Abstract
Extreme gradient boosting methods outperform conventional machine-learning models. Here, we have developed the LEukemia Artificial intelligence Program (LEAP) with the extreme gradient boosting decision tree method for the optimal treatment recommendation of tyrosine kinase inhibitors (TKIs) in patients with chronic myeloid leukemia in chronic phase (CML-CP). A cohort of CML-CP patients was randomly divided into training/validation (N = 504) and test cohorts (N = 126). The training/validation cohort was used for 3-fold cross validation to develop the LEAP CML-CP model using 101 variables at diagnosis. The test cohort was then applied to the LEAP CML-CP model and an optimum TKI treatment was suggested for each patient. The area under the curve in the test cohort was 0.81899.Backward multivariate analysis identified age at diagnosis, the degree of comorbidities, and TKI recommended therapy by the LEAP CML-CP model as independent prognostic factors for overall survival. The bootstrapping method internally validated the association of the LEAP CML-CP recommendation with overall survival as an independent prognostic for overall survival. Selecting treatment according to the LEAP CML-CP personalized recommendations, in this model, is associated with better survival probability compared to treatment with a LEAP CML-CP non-recommended therapy. This approach may pave a way of new era of personalized treatment recommendations for patients with cancer.
1 |. INTRODUCTION
The survival of patients with chronic myeloid leukemia in chronic phase (CML-CP) is approaching that of the general population with the use of tyrosine-kinase inhibitors (TKI), particularly in younger patients who achieve remission within 1 year of TKI therapy.1–6 The current guidelines recommend treating patients with a TKI in the frontline aiming for response milestones at specific time points.7,8 However, even with effective TKI therapy, most trials suggest that at least 30%−40% of patients will require a change to another TKI.9,10 The decision regarding what is the optimal initial TKI for a given patient is frequently based on comorbidities, the biological features of CML-CP, and social geodemographic features. However, most of the treatment decisions are based on indirect comparisons of unrelated studies and personal preferences.
Recently developed machine learning algorithms outperform conventional statistical models for the accuracy of prediction.11–13 Randomized clinical trials can compare the efficacy of treatment between patient groups. However, selection of the best treatment decision for an individual patient, with their own clinical and biological features, and in the context of highly effective treatment options, is more complex and frequently based on subjective criteria. A machine-learning-assisted approach may help with decision-making in complex clinical situations. Gradient tree boosting is a machine learning model through ensemble learning with weak prediction models.14 Gradient boosting decision tree models have outperformed other machine learning methods for classification and ranking problems.15,16 Extreme gradient boosting (XGBoost) is an advanced implementation of gradient boosting, which enables regularization to reduce overfitting and to improve model performance. Among machine learning algorithms with gradient tree boosting, the XGBoost approach consistently wins international data analysis challenges.17 Among 29 data analysis challenges at Kaggle, XGBoost was utilized for 17 solutions and continues to be the dominant state-of-the-art method for data analysis competitions. Thus, the state-of-art gradient boosting method might improve overall survival by selecting the optimal frontline TKI with accurate prediction. The aim of this study is to develop the LEukemia Artificial intelligence Program (LEAP) to assist with treatment selection for patients with CML-CP.
2 |. METHODS
2.1 |. Patients
From 30 July 2000 to 25 November 2014, 630 consecutive patients with newly diagnosed CML-CP were enrolled in seven consecutive or parallel prospective clinical trials at a single institution and were included in this analysis. Therapy consisted of imatinib (starting dose of 400 or 800 mg daily, alone or with pegylated interferon after 6 months of imatinib), dasatinib (50 mg orally twice daily or 100 mg orally once daily), nilotinib (400 mg orally twice daily), or ponatinib (45 mg orally daily). All of the patients who enrolled in these clinical trials were analyzed in this study. These trials were registered at www.clinicaltrials.gov as NCT00038649, NCT00048672, NCT00333840, NCT00050531, NCT00254423, NCT00129740, and NCT01570868. All protocols and the development of the LEAP CML-CP were approved by the institutional review board and informed consent was obtained in accordance with the Declaration of Helsinki.
Diagnosis of CML in early chronic phase was defined as the presence of Philadelphia chromosome or BCR-ABL1 rearrangement with the presence in the peripheral blood of <15% blasts, <20% basophils, <30% blasts and promyelocytes, and platelets >100 × 109/L, with a time interval from diagnosis to enrollment of 12 months or less. The inclusion criteria were similar for all trials, including age equal to or older than age 16, adequate heart, liver and renal function, and Eastern Cooperative Oncology Group (ECOG) performance status of 0–2. Patients with clonal evolution at the time of diagnosis were eligible for these studies.
We evaluated the severity of comorbidities by Adult Comorbidity Evaluation 27 (ACE-27), a 27-item validated comorbidity index for patients with cancer.18 Briefly, the ACE-27 grades the specific conditions into one of three levels including grade one (mild), grade two (moderate), or grade three (severe) according to the functionality of individual organ. The overall comorbidity score of ACE-27 (none, mild, moderate, or severe) is assigned based on the highest rank of single organ system function. A patient with two or more moderate comorbidity in different systems is designated as severe overall comorbidity.
2.2 |. Endpoints and assessment
The cytogenetic and molecular response criteria were previously described.19 Briefly, cytogenetic response was assessed by conventional cytogenetic analysis performed in bone marrow cells with the G-banding technique, with at least 20 metaphases analyzed. Fluorescent in-situ hybridization (FISH) on peripheral blood was used to assess response only when routine cytogenetic analysis was insufficient number of metaphases. Complete cytogenetic response was defined as the absence of Philadelphia chromosome by conventional cytogenetic analysis or FISH. Molecular response was assessed by reverse transcription-polymerase chain reaction (RT-PCR) and expressed as the BCR-ABL/ABL ratio on the international scale. A major molecular response (MMR) was defined as BCR-ABL/ABL transcript ratio less than or equal to 0.1%, and MR4.0 and MR4.5 as a 4.0 and 4.5 log reduction or greater in BCR-ABL transcripts with a ratio of less than or equal to 0.01% and 0.0032%, respectively. Sustained MR4.5 was defined as the duration of MR4.5 or deeper for at least 2 years consistently.
Event-free survival (EFS) was calculated from the start of therapy to loss of complete hematologic response, loss of major cytogenetic response, transformation to accelerated (AP) or blast phase (BP), or death from any cause during study therapy. Transformation-free survival (TFS) was calculated from the start of therapy to transformation to AP or BP, or death during study therapy. Failure-free survival (FFS) was calculated from the start of TKI to an event (as defined above), discontinuation of therapy for any reason, or death. Patients who were alive at the end of the study period were censored at the date of last follow-up. Overall survival (OS) was dated from the start of therapy until death from any cause at any time regardless of the termination of the study.
2.3 |. Statistical analysis
We developed the machine learning model using 101 variables that included age at diagnosis of CML-CP, gender, primary ethnicity, Hispanic, primary language, international patients, marital status, distance in kilometers from the home zone improvement plan (ZIP) code to our institution, height, weight, body mass index, body surface area, palpable spleen size below left costal margin on physical examination, the time period from the diagnostic date of CML-CP to the start date of TKI, white blood cell count, red blood cell count, hemoglobin, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, red cell distribution width, platelet count, mean platelet volume, the percentage of neutrophils, lymphocytes, monocytes, eosinophils. Basophils, blasts, promyelocytes, myelocytes, metamyelocytes, and band neutrophils in peripheral blood, the percentage of blasts, promyelocytes, myelocytes, metamyelocytes, neutrophils, eosinophils, basophils, lymphocytes, monocytes, plasma cells, pronormoblasts, and normoblasts in bone marrow, myeloid:erythroid ratio in bone marrow, albumin, blood urea nitrogen, lactate dehydrogenase, total bilirubin, alanine transaminase, Sokal risk score, Sokal risk classification, Hasford score, Hasford risk classification, European Treatment and Outcome Study (EUTOS) score, EUTOS risk classification, EUTOS long-term survival (ELTS) score, ELTS risk classification, the presence of clonal evolution, cryptic, and variant Philadelphia chromosome abnormality, transcript type of BCR-ABL1, the percentage of Philadelphia chromosome by conventional cytogenetic karyotype analysis and fluorescence in situ hybridization, BCR-ABL1 levels by reverse transcription polymerase chain reaction, and the daily dose of frontline TKI therapy including imatinib, dasatinib, nilotinib, and ponatinib, and the severity of comorbidity. We assessed the severity of each organ system by ACE-27 for the assessment of the degree of comorbidities for cardiovascular system (myocardial infarction, angina/coronary artery disease, congestive heart failure, arrhythmias, hypertension, venous disease, and peripheral arterial disease), respiratory system, gastrointestinal system (hepatic function, stomach/intestine, and pancreas), renal system (end-stage renal disease), endocrine system (diabetes mellitus), neurological system (stroke, dementia, paralysis, and neuromuscular disease), psychiatric disease, rheumatologic disease, immunological system, malignancy (solid tumor including melanoma, leukemia and myeloma, and lymphoma), substance abuse (alcohol, and illicit drugs), and body weight (obesity). White blood cell count, red blood cell count, hemoglobin, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, red cell distribution width, platelet count, mean platelet volume, the percentage of neutrophils, lymphocytes, monocytes, eosinophils. Basophils, blasts, promyelocytes, myelocytes, metamyelocytes, and band neutrophils in peripheral blood, the percentage of blasts, promyelocytes, myelocytes, metamyelocytes, neutrophils, eosinophils, basophils, lymphocytes, monocytes, plasma cells, pronormoblasts, and normoblasts in bone marrow, myeloid:erythroid ratio in bone marrow, albumin, blood urea nitrogen, lactate dehydrogenase, total bilirubin, alanine transaminase, Sokal risk score, Sokal risk classification, Hasford score, Hasford risk classification, European Treatment and Outcome Study (EUTOS) score, EUTOS risk classification, EUTOS long-term survival (ELTS) score, ELTS risk classification, the presence of clonal evolution, cryptic, and variant Philadelphia chromosome abnormality, transcript type of BCR-ABL1, the percentage of Philadelphia chromosome by conventional cytogenetic karyotype analysis and fluorescence in situ hybridization, BCR-ABL1 levels by reverse transcription polymerase chain reaction, and the daily dose of frontline TKI therapy including imatinib, dasatinib, nilotinib, and ponatinib. We included variables partly related to CML, and clinical factors which potential affect prediction on survival. The proposed LEAP CML-CP model was intended for an optimal TKI selection at the time of diagnosis and response after therapy was not included to avoid potential bias in the development of the LEAP CML-CP model.
We developed an extreme gradient boosting decision tree model with the XGBoost package through ensemble learning which combines weak decision tree models after random sampling and random variable sampling. Hyperparameter optimization was performed by Python with Stampede2, a supercomputer located at Texas Advanced Computing Center, which was ranked at the 19th fastest supercomputer in the world in June 2019 by TOP500. The hyperparameter tuning included maximal depth, minimal child weight, learning rate, subsample ratio of variables by tree, regularization parameter (alpha and lambda). The accuracy of prediction was measured by area under the curve in the training/validation and test cohorts. The extreme gradient boosting decision tree model estimated hazard ratios for overall survival using only the training/validation cohort. The final performance was evaluated with the independent test cohort that was not used for the development of the machine learning model.
In the test cohort, expected hazard ratios were calculated with potential treatment options. The treatment option with the lowest hazard ratio was considered the best treatment option for individual patients. A difference in hazard ratios of less than 0.005 was considered as comparable treatment options. We considered the best and comparable treatment options as the LEAP CML-CP recommendations. The test cohort was divided into the LEAP CML-CP recommendation and the LEAP CML-CP non-recommendation cohorts.
In the test cohort, categorical variables were compared with a Fisher exact or Pearson χ2 test. Continuous variables were analyzed by a Mann-Whitney U test. A Kaplan-Meier plot was used to visualize survival distributions between the LEAP CML-CP recommendation and the LEAP CML-CP non-recommendation cohorts. Differences in survival between groups were assessed by a log-rank test. Multiple imputation was performed for missing variables to reduce potential bias. To evaluate the association of the LEAP CML-CP recommendation with overall survival, we built a multivariate Cox proportional hazards with backward elimination after the initial feature selection with the P value cutoff of .100 by univariate Cox regression analysis. For the internal validation, bootstrapping method was performed with 2000 bootstrap sampling. To evaluate the causation of the LEAP CML-CP recommendation with survival, we performed inverse probability of treatment weighing (IPTW) to balance baseline difference of covariates. We selected well-validated prognostic covariates to verify the significance of the LEAP CML-CP recommendations. Covariates for the calculations of propensity scores included age at diagnosis, Sokal score, Hasford score, EUTOS score, ELTS score, and ACE-27. Logistic regression was performed to calculate propensity score before IPTW analysis. To validate the results of crude IPTW analysis, we performed adjusted IPTW analyses: IPTW with removal of subjects whose propensity scores below the second percentile of the LEAP CML-CP recommendation cohort and above 98th percentile of the LEAP CML-CP non-recommendation cohort; IPTW analysis with the maximum weight cap at 100. The P values < .05 were considered as statistically significant.
We calculated SHapley Additive exPlanations (SHAP) values to interpret the black box of the LEAP CML-CP recommendation.20 Shapley values were calculated from cooperative game therapy to explain the contribution of each player.21 The SHAP values were modified from the Shapely values for the intuitive understanding of machine learning models with additive feature attribution. The SHAP values demonstrated the positive and negative impact of each feature, and the sum of SHAP values determined the final prediction. Statistical analysis was performed using Statistical Package for Social Sciences (SPSS) software (version 24, SPSS, Inc, Chicago, Illinois), R (version 3.5.0), and Python (version 3.7.3).
3 |. RESULTS
3.1 |. Patient cohort
From 30 July 2000 to 25 November 2014, 630 patients with newly diagnosed CML-CP were treated with frontline TKIs in prospective clinical trials were included in this analysis. Therapy consisted of imatinib (starting dose of 400 or 800 mg daily, alone or with pegylated interferon after 6 months of imatinib), dasatinib (50 mg orally twice daily or 100 mg orally once daily), nilotinib (400 mg orally twice daily), or ponatinib (45 mg orally daily). These were sequential or parallel clinical trials (clinicaltrials.gov numbers NCT00038649, NCT00048672, NCT00333840, NCT00050531, NCT00254423, NCT00129740, and NCT01570868, respectively) and all enrolled patients were included in this analysis. Patient characteristics are summarized in Table 1.
TABLE 1.
Main patient characteristics
| Test Cohort: LEAP CML-CP recommendation | ||||
|---|---|---|---|---|
| Median (range)/No. (%) | Training/validation Cohort N = 504 | Recommended N = 94 | Not recommended N = 32 | P recommended vs. not recommended |
| Age (y) | 49 (15.1–86.5) | 45 (17.0–72.7) | 63 (23.4–81.6) | <.001 |
| Adult comorbidity evaluation-27 | ||||
| None | 280 (56) | 54 (57) | 8 (25) | <.001 |
| Mild | 158 (31) | 35 (37) | 10 (31) | |
| Moderate | 55 (11) | 4 (4) | 12 (38) | |
| Severe | 11 (2) | 1 (1) | 2 (6) | |
| Sokal risk | ||||
| Low | 328 (65) | 63 (67) | 14 (44) | .004 |
| Intermediate | 132 (26) | 21 (22) | 17 (53) | |
| High | 44 (9) | 10 (11) | 1 (3) | |
| Hasford risk | ||||
| Low | 309 (61) | 60 (64) | 14 (44) | .101 |
| Intermediate | 181 (36) | 30 (32) | 17 (53) | |
| High | 14 (3) | 4 (4) | 1 (3) | |
| EUTOS risk | ||||
| Low | 447 (89) | 80 (85) | 28 (88) | .498 |
| High | 57 (11) | 14 (15) | 4 (13) | |
| ELTS risk | ||||
| Low | 447 (89) | 82 (87) | 30 (94) | .443 |
| Intermediate | 39 (8) | 8 (9) | 2 (6) | |
| High | 18 (4) | 4 (4) | 0 | |
| TKI therapy, No. (%) | ||||
| Imatinib 400 mg/d | 62 (12) | 7 (7) | 4 (13) | .128 |
| Imatinib 800 mg/d | 165 (33) | 32 (34) | 11 (34) | |
| Nilotinib 800 mg/d | 111 (22) | 24 (26) | 13 (41) | |
| Dasatinib 100 mg/d | 123 (24) | 25 (27) | 2 (6) | |
| Ponatinib 30 mg/d | 7 (1) | 0 | 0 | |
| Ponatinib 45 mg/d | 36 (7) | 6 (6) | 2 (6) | |
Abbreviations: EUTOS, European Treatment and Outcome Study; ELTS, EUTOS long-term survival; LEAP CML-CP, LEukemia Artificial intelligence Program for chronic myeloid leukemia in chronic phase; TKI, tyrosine kinase inhibitor.
3.2 |. Development of LEAP CML-CP
The process of the LEAP CML-CP development is summarized in Figure 1. The whole cohort was randomly divided into training/validation (N = 504) and test cohorts (N = 126) at a 4:1 ratio. The training/validation cohort was used for 3-fold cross validation to develop the LEAP CML-CP model. Hyperparameter tuning was performed for colsample by tree, learning rate, maximal depth of decision trees, and minimal number of patients in each leaf of decision trees to optimize the development of the LEAP CML-CP. Each hyperparameter tuning was performed until no further improvement of the area under the curve (AUC) in the validation cohort was observed after 200 rounds of adjustment. The best hyperparameter was selected among 50 000 evaluations (Figure S1). Hyperparameter tuning identified colsample by tree of 0.8893679364029201, learning rate of 0.00023388082385954239, maximal depth of nine, minimal child weight of 28, regularization alpha of 3.316555622922197, regularization lambda of 1.3127892578303628, and subsample of 0.9996679829232776. The number of decision trees was 8417, 14 659, and 14 190 in the first, second, and third validation cohort, respectively.
FIGURE 1.
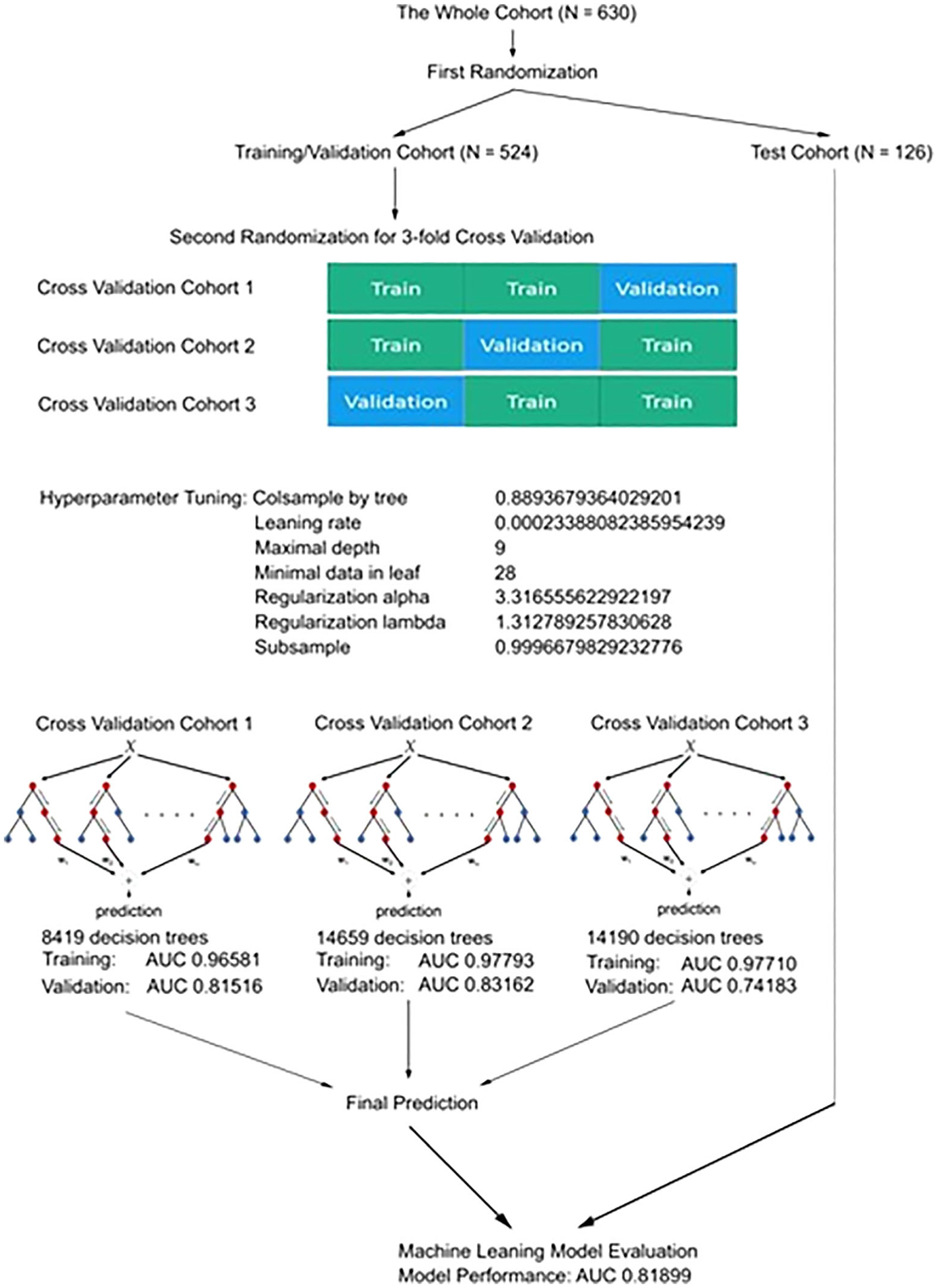
The development process of the LEAP CML-CP model. Abbreviations: LEAP CML-CP, Leukemia Artificial intelligence Program for chronic myeloid leukemia in chronic phase
The AUC of the training in the first, second, and third cross validation cohort was 0.9658104824713641, 0.9779276025363192, and 0.9771049983227105, respectively; the AUC of the validation in the first, second, and third cross validation cohort was 0.8151599875737807, 0.8316176470588236, and 0.7418278852568378, respectively (Figure S2). The AUC in the test cohort was 0.81899. We calculated concordance index for the prediction on survival by well-validated conventional statistical model with the ELTS risk classification and ACE27 classification. The concordance index was 0.5513 and 0.7148 by the ELTS and ACE27 risk classification in the training/validation cohort; the concordance index was 0.4876 and 0.8044 by the ELTS and ACE27 risk classification in the test cohort.
3.3 |. Treatment recommendation by LEAP CML-CP
The performance of the LEAP CML-CP model was evaluated by calculating the hazard ratios for overall survival by treatment in the test cohort. A difference in predicted hazard ratios of less than 0.005 was considered as comparable treatment options. We considered the best and comparable treatment options as LEAP CML-CP recommendations. The test cohort (N = 126) was divided into those that, in retrospect, were treated in the frontline with the TKI that would have been recommended by LEAP CML-CP (ie, the LEAP CML-CP recommendation cohort; N = 94, 75%) and those that were treated with a TKI that was not the one recommended by LEAP CML-CP (ie, the LEAP CML-CP non-recommendation cohort; N = 32, 25%). The median follow-up for the total population was 139 months (range, 3.7–216.1), and was 127 and 148 months in the LEAP CML-CP recommendation and LEAP CML-CP non-recommendation cohorts, respectively (P = .902). The median age at diagnosis was 43 (17.0–72.7) and 63 (23.4–81.6) in patients with the LEAP CML-CP recommendation and LEAP CML-CP non-recommendation cohorts, respectively (P < .001). The degree of comorbidity was more severe in patients in the LEAP CML-CP non-recommendation cohort (P < .001); 57% of patients in the LEAP CML-CP recommendation cohort had no comorbidity, and 44% of patients in the LEAP CML-CP non-recommendation cohort had moderate or severe comorbidities by Adult Comorbidity Evaluation 27 (ACE-27), a 27-item validated comorbidity index for patients with cancer.18 Intermediate and high Sokal risk classification, a prognostic classification from four clinical features (spleen size on physical examination, blast percentage in peripheral blood, platelet count, and age at diagnosis), was observed in 33% and 56% of patients in the LEAP CML-CP recommendation and LEAP CML-CP no-recommendation cohorts, respectively (P = .004). The type and dosage of TKI did not differ significantly between cohorts (P = .128). Overall survival did not differ significantly by the type and dose of TKI (P = .472) (Figure S3).
In the test cohort, the overall rates of complete cytogenetic response (CCyR), major molecular response (MMR), molecular response by a 4.0 log reduction or BCR-ABL transcripts with a ratio of less than or equal to 0.01% on the international scale (MR4), molecular response by a 4.5 log reduction or BCR-ABL transcripts with a ratio of less than or equal to 0.0032% on the international scale (MR4.5), and sustained MR4.5 for the LEAP CML-CP recommendation and LEAP CML-CP non-recommendation cohorts were 89% and 81%, 82% and 75%, 73% and 53%, 70% and 47%, and 39% and 16%, respectively (P = .186; P = .397; P = .033; P = .017; P = .014). Similarly, the rates of 5-year failure-free survival, transformation-free survival, event-free survival, and overall survival were 63% and 28%, 98% and 76%, 92% and 58%, and 98% and 77% in the LEAP CML-CP recommendation and LEAP CML-CP non-recommendation cohorts, respectively (P < .001; P = .002; P < .001; P < .001) (Table 2); the median failure-free survival was not reached and 3 months, respectively (P < .001) (Figure 2A); the median transformation-free survival was not reached in either cohort (P = .002) (Figure 2B); the median event-free survival was not reached and 98 months, respectively (P < .001) (Figure 2C); the median overall survival was 210 and 150 months, respectively (P < .001) (Figure 2D).
TABLE 2.
Response and outcomes
| Training/validation Cohort N = 504 | Test Cohort: LEAP CML-CP recommendation | P recommended vs. not recommended | ||
|---|---|---|---|---|
| Recommended N = 94 | Not recommended N = 32 | |||
| Response within 1 y of TKI therapy, No. (%) | ||||
| CCyR | 440 (87) | 83 (88) | 25 (78) | .155 |
| MMR | 367 (73) | 68 (72) | 19 (59) | .171 |
| MR4 | 237 (47) | 38 (40) | 12 (38) | .770 |
| MR4.5 | 190 (38) | 31 (33) | 9 (28) | .610 |
| Overall response, No. (%) | ||||
| CCyR | 452 (89) | 84 (89) | 26 (81) | .186 |
| MMR | 425 (84) | 77 (82) | 24 (75) | .397 |
| MR4 | 364 (72) | 69 (73) | 17 (53) | .033 |
| MR4.5 | 342 (68) | 66 (70) | 15 (47) | .017 |
| Sustained MR4.5 | 69 (14) | 37 (39) | 5 (16) | .014 |
| Five-year outcome, (%) | ||||
| FFS | 67 | 63 | 28 | <.001 |
| TFS | 93 | 98 | 76 | .002 |
| EFS | 85 | 92 | 58 | <.001 |
| OS | 93 | 98 | 77 | <.001 |
Abbreviations: CCyR, complete cytogenetic response; MMR, major molecular response; MR4.5, molecular response by a 4.5 log reduction on the international scale; TKI, tyrosine kinase inhibitor.
FIGURE 2.
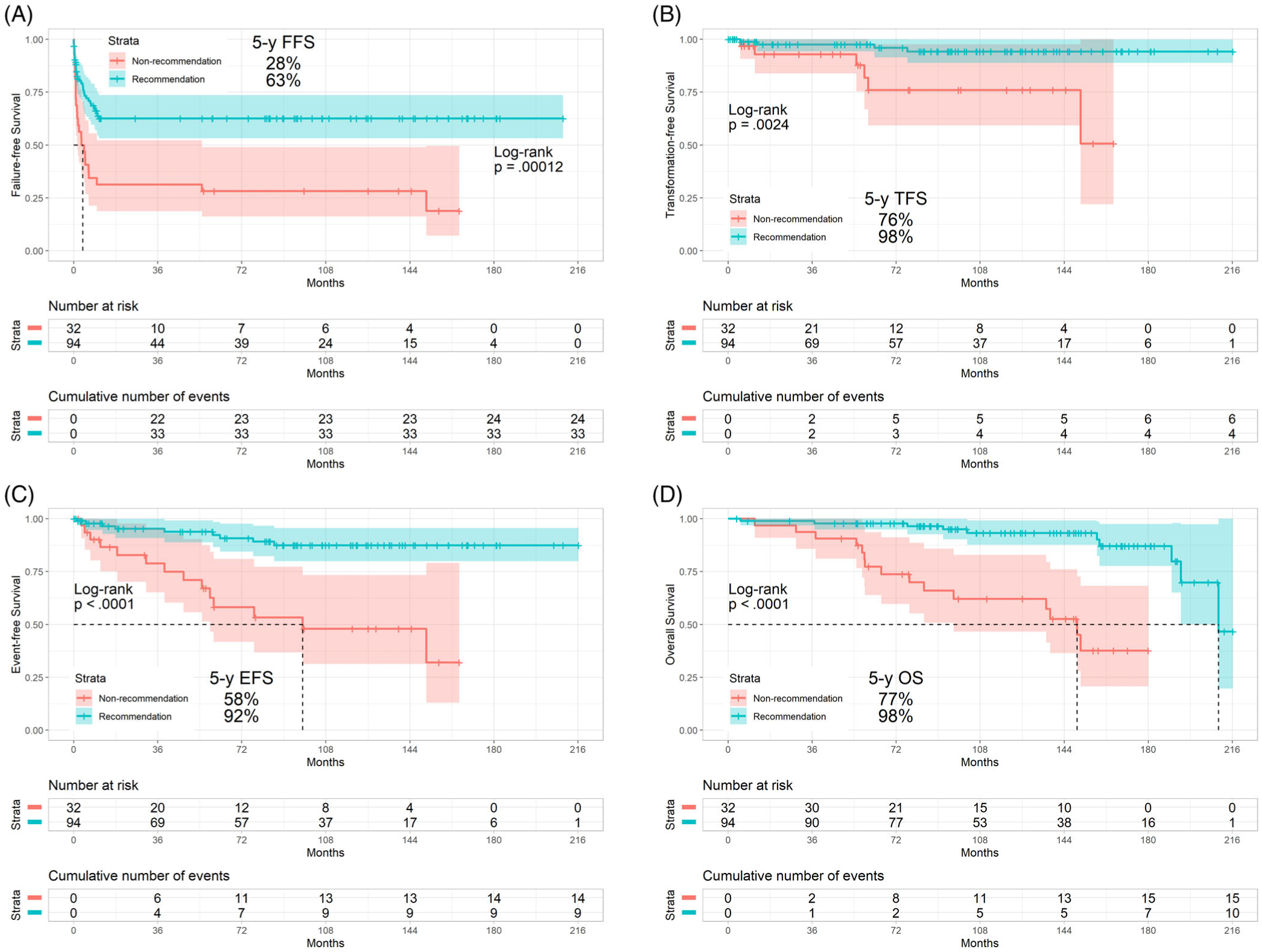
Survival by the LEAP CML-CP recommendation in the test cohort: A, failure-free survival, B, transformation-free survival, C, event-free survival, D, overall survival. Abbreviations: LEAP, LEukemia Artificial intelligence Program for chronic myeloid leukemia in chronic phase; FFS, failure-free survival; TFS, transformation-free survival; EFS, event-free survival; OS, overall survival
Backward multivariate analysis identified age at diagnosis (P = .045; hazard ratio [HR], 1.041; 95% confidence interval [CI], 1.001–1.083), the degree of ACE-27 (P = .032; HR, 1.742; 95% CI, 1.012–2.997), and recommended TKI therapy by the LEAP CML-CP model (P = .032; HR, 0.280; 95% CI, 0.087–0.895) as independent prognostic factors for OS (Table S1). The bootstrapping method internally validated the association of the LEAP CML-CP recommendation with overall survival (P = .007; HR, 0.234; 95% CI, 0.384–0.768) as an independent prognostic for overall survival. Inverse probability of treatment weighing (IPTW) methods with truncation by propensity score and with weight capping at 100 confirmed that the recommendation by the LEAP CML-CP model improves failure-free survival, transformation-free survival, event-free survival, and overall survival in patients with CML-CP (P < .001 and P < .001; P < .001 and P < .001; P < .001 and P < .001; P < .001 and P < .001) (Table S2).
We observed 25 deaths in the test cohort (10 deaths and 15 deaths in the LEAP CML-CP recommendation and LEAP CML-CP non-recommendation cohorts, respectively). In the LEAP CML-CP recommendation cohort, six patients died of unknown causes; one, CML-CP progression; one, influenza B, one, fall; one, suicide; and there were no deaths due to cardiovascular events. In the LEAP CML-CP non-recommendation cohort, four patients died of cardiovascular events; four, unknown causes; two, second malignancies (non-small cell lung cancer; Philadelphia chromosome negative acute myeloid leukemia); two, pneumonia; one, complications from stem cell transplantation following disease progression; one, car accident; and one, fall.
We calculated SHapley Additive exPlanations (SHAP) values to interpret the black box of the LEAP CML-CP recommendation (Figure S4). SHAP values attribute to each variable the change in the expected model prediction when conditioning on that variable. The SHAP values enable interpretation of LEAP CML-CP predictions. The summary of SHAP values identifies the order of variable importance for prediction. The presence and degrees of comorbidities were the first and third most important variables for the prediction of overall survival in patients with CML-CP. The European Treatment and Outcome Study (EUTOS) long-term survival (ELTS) score was the second important prognostic factor for overall survival. Though basophilia was not a part of ELTS score, the percentage of basophils was the seventh most important prognostic factor, with less predictive significance compared to comorbidity and ELTS scores.
4 |. DISCUSSION
In the current study, we show that treatment recommendations by the LEAP CML-CP have the potential to improve the clinical outcomes of patients with CML-CP. The LEAP CML-CP incorporated 101 variables including comorbidity by ACE-27, Sokal risk, Hasford risk, EUTOS risk, and ELTS risk for the prediction of overall survival. We hypothesized that a machine learning model could support decision making among patients, caregivers, and physicians for the optimal TKI recommendation in patients with CML-CP, and we demonstrated that patients who received the TKI that the LEAP CML-CP would have recommended improved survival compared to that of patients who did not receive the TKI that the LEAP CML-CP would have recommended. The LEAP CML-CP recommendation was not associated with 1-year cytogenetic and molecular response but was associated with overall deep response, including MR4, MR4.5, and sustained MR4.5. Given failure-free survival and transformation-survival improved with the LEAP CML-CP recommendation, the LEAP CML-CP recommendation evaluated tolerance and resistance to TKI in CML-CP. The expected higher tolerance and lower resistance translated into improved event-free survival and overall survival. The difference of age at diagnosis, the degrees of ACE-27, and the Sokal risk scores between the two cohorts suggested that older patients with moderate/severe comorbidities and high risk CML-CP required cautious selection of frontline TKI. These elements are informally incorporated into the decision process for most patients by their treating physicians. Note, LEAP CML-CP incorporates these elements in a more formal and structured manner. Of note, LEAP CML-CP did not always recommend one particular TKI. To better understand how LEAP CML-CP recommendations may lead to improved survival, we balanced baseline difference of age, comorbidities, and risk classification with inverse probability of treatment weighing with propensity score analysis. We confirmed the LEAP CML-CP recommendation was associated with improved overall survival (Figure 2D). Given the impracticalities of conducting randomized clinical trials in specific subsets of patients with a relatively rare disease such as CML-CP, this approach may pave the way for a new era of personalized treatment recommendations for individual patients based on their unique clinical, social geodemographic, biological, chromosomal, and molecular features.
There appears to be a higher discordance between the TKI that patients actually received and the TKI that was recommended by LEAP CML-CP among older patients. This high rate of discordance in older patients suggests that older patients are likely to receive the greatest benefit of the LEAP CML-CP recommendation. With adequate access to TKI, survival in young patients with CML-CP who achieved at least a complete cytogenetic response (which is the overwhelming majority) is similar to that of general population.1 However, the presence of comorbidities, more commonly seen in older patients, influences overall survival expectations.22 Each TKI has its own specific safety profile.8 Importantly, several TKIs increase the risk of arterial and venous vascular events in patients with CML-CP.23,24 TKIs may also induce pulmonary arterial hypertension, which may persist after discontinuation of TKI treatment.25 Also, the incidence of second malignancy has increased in patients with CML-CP compared to that of general population since the advent of TKIs.26 Clearly, treatment recommendation requires a holistic review of patient characteristics including comorbidities in all organ systems. We used ACE-27, a well-validated comorbidity scoring system in patients with cancer, to assess the degrees of comorbidities.27 Multimorbidity is increasingly common with aging, and approximately half of patients over age 65 had at least three morbidities.28 Given that the reported median age of CML patients is 67 years,29 the assessment of multimorbidity must be a part of treatment recommendations to determine the optimal TKI and avoid adverse events. To incorporate the clinical, biological, chromosomal, and molecular features of CML-CP into a prognostic model, we used the state-of-art machine-learning model with the extreme gradient boosting decision tree model. Machine learning is starting to be utilized in medical fields, and outperforms conventional statistical models.30–32 Our machine learning model can support the process of decision making for the selection of optimal treatment among available options. Importantly, LEAP CML-CP does not replace physician expertise and proper care. Human experts are vital for the supervision and monitoring of response and adverse events over the course of TKI therapy, and to ensure that the treatment recommendation is acceptable based on published medical literature for patient safety and the results being observed on individual patients. However, with the supervision of machine learning models by human experts, the LEAP CML-CP model can achieve improved patient outcomes.
We calculated SHAP values to estimate the impact of each variable on the prediction by the LEAP CML-CP model (Figure S4). The summary of SHAP values suggested the presence and degree of comorbidities and ELTS scores (which included age at diagnosis, spleen size, the percentage of blasts in peripheral blood, and platelet count) as the most important contributors.33 The ELTS classification was developed based on survival data of patients in the TKI era, and has been suggested to be a better survival predictor for CML-specific death than the Sokal, Hasford, and EUTOS scores.34–36 The summary of SHAP values in the LEAP CML-CP has successfully captured the importance of ELTS scores and the degrees of comorbidity among 101 variables in the era of TKI.
There are several limitations to our study. First, the clinical data for frontline bosutinib was not available, thus precluding evaluation of one of the four approved treatment options for frontline therapy, while including ponatinib which is not approved as frontline therapy. However, the LEAP CML-CP included long-term follow-up data over a decade with frontline imatinib, dasatinib, and nilotinib. Given that the LEAP CML-CP recommendations retrospectively improved overall survival through improving tolerance and overcoming resistance for long-term TKI, we plan to incorporate long-term frontline bosutinib for the next future version update. Second, the LEAP CML-CP model is intended for the recommendation of frontline TKI selection. The LEAP CML-CP recommendation could potentially be used for selection of the optimal TKI therapy after frontline failure, however, a different dataset of patients who failed front-line TKI therapy is ideally required to develop another LEAP CML-CP model for the optimal decision of treatment after frontline failure. Our current LEAP CML-CP model focused on the treatment decision at diagnosis, which leads to higher rates of sustained MR4.5. Ideally, patients who received an optimal TKI treatment by the LEAP CML-CP would achieve higher rates of treatment-free remission without the need of second line therapy. Third, the LEAP CML-CP was designed based on patients treated in a dedicated CML clinic and with patients enrolled in clinical trials. Although we believe the model applies to patients regardless of whether they are treated on clinical trials or not and in various settings, this would have to be confirmed in additional testing. Fourth, we analyzed patients on seven consecutive clinical trials that required compliance on TKI therapies. Given non-compliance adversely affects outcome in patients with CML-CP, the application of the LEAP CML-CP model can be limited to compliant patients. The incorporation of socioeconomic factors at diagnosis might be considered to predict for compliance after starting TKI therapy in an updated version of the LEAP CML-CP model.
In conclusion, the LEAP CML-CP model with the extreme gradient boosting decision tree method retrospectively improved overall survival in patients with CML-CP through optimal selection of frontline TKI therapy among available treatment options. The LEAP CML-CP has the potential to support patients, caregivers, and physicians for personalized treatment recommendations, and contribute to further improvement of clinical outcome in patients with CML-CP.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients and their families who enrolled in the original trials.
Funding information
This work is supported in part by the University of Texas MD Anderson Cancer Center Support Grant from the National Institutes of Health P30 CA016672, the National Institutes of Health/National Cancer Institute under award P01 CA049639, the University of Texas MD Anderson MDS/AML Moon Shot, and Leukemia Texas.
CONFLICT OF INTEREST
K.S. declares honoraria from Otsuka Pharma, and a consultancy fee from Pfizer Japan. E.J.J. received consultancy fees from Ariad, Bristol-Myers Squibb, Teva, and Pfizer. F.R. received research funding from Novartis and Bristol-Myers Squibb. M.K. declares consulting and honoraria from AbbVie, Genentech, F. Hoffman La-Roche, Stemline Therapeutics, Amgen, Forty-Seven, and Kisoji; research funding and/or clinical trial support from AbbVie, Genentech, F. Hoffman La-Roche, Eli Lilly, Cellectis, Calithera, Ablynx, Stemline Therapeutics, Agios, Ascentage, and Astra Zeneca; and stock options/royalties from Reata Pharmaceutical. G.G-M. declares research support from Amphivena, Helsinn, Novartis, Abbvie, Celgene, Astex, Onconova, and Merck. J.E.C. declares support (to the institution) from and consultancy for BMS, Novartis, Astellas, Daiichi-Sankyo, Pfizer, Jazz Pharma. C.D. declares consultancy fee for Abbvie, Agios, Celgene and honoraria from Medimmune, Daiichi Sankyo, Abbvie, Agios, Jazz, Celgene, and Syros. G.B., W.G.W., N.D., K.T., K.N., G.M.B., R.K.-S., G.I., P.J., J.K., M.B.R., S.P., K.A.S., and J.S. have no relevant conflict of interest. The rest of authors declare no competing financial interests.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
The University of Texas MD Anderson Cancer Center is committed to providing qualified scientific collaborators access to anonymized patient-level data for the purpose of conducting legitimate clinical research upon the approval of institutional review board at MD Anderson Cancer Center. To protect the rights and privacy of patients on clinical trials, external researchers will be evaluated for the ability of fulfilling suggested collaborative projects for the qualification of data access.
REFERENCES
- 1.Sasaki K, Strom SS, O’Brien S, et al. Relative survival in patients with chronic-phase chronic myeloid leukaemia in the tyrosine-kinase inhibitor era: analysis of patient data from six prospective clinical trials. Lancet Haematol. 2015;2:e186–e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortes JE, Gambacorti-Passerini C, Deininger MW, et al. Bosutinib vs imatinib for newly diagnosed chronic myeloid leukemia: results from the randomized BFORE trial. J Clin Oncol. 2018;36:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes JE, Saglio G, Kantarjian HM, et al. Final 5-year study results of DASISION: the dasatinib vs imatinib study in treatment-naive chronic myeloid leukemia patients trial. J Clin Oncol. 2016;34:2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochhaus A, Larson RA, Guilhot F. et al. ; IRIS InvestigatorsLong-term outcomes of imatinib treatment for chronic myeloid leukemia. N Engl J Med. 2017;376:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochhaus A, Saglio G, Hughes TP, et al. Long-term benefits and risks of frontline nilotinib vs imatinib for chronic myeloid leukemia in chronic phase: 5-year update of the randomized ENESTnd trial. Leukemia. 2016;30:1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain P, Kantarjian H, Alattar ML, et al. Long-term molecular and cytogenetic response and survival outcomes with imatinib 400 mg, imatinib 800 mg, dasatinib, and nilotinib in patients with chronic-phase chronic myeloid leukaemia: retrospective analysis of patient data from five clinical trials. Lancet Haematol. 2015;2:e118–e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radich JP, Deininger M, Abboud CN, et al. Chronic myeloid leukemia, version 1.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16:1108–1135. [DOI] [PubMed] [Google Scholar]
- 9.Jabbour E, Kantarjian HM, Saglio G, et al. Early response with dasatinib or imatinib in chronic myeloid leukemia: 3-year follow-up from a randomized phase 3 trial (DASISION). Blood. 2014;123:494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jabbour E, Kantarjian H, O’Brien S, et al. The achievement of an early complete cytogenetic response is a major determinant for outcome in patients with early chronic phase chronic myeloid leukemia treated with tyrosine kinase inhibitors. Blood. 2011;118:4541–4546; quiz 4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hale AT, Stonko DP, Brown A, et al. Machine-learning analysis outperforms conventional statistical models and CT classification systems in predicting 6-month outcomes in pediatric patients sustaining traumatic brain injury. Neurosurg Focus. 2018;45:E2. [DOI] [PubMed] [Google Scholar]
- 12.Singal AG, Mukherjee A, Elmunzer JB, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013; 108:1723–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamshidi A, Pelletier JP, Martel-Pelletier J. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nat Rev Rheumatol. 2019;15:49–60. [DOI] [PubMed] [Google Scholar]
- 14.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–1232. [Google Scholar]
- 16.Burges CJC. From RankNet to LambdaRank to LambdaMART: an overview. Learning. 2010;11:23–581. [Google Scholar]
- 17.Chen Tianqi, Guestrin Carlos. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. August 13–17, 2016, San Francisco, CA. [Google Scholar]
- 18.Piccirillo JF, Tierney RM, Costas I, Grove L, Spitznagel EL Jr. Prognostic importance of comorbidity in a hospital-based cancer registry. JAMA. 2004;291:2441–2447. [DOI] [PubMed] [Google Scholar]
- 19.Baccarani M, Cortes J, Pane F. et al. ; European LeukemiaNetChronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27:6041–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Proceedings of the 31st International Conference on Neural Information Processing Systems. Long Beach, CA: Curran Associates Inc.; 2017:4768–4777. [Google Scholar]
- 21.Shapley LS. A value for n-person games. Ann Mathemat Stud. 1953; 28:307–317. [Google Scholar]
- 22.Saussele S, Krauss MP, Hehlmann R. et al. ; Schweizerische Arbeitsgemeinschaft für Klinische Krebsforschung and the German CML Study GroupImpact of comorbidities on overall survival in patients with chronic myeloid leukemia: results of the randomized CML study IV. Blood. 2015;126:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahlen T, Edgren G, Lambe M, et al. Cardiovascular events associated with use of tyrosine kinase inhibitors in chronic myeloid leukemia: a population-based cohort study. Ann Intern Med. 2016;165:161–166. [DOI] [PubMed] [Google Scholar]
- 24.Jain P, Kantarjian H, Boddu PC, et al. Analysis of cardiovascular and arteriothrombotic adverse events in chronic-phase CML patients after frontline TKIs. Blood Adv. 2019;3:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weatherald J, Chaumais MC, Montani D. Pulmonary arterial hypertension induced by tyrosine kinase inhibitors. Curr Opin Pulm Med. 2017;23:392–397. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki K, Kantarjian HM, O’Brien S, et al. Incidence of second malignancies in patients with chronic myeloid leukemia in the era of tyrosine kinase inhibitors. Int J Hematol. 2019;109:545–552. [DOI] [PubMed] [Google Scholar]
- 27.Binder PS, Peipert JF, Kallogjeri D, et al. Adult comorbidity evaluation 27 score as a predictor of survival in endometrial cancer patients. Am J Obstet Gynecol. 2016;215:766.e761–766.e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonelli M, Wiebe N, Straus S. et al. ; for the Alberta Kidney Disease NetworkMultimorbidity, dementia and health care in older people:a population-based cohort study. CMAJ Open. 2017;5:E623–e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 30.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ting DSW, Cheung CYL, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 33.Pfirrmann M, Baccarani M, Saussele S, et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia. 2016;30:48–56. [DOI] [PubMed] [Google Scholar]
- 34.Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood. 1984;63:789–799. [PubMed] [Google Scholar]
- 35.Hasford J, Pfirrmann M, Hehlmann R, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst. 1998;90:850–858. [DOI] [PubMed] [Google Scholar]
- 36.Hasford J, Baccarani M, Hoffmann V, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: the EUTOS score. Blood. 2011;118:686–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The University of Texas MD Anderson Cancer Center is committed to providing qualified scientific collaborators access to anonymized patient-level data for the purpose of conducting legitimate clinical research upon the approval of institutional review board at MD Anderson Cancer Center. To protect the rights and privacy of patients on clinical trials, external researchers will be evaluated for the ability of fulfilling suggested collaborative projects for the qualification of data access.


