Abstract
Context.—
Evaluation of HER2 gene amplification by fluorescence in situ hybridization (FISH) was changed by recent American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) guidelines.
Objective.—
To determine frequencies and assess patterns of HER2 protein expression for each ASCO-CAP guideline FISH category among 7526 breast cancers accrued to our consultation practice.
Design.—
We retrospectively reevaluated the HER2 FISH status of breast cancers in our consultation practice according to ASCO-CAP FISH guidelines, and documented HER2 protein levels in each category.
Results.—
According to new guidelines, 17.7% of our consultation breast cancers were “ISH-positive” with HER2:CEP17 FISH ratios ≥2.0 and average HER2 gene copies per cell ≥4.0 (group 1); 0.4% were “ISH-positive” with ratios ≥2.0 and average copies <4.0 (group 2); 0.6% were “ISH-positive” with ratios <2.0 and average copies ≥6.0 (group 3); 4.6% were “ISH-equivocal” with ratios <2.0 and average copies ≥4.0 and <6.0 (group 4); and 76.7% were “ISH-negative” with ratios <2.0 and average copies <4.0 (group 5). However, only groups 1 (HER2 amplified) and 5 (HER2 not amplified) agreed with our previously reported status, and only these groups demonstrated the expected immunohistochemistry status, over-expression and low expression, respectively. Groups 2 and 4 breast cancers lacked overexpression, whereas group 3 was not significantly associated with either increased or decreased HER2 expression.
Conclusions.—
Although the status of approximately 95% of our cases (groups 1 and 5) is not affected by the new guidelines, those of the other 5% (groups 2–4) conflict with previous HER2 gene amplification status and with HER2 status by immunohistochemistry.
The human epidermal growth factor receptor type 2 (HER2) gene, also known as the neu oncogene and avian erythroblastosis oncogene B2 (ERBB2), was first shown to be amplified in approximately 20% to 30% of human breast cancer specimens by Southern hybridization using arginase (ARG1) as an internal control gene and with a HER2:ARG1 ratio greater than or equal to 2.0 defined as gene amplification.1 Those women whose breast cancers contained HER2 amplification, especially those whose cancers had greater than 5-fold amplification, had worse disease-free survival (DFS) and overall survival (OS).1 Additional studies with other control genes—for example, myeloperoxidase (MPO)—used the same ratio of 2.0 or greater for amplification, and also found an association with worse DFS and OS.2 In addition, a direct relationship between HER2 amplification and increased expression (overexpression) of mRNA and protein was demonstrated.2,3 These early studies of HER2 gene amplification, performed with Southern hybridization, required frozen tissue. Because clinical specimens are available predominantly as fixed, paraffin-embedded tissue blocks, the development of alternative techniques, such as fluorescence in situ hybridization (FISH), was highly desirable for evaluation of this alteration in clinical samples. Early studies of HER2 by FISH3–6 compared the number of HER2 gene signals per tumor cell nucleus with the number of copies of an internal control on the same chromosome, predominantly chromosome 17 centromere (CEP17), to assess amplification using the same 2.0 HER2 ratio, as previously established by Southern hybridization, which similarly showed the correlation of amplification with shorter DFS and OS in breast cancer patients.6
Clinical trials of HER2-targeted therapies, including initial studies of trastuzumab7–11 and lapatinib12 as well as subsequent trials of pertuzumab13,14 and trastuzumab emtansine,15 selected women whose breast cancers had HER2 overexpression, as determined by immunohistochemistry (IHC 3+), or HER2 gene amplification, as determined by FISH (HER2:CEP17 ratio ≥2.0), as eligible for trial enrollment. These trials all demonstrated significant improvements in progression-free survival or DFS/OS for trial participants receiving targeted therapy in addition to standard chemotherapy regimens.
New guidelines for HER2 testing have been recently formulated by a committee that represents the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP). Although there were few recommendations for changes in the established standards for IHC testing, significant changes were made in guidelines for HER2 testing by FISH. However, no data were provided with these guidelines regarding the impact of these changes or the proportion of breast cancer patients whose status could be affected by these changes. We retrospectively reevaluated HER2 gene amplification and expression status in our consultation practice to determine what proportion of breast cancers are in each of these ASCO-CAP guidelines’ categories and to assess the patterns of protein overexpression for each of these categories.
MATERIALS AND METHODS
Study Cohort
All primary breast cancer cases accrued to our University of Southern California (Los Angeles, California) Breast Cancer Analysis Laboratory consultation practice from April 1999 until September 2015 that had both HER2 gene amplification status determined by FISH and HER2 protein level determined by IHC were eligible for inclusion in this study. Metastatic breast cancers and other primary malignancies analyzed for HER2 status were excluded. We also excluded primary breast cancer cases that were not assigned a unique HER2 gene amplification status, particularly 86 breast cancers with HER2 genomic heterogeneity.16 These 86 cases with HER2 genomic heterogeneity (1.1%) in our practice during this period were not included because they could not be uniquely assigned to an individual gene amplification category. These cases were considered to have both HER2-amplified and HER2–not amplified geographically separate areas of breast carcinoma present in the same biopsy specimen, as we have described elsewhere.16,17 The University of Southern California Institutional Research Board approved analyses of the laboratory data. All HER2 FISH and IHC results were evaluated by a board-certified pathologist.
HER2 Gene Amplification by FISH
HER2 gene amplification was determined using the Abbott-Molecular Inc (Des Plaines, Illinois), formerly Vysis Inc (Downers Grove, Illinois), PathVysion HER2 FISH assay, according to the package insert as described.16–18 Our laboratory’s interpretation of these FISH results was initially based on an approach submitted to the US Food and Drug Administration (FDA) as part of the approval process for the Oncor INFORM HER FISH assay using a ratio of average HER2 gene copy number divided by the average CEP17 determined in at least 20 nonoverlapping tumor cell nuclei, which was a modification permitted from an original requirement of 100 tumor cell nuclei.6
Those breast cancers with a ratio greater than or equal to 2.0 were considered “amplified,” with one exception. Because patients with primary, node-negative invasive breast cancers having an average HER2 gene copy number per nucleus less than 4.0 had an OS and a DFS indistinguishable from those whose breast cancers lacked HER2 gene amplification,6 we also required an average HER2 gene copy number greater than or equal to 4.0 to be classified as “amplified” as described elsewhere.19
Breast cancers with an average HER2 copy number per tumor cell to CEP17 ratio that was lower than 2.0 were considered “not amplified.”17,19 However, breast cancers that had a HER2 FISH ratio lower than 2.0 but an average HER2 gene copy number of 6.0 or greater, especially those greater than 8.0, were evaluated with alternative control probes to exclude the possibility that CEP17 alpha satellite DNA had become part of the HER2 amplicon, leading to increased CEP17 copy number with a consequential reduced ratio. Such cases were considered to have actual HER2 amplification, if use of an alternative control gene on the same chromosome led to a HER2 FISH ratio greater than or equal to 2.0. A formalized strategy using alternative control probes on the same chromosome, described by Troxell and others,20 is the approach we prefer.21
We do not have an “equivocal” interpretation.19 We resolve the status of breast cancers with HER2 FISH ratios within 10% of the 2.0 cutoff as required by the FDA and the manufacturer’s package insert. This involves the scoring at least 40 additional tumor cell nuclei by each of two different clinical laboratory scientists with reassessment of the FISH ratio; those having a ratio lower than 2.0 in both separate assessments are considered not amplified.
Our original HER2 status was compared with the HER2 status required by the new ASCO-CAP guidelines,22 and also with expression of HER2 protein determined by IHC.
HER2 Protein Expression by IHC
HER2 protein expression was determined with one of two different immunohistochemical assays: our laboratory-developed 10H8 HER2 assay17,18 or the FDA-approved HercepTest (Dako, Carpinteria, California).16,18 The HercepTest is used for those referred cases requesting a determination of HER2 protein by IHC, whereas the 10H8-HER2 IHC assay is used routinely for most cases. Although HER2 expression by 10H8-IHC is significantly more accurate for assessment of HER2 status as determined with molecularly characterized specimens,18 these two IHC assays provide similar categorization of expression levels—with 10H8-IHC 3+ showing a stronger association with HER2 gene amplification by FISH—but a slightly higher false-negative (IHC 0 or 1+ with gene amplification) rate, as described elsewhere.16–19 The subjective microscopic scoring of tumor cells is interpreted similarly, with both assays scored 0, 1+, 2+, and 3+, depending on the proportion of tumor cells and the intensity of immunostaining, as described in the package insert and elsewhere.16–19
Statistical Methods
Counts and percentages were used to summarize the study data. Within each of the 5 ASCO-CAP FISH groups, the χ2 test for goodness of fit was used to test the hypothesis of equal proportions in each of the 4 IHC categories, that is, 0, 1+, 2+, and 3+. Asymptotic distribution of the Wald statistic for the Spearman correlation coefficient23 was used to test the association of HER2 FISH ratio and average HER2 gene copy number per tumor cell.23
RESULTS
Distribution of Primary Breast Cancers According to ASCO-CAP FISH Groups
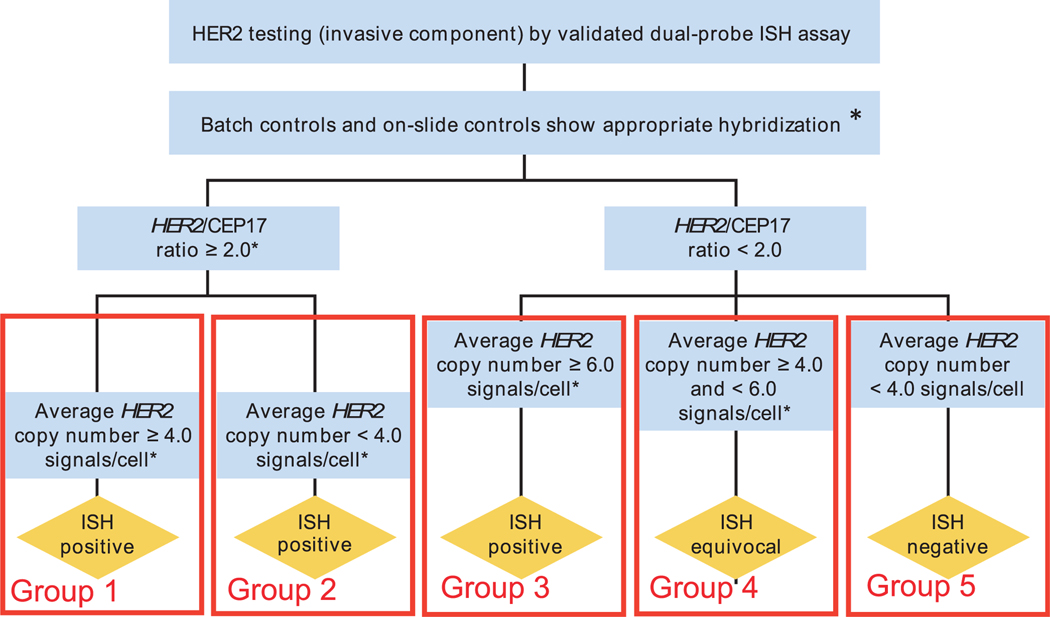
The current ASCO-CAP guidelines22 assess HER2 gene amplification status by using a formalized separation of breast cancer into 5 different groups, with 3 of these groups designated as “in situ hybridization (ISH) positive,” 1 as “ISH equivocal,” and 1 as “ISH negative” (Figure 1). Breast cancers with HER2:CEP17 ratios greater than or equal to 2.0 are divided in two groups: one with an average HER2 gene copy number per tumor cell of 4.0 or greater (group 1), and one with an average HER2 gene copy number per tumor cell lower than 4.0 (group 2)—both classified as “ISH positive.” Breast cancers with HER2:CEP17 ratios lower than 2.0 are separated into 3 additional groups: one with an average HER2 gene copy number per tumor cell of 6.0 or greater (group 3), also classified as “ISH positive”; another with an average HER2 gene copy number per tumor cell of 4.0 or greater but lower than 6.0 (group 4), classified as “ISH equivocal”; and one with breast cancers containing an average HER2 gene copy number per tumor cell lower than 4.0 (group 5), classified as “ISH negative.” Therefore, according to the ASCO-CAP guidelines,22 breast cancers in groups 1, 2, and 3 are interpreted as “ISH positive,” group 4 as “ISH equivocal,” and group 5 as “ISH negative” (Figure 1). Breast cancers in ISH group 4 must be subjected to a reflex test, such as IHC (same specimen), or tested with an alternative ISH chromosome 17 probe, such as retinoic acid receptor alpha (RARA),20 or a new test should be performed with a new specimen, if available, to complete assessment of HER2 status.22
Figure 1.
Schematic diagram of the “Algorithm for evaluation of human epidermal growth factor receptor 2 (HER2) gene amplification by in situ hybridization (ISH) assay of the invasive component of a breast cancer specimen using a dual-signal (HER2 gene) assay (dual-probe ISH)” as published by the American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) guidelines committee,22 modified here by introduction of the groups 1 to 5 to identify the various ASCO-CAP fluorescence in situ hybridization (FISH) categories used in this analysis. Breast cancers with HER2:CEP17 ratios of 2.0 or greater are divided into two groups: one with an average HER2 gene copy number per tumor cell greater than/equal to 4.0 (ISH-positive, or our group 1) and one with an average HER2 gene copy number per tumor cell lower than 4.0 (ISH-positive, or our group 2). Breast cancers with HER2:CEP17 ratios lower than 2.0 are separated into three additional groups: one with an average HER2 gene copy number per tumor cell of 6.0 or greater (ISH-positive, or our group 3 [N or A]); another with an average HER2 gene copy number per tumor cell of 4.0 or greater but less than 6.0 (ISH-equivocal, or our group 4); and one with breast cancers containing an average HER2 gene copy number per tumor cell lower than 4.0 (ISH-negative, or our group 5). Therefore, according to the ASCO-CAP guidelines,22 breast cancers in groups 1, 2, and 3 are interpreted as “ISH positive,” group 4 as “ISH equivocal,” and group 5 as “ISH negative.” This figure has been modified from Figure 3 of the previously published article by Wolff et al,22 Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256, with permission from the Archives of Pathology & Laboratory Medicine. Copyright 2014 College of American Pathologists. (*) Observed in a homogeneous and contiguous population.
Accordingly, of the 7526 primary breast cancers analyzed in this report, 1328 had a HER2 FISH ratio of 2.0 or higher and an average HER2 gene copy number per tumor cell of 4.0 or greater (1328 of 7526; 17.7%); 31 had a HER2 FISH ratio of 2.0 or higher but an average HER2 gene copy number per tumor cell lower than 4.0 (31 of 7526; 0.4%); 48 had a HER2 FISH ratio lower than 2.0 and an average HER2 gene copy number per tumor cell of 6.0 or greater (48 of 7526; 0.6%); 345 had a HER2 FISH ratio lower than 2.0 but an average HER2 gene copy number per tumor cell of 4.0 or greater but lower than 6.0 (345 of 7526; 4.6%); and 5774 had a HER2 FISH ratio lower than 2.0 but an average HER2 gene copy number per tumor cell lower than 4.0 (5774 of 7526; 76.7%; Figures 2 through 4; Table 1). Because HER2 gene amplification is correlated with HER2 protein overexpression, we also characterized each ASCO-CAP FISH group according to the distribution of HER2 protein expression determined by IHC with two different IHC assays used routinely in our laboratory.
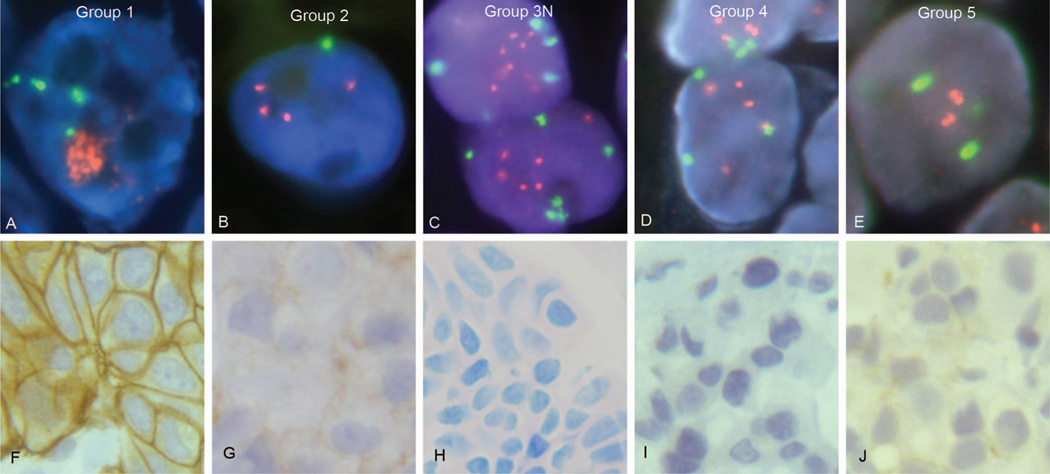
Figure 2.
Fluorescence in situ hybridization (FISH) for HER2 gene status (A through E) and immunohistochemical staining for HER2 protein status (F through J) are illustrated with cases representing each of the American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) guidelines for in situ hybridization (ISH) algorithm groups 1 to 5. The ASCO-CAP guidelines’ algorithm ISH groups are compared with observed HER2 gene amplification status by FISH, and HER2 protein expression status by immunohistochemical staining with either the Dako HercepTest or our laboratory-developed 10H8 immunohistochemistry (IHC) assay. A, ASCO-CAP group 1 breast cancer with HER2 gene amplification by FISH, consistent with the ASCO-CAP guidelines’ designation of “ISH positive” (and consult practice designation of “HER2 amplified”). The average HER2 gene copy number for this case was 28.8 copies per tumor cell, with an average of 4.45 chromosome 17 centromere (CEP17) copies per tumor cell and a HER2:CEP17 FISH ratio of 6.47. The HER2 signals are sufficiently numerous and are not captured in a single plane of focus in this photomicrograph, so they appear here as orange clusters of intranuclear signals. Consultation case number C17984. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. B, ASCO-CAP group 2 breast cancer, previously reported in consultation with a lack of HER2 gene amplification by FISH, contradicts the ASCO-CAP guidelines’ designation of “ISH positive.” The average HER2 gene copy number for this breast cancer was 3.4 copies per tumor cell, with an average of 1.2 CEP17 copies per tumor cell and a HER2:CEP17 FISH ratio of 2.8. Consultation case number C20890. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. C, ASCO-CAP group 3 breast cancer, one of our “group 3N” cases, was reported to have a lack of HER2 gene amplification by FISH in consultation, contrary to the ASCO-CAP guidelines’ designation of “ISH positive.” This breast cancer had an average of 6.6 HER2 gene copies per tumor cell and an average of 3.9 CEP17 copies, with a HER2:CEP17 FISH ratio of 1.69. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. Consultation case number C18756. D, ASCO-CAP group 4 breast cancer, designated as “ISH equivocal” by ASCO-CAP but reported in our consultation practice as “HER2 not amplified” by FISH. This breast cancer had an average HER2 gene copy number of 5.3 HER2 gene copies per tumor cell, an average CEP17 copy number of 3.0 per tumor cell, and therefore had a HER2:CEP17 FISH ratio of 1.77. The use of retinoic acid receptor alpha (RARA) gene probe as an alternative CEP17 control demonstrated an average of 3.3 RARA copies per tumor cell, providing a HER2:RARA ratio of 1.6. Similarly, using the Smith-Magenis Syndrome (SMS) region FISH probe as an alternative control, there were 2.9 copies per tumor cell, providing a HER2:SMS ratio of 1.8. Consultation case number C18137. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. E, ASCO-CAP group 5 breast cancer, consistent with the guidelines’ designation of “ISH negative,” which was reported as “HER2 not amplified” by FISH in our consultation practice. The case had an average HER2 gene copy number of 2.65 per tumor cell, a CEP17 average of 2.05 copies per tumor cell, and a HER2:CEP17 ratio of 1.29. Consultation case number C18066. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. F, ASCO-CAP group 1 breast cancer case with HER2 protein overexpression, IHC 3+, both by the Dako HercepTest (illustrated) and our laboratory-developed 10H8-HER2 (data not shown) immunohistochemical assays. This breast cancer, corresponding to A above, is an ASCO-CAP FISH group 1 case consistent with the ASCO-CAP guidelines’ designation of “ISH positive.” Consultation case number C17984. G, ASCO-CAP group 2 breast cancer, corresponding to the breast cancer in B above, with HER2 protein expression determined as IHC 1+ with the HercepTest (illustrated) and the 10H8-HER2 IHC assay (not shown), contradicts the ASCO-CAP guidelines’ designation of “ISH positive.” Consultation case number C20890. H, ASCO-CAP group 3 breast cancer with low HER2 protein expression by the HER2 10H8-IHC (IHC 0) immunohistochemical assay. This breast cancer, corresponding to C above, was reported as not amplified, contrary to the ASCO-CAP guidelines’ designation of “ISH positive.” Consultation case number C18756. I, ASCO-CAP group 4 breast cancer, corresponding to D above, had low HER2 protein expression by both the 10H8-IHC HER2 assay (IHC 0, data not shown) and the Dako HercepTest (IHC 1+), as illustrated. Consultation case number C18137. J, ASCO-CAP group 5 breast cancer, corresponding to E above, with low HER2 protein expression by IHC with both the Dako HercepTest (IHC 1+, as illustrated) and 10H8-IHC (IHC 0, data not shown), consistent with the ASCO-CAP guidelines’ designation of “ISH negative.” Consultation case number C18066. A normal immunoglobulin G (IgG)–negative control was performed for each of the immunohistochemical assays used in F through J and showed a lack of any staining; however, these have not been illustrated (original magnifications ×1000 [A through E] and ×400 [F through J]).
Table 1.
Consultation Cases Arranged According to American Society of Clinical Oncology/College of American Pathologists Guidelines’ Fluorescence In Situ Hybridization (FISH) Groups
| Group | Description of FISH Category | No. (%) of Cases |
|---|---|---|
|
| ||
| 1 | Ratio ≥2.0, HER2 average ≥4.0 | 1328 (17.7) |
| 2 | Ratio ≥2.0, HER2 average <4.0 | 31 (0.4) |
| 3 | Ratio <2.0, HER2 average ≥6.0 | 48 (0.6) |
| 4 | Ratio <2.0, HER2 average ≥4.0 and <6.0 | 345 (4.6) |
| 5 | Ratio <2.0, HER2 average <4.0 | 5774 (76.7) |
| Total | 7526 (100.0) | |
Association of Average HER2 Gene Copy Number per Tumor Cell as Well as HER2 FISH Ratios With HER2 Protein Expression
As expected, among the 7526 breast cancers in this study there was a significant association between increasing average HER2 gene copy number per tumor cell determined by FISH and increasing HER2 protein expression determined by IHC (P=.01; Table 2). Likewise, there was also a significant association between increasing HER2 FISH ratios (ratio of average HER2 gene copy number per tumor cell to CEP17) and increasing HER2 protein expression determined by IHC (P= .009; Table 2).
Table 2.
Comparison of Average HER2 Gene Copy Numbers per Tumor Cell and HER2 Fluorescence In Situ Hybridization (FISH) Ratios With HER2 Protein Expression by Either HercepTest or 10H8 Immunohistochemistry (IHC) Scores in 7526 Breast Cancers
| Association of Average HER2 Gene Copy Number and HER2 FISH Ratios With HER2 Protein by IHC Scores | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| HER2 FISH Assay Results | IHC Score, No. (%)a | ||||||
|
|
|
||||||
| HER2 FISH Ratios | Average HER2 Copy Number per Cell | IHC 0 | IHC 1+ | IHC 2+ | IHC 3+ | Totals, No. (%) | P Valueb |
|
| |||||||
| — | <4.0 | 3612 (62.2) | 1977 (34.1) | 204 (3.5) | 12 (0.2) | 5805 (77.1) | .01 |
| — | 4.0–5.9 | 202 (37.2) | 229 (42.2) | 102 (18.8) | 10 (1.8) | 543 (7.2) | |
| — | 6.0–7.9 | 44 (26.4) | 46 (27.5) | 60 (35.9) | 17 (10.2) | 167 (2.2) | |
| — | 8.0–9.9 | 15 (12.9) | 28 (24.1) | 47 (40.5) | 26 (22.4) | 116 (1.5) | |
| — | ≥10.0 | 16 (1.8) | 54 (6.0) | 259 (28.9) | 566 (63.2) | 895 (11.9) 7526 | |
| <2.0 | — | 3753 (60.9) | 2157 (35.0) | 238 (3.9) | 19 (0.3) | 6167 (81.9) | .009 |
| 2.00–5.0 | — | 119 (22.1) | 122 (22.6) | 213 (39.5) | 85 (15.8) | 539 (7.2) | |
| 5.01–10.0 | — | 17 (2.8) | 40 (6.6) | 177 (29.2) | 372 (61.4) | 606 (8.1) | |
| >10.0 | — | 0 (0) | 15 (7.0) | 44 (20.6) | 155 (72.4) | 214 (2.8) | |
When data from both HER2 immunohistochemical assays, 10H8 and HercepTest, were available, the 10H8 assay result was used.
P value based on the asymptotic distribution of the Wald statistic for the Spearman correlation coefficient.24
HER2 Protein Expression by IHC According to ASCO-CAP FISH Groups
At least one immunohistochemical assay result was available for 7526 breast cancers, 7029 by 10H8 IHC, 693 by HercepTest IHC, and 196 by both IHC assays. Similar significant associations were demonstrated with each IHC assay individually or with both IHC assays combined as presented here. The distribution of combined IHC results by ASCO-CAP guidelines group is summarized below and in Table 3.
Table 3.
American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) Fluorescence In Situ Hybridization Groupings Compared with HER2 Protein by Immunohistochemistry (IHC) Scores
| ASCO-CAP Group | HER2/CEP17 Ratio | Average HER2 Copy Number per Cell | HER2 Protein Expression by 10H8 or HercepTest IHCa | Totals of Each Group, No. (% of the Total Cases) | P Valueb | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| IHC 0, No. (%) | IHC 1+, No. (%) | IHC 2+, No. (%) | IHC 3+, No. (%) | |||||
|
| ||||||||
| Group 1 | ≥2.0 | ≥4.0 | 123 (9.3) | 167 (12.6) | 427 (32.2) | 611 (46.0) | 1328 (100/17.6) | <.001 |
| Group 2 | ≥2.0 | <4.0 | 13 (41.9) | 10 (32.3) | 8 (25.8) | 0 (0) | 31 (100/0.4) | .02 |
| Group 3 | <2.0 | ≥6.0 | 15 (31.3) | 22 (45.8) | 7 (14.6) | 4 (8.3) | 48 (100/0.6) | <.001 |
| Group 4 | <2.0 | ≥4.0 and <6.0 | 139 (40.3) | 168 (48.7) | 35 (10.1) | 3 (0.9) | 345 (100/4.6) | <.001 |
| Group 5 | <2.0 | <4.0 | 3599 (62.3) | 1967 (34.0) | 197 (3.4) | 11 (0.2) | 5774 (100/76.7) 7526 | <.001 |
| Average, Mean ± SD | ||||||||
|
| ||||||||
| Group 3A | <2.0 | 12.3 ± 6.6 | 1 (12.5) | 1 (12.5) | 2 (25.0) | 4 (50.0) | 8 (100/16.7) | .39 |
| Group 3N | <2.0 | 6.8 ± 0.9 | 14 (35.0) | 21 (52.5) | 5 (12.5) | 0 (0.0) | 40 (100/83.3) 48 | <.001 |
When data from both HER2 immunohistochemical assays, 10H8 and HercepTest, were available, the HercepTest assay result was used.
P value based on χ2 test for goodness of fit test of the hypothesis of equal proportions in each of the 4 IHC categories.
Group 1 (Ratio .>2.0, Average HER2 ≥4.0; “ISH Positive”).—
The vast majority (1038 of 1328; 78.2%) of breast cancers in this group had IHC 2+ or IHC 3+ immunostaining when analyzed by either 10H8 or Her-cepTest IHC (Table 2). Breast cancers with IHC 0/1+ were less frequent (290 of 1328; 21.8%). Among these breast cancers, previously reported as “HER2 amplified” in our consultation practice, there was a statistically significant increased frequency of tumors with IHC 2+/3+ status (P < .001; group 1 in Figures 1 and 2; Table 3).
Group 2 (Ratio .>2.0, Average HER2 <4.0; “ISH Positive”).—
In contrast to ASCO-CAP group 1 above, most group 2 breast cancers had IHC 0 or IHC 1+ immunostaining (23 of 31; 74.2%), with relatively few showing IHC 2+ (8 of 31; 25.8%) and none (0%) showing IHC 3+ immunostaining (Table 3). In these breast cancers, previously reported as “HER2 not amplified” through our consultation practice, there was a statistically significant increased frequency of tumor with IHC 0/1+low-expression status (P= .02; group 2 in Figures 1 and 2; Table 3).
Group 3 (Ratio <2.0, Average HER2 >6.0; “ISH Positive”).—
Among the 48 breast cancers in this group, most (37; 77.1%) had IHC 0 or 1+ low HER2 expression; however, 4 (8.3%) were IHC 3+(P, .001; Figure 3; Table 3). Breast cancers in this group had been reported as either “HER2 amplified” (group 3A; 8 of 48; 16.7%; Table 3) or as “HER2 not amplified” (group 3N; 40 of 48; 83.3%; Table 3). Separation of these cases according to the reported HER2 amplification status by FISH demonstrated that those reported as “amplified” (16.7%) had on average 12.3 HER2 gene copies per tumor cell, whereas those reported as “not amplified” (83.3%) had on average only 6.8 HER2 gene copies per tumor cell. In addition, the 8 breast cancers originally reported as HER2 amplified (Figure 3; Table 3, group 3A) were predominantly (75%) IHC 2+ or 3+, whereas the 40 reported as HER2 not amplified (Table 3, group 3N), using alternative control gene probes on chromosome 17 with previously described criteria,17,20,21 were predominantly (87.5%) IHC 0/1þ, with no IHC 3+cases (Figures 1 and 2, C and H [group 3N]). These subgroup distributions were different from one another in terms of HER2 protein expression by IHC (P <.001; Table 3). Although the “amplified” (group 3A) breast cancers were predominantly (75%) associated with IHC 2+/3+ immunostaining, the association was not statistically significant from the perspective of a deviation from an equal distribution across all 4 IHC categories, probably related to the small sample size (n = 8). This was consistent with the reported status of these two different subgroups but was inconsistent with the ASCO-CAP guidelines’ assignment of all of these cases to a single “ISH-positive” category (group 3).
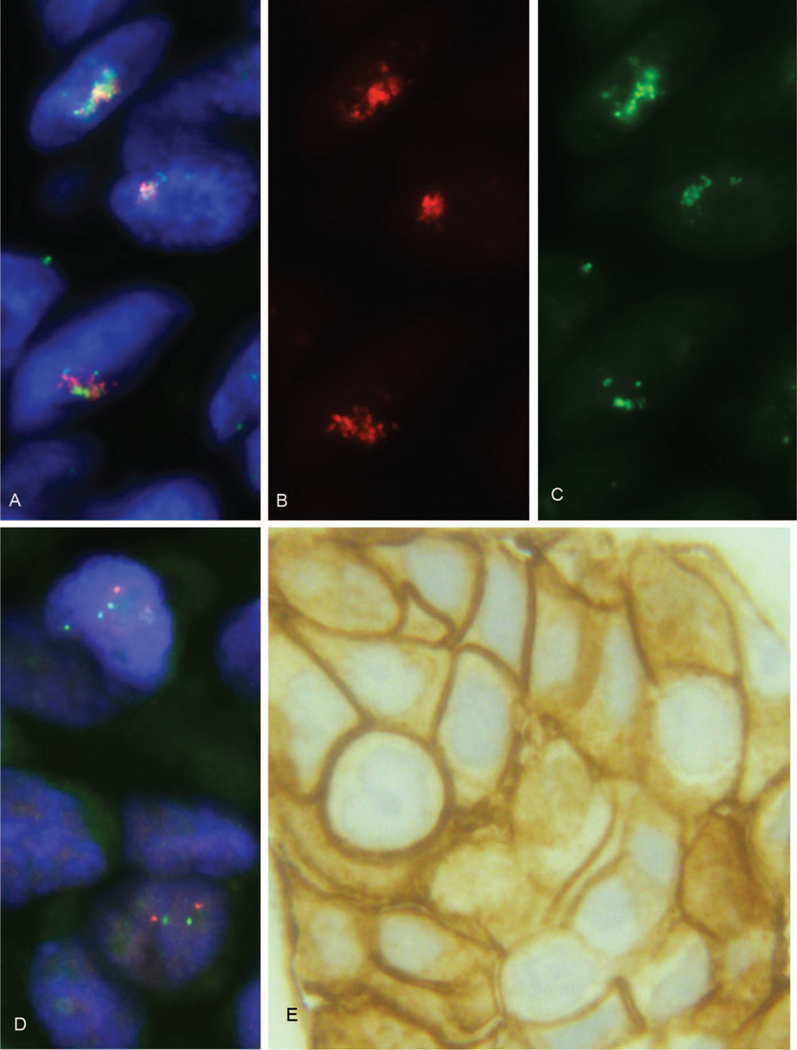
Figure 3.
A minority of American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) group 3 breast cancers, referred to here as “group 3A,” show HER2 gene amplification and HER2 protein overexpression. A, ASCO-CAP group 3 breast cancer, one of our group 3A cases. This breast cancer has an average HER2 gene copy number of 23.2 per tumor cell and an average chromosome 17 centromere (CEP17) copy number of 15.75 per tumor cell, and it therefore has a HER2 FISH ratio of only 1.47. This triple bandpass image shows the composite (blue/orange/green) image with HER2 gene copies (orange) and CEP17 copies (green) arranged together in a limited geographic area of tumor cell nuclei (blue). Please note that the HER2 gene signals (orange) and CEP17 signals (green) are aggregated together in this same limited geographic area of the nuclei, making assessment of individual signals challenging without the aid of single bandpass filters, as illustrated in B and C. HER2 gene (orange) and CEP17 (green) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. Consultation case number C20906. B, This single bandpass image (orange filter) shows the distribution of HER2 gene copies in the same tumor cell nuclei illustrated in A. HER2 gene copies (orange) are identified using the Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. Consultation case number C20906. C, This single bandpass image (green filter) shows the distribution of alpha satellite DNA of CEP17 (green) in the same tumor cell nuclei illustrated in A. Abbott-Molecular PathVysion HER2 DNA probe kit (Vysis LSI HER-2/neu SpectrumOrange/CEP17 SpectrumGreen) FISH assay. Consultation case number C20906. D, FISH of alternative control probes located on chromosome 17 remote from the HER2 locus. The use of retinoic acid receptor alpha (RARA) gene probe (green) in this breast cancer demonstrated an average of 2.55 RARA copies per tumor cell by FISH, providing a HER2:RARA ratio of 9.1. Similarly, using the Smith-Magenis Syndrome (SMS) region FISH probe (orange) as an alternative control gene probe there were 1.85 copies per tumor cell, providing a HER2:SMS ratio of 12.54. This breast cancer was reported as “HER2 amplified” in our consultation practice, consistent with the ASCO-CAP guidelines’ designation of “ISH positive.” RARA gene (green) and SMS (orange) are identified using the Abbott-Molecular Vysis Smith-Magenis Region LSI SMS SpectrumOrange/RARA SpectrumGreen probes. Consultation case number C20906. E, ASCO-CAP group 3 breast cancer, corresponding to our “group 3A” cases, with HER2 protein overexpression by immunohistochemistry (IHC; IHC 3+by HercepTest) consistent with the ASCO-CAP guidelines’ designation of “ISH positive.” Similar results were obtained with our 10H8-IHC assay (IHC 3+, data not shown). Consultation case number C20906 (original magnifications ×1000 [A through D] and ×400 [E]).
Group 4 (Ratio <2.0, Average HER2 .>4.0 and <6.0; “ISH Equivocal”).—
Similar to group 2 above and group 5 below, few breast cancers in ASCO-CAP group 4 had IHC 2+(n=35; 10.1%) or IHC 3+(n=3; 0.9%) immunostaining, whereas most (n = 307; 89%) had IHC 1+ or IHC 0 immunostaining (group 4 in Figures 1 and 2, D and I; Table 3). This group, like groups 2 and 5, was significantly (P =.001) associated with IHC 0/1+ low–HER2 expression status (Figures 1 [group 4] and 2, D and I). None of the breast cancers in this ISH group were previously reported by us to be HER2 “equivocal” by FISH in our consultation practice. All were reported as “HER2 not amplified.”
Group 5 (Ratio <2.0, Average HER2 <4.0; “ISH Negative”).—
As expected, most breast cancers in this group had IHC 0 or IHC 1+ immunohistochemical staining status (n = 5566; 96.4%; Table 3), whereas only 197 (3.4%) showed IHC 2+ immunostaining and fewer (n = 11; <1%) had IHC 3+ scores (Figures 1 [group 5] and 2, E and J). This ASCO-CAP FISH group of breast cancers had all been reported as “HER2 not amplified” in our practice and were significantly associated with low expression (IHC 0/1+; P <.001; Table 3).
DISCUSSION
Amplification and overexpression of HER2 is an established therapeutic target in breast carcinomas. Although mutations24 in the HER2 gene may prove to be responsive to small-molecule kinase inhibitors, currently patients whose breast carcinomas have HER2 amplification/overexpression are the only patients approved by regulatory authorities to be selected for HER2-targeted therapies. Several companion diagnostic assays have been approved to characterize breast cancers’ HER2 status. Some companion diagnostic tests, especially IHC assays, are reported to have variability in terms of test accuracy.18,25 This variability led the ASCO and the CAP to convene a panel to make recommendations related to HER2 testing, including the interpretation of assay results. The ASCO-CAP guidelines, initially published in 2007,25 have been recently revised.22
Previously, the ASCO-CAP guidelines required a change in the HER2:CEP17 ratio from the FDA-approved FISH ratio of 2.0 to 2.2 for the establishment of HER2 gene amplification status. Although the acceptable FISH ratio in the revised, current guideline is, once again, 2.0, the ASCO-CAP committee has now recommended separation of HER2 status into 5 FISH groups based on a combination of the 2.0 FISH ratio and the average HER2 gene copy number per tumor cell. These 5 HER2 FISH groups are then each categorized as HER2 “ISH positive,” HER2 “ISH negative,” or HER2 “ISH equivocal” for patient selection to targeted therapies.22 Because these definitions differ from our own previous decision-making process related to HER2 gene amplification status, we decided to reevaluate the HER2 gene status of patients referred to our consultation practice for determination of HER2 status by FISH. We compared HER2 gene amplification status determined by FISH according to the requirements of the FDA, as modified6,19–21 and summarized in the Materials and Methods, with the current 2013/2014 ASCO-CAP guidelines,22 using 7526 breast cancers from our consultation practice to determine the impact on patient classifications. We found that only 2 of the 5 ASCO-CAP ISH groups—group 1 and group 5—had clear HER2 protein expression patterns in the manner that would be predicted for HER2 “ISH-positive” (overexpression) and HER2 “ISH-negative” (low expression) groups, respectively (Table 4).
Table 4.
Summary of Associations Between American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) In Situ Hybridization (ISH) Guidelines Groups, HER2 Protein Expression, and Diagnostic Status in Our Consult Practice
| ASCO-CAP FISH Group | ASCO-CAP Designation | Predominant Pattern of HER2 Protein Expression | Consult Practice Diagnoses |
|---|---|---|---|
|
| |||
| 1. Ratio ≥2.0, HER2 copies ≥4.0 | ISH positive | HER2 overexpression (IHC 2+/3+); P < .001 | HER2 amplified |
| 2. Ratio ≥2.0, HER2 copies <4.0 | ISH positive | HER2 low expression (IHC 0/1+); P = .02 | HER2 not amplified |
| 3. Ratio <2.0, HER2 copies ≥6.0 | ISH positive | HER2 mixed protein expression | Mixed category of HER2 not amplified and HER2 amplified cases |
| 4. Ratio <2.0, HER2 copies ≥4.0 and <6.0 | ISH equivocal | HER2 low expression (IHC 0/1+); P < .001 | HER2 not amplified |
| 5. Ratio <2.0, HER2 copies <4.0 | ISH negative | HER2 low expression (IHC 0/1+); P < .001a | HER2 not amplified |
Abbreviations: FISH, fluorescence in situ hybridization; IHC, immunohistochemistry.
P-value is based on the chi-square statistic that tests the hypothesis that equal proportions of tumors (0.25 in each) fall into the 4 IHC expression groups.
Group 2, characterized as “ISH-positive” by ASCO-CAP guidelines, was significantly associated with low HER2 expression, as was group 4 (“ISH-equivocal”), which strongly suggests neither of these groups contains cases with HER2 gene amplification, as previously reported6,19 (Table 4).
Group 3—also “ISH-positive” according to the ASCO-CAP guidelines—although significantly associated with low HER2 protein expression (Table 3), contains a small subgroup (approximately 17%) of breast cancers that we have previously characterized as HER2 amplified. This ASCO-CAP FISH group is composed of breast cancers that were previously reported in our consultation practice as HER2 not amplified (40 of 48; 83.3%) as well as others that were HER2 amplified (8 of 48; 16.7%). Because we have considered cases in this group with high HER2 gene copy number and high CEP17 copy number to have a HER2 amplicon that includes alpha-satellite DNA of CEP17, we have routinely used an approach for characterization of HER2 status, described by Troxell et al,20 that involves the use of alternative control gene loci (RARA and Smith-Magenis Syndrome) on chromosome 17 considered sufficiently remote from the HER2 locus to likely be uninvolved in the HER2 amplicon. This strategy leads to designations of HER2 not amplified breast cancers that are significantly correlated with low HER2 protein expression, supporting our interpretation that the ASCO-CAP guidelines’ ISH group 3 is composed of two different subgroups of breast cancer, one HER2 amplified and the other HER2 not amplified.
Although our findings conflict with the ASCO-CAP guidelines for 3 of the 5 FISH groupings (Table 4), the 2 groups for which our FISH and IHC results are concordant represent nearly 95% of breast cancers in our consultation practice, indicating disagreements between our consultation practice diagnoses and the ASCO-CAP guidelines are limited to approximately 5% of breast cancer patients. Because one of the limitations of this study is the lack of long-term clinical follow-up information, additional studies will be required to confirm that the clinical behaviors of groups 2 and 4 are those of HER2 not amplified, HER2 low-expression breast cancer patients, with regard to both prognosis and response to targeted therapy. Likewise, outcome information for group 3 will be required to confirm that this group of breast cancer patients represents both HER2 not amplified, low-expression breast cancer patients and a smaller subpopulation of HER2 amplified, HER2 overexpression breast cancer patients. Nevertheless, our findings raise serious questions about the validity of 3 of the 5 ASCO-CAP guidelines’ FISH groupings newly instituted—contrary to previous classification schemes established by the manufacturers and the FDA—without prior published data.
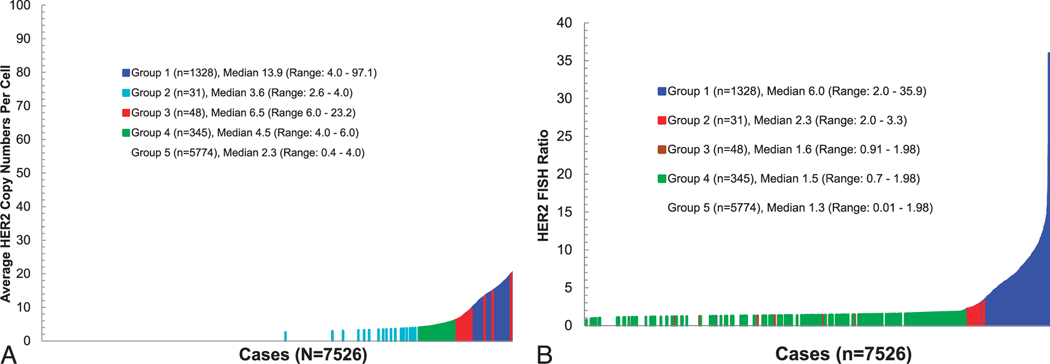
Figure 4.
Distribution of average HER2 gene copy number and HER2 fluorescent in situ hybridization (FISH) ratios among 7526 breast cancers evaluated in an academic private practice. A, Plot of average HER2 gene copy number per tumor cell nucleus from lowest to highest, but with cases identified according to the American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) guidelines as groups 1 (blue), 2 (turquoise), 3 (red), 4 (green), and 5 (no color). Please note that no color was assigned to group 5 cases because doing so in such a large group obscures the colors of some cases from less frequent groups, especially group 2. B, Plot of HER2 FISH ratios arranged from lowest to highest with identification of ASCO-CAP groups 1 (blue), 2 (red), 3 (brown), 4 (green), and 5 (no color). Please note that no color was assigned to group 5 cases because doing so in such a large group obscures the colors of some cases from less frequent groups, especially groups 2, 3 and 4.
Acknowledgments
This study was supported in part by a grant from the Breast Cancer Research Foundation (Dr Press), a grant from the Entertainment Industry Foundation (Dr Press), and a gift from Dr Richard Blach (Dr Press) as well as an endowed chair, the Harold E. Lee Chair for Cancer Research (Dr Press). The project was also supported in part by award number P30CA014089 from the National Cancer Institute (Dr Press, Ms Tsao-Wei, and Dr Groshen). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Note: Another paper addressing the ASCO-CAP guidelines in the HERA trial was published while this paper was in review (Mod Pathol.2015;28:1528-1534).
The authors have no relevant financial interest in the products or companies described in this article. However, Dr Press has collaborated with a competing company on an unrelated project with placement of a loaned machine in his laboratory, the Ventana Benchmark Ultra (Ventana Medical Systems Inc, Tucson, Arizona).
References
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL.Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu protooncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. [DOI] [PubMed] [Google Scholar]
- 3.Pauletti G, Godolphin W, Press MF, Slamon DJ. Detection and quantitationof HER-2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene. 1996;13(1):63–72. [PubMed] [Google Scholar]
- 4.Press MF, Pike MC, Hung G, et al. Amplification and overexpression ofHER-2/neu in carcinomas of the salivary gland: correlation with poor prognosis. Cancer Res. 1994;54(21):5675–5682. [PubMed] [Google Scholar]
- 5.Saffari B, Jones LA, el-Naggar A, Felix JC, George J, Press MF. Amplificationand overexpression of HER-2/neu (c-erbB2) in endometrial cancers: correlation with overall survival. Cancer Res. 1995;55(23):5693–5698. [PubMed] [Google Scholar]
- 6.Press MF, Bernstein L, Thomas PA, et al. HER-2/neu gene amplificationcharacterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol. 1997;15(8):2894–2904. [DOI] [PubMed] [Google Scholar]
- 7.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus amonoclonal antibody against HER2 for metastatic breast cancer that over-expresses HER2. N Engl J Med. 2001;344(11):783–792. [DOI] [PubMed] [Google Scholar]
- 8.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. ; Herceptin Adjuvant(HERA) Trial Study Team. Trastuzumab after adjuvant chemotherapy in HER2positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. [DOI] [PubMed] [Google Scholar]
- 9.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005; 353(16):1673–1684. [DOI] [PubMed] [Google Scholar]
- 10.Slamon D, Eiermann W, Robert N, et al. ; Breast Cancer InternationalResearch Group. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mass RD, Press MF, Anderson S, et al. Evaluation of clinical outcomesaccording to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer. 2005;6(3): 240–246. [DOI] [PubMed] [Google Scholar]
- 12.Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine forHER2-positive advanced breast cancer. N Engl J Med. 2006;355(26):2733–2743. [DOI] [PubMed] [Google Scholar]
- 13.Baselga J, Cortes J, Kim SB, et al. ; CLEOPATRA Study Group. Pertuzumabplus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012; 366(2):109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swain SM, Kim SB, Cortes J, et al. Pertuzumab, trastuzumab, and docetaxelfor HER2-positive metastatic breast cancer (CLEOPATRA study): overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 2013;14(6):461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2positive advanced breast cancer. N Engl J Med. 2012;367(19):1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Press MF, Finn RS, Cameron D, et al. HER-2 gene amplification, HER-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin Cancer Res. 2008;14(23): 7861–7870. [DOI] [PubMed] [Google Scholar]
- 17.Press MF, Sauter G, Bernstein L, et al. Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res. 2005; 11(18):6598–6607. [DOI] [PubMed] [Google Scholar]
- 18.Press MF, Slamon DJ, Flom KJ, Park J, Zhou JY, Bernstein L. Evaluation ofHER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol. 2002;20(14):3095–3105. [DOI] [PubMed] [Google Scholar]
- 19.Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for humanepidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27(8):1323–1333. [DOI] [PubMed] [Google Scholar]
- 20.Troxell ML, Bangs CD, Lawce HJ, et al. Evaluation of Her-2/neu status incarcinomas with amplified chromosome 17 centromere locus. Am J Clin Pathol. 2006;126(5):709–716. [DOI] [PubMed] [Google Scholar]
- 21.Press MF. How is Her-2/neu status established when Her-2/neu andchromosome 17 centromere are both amplified? Am J Clin Pathol. 2006;126(5): 673–674. [DOI] [PubMed] [Google Scholar]
- 22.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for humanepidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31(31):3997–4013. Arch Pathol Lab Med. 2014; 138(2):241–256. [DOI] [PubMed] [Google Scholar]
- 23.Brown MB, Benedetti JK. Sampling behavior of tests for correlation in twoway contingency tables. J Am Stat Assoc. 1977;72(358):309–315. [Google Scholar]
- 24.Wen W, Chen W, Xiao N, et al. Mutations in the kinase domain of theHER2/ERBB2 gene identified in a wide variety of human cancers J Mol Diagn. 2015;17(5):487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. Arch Pathol Lab Med. 2007;131(1):18–43. [DOI] [PubMed] [Google Scholar]