Abstract
Interactions between the neonate host and its gut microbiome are central to the development of a healthy immune system. However, the mechanisms by which animals alter early colonization of microbiota for their benefit remain unclear. Here, we investigated the role of early-life expression of the α2,6-sialyltransferase ST6GAL1 in microbiome phylogeny and mucosal immunity. Fecal, upper respiratory, and oral microbiomes of pups expressing or lacking St6gal1 were analyzed by 16S rRNA sequencing. At weaning, the fecal microbiome of St6gal1-KO mice had reduced Clostridiodes, Coprobacillus, and Adlercreutzia, but increased Helicobacter and Bilophila. Pooled fecal microbiomes from syngeneic donors were transferred to antibiotic-treated wild-type mice, before analysis of recipient mucosal immune responses by flow cytometry, RT-qPCR, microscopy, and ELISA. Transfer of St6gal1-KO microbiome induced a mucosal Th17 response, with expression of T-bet and IL-17, and IL-22-dependent gut lengthening. Early life intestinal sialylation was characterized by RT-qPCR, immunoblot, microscopy, and sialyltransferase enzyme assays in genetic mouse models at rest or with glucocorticoid receptor modulators. St6gal1 expression was greatest in the duodenum, where it was mediated by the P1 promoter and efficiently inhibited by dexamethasone. Our data show that the inability to produce α2,6-sialyl ligands contributes to microbiome-dependent Th17 inflammation, highlighting a pathway by which the intestinal glycosylation regulates mucosal immunity.
Keywords: sialic acid, ST6GAL1, neonatal microbiome, Helicobacter, Th17
1. Introduction
The mammalian gastrointestinal tract is densely colonized by microbes that engage the host in a complex symbiotic relationship. During neonatal life, the gut microbiome transitions from the relative sterility of the womb to a diverse and stable community, reflecting increasing exposure to the environment (Palmer et al. 2007). Disruption of this process by antibiotics, caesarean delivery, or formula feeding results in long-lasting changes to the microbiome, the consequences of which can manifest both locally and systemically (Salminen et al. 2004; Dominguez-Bello et al. 2010; Koenig et al. 2011; Guaraldi and Salvatori 2012). Commensal species are of particular importance, as their colonization directly competes with the growth of disease-causing pathogens and educates host immune cells to mount effect responses against viral, bacterial, and fungal pathogens throughout the body (Koenig et al. 2011; Abt et al. 2012; Madan et al. 2012; Deshmukh et al. 2014; McAleer et al. 2016; Schuijt et al. 2016; Budden et al. 2017; Stewart et al. 2017). However, dysregulated mucosal immune responses can also fuel the pathogenesis of autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, and inflammatory bowel disease (Devkota et al. 2012; Zhang et al. 2015; Berer et al. 2017; van den Hoogen et al. 2017). Thus, the determinants of colonization of the neonatal microbiome are integral to understanding health in early life and beyond.
Considerable variation in gut microbiota exists between individuals in different geographic locations, with multiple stable community structures conducive to human health (Human Microbiome Project 2012; Yatsunenko et al. 2012; Lloyd-Price et al. 2016). Environmental flora and dietary intake are major contributors to the microbiome and can rapidly alter its structure and function (Wu et al. 2011; David et al. 2014; Singh et al. 2017; Tun et al. 2017). However, twin studies have demonstrated that microbiome phylogeny is also at least partially heritable (Goodrich et al. 2014). The mechanisms of gene–microbe cross talk by which the host might influence bacterial colonization to safeguard its associated benefits remain poorly understood (Kurilshikov et al. 2017).
In the gastrointestinal tract, host-derived carbohydrates serve both as metabolic substrates and adhesive receptors for bacteria (Sonnenburg et al. 2005; Martens et al. 2008). Whereas certain commensals independently catabolize host mucins, the liberation of sugars from host cells can also fuel the outgrowth of pathogenic Salmonella and Clostridiodes species (Ng et al. 2013). Conversely, specific monosaccharides such as fucose, present within the host glycocalyx, can be upregulated by the host to resist colonization by Salmonella and Enterococcus (Goto et al. 2014; Pham et al. 2014). The incorporation of fucose by the FUT2 fucosyltransferase is integrated into mucosal immune signaling, being directly regulated by IL-22 released by lamina propria innate lymphoid cells (Goto et al. 2016). Aside from fucose, sialic acid, a nine-carbon terminal monosaccharide on deuterostome glycans, is a viable metabolic substrate for many bacterial species (Vimr 2013; Charbonneau et al. 2016). An extensive body of literature demonstrates the functional role of leukocyte sialic acid in adhesive interactions necessary for tissue infiltration, outside–in signaling triggered by cytokines and growth factors, and cellular differentiation, underscoring its broad relevance in the immune system (Anderson and Anderson 1976; Bistrup et al. 1999; Hernandez and Baum 2002; Ghosh et al. 2006; Schmidt et al. 2013; Dougher et al. 2017; Leney et al. 2017; Irons and Lau 2018). Sialic acid moieties are present predominantly as terminal α2,6- or α2,3-linked structures on glycoconjugates. α2,3-linked sialic acid, acquired by neonates as oligosaccharides in breastmilk, promotes intestinal inflammation and the adhesion of pathogens such as Helicobacter pylori (Aspholm et al. 2006; Fuhrer et al. 2010). Although less is known about α2,6-linked sialic acids in host–microbe dynamics, genetic variation in the α2,6-sialyltransferase ST6GAL1 is associated with changes in the human gut microbiome (Snijders et al. 2016). Elsewhere, sialylation by ST6GAL1 is implicated in a plethora of physiologic functions including inflammation (Su et al. 2010), humoral immunity (Irons et al. 2020), cancer (Schultz et al. 2016; Holdbrooks et al. 2018; Jones et al. 2018; Dorsett et al. 2019), cellular survival (Britain et al. 2017, 2018), and chemo-/radio-resistance (Chakraborty et al. 2018; Punch et al. 2020). Emerging evidence continues to link glycosylation of intestinal epithelium and its associated mucins with the severity of gastrointestinal diseases, including inflammatory bowel disease and colorectal cancer (Swindall et al. 2013; Theodoratou et al. 2014; Bergstrom et al. 2016; Jiang et al. 2018; Earley et al. 2019; Cornelissen et al. 2020). However, the regulation and functional importance of host gut sialylation in mediating microbial colonization and mucosal immunity remain unknown.
In this study, we utilized genetically engineered mouse models to determine the relevance of α2,6-sialic acids synthesized by ST6GAL1 to microbiome colonization in early life. We found that ST6GAL1 expression profoundly altered microbiome composition at the time of weaning, enriching for Clostridiodes, Coprobacillus, and Adlercreutzia species while inhibiting the colonization of Helicobacter and Bilophila. Fecal microbiome transfer experiments demonstrated that the disrupted microbiota of St6gal1-KO animals triggered local and systemic Th17 immune responses, which were partly dependent on activation of the aryl hydrocarbon receptor and promoted epithelial hyperplasia downstream of IL-22. Consistent with its role in neonatal microbiome formation, we find ST6GAL1 is expressed within duodenal epithelium between birth and weaning due to glucocorticoid disinhibition of the P1 promoter during the neonatal stress hyporesponsive period (SHRP) (Matthews 2002; van Bodegom et al. 2017). Our results demonstrate a link between a developmentally programmed period in neonatal life and microbiome colonization that is mediated by host induction of an intestinal sialyltransferase and shed light on possible implications of this process in mucosal immunity.
2. Results
2.1 ST6GAL1 modifies the postnatal gut microbiome
Comprehensive changes in the glycosylation of intestinal epithelium occur during neonatal life (Chu and Walker 1986; Jaswal et al. 1988). In particular, α2,6-sialylation of the small intestinal epithelium reaches a maximum during early postnatal life, followed by a sweeping transition from terminal sialylation to fucosylation of glycans after weaning (Torres-Pinedo and Mahmood 1984; Mahmood and Torres-Pinedo 1985). However, the functional importance of this characteristic expression of sialic acid in the preweaned animal remains elusive.
We hypothesized that the presence of α2,6-linked sialic acid in the neonatal gastrointestinal tract, either due to endogenous enterocyte expression or the ingestion of breast milk ST6GAL1 and sialylated oligosaccharides, alters the microbiome at weaning. To test this, we performed 16S rRNA sequencing of fecal pellets collected immediately upon weaning. To control for mouse genomic background and genetic drift, we performed parallel comparisons of St6gal1+/+ and St6gal1−/− mice on wild-type (C57BL/6J) and B cell–deficient Ighm−/− (μMT.B6) backgrounds. To control for environmental factors, mice were provided identical cages, food, and water. Poisson analysis and principal component analysis (PCA) demonstrated distinct clustering of mice microbiomes by genotype, suggesting that community heterogeneity could be partially attributed to host genotype at the Ighm and St6gal1 loci (Fig. 1A). In particular, variation due to ST6GAL1 status was captured in the first principal component, representing 31.9% of total variation (Fig. 1A). Original taxonomic units (OTUs) were assigned to known taxonomic groups based on sequence identity (Supplementary Fig. 1). Statistically significant (Padj < 0.05) differences with a minimum twofold difference in means (FC > 2) were identified between WT and St6gal1-KO, or μMT and μMT/St6gal1-DKO microbiomes at the class, family, and genus taxonomic levels (full comparisons in Supplementary Fig. 2). Although numerous specific differences in microbial phylogeny were observed, ST6GAL1 expression did not significantly alter global microbiome diversity (Fig. 1B). Parallel analyses of the upper respiratory tract microbiome (obtained by caudocephalad saline flush of trachea and nasopharynx) and oral microbiome (obtained by tongue tissue) from mice at the same age revealed only weak and inconsistent clustering of samples by host genotype, highlighting that the influence of ST6GAL1 on the microbiome was restricted to the gastrointestinal system (Supplementary Figs 3 and 4).
Fig. 1.
ST6GAL1 influences neonatal Fecal microbiome composition. Fecal pellets were collected from mice of indicated genotypes upon weaning and extracted DNA was subjected to 16S rRNA sequencing. A) Principal components analysis of 16S sequencing data for ST6GAL1-sufficient and deficient mice on C57BL/6 J and B6.μMT backgrounds. B) Simpson diversity score for microbiomes derived from indicated genotypes. C) Relative abundances of Bacteroides, Prevotella, and Ruminococcus genera, indicative of enterotypes observed in the human microbiome. D) OTUs assigned to Clostridioides, Coprobacillus, and Adlercreutzia genera were enriched in ST6GAL1-expressing animals. E) OTUs assigned to Helicobacter and Bilophila were depleted in ST6GAL1-expressing animals. Data shown as mean ± SD of n = 5 mice per group. *P < 0.05.
Previous studies have identified multiple fecal microbiome enterotypes in healthy adult humans, each representing stable community structures characterized by the high abundance of a single bacterial genus adapted to specific host and environmental factors (Arumugam et al. 2011). To understand whether expression of ST6GAL1 was associated with the described microbiome enterotypes, we analyzed the levels of enterotype-defining genera Bacteroides, Prevotella, and Ruminococcus in WT and St6gal1-KO mice. St6gal1-KO mice harbored significantly elevated levels of Ruminococcus OTUs, with a corresponding trending increase in Bacteroides and trending decrease in Prevotella OTUs (Fig. 1C). These results are consistent with a potential role for ST6GAL1 in promoting the type 2 enterotype, characterized by a higher abundance of Prevotella species, efficient thiamine biosynthesis, and catabolism of fiber and host-derived mucins (Wu et al. 2011; Gorvitovskaia et al. 2016).
To identify reproducible changes in specific OTUs associated with ST6GAL1 expression, we identified statistically significant (FC > 2, P < 0.05) differences between comparisons of WT and St6gal1-KO mice on C57BL/6J and B6.μMT backgrounds and limited our analysis to OTUs that exhibited similar changes in both comparisons (Supplementary Fig. 5). We first noted that ST6GAL1 expression was strongly associated with the presence of two Firmicutes genera, Coprobacillus and Clostridiodes (Fig. 1D, left). Mice lacking ST6GAL1 had virtually no detectable reads assigned to either of these closely related OTUs. We also observed that abundance of the equol-producing Actinobacteria genus Adlercreutzia was reduced in ST6GAL1-deficient animals (Fig. 1D, right). In contrast, several Proteobacteria genera were expanded in ST6GAL1-deficient animals, including Helicobacter and Bilophila OTUs (Fig. 1E).
2.2 ST6GAL1 deficiency triggers a Th17 response via the microbiome
Commensal and pathogenic gastrointestinal microbes can have profound effects on the education and activation of mucosal immune cells. The neonatal period is hypothesized to be a critical window during which both protective antimicrobial responses and tolerance of gut commensals develop (Palmer et al. 2007; Koenig et al. 2011). Given the influence of ST6GAL1 on microbiome phylogeny during a period of early bacterial colonization, we hypothesized that mice lacking ST6GAL1 expression would harbor a microbiome that changes the population of leukocytes in the intestinal lamina propria; expression of ST6GAL1 may alter the mucosal immune response by selective pressure on certain bacterial species.
To test this, we reconstituted antibiotic-depleted, genetically identical neonatal wild-type mice with fecal microbiome from either WT or St6gal1-KO donors. To deplete endogenous microbiota, neonatal WT mice were administered a broad-spectrum antibiotic mix (vancomycin, neomycin, ampicillin, metronidazole) starting at postnatal day 7–10 for 2 weeks to deplete intestinal colonization, as has been reported elsewhere (Rakoff-Nahoum et al. 2004). One day after the cessation of antibiotics, mice were given 50 μL of homogenized fecal pellets (normalized to 40 mg/mL) in sterile PBS by oral gavage, once per day for three consecutive days (Fig. 2A). In our hands, antibiotic treatment depleted detectable fecal bacterial DNA to ~1% of native levels, and fecal microbiome transplantation restored predepletion levels, with no significant difference between mice receiving WT or St6gal1-KO feces (Fig. 2B). qPCR analysis of reconstituted fecal pellets demonstrated that KO to WT fecal transplantation reproduced the elevated levels of Helicobacter seen in St6gal1-KO mice (Fig. 2C). Overall mouse weights remained unchanged after fecal transplantation (Supplementary Fig. 6A).
Fig. 2.
ST6GAL1 deficiency promotes microbiome-dependent local and systemic Th17 responses. A) Wild-type mice were treated for 7 days with antibiotics (ampicillin, vancomycin, metronidazole, neomycin) in drinking water and then given fecal microbiome transplants (FMT) with either WT or St6gal1-KO feces from postnatal day 20–35 donor mice. After 4–8 days, mice were sacrificed for analysis. B) Depletion of fecal microbiome by antibiotics and restoration by FMT. C) Reconstitution of elevated Helicobacter prevalence in mice receiving St6gal1-KO microbiome. D) Quantitation of frequency of CD4+ T cell subsets with indicated fecal microbiome transfer donor genotype. E) Treatment with CH-233191 depletes Th17 cell increase induced by St6gal1-KO microbiome transfer. F) qPCR analysis of total lamina propria cells between mice receiving WT or St6gal1-KO fecal microbiome. G) Immunofluorescence staining of mesenteric lymph nodes for CD4 (red) and IL-17 (green). White arrows point to some double CD4/IL-17 positive cells (yellow). H) qPCR analysis of IL-17A expression within the spleen. I) Serum analysis of indicated cytokines after FMT. Main data are collated from two experiments with n = 3–5 per group. Data shown as mean ± SD. *P < 0.05, **P < 0.01, ****P < 0.0001.
Given the well-described role of Helicobacter species as pathogens in both humans and rodents, we hypothesized that transplantation of the fecal microbiome of St6gal1-KO mice would induce an antibacterial immune response at the mucosa. In order to capture changes in immune cell polarization and cytokine production induced by fecal transplantation, mice were euthanized 1–2 days posttransplantation and small intestinal lamina propria cells isolated for flow cytometry. Our analysis of several major hematopoietic cell types, including total myeloid cells, neutrophils, total T cells, as well as CD4+ and CD8+ T cell subsets, revealed no significant differences in abundance (Supplementary Fig. 6B and C). However, we noted a striking enrichment in IL-17-producing CD4+ T cells in mice receiving St6gal1-KO microbiota (Fig. 2D). In contrast, levels of FoxP3+ Tregs or IFN-γ-producing Th1 cells were not significantly altered. The induction of a Th17 phenotype is thought to result from production of IL-1β and IL-6, which skew undifferentiated CD4+ T cells toward expression of ROR-γt, resulting in “nonpathogenic” Th17 cells that express IL-17 and IL-10 (Wu et al. 2018). Antigen-presenting dendritic cells produce proinflammatory IL-23, leading to further differentiation of Th17 cells into a “pathogenic” phenotype (Haines et al. 2013; Jain et al. 2016). A major mechanism by which IL-17 production and the Th17 phenotype is stabilized in the gut lamina propria is by dietary and microbial tryptophan metabolites that engage the aryl hydrocarbon receptor (Gutierrez-Vazquez and Quintana 2018; Rothhammer and Quintana 2019). When we administered the AhR inhibitor CH-233191 concurrently with fecal microbiome transplantation, the enrichment of Th17 cells within the lamina propria of St6gal1-KO FMT recipients was reversed, suggesting a role for AhR in the maintenance of this Th17 response (Fig. 2E). In order to gain a comprehensive understanding of the immune pathways activated by St6gal1-KO microbiota, we analyzed small intestinal lamina propria RNA by qPCR. In our analysis, Treg-associated immunosuppressive genes (FoxP3, Il10, Tgfb1) and Th2-associated Gata3 were unaltered by the St6gal1-KO microbiome (Fig. 2F). Meanwhile, several Th1- and Th17-associated genes (Tbx21, Il17a, Il22) were markedly upregulated, with related genes trending but not significantly increased (Rorc, Il23) (Fig. 2F). Downstream epithelial genes associated with Th17 pathways (Reg3b, Reg3g) and inflammatory cell recruitment (Il6, Cxcr2, Nos2) were not altered in our experiments (Fig. 2F). Collectively, these data suggest that St6gal1-KO microbiota induce a local Th17 immune response, in part by activating the aryl hydrocarbon receptor.
To assess whether this Th17 response was a localized or a systemic change, we analyzed the draining mesenteric lymph nodes, spleen, and blood of recipient animals. Within the mesenteric lymph nodes, an increase in CD4+ T cells staining positive for IL-17 in mice receiving St6gal1-KO microbiota was noted, suggesting a parallel process to the mucosa was occurring in a secondary lymphoid organ (Fig. 2G, yellow cells). To understand if the IL-17 response had spread systemically, we analyzed the RNA levels of Il17a within the spleen. Here as well, mice receiving St6gal1-KO microbiota expressed significantly increased levels of IL-17a mRNA (Fig. 2H). Finally, analysis of serum cytokines indicated that although KO recipients did not exhibit increased systemic inflammation, as evidenced by proinflammatory cytokines IL-1β and IL-6, or anti-inflammatory cytokine IL-10, they did have greatly increased blood IL-17A (Fig. 2I). Other tested serum factors are shown in Supplementary Fig. 6E and F. Collectively, these results demonstrate that a genetic inability to express the sialyltransferase ST6GAL1 results in gut microbiome changes, which induce a systemic enrichment of IL-17- and IL-22-producing Th17 cells.
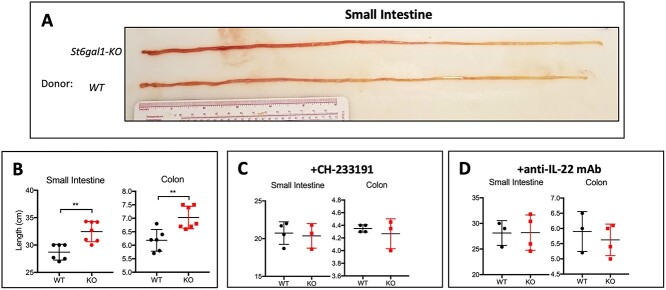
In contrast to IL-17, which primarily functions to recruit neutrophils and stimulate the production of IL-6, GM-CSF, G-CSF, and IL-1, IL-22 primarily acts upon epithelial cells and can have a strong prosurvival and proproliferative effect (Lindemans et al. 2015; Neumann et al. 2019; Kim et al. 2020). In line with this, we also observed that mice receiving St6gal1-KO microbiota developed a longer gastrointestinal tract within days of treatment (Fig. 3A). This difference in length was present both in the small intestine and colon (Fig. 3B). Consistent with a microbiome-dependent mechanism, administration of the AhR inhibitor CH-233191 equalized intestinal lengths between treatment groups (Fig. 3C). Finally, to test whether gut lengthening was dependent on Th17-dependent production of IL-22, we administered an anti-IL-22 neutralizing antibody during microbiome transplantation, which was also able to normalize the changes in gut length (Fig. 3D). These results suggest that the microbiome of ST6GAL1-deficient mice induces a Th17 response dependent on AhR activation, which promotes a lengthening of the gastrointestinal tract by IL-22-dependent epithelial proliferation.
Fig. 3.
ST6GAL1-deficient microbiome promotes gut lengthening via an AhR/IL-22 pathway. A) Representative image of small intestine from FMT recipients at time of sacrifice. B) Length of small intestine and colon between mice receiving WT or KO FMT, with or without concurrent administration of C) CH-233191 or D) neutralizing anti-IL-22 mAb. Representative data from multiple experiments are shown, with n = 3–5 per group. Data shown as mean ± SD. **P < 0.01.
2.3 Expression of ST6GAL1 in the neonatal intestine
Our findings so far demonstrate a previously unknown function of ST6GAL1 in altering the colonization of bacteria in the preweaning period. To assess the responsible cell types, as well as the mechanisms driving ST6GAL1 expression during the preweaning period, we analyzed the expression of St6gal1 mRNA in various segments of intestinal tract in preweaned (postnatal days 10 and 19) and weaned (postnatal day 44) mice. In suckling mice, St6gal1 mRNA in the duodenum was significantly higher than in the ileum and colon (Fig. 4A). In contrast, intestinal St6gal1 expression shifted distally to the jejunum and ileum in adult mice (Fig. 4A). At all times examined, there was minimal evidence for St6gal1 expression in the colon. Using primers specific for the 5′-untranslated regions (5’UTR) of St6gal1 (Wuensch et al. 2000), we found that both expected tissue-specific P1- and P3-dependent transcripts contributed to duodenal St6gal1 expression (primer sequences in Supplementary Fig. 7). Notably, P1-dependent St6gal1 transcripts were limited to the duodenum, where they were upregulated over 20-fold compared to more distal portions of the intestine. P3-dependent transcripts, thought to exist constitutively at low levels in most cells and tissues, were similarly present in the duodenum to a lesser extent (Fig. 4B) (Wuensch et al. 2000). Cell lysates were collected from duodenum, jejunum, ileum, and colon at similar times and subjected to immunoblot analysis for ST6GAL1 protein. At postnatal day 13 (PND13), ST6GAL1 protein levels were highest in the colon, possibly due to the ingestion of enzyme from maternal-derived milk and its accumulation in the distal GI tract, as ST6GAL1 is abundantly expressed in colostrum from lactating mammary glands (Dalziel et al. 2001). By postnatal day 19, an ST6GAL1 surge was noted in the duodenum, jejunum, and ileum, consistent with endogenous mRNA levels (Fig. 4C). Interestingly and quite unexpectedly, by adulthood (postnatal day 44), the vast majority of ST6GAL1 protein was found in colon (Fig. 4C).
Fig. 4.

ST6GAL1 expression in the neonatal duodenum is mediated by the P1 promoter and inhibited by glucocorticoids. A) Relative expression of St6gal1 transcripts within the duodenum, jejunum, ileum, and colon on postnatal days (PND) 10, 19, and 44. B) Relative abundance of P1-dependent and P3-dependent St6gal1 transcripts on postnatal day 10. C) Immunoblot for ST6GAL1 from total tissue of duodenum, jejunum, ileum, and colon on postnatal days 13, 19, and 44. D) Frozen sections of mouse total small intestine tissue at postnatal days 1, 12, and 24 were stained for ST6GAL1 (red) and with Sambucus nigra lectin (green). Comparison of wild-type (WT), P1 promoter conditional knockout (dP1), and global St6gal1 KO mice is shown. E) α2,6-sialyltransferase activity in indicated WT gastrointestinal tissues at indicated ages (above) and in fecal pellets at 10 days of age (below). F) Postnatal day 10 mice were given a single bolus of intraperitoneal dexamethasone and then sacrificed after 24 h and total RNA levels of total, P1-specific and P3-specific St6gal1 transcripts quantified in duodenum and jejunum (left). Proximal small intestine was stained for ST6GAL1 protein in vehicle and dexamethasone-treated PND15 mice (right). G) Adult WT mice were treated with intraperitoneal RU-486 or vehicle control for 3 days and then abundance total, P1-specific and P3-specific St6gal1 transcripts quantified in duodenum and jejunum. Major results are representative of multiple experiments. Data shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
ST6GAL1 is expressed in diverse cell types, including those of the epithelial, endothelial, mesenchymal, and hematopoietic lineages, all of which are present within the small intestine (Zhang et al. 2017; Imamaki et al. 2018; Irons et al. 2019). To understand which cell types were responsible for the neonatal spike of St6gal1 transcripts, a histology approach was used to compare the small intestines of wild-type, global St6gal1-KO, and dP1 pups (which lack the P1 promoter for St6gal1). To analyze the morphologic distribution of sialic acid, we used SNA, a lectin from Sambucus nigra that specifically recognizes the α2,6-sialyl product of ST6GAL1. Consistent with our previous observations, we noted very high SNA reactivity within the submucosa, muscularis layers, and adventitia, which was partially attenuated in dP1 mice and virtually absent in global St6gal1-KO mice (Fig. 4D). This SNA reactivity increased between postnatal day 1 and 12 but remained constant in intensity thereafter. In contrast, the small intestinal epithelium exhibited maximal ST6GAL1 staining at postnatal day 12, with corresponding SNA reactivity, which was not detected after weaning on postnatal day 24 (Fig. 4D). Epithelial ST6GAL1 was not evident in St6gal1-KO mice and was reduced in mice lacking the P1 promoter (Fig. 4D). However, epithelial SNA reactivity in dP1 pups was similar to full knockouts, confirming that P1-dependent ST6GAL1 expression is a major determinant of epithelial sialylation (Fig. 4D).
α2,6-sialyltransferase activity toward Gal-β1,4-GlcNAc acceptors, indicative of ST6GAL1 and ST6GAL2, was quantified in total intestinal cell lysates. To control for potential ST6GAL2 activity, St6gal1-KO tissues were used as a negative control for all samples (not shown). In contrast to the RNA and protein expression data, we were unable to detect any spike in sialyltransferase activity in the neonatal duodenum of suckling pups, with detectable but minimal activity downstream in the jejunum and ileum. However, the colon exhibited strikingly high α2,6-sialyltransferase activity, increasing nearly fivefold between postnatal day 13 and 44 (Fig. 4E, top). Furthermore, α2,6-sialyltransferase activity was detectable in the homogenized fecal pellets of suckling WT, but not age-matched St6gal1-KO or dP1 mice, nor adult WT mice (Fig. 4E, bottom). Together, these observations further suggest that ST6GAL1, though expressed proximally in the duodenum, accumulates in the colon of suckling mice.
Rodents undergo a period of stress hyporesponsiveness between birth and weaning, during which time blood adrenocorticotropic hormone (ACTH) and glucocorticoid levels remain low and unresponsive to most stressors. Upon weaning, the stress of maternal separation triggers a surge of glucocorticoid production that drives the maturation of both pulmonary and gastrointestinal tissues (De Kloet et al. 1988; van Bodegom et al. 2017). Given the significant downregulation of ST6GAL1 expression in the proximal small intestine upon weaning, we hypothesized that glucocorticoid sensitivity was involved in its expression. First, we attempted to inhibit neonatal ST6GAL1 expression by providing an early bolus of systemic glucocorticoids to neonatal mice. Postnatal day 10 pups were given a single dose of intraperitoneal dexamethasone and analyzed after one day for changes in expression of St6gal1 at the RNA level. We observed that dexamethasone reduced duodenal transcripts ~fourfold, attributable to a reduction of transcripts expressed under the P1, but not P3 promoter (Fig. 4F, left). Downregulation of ST6GAL1 in the proximal small intestine was also observed by immunofluorescence microscopy (Fig. 4F, right). To understand if endogenous glucocorticoid-mediated repression was involved in the low levels of intestinal ST6GAL1 expression in adults, we treated 6- to 8-week-old adult mice with the steroid receptor inhibitor RU-486 for three successive days and then analyzed ST6GAL1 expression within intestinal tissues. Total St6gal1 transcripts were elevated within the duodenum and jejunum, largely attributable to an increase in P1-dependent transcripts (Fig. 4G). Although the inherent variability in this system precluded statistical significance, our results indicate that ST6GAL1 expression in the neonatal period is likely secondary to glucocorticoid disinhibition during the stress hyporesponsive period.
3. Discussion
The neonatal period represents a time of rapid evolution for the microbiome. Central to these changes is the balance between symbionts and pathogens, mediated by complex host and environmental factors that are poorly understood. Our results implicate ST6GAL1 as a previously unrecognized genetic factor facilitating the colonization of specific microbes of the Firmicutes phylum. The scavenging of host sialic acid by pathogenic and commensal bacteria for catabolism relies on the expression of three genes of the Nan cluster (NanA, NanK, and NanE), as well as the deacetylase NagA and deaminase NagB (Vimr et al. 2004; Vimr 2013). The presence of the Nan cluster within bacterial genomes is limited to species of the Gamma-Proteobacteria and Fusobacteria phyla, as well as Bacillales, Clostridioides, and Lactobacillales of the Firmicutes phylum (Almagro-Moreno and Boyd 2009; McDonald et al. 2016). In our data, colonization by bacteria of the genus Clostridiodes was completely dependent on host expression of ST6GAL1, consistent with the expansion of pathogenic Clostridioides species in response to sialic acid liberated by other environmental microbes observed by others (Ng et al. 2013).
Spore-forming bacteria such as Clostridia dramatically expanded in abundance in the human gut after the first year of life, consistent with their ability to tolerate the increasingly anoxic conditions of the developing colon (Guittar et al. 2019). In contrast to adults, wherein certain Clostridiodes species may cause infection, the accumulation of Clostridioides in the early postnatal period directly competes with adhering/effacing pathogens, protecting mice from life-threatening infection (Kim et al. 2017).
Our findings also indicate that ⍺2,6-sialyl ligands support the key isoflavone-producing species Adlercreutzia, the presence of which is associated with improved blood lipids and reduced diet-induced obesity (Zietak et al. 2016; Zheng et al. 2019). To the best of our knowledge, there have not been any published studies of the ability of the genus Adlercreutzia to metabolize sialic acid (Maruo et al. 2008; Danylec et al. 2019; Florez et al. 2019). We also show an association between ST6GAL1 expression and reduced colonization of Helicobacter and Bilophila species, which are collectively implicated in the pathogenesis of infection, cancer, and metabolic dysfunction (Feng et al. 2017; Dahmus et al. 2018; Natividad et al. 2018). Interestingly, free sialic acid has been shown to have anti-Helicobacter properties both in vitro and in vivo (Yang et al. 2008, 2013; Salcedo et al. 2013; Rhee et al. 2016; Noh et al. 2017; Benktander et al. 2018). Bile-metabolizing bacteria of the Bilophila genus, which expand in response to dietary fat intake, were recently identified as pathobionts, capable of promoting colitis (Feng et al. 2017; Natividad et al. 2018).
Overall, our data support a model that both agrees with and expands upon the existing literature on milk-derived glycans, highlighting a health-promoting role for ST6GAL1 in altering the balance of commensals and pathogens colonizing the neonatal gut (Newburg and Morelli 2015). Our microbiome data do not distinguish between the effects of ⍺2,6-sialyl glycans in milk oligosaccharides and gut glycocalyx, but future experiments using a combination of cross-fostering and tissue-specific knockouts may shed light on the relative importance of these 2 pathways. Neither do we here investigate whether the described microbiome changes persist into adult life. However, our previous findings of neutrophilia and inflammatory tissue damage in St6gal1-deficient mice allow for the possibility that microbiome-related processes may still contribute to a tendency toward inflammation in these animals (Appenheimer et al. 2003; Nasirikenari et al. 2006, 2010, 2014, 2019; Jones et al. 2012; Dougher et al. 2017).
As a fundamental metabolic substrate, carbohydrates shape microbial fitness within the gastrointestinal ecosystem (Koropatkin et al. 2012; Poole et al. 2018). Host-derived glycans exist both on the plasma membrane glycocalyx and mucins secreted into the extracellular space (Koropatkin et al. 2012). The glycosylation of major mucins such as Muc2, along with host expression of associated glycosyltransferases, varies along the intestinal tract in a regiospecific and microbiome-dependent manner (Arike et al. 2017). Complete normalization of intestinal mucus layers requires at least 6 weeks of microbial colonization, with stereotyped shifts in major bacterial populations that may mimic those seen during initial colonization of the neonatal intestine (Johansson et al. 2015). At weaning, a surge in microbial exposure activates IL-22 production, which promotes expression of glycocalyx-associated MUC17, along with a global transition from sialylated to fucosylated glycans in the intestinal mucosa (Chu and Walker 1986; Layunta et al. 2021). Fucosylation of enterocytes prevents the expansion of disease-causing pathobionts, as part of a dynamic paracrine circuit regulated by type 3 innate lymphoid cells (Goto et al. 2014; Pham et al. 2014). As the immune system matures, infiltrating adaptive CD4+ Treg and Th17 cells gradually dominate the symbiotic relationship with commensal species, dampening IL-22- and IL-23-mediated STAT3 activation, counteracting ILC3s to normalize lipid metabolism in enterocytes (Mao et al. 2018). Our findings add to the understanding of the development of IL-17-dependent responses in the neonate, underscoring a role for ST6GAL1 in preventing Th17 accumulation in the gut prior to weaning. This is achieved by the careful calibration of bacterial colonizers during the suckling period, prior to the wholesale changes brought about by an adult diet. These findings naturally beg the question of whether the bacterial colonization that accompanies weaning is itself regulating the expression of ST6GAL1, as has been demonstrated for other glycosyltransferases. In this regard, our preliminary data suggest that ST6GAL1-associated sialic acid may be sensitive to antibiotic depletion and increases with fecal transplant, consistent with a study of glycosylation changes in germ-free mice (Arike et al. 2017). Surprisingly, despite high ST6GAL1 expression in the duodenum, the majority of ST6GAL1 protein and ⍺2,6-sialyltransferase activity accumulates distally within the colon, consistent with previous studies showing detectable ST6GAL1 protein in the colon (Arike et al. 2017). These observations are important in explaining how neonatal ST6GAL1 expression in the proximal gastrointestinal tract might alter microbiome composition in the distal gastrointestinal tract, potentially by direct release into the lumen and distal transport. Furthermore, the disproportionately high ⍺2,6-sialyltransferase activity in the colon of adult mice (~100-fold higher than upstream tissues) may indeed be attributable to a previously reported protein cofactor of ST6GAL1 in that tissue (Nagpurkar et al. 1996).
Glucocorticoids are widely recognized for their importance in lung and gut epithelium maturation (Nanthakumar, Young, et al. 2005b; Roberts et al. 2017). Previous studies documented the ability of glucocorticoids to upregulate enzymes involved in dietary carbohydrate metabolism, as well as Fut2, affecting an increase in fucosylation during the postweaning period (Solomon et al. 2001; Nanthakumar, Dai, et al. 2005a; Nanthakumar et al. 2013). In early works, we noted that transcription of St6gal1 was mediated by a number of distinct promoter/transcription initiation regions (Wang et al. 1990, 1993; Dalziel et al. 2001). Others have shown that intestinal ST6GAL1 is upregulated in the presence of microbes and downregulated during colitis and in response to a high-protein diet (Ilott et al. 2016; Arike et al. 2017). In our previous studies, transcription of St6gal1 in the neonatal intestine was associated with expression from the P1 promoter, previously known only to be utilized by the liver (Vertino-Bell et al. 1994). Here, our data expand on earlier findings and indicate that glucocorticoids also potently inhibit ST6GAL1 expression via the P1 promoter, placing ST6GAL1 among a suite of other investigated intestinal enzymes whose expression levels are linked to serum glucocorticoids (Rudman 1973; Yeh, Yeh, Holt 1991a; Yeh, Yeh, Montgomery, et al. 1991b). Ultimately, glucocorticoids likely play a central role in the transition from sialylated to galactosylated and fucosylated glycans in the intestinal epithelium that occurs with weaning (Biol-N'garagba et al. 2003). Although not tested in this study, we hypothesize that this surge of glucocorticoid production and sensitivity is most likely triggered by the maternal separation of weaning, terminating the neonatal period of stress hyporesponsiveness (van Bodegom et al. 2017).
In humans, Helicobacter pylori is regarded as a carcinogen for its role in the pathogenesis of gastric adenocarcinoma and MALT lymphoma (Lee et al. 2016). In mice, Helicobacter hepaticus is a pathogen that causes hepatitis, colitis, and colorectal cancer (Fox et al. 2011). Several lines of evidence point to a central role for bacteria of the Helicobacter genus in our data. A greater abundance of Helicobacter in ST6GAL1-deficient animals is consistent with a number of previous reports demonstrating anti-Helicobacter activity of soluble sialic acid preparations by at least two independent mechanisms. Firstly, sialic acid has direct antibacterial properties toward Helicobacter species both in vitro and in vivo, an effect that is augmented by coadministration of antioxidant catechins (Yang et al. 2008, 2013; Rhee et al. 2016; Noh et al. 2017). Secondly, sialylated gastrointestinal mucins, particularly those of the stomach, act as decoys for Helicobacter strains expressing the sialic acid–binding SabA adhesin by competing with sialylated structures in the gastric epithelial surface necessary for initiating infection (Mentis et al. 1990; Simon et al. 1997; Valkonen et al. 1997; Hirmo et al. 1998). Physiologically, it remains a matter of speculation whether neonate-derived or maternal-derived free sialic acid is more important in preventing Helicobacter infection. Although a physiological role for milk-derived sialic acid in protecting the neonate from Helicobacter colonization is plausible, human studies in multiple populations have failed to demonstrate any association between breast-feeding and Helicobacter colonization (Rothenbacher et al. 2002; Rodrigues et al. 2006; Senbanjo et al. 2014; Soltani et al. 2014).
Helicobacter infection induces a robust but plastic CD4+ T cell response with both Th1 and Th17 characteristics, characterized by expression of T-bet and ROR-γt, as well as by production of IFN-γ, IL-17, and IL-22 (Morrison et al. 2013). Ensuing inflammation often targets other bacterial species and is associated with increased tissue damage in both humans and mice (Gomes-Neto et al. 2017; Bagheri et al. 2018). In our experiments, fecal transplantation of St6gal1-KO microbiome recapitulated an elevated abundance of Helicobacter DNA and provoked a local and systemic Th17-mediated immune response within days, consistent with the initial stages of Helicobacter infection. Interestingly, we show that the Th17 response is highly dependent on AhR engagement in our model, paralleling findings that Ahr-deficient mice develop rectal prolapse associated with uncontrolled H. hepaticus infection (Fernandez-Salguero et al. 1997). During persistent infection, Helicobacter pylori often achieves immune escape by inducing tolerance after an early failure of Th17-mediated pathogen clearance, a process dependent on dendritic cells and c-Maf+ regulatory T cells (Zagon et al. 2010; Xu et al. 2018). Our findings add to existing literature by demonstrating the importance of early life expression of ST6GAL1 in reducing the colonization of Helicobacter pathogens. In addition to the possibility of using oral sialic acid as a treatment for Helicobacter infections, our data hint at the importance of the neonatal stress hyporesponsive period in ensuring appropriate microbial colonization in the gut.
Intestinal sialylation by the sialyltransferase ST6GAL1 in the neonatal period is a developmentally regulated host mechanism coordinating bacterial colonization in the early gut microbiome. The inability to produce α2,6-sialyl ligands predisposes animals toward a microbiome-dependent Th17 responses, highlighting a pathway by which intestinal epithelium regulates mucosal immunity. Considering the prevalence of intestinal fucosylation in adult animals, sialic acid may promote an early stage of microbial ecological succession in the developing gut.
4. Materials and methods
4.1 Animal models
All animal usage in this study were approved by Roswell Park Institutional Animal Care and Use Committee under protocol 1071M. Wild-type mice (C57BL/6J) were purchased from Jackson Laboratory and regularly replenished by backcrossing. St6gal1-KO mice were generated as described previously and backcrossed onto a C57BL/6J background for at least 15 generations (Hennet et al. 1998). dP1 mice were generated in the laboratory and extensively validated to have undetectable P1 transcripts of ST6GAL1 in the liver and reduced circulatory ST6GAL1 activity (Appenheimer et al. 2003). μMT mice were purchased from Jackson Laboratory, and μMT/St6gal1-DKO double knockouts were generated in multiple crossings between single knockouts, followed by genotyping and phenotyping analyses, as previously reported (Irons and Lau 2018). Standard housing conditions, which do not guarantee exclusion of specific pathogens, were used unless otherwise indicated.
4.2 Antibodies
Anti-ST6GAL1 (AF5924, R&D Biosystems), SNA-FITC (FL-1301-2, Vector Labs), anti-goat-Cy3 (111–165-003, Jackson ImmunoResearch), anti-IL-17A-AlexaFluor488 (eBio17B7, Thermo Fisher), anti-CD4-PE (GK1.5, Invitrogen), anti-CD3-PE (17A2, BioLegend), anti-CD4-BV510 (GK1.5, BioLegend), anti-CD8-biotin (53-6.7, eBioscience), streptavidin-PerCP/Cy5.5 (45-4317, eBioscience), anti-FoxP3-AlexaFluor488 (MF-14, BioLegend), anti-IFN-g-APC (XMG1.2, BioLegend), anti-IL17A-PE/Cy7 (TC11-18410.1, BioLegend), anti-IL-22 mAb functional grade (IL22JOP, eBioscience), anti-CD11b-BV711 (M1/70, BioLegend), anti-Ly6G-APC (1A8, BioLegend).
4.3 Immunofluorescence microscopy
Intestinal tissue was separated into small intestine (gastric pylorus to cecum) and colon (cecum to rectum). Mesenteric lymph nodes were isolated by careful dissection of mesenteric fat adjacent to the descending colon. All tissues were snap frozen before sectioning at 10 μm thickness onto charged glass microscope slides. Tissue sections were fixed at −20 °C in acetone and then hydrated in PBS before blocking in 1% BSA for 1 h. Sections were stained with anti-ST6GAL1 primary antibody and FITC-conjugated SNA lectin overnight, washed thoroughly with PBS, and stained with donkey–anti-goat-Cy3 secondary for 1 h. For mesenteric lymph nodes, tissues were incubated with anti-IL-17-FITC and anti-CD4-PE overnight. All slides were washed extensively, rinsed with DAPI, air-dried, and then mounted with cover slips in 10% glycerol. Images were captured with a Nikon Eclipse E600 microscope with EXFO X-cite 120 light source, Spot RT3 camera, and Spot Software.
4.4 Sialyltransferase assay
Sialyltransferase activity within mouse tissues was determined using an artificial O-benzyl conjugated Gal-β1,4-GlcNAc acceptor, as has been described before (Lee et al. 2014). Briefly, serum or lysed cells were incubated at 37 C with artificial acceptor and tritium-labeled CMP–sialic acid for 1 h. The resulting reaction mix was applied to a SepPak column and extensively washed and then eluted with methanol. Radioactive counts in the sample were quantified using a Beckman Coulter LS 6500 scintillation counter. α2,6-sialylated product was precipitated with SNA-agarose beads, and SNA-reactive fraction once again counted to quantify α2,6-sialyltransferase activity. Remaining α2,3-sialyltransferase activity was inferred.
4.5 Glucocorticoid experiments
To test the effect of exogenous glucocorticoids, mice at postnatal day 10 were given a single intraperitoneal injection of 5 μg dexamethasone (Sigma) in mineral oil, or vehicle only control. About 24 h after treatment, mice were euthanized, and tissues collected for histologic and RNA analysis. In other experiments, glucocorticoid receptor was blocked in adult mice (age 8–12 weeks) with three consecutive days of intraperitoneal injections of 40 μg RU-468 (Sigma) in 50 μL mineral oil or vehicle only. On day 4, mice were euthanized, and tissues collected for histologic and RNA analysis.
4.6 RNA analysis
Tissue was preserved in Tri Reagent (MRC Inc.) at −80 °C prior to extraction. RNA extraction was performed in accordance with manufacturer instructions, and RNA concentration and purity were quantified immediately. cDNA synthesis was performed with iSCRIPT cDNA synthesis kit (Bio-rad) on normalized amounts (0.5–2 μg) of RNA, and RT-qPCR analysis performed with iQ SYBR-Green kit (Bio-rad). Primer sequences, melting temperatures, and references can be found in Supplementary Fig. 7. For liver and intestinal ST6GAL1 expression, cycles of amplification were normalized to β2-microglobulin or β-actin control, and relative expression (2dCT) was presented. For lamina propria gene expression, relative expression was normalized to WT microbiome transplant recipients (2ddCT).
4.7 16S rRNA sequencing
Mouse fecal pellets were collected fresh in sterile tubes. Oral microbiome samples represent anterior tongue tissue, collected immediately after sacrifice. Upper respiratory tract microbiome samples were collected immediately postmortem by anterior dissection to the trachea and caudocephalad sterile saline lavage of the nasopharynx and nasal cavities, with the mouse being held in a supine position. About 1 mL of sterile saline was used for lavage, and output collected from the nares. All samples were collected within 2 days of weaning and immediately stored at −80 °C. The sequencing libraries were prepared using a two-step PCR method for targeting an ~500 bp region of the 16S V3 and V4 rDNA. The first PCR (25-cycle) used 25 ng of DNA to amplify the target region, where the PCR primers have overhang adapter sequence necessary for the second PCR step. After purification, the amplicon from the first stem is amplified with 8 cycles of PCR using the Nextera Index Kit (Illumina Inc.), which uses primers that target the overhang adaptor sequence added during the first round of PCR. The second round of PCR adds one of 384 different combinations of indexed tags to each sample, which allows pooling of libraries and multiplex sequencing. Prior to pooling, each individual sample’s amplified DNA is visualized on a TapeStation 4200 D1000 tape (Agilent Technologies) for expected amplicon size, purity, and concentration. Validated libraries are pooled equal molar in a final concentration of 4 nM in Tris–HCl 10 mM, pH 8.5, before 2 × 300 cycle sequencing on a MiSeq (Illumina, Inc.).
Paired-end fastq reads were demultiplexed, processed, and analyzed using QIIME v1.9.1. OTUs were assigned using QIIME’s uclust-based open-reference OTU-picking pipeline using Greengenes bacterial 16S rRNA reference (v13.8); bacterial sequences were aligned using PyNAST. These alignments were refined by removing chimeric sequences using ChimeraSlayer. OTUs with less than 0.001% assigned sequences were removed from each sample to avoid biased and inflated diversity estimates. Positive and negative control samples were checked against the whole batch and then removed from the data. Relative abundance bar plots were generated at the Class, Family, and Genus levels. Results were summarized estimating alpha-diversity scores using inverse Simpson’s diversity index. Phylogenetic composition plots at different taxonomic levels, sample-to-sample heatmaps, and PCA plots were also generated.
Statistical analyses and comparisons were carried out using DESeq2 (v1.20.0) and phyloseq (v1.26.0) packages from R (v3.5.0). This methodology implements a likelihood ratio test using a generalized linear model assuming the outcome variable is negative-binomial distributed. Bi-taxa plots were used to examine relevant OTUs, subsetting those having absolute fold-change values greater than 2 (absFC > 2) and P value < 0.05. Bi-taxa plots are displayed pairing Phylum (L2) vs Class (L3), Family (L4) and Genus (L5). K vs W and D vs U were compared to find differentially abundant OTUs, accounting for animal sex, and summarized in a Venn Diagram at Class, Family, and Genus levels. Although initially excluded due to high sample variation, Clostridiodes genus was significantly abundant in condition W compared to K (P value < 3.4e−8) and U compared to D (P value < 1.3e−7).
4.8 Bacterial quantification
At 21 days of age, fresh fecal pellets were collected in sterile tubes. Pellets were digested in proteinase K–containing buffer at 55 °C overnight, precipitates removed in 6 M NaCl solution, and remaining DNA isolated by ethanol precipitation (Miller et al. 1988). DNA yields were quantified and a normalized 0.5 ng of DNA was used as template in qPCR analysis (SYBR Green, Bio-rad). Eubacteria-specific primers were used to quantify total bacterial DNA, which is presented relative to amplification of an unrelated host gene.
4.9 Fecal microbiome transplantation
At age 7–10 days, wild-type C57BL/6J mice were administered a combination of vancomycin (0.5 g/L, VWR), ampicillin (1 g/L, Sigma), neomycin (1 g/L, VWR), and metronidazole (1 g/L; Beantown Chemical) in acidified drinking water for 14 days. On day 15–17, fecal pellets from WT or St6gal1-KO mice of age 20–35 days were collected (minimum n = 7), vortexed for 3 min in sterile PBS, and administered to antibiotic-pretreated mice by 50 μL gastric gavage at 0.4 mg/mL concentration. After the final transfer, mice were euthanized between days 18 and 20 for analysis. Where indicated, mice were also intraperitoneally administered 200 μg AhR inhibitor CH-233191 (Sigma) in 25 μL of mineral oil (or vehicle control) prior to each FMT gavage. In IL-22 neutralization experiments, mice were intraperitoneally administered 1.25 μg of anti-IL22 neutralizing mAb per day (or isotype control) between the initiation of transplantation and analysis.
4.10 Cytokine quantification
Serum samples were diluted according to manufacturer’s instructions to fall within dynamic range of the specific ELISA assay. In all cases, recombinant cytokine standards were utilized in duplicate to quantify the analyte within serum. Kits used include IL-1β, IL-6, IL-10, IL-17 (all Invitrogen) and soluble RAGE (R&D).
4.11 Lamina propria isolation and flow cytometry
Small intestines were flushed of fecal matter and then carefully stripped of adipose tissue and Peyer’s patches. Guts were separated into segments, inverted, and epithelial layer dissociated at 37 °C for 15 min using EDTA and DTT containing buffer. Remaining submucosal tissue was further digested in collagenase-containing solution at 37 °C for 30 min, filtered, washed, and cells enumerated. Cells were seeded at 4 × 106 cells/mL in 100 μL RPMI with 10% FBS and stimulated for 5–6 h with PMA and ionomycin-containing cell activation cocktail (BioLegend). Cells were then collected and stained for cell surface markers (CD3, CD4, CD8), and fixed for 20 min in 2% formalin. Washed cells were permeabilized either with saponin-containing buffer (BD Cytoperm, BD Biosciences) with antibodies for cytokines (anti-IFN-γ, anti-IL-17A) or with 0.1% Triton-X 100, followed by anti-FoxP3 antibody in staining buffer. Cells were analyzed within 24 h of animal euthanasia.
Supplementary Material
Acknowledgments
We acknowledge Dr. Tianxin Yu for her assistance in performing some of the experiments reported in this paper.
Contributor Information
Eric E Irons, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Eduardo Cortes Gomez, Department of Biostatistics and Bioinformatics, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Valerie L Andersen, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Joseph T Y Lau, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Funding
This work was supported by NIH National Institute of Allergy and Infectious Diseases (NIAID) R01AI140736. The core facilities of Roswell Park Comprehensive Cancer Center used in this work, including the Genomics Shared Resource and Flow and Image Cytometry Shared Resource, were supported in part by NIH National Cancer Institute Cancer Center Support Grant 5P30 CA016056.
Conflict of interest statement
None declared.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Ethics approval
This manuscript does not report studies involving human participations, human data, or human tissues.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 37:158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S, Boyd EF. 2009. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AO, Anderson ND. 1976. Lymphocyte emigration from high endothelial venules in rat lymph nodes. Immunology. 31:731–748. [PMC free article] [PubMed] [Google Scholar]
- Appenheimer MM, Huang RY, Chandrasekaran EV, Dalziel M, Hu YP, Soloway PD, Wuensch SA, Matta KL, Lau JT. 2003. Biologic contribution of P1 promoter-mediated expression of ST6Gal I sialyltransferase. Glycobiology. 13:591–600. [DOI] [PubMed] [Google Scholar]
- Arike L, Holmen-Larsson J, Hansson GC. 2017. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. 27:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. 2011. Enterotypes of the human gut microbiome. Nature. 473:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspholm M, Olfat FO, Norden J, Sonden B, Lundberg C, Sjostrom R, Altraja S, Odenbreit S, Haas R, Wadstrom T, et al. 2006. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri N, Razavi A, Pourgheysari B, Azadegan-Dehkordi F, Rahimian G, Pirayesh A, Shafigh M, Rafieian-Kopaei M, Fereidani R, Tahmasbi K, et al. 2018. Up-regulated Th17 cell function is associated with increased peptic ulcer disease in Helicobacter pylori-infection. Infect Genet Evol. 60:117–125. [DOI] [PubMed] [Google Scholar]
- Benktander J, Barone A, Johansson MM, Teneberg S. 2018. Helicobacter pylori SabA binding gangliosides of human stomach. Virulence. 9:738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE, et al. 2017. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 114:10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K, Liu X, Zhao Y, Gao N, Wu Q, Song K, Cui Y, Li Y, McDaniel JM, McGee S, et al. 2016. Defective intestinal mucin-type O-glycosylation causes spontaneous colitis-associated cancer in mice. Gastroenterology. 151:152–164 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biol-N'garagba MC, Niepceron E, Mathian B, Louisot P. 2003. Glucocorticoid-induced maturation of glycoprotein galactosylation and fucosylation processes in the rat small intestine. J Steroid Biochem Mol Biol. 84:411–422. [DOI] [PubMed] [Google Scholar]
- Bistrup A, Bhakta S, Lee JK, Belov YY, Gunn MD, Zuo FR, Huang CC, Kannagi R, Rosen SD, Hemmerich S. 1999. Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J Cell Biol. 145:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britain CM, Dorsett KA, Bellis SL. 2017. The glycosyltransferase ST6Gal-I protects tumor cells against serum growth factor withdrawal by enhancing survival Signaling and proliferative potential. J Biol Chem. 292:4663–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britain CM, Holdbrooks AT, Anderson JC, Willey CD, Bellis SL. 2018. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J Ovarian Res. 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. 2017. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 15:55–63. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Dorsett KA, Trummell HQ, Yang ES, Oliver PG, Bonner JA, Buchsbaum DJ, Bellis SL. 2018. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J Biol Chem. 293:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, et al. 2016. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 164:859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu SH, Walker WA. 1986. Developmental changes in the activities of sialyl- and fucosyltransferases in rat small intestine. Biochim Biophys Acta. 883:496–500. [DOI] [PubMed] [Google Scholar]
- Cornelissen LAM, Blanas A, Zaal A, van der Horst JC, Kruijssen LJW, O'Toole T, van Kooyk Y, van Vliet SJ. 2020. Tn antigen expression contributes to an immune suppressive microenvironment and drives tumor growth in colorectal cancer. Front Oncol. 10:1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus JD, Kotler DL, Kastenberg DM, Kistler CA. 2018. The gut microbiome and colorectal cancer: a review of bacterial pathogenesis. J Gastrointest Oncol. 9:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel M, Huang RY, Dall'Olio F, Morris JR, Taylor-Papadimitriou J, Lau JT. 2001. Mouse ST6Gal sialyltransferase gene expression during mammary gland lactation. Glycobiology. 11:407–412. [DOI] [PubMed] [Google Scholar]
- Danylec N, Stoll DA, Huch M. 2019. Draft genome sequences of type strains of Adlercreutzia muris and Ellagibacter urolithinifaciens, belonging to the family Eggerthellaceae. Microbiol Resour Announc. 8:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kloet ER, Rosenfeld P, Van Eekelen JA, Sutanto W, Levine S. 1988. Stress, glucocorticoids and development. Prog Brain Res. 73:101–120. [DOI] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE, Oliver PM, Kolls JK, Weiser JN, Worthen GS. 2014. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 20:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 487:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett KA, Jones RB, Ankenbauer KE, Hjelmeland AB, Bellis SL. 2019. Sox2 promotes expression of the ST6Gal-I glycosyltransferase in ovarian cancer cells. J Ovarian Res. 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougher CWL, Buffone A Jr, Nemeth MJ, Nasirikenari M, Irons EE, Bogner PN, Lau JTY. 2017. The blood-borne sialyltransferase ST6Gal-1 is a negative systemic regulator of granulopoiesis. J Leukoc Biol. 102:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley H, Lennon G, Balfe A, Coffey JC, Winter DC, O'Connell PR. 2019. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci Rep. 9:15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Long W, Hao B, Ding D, Ma X, Zhao L, Pang X. 2017. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. 9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. 1997. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet Pathol. 34:605–614. [DOI] [PubMed] [Google Scholar]
- Florez AB, Vazquez L, Rodriguez J, Redruello B, Mayo B. 2019. Transcriptional regulation of the equol biosynthesis gene cluster in Adlercreutzia equolifaciens DSM19450(T). Nutrients. 11:8–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. 2011. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 4:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer A, Sprenger N, Kurakevich E, Borsig L, Chassard C, Hennet T. 2010. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med. 207:2843–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Bandulet C, Nitschke L. 2006. Regulation of B cell development and B cell signalling by CD22 and its ligands alpha2,6-linked sialic acids. Int Immunol. 18:603–611. [DOI] [PubMed] [Google Scholar]
- Gomes-Neto JC, Kittana H, Mantz S, Segura Munoz RR, Schmaltz RJ, Bindels LB, Clarke J, Hostetter JM, Benson AK, Walter J, et al. 2017. A gut pathobiont synergizes with the microbiota to instigate inflammatory disease marked by immunoreactivity against other symbionts but not itself. Sci Rep. 7:17707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. 2014. Human genetics shape the gut microbiome. Cell. 159:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvitovskaia A, Holmes SP, Huse SM. 2016. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, Takeyama N, Kamioka M, Sakamoto M, Matsuki T, et al. 2014. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 345:1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Uematsu S, Kiyono H. 2016. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. 17:1244–1251. [DOI] [PubMed] [Google Scholar]
- Guaraldi F, Salvatori G. 2012. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol. 2:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guittar J, Shade A, Litchman E. 2019. Trait-based community assembly and succession of the infant gut microbiome. Nat Commun. 10:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Vazquez C, Quintana FJ. 2018. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 48:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines CJ, Chen Y, Blumenschein WM, Jain R, Chang C, Joyce-Shaikh B, Porth K, Boniface K, Mattson J, Basham B, et al. 2013. Autoimmune memory T helper 17 cell function and expansion are dependent on interleukin-23. Cell Rep. 3:1378–1388. [DOI] [PubMed] [Google Scholar]
- Hennet T, Chui D, Paulson JC, Marth JD. 1998. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 95:4504–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JD, Baum LG. 2002. Ah, sweet mystery of death! Galectins and control of cell fate. Glycobiology. 12:127R–136R. [DOI] [PubMed] [Google Scholar]
- Hirmo S, Kelm S, Iwersen M, Hotta K, Goso Y, Ishihara K, Suguri T, Morita M, Wadstrom T, Schauer R. 1998. Inhibition of Helicobacter pylori sialic acid-specific haemagglutination by human gastrointestinal mucins and milk glycoproteins. FEMS Immunol Med Microbiol. 20:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdbrooks AT, Britain CM, Bellis SL. 2018. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 293:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project, C . 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE, Bollrath J, Danne C, Schiering C, Shale M, Adelmann K, Krausgruber T, Heger A, Sims D, Powrie F. 2016. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 10:2389–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamaki R, Ogawa K, Kizuka Y, Komi Y, Kojima S, Kotani N, Honke K, Honda T, Taniguchi N, Kitazume S. 2018. Glycosylation controls cooperative PECAM-VEGFR2-beta3 integrin functions at the endothelial surface for tumor angiogenesis. Oncogene. 37:4287–4299. [DOI] [PubMed] [Google Scholar]
- Irons EE, Lau JTY. 2018. Systemic ST6Gal-1 is a pro-survival factor for murine transitional B cells. Front Immunol. 9:2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons EE, Lee-Sundlov MM, Zhu Y, Neelamegham S, Hoffmeister KM, Lau JT. 2019. B cells suppress medullary granulopoiesis by an extracellular glycosylation-dependent mechanism. Elife. 8:7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons EE, Punch PR, Lau JTY. 2020. Blood-borne ST6GAL1 regulates immunoglobulin production in B cells. Front Immunol. 11:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Chen Y, Kanno Y, Joyce-Shaikh B, Vahedi G, Hirahara K, Blumenschein WM, Sukumar S, Haines CJ, Sadekova S, et al. 2016. Interleukin-23-induced transcription factor Blimp-1 promotes pathogenicity of T helper 17 cells. Immunity. 44:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaswal VM, Babbar HS, Mahmood A. 1988. Changes in sialic acid and fucose contents of enterocytes across the crypt-villus axis in developing rat intestine. Biochem Med Metab Biol. 39:105–110. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liu Z, Xu F, Dong X, Cheng Y, Hu Y, Gao T, Liu J, Yang L, Jia X, et al. 2018. Aberrant O-glycosylation contributes to tumorigenesis in human colorectal cancer. J Cell Mol Med. 22:4875–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Backhed F, et al. 2015. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 18:582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Nasirikenari M, Lugade AA, Thanavala Y, Lau JT. 2012. Anti-inflammatory IgG production requires functional P1 promoter in beta-galactoside alpha2,6-sialyltransferase 1 (ST6Gal-1) gene. J Biol Chem. 287:15365–15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Dorsett KA, Hjelmeland AB, Bellis SL. 2018. The ST6Gal-I sialyltransferase protects tumor cells against hypoxia by enhancing HIF-1alpha signaling. J Biol Chem. 293:5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG 3rd, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, et al. 2017. Neonatal acquisition of clostridia species protects against colonization by bacterial pathogens. Science. 356:315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Ahn JB, Kim DH, Kim S, Ma HW, Che X, Seo DH, Kim TI, Kim WH, Cheon JH, et al. 2020. Glutathione S-transferase theta 1 protects against colitis through goblet cell differentiation via interleukin-22. FASEB J. 34:3289–3304. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 108(Suppl 1):4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 10:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. 2017. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 38:633–647. [DOI] [PubMed] [Google Scholar]
- Layunta E, Javerfelt S, Dolan B, Arike L, Pelaseyed T. 2021. IL-22 promotes the formation of a MUC17 glycocalyx barrier in the postnatal small intestine during weaning. Cell Rep. 34:108757. [DOI] [PubMed] [Google Scholar]
- Lee MM, Nasirikenari M, Manhardt CT, Ashline DJ, Hanneman AJ, Reinhold VN, Lau JT. 2014. Platelets support extracellular sialylation by supplying the sugar donor substrate. J Biol Chem. 289:8742–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. 2016. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 150:1113–1124 e1115. [DOI] [PubMed] [Google Scholar]
- Leney AC, El Atmioui D, Wu W, Ovaa H, Heck AJR. 2017. Elucidating crosstalk mechanisms between phosphorylation and O-GlcNAcylation. Proc Natl Acad Sci U S A. 114:E7255–E7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, et al. 2015. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 528:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Abu-Ali G, Huttenhower C. 2016. The healthy human microbiome. Genome Med. 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan JC, Farzan SF, Hibberd PL, Karagas MR. 2012. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr Opin Pediatr. 24:753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood A, Torres-Pinedo R. 1985. Effect of hormone administration on the sialylation and fucosylation of intestinal microvillus membranes of suckling rats. Pediatr Res. 19:899–902. [DOI] [PubMed] [Google Scholar]
- Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, Huang Y, Gerner MY, Belkaid Y, Germain RN. 2018. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 554:255–259. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 4:447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. 2008. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 58:1221–1227. [DOI] [PubMed] [Google Scholar]
- Matthews SG. 2002. Early programming of the hypothalamo-pituitary-adrenal axis. Trends Endocrinol Metab. 13:373–380. [DOI] [PubMed] [Google Scholar]
- McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, Armentrout RA, Pociask DA, Hein A, Yu A, et al. 2016. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J Immunol. 197:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald ND, Lubin JB, Chowdhury N, Boyd EF. 2016. Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo. MBio. 7:e02237–e02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis A, Tzouvelekis L, Spiliadis C, Blackwell CC, Weir DM. 1990. Inhibition of Helicobacter pylori haemagglutination activity by human salivary mucins. FEMS Microbiol Immunol. 2:125–127. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison PJ, Bending D, Fouser LA, Wright JF, Stockinger B, Cooke A, Kullberg MC. 2013. Th17-cell plasticity in helicobacter hepaticus-induced intestinal inflammation. Mucosal Immunol. 6:1143–1156. [DOI] [PubMed] [Google Scholar]
- Nagpurkar A, Hunt D, Mookerjea S. 1996. Specific stimulation of alpha 2-6 sialyltransferase activity by a novel cytosolic factor from rat colon. Int J Biochem Cell Biol. 28:1337–1348. [DOI] [PubMed] [Google Scholar]
- Nanthakumar NN, Dai D, Meng D, Chaudry N, Newburg DS, Walker WA. 2005a. Regulation of intestinal ontogeny: effect of glucocorticoids and luminal microbes on galactosyltransferase and trehalase induction in mice. Glycobiology. 15:221–232. [DOI] [PubMed] [Google Scholar]
- Nanthakumar NN, Young C, Ko JS, Meng D, Chen J, Buie T, Walker WA. 2005b. Glucocorticoid responsiveness in developing human intestine: possible role in prevention of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 288:G85–G92. [DOI] [PubMed] [Google Scholar]
- Nanthakumar NN, Meng D, Newburg DS. 2013. Glucocorticoids and microbiota regulate ontogeny of intestinal fucosyltransferase 2 requisite for gut homeostasis. Glycobiology. 23:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Segal BH, Ostberg JR, Urbasic A, Lau JT. 2006. Altered granulopoietic profile and exaggerated acute neutrophilic inflammation in mice with targeted deficiency in the sialyltransferase ST6Gal I. Blood. 108:3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Chandrasekaran EV, Matta KL, Segal BH, Bogner PN, Lugade AA, Thanavala Y, Lee JJ, Lau JT. 2010. Altered eosinophil profile in mice with ST6Gal-1 deficiency: an additional role for ST6Gal-1 generated by the P1 promoter in regulating allergic inflammation. J Leukoc Biol. 87:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Veillon L, Collins CC, Azadi P, Lau JT. 2014. Remodeling of marrow hematopoietic stem and progenitor cells by non-self ST6Gal-1 sialyltransferase. J Biol Chem. 289:7178–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Lugade AA, Neelamegham S, Gao Z, Moremen KW, Bogner PN, Thanavala Y, Lau JTY. 2019. Recombinant sialyltransferase infusion mitigates infection-driven acute lung inflammation. Front Immunol. 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, da Costa G, van Hylckama Vlieg J, Sovran B, Chamignon C, et al. 2018. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun. 9:2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C, Blume J, Roy U, Teh PP, Vasanthakumar A, Beller A, Liao Y, Heinrich F, Arenzana TL, Hackney JA, et al. 2019. C-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol. 20:471–481. [DOI] [PubMed] [Google Scholar]
- Newburg DS, Morelli L. 2015. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr Res. 77:115–120. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 502:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh HJ, Koh HB, Kim HK, Cho HH, Lee J. 2017. Anti-bacterial effects of enzymatically-isolated sialic acid from glycomacropeptide in a Helicobacter pylori-infected murine model. Nutr Res Pract. 11:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP, Keane JA, Page AJ, Kumasaka N, Kane L, et al. 2014. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 16:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole J, Day CJ, von Itzstein M, Paton JC, Jennings MP. 2018. Glycointeractions in bacterial pathogenesis. Nat Rev Microbiol. 16:440–452. [DOI] [PubMed] [Google Scholar]
- Punch PR, Irons EE, Manhardt CT, Marathe H, Lau JTY. 2020. The sialyltransferase ST6GAL1 protects against radiation-induced gastrointestinal damage. Glycobiology. 30:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 118:229–241. [DOI] [PubMed] [Google Scholar]
- Rhee YH, Ku HJ, Noh HJ, Cho HH, Kim HK, Ahn JC. 2016. Anti-Helicobacter pylori effect of CaG-NANA, a new sialic acid derivative. World J Gastrointest Pathophysiol. 7:300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Brown J, Medley N, Dalziel SR. 2017. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst Rev. 3:CD004454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MN, Queiroz DM, Braga AB, Rocha AM, Eulailo EC, Braga LL. 2006. History of breastfeeding and Helicobacter pylori infection in children: results of a community-based study from northeastern Brazil. Trans R Soc Trop Med Hyg. 100:470–475. [DOI] [PubMed] [Google Scholar]
- Rothenbacher D, Bode G, Brenner H. 2002. History of breastfeeding and Helicobacter pylori infection in pre-school children: results of a population-based study from Germany. Int J Epidemiol. 31:632–637. [DOI] [PubMed] [Google Scholar]
- Rothhammer V, Quintana FJ. 2019. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 19:184–197. [DOI] [PubMed] [Google Scholar]
- Rudman D. 1973. Cortisone induction of small bowel sucrase activity. Nutr Rev. 31:158–159. [DOI] [PubMed] [Google Scholar]
- Salcedo J, Barbera R, Matencio E, Alegria A, Lagarda MJ. 2013. Gangliosides and sialic acid effects upon newborn pathogenic bacteria adhesion: an in vitro study. Food Chem. 136:726–734. [DOI] [PubMed] [Google Scholar]
- Salminen S, Gibson GR, McCartney AL, Isolauri E. 2004. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 53:1388–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Moser M, Sperandio M. 2013. The molecular basis of leukocyte recruitment and its deficiencies. Mol Immunol. 55:49–58. [DOI] [PubMed] [Google Scholar]
- Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, et al. 2016. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 65:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, Conner MG, Arend RC, Yoon KJ, Klug CA, et al. 2016. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 76:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senbanjo IO, Oshikoya KA, Njokanma OF. 2014. Helicobacter pylori associated with breastfeeding, nutritional status and recurrent abdominal pain in healthy Nigerian children. J Infect Dev Ctries. 8:448–453. [DOI] [PubMed] [Google Scholar]
- Simon PM, Goode PL, Mobasseri A, Zopf D. 1997. Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect Immun. 65:750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, et al. 2017. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Langley SA, Kim YM, Brislawn CJ, Noecker C, Zink EM, Fansler SJ, Casey CP, Miller DR, Huang Y, et al. 2016. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat Microbiol. 2:16221. [DOI] [PubMed] [Google Scholar]
- Solomon NS, Gartner H, Oesterreicher TJ, Henning SJ. 2001. Development of glucocorticoid-responsiveness in mouse intestine. Pediatr Res. 49:782–788. [DOI] [PubMed] [Google Scholar]
- Soltani J, Nikkhoo B, Khormehr J, Ataee P, Hakhamaneshi MS, Gharibi F. 2014. Breastfeeding and Helicobacter pylori infection in early childhood: a continuing dilemma. Iran J Pediatr. 24:745–752. [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 307:1955–1959. [DOI] [PubMed] [Google Scholar]
- Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, Skeath T, Perry JD, Petrosino JF, Berrington JE, et al. 2017. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, You P, Li WL, Tao XR, Zhu HY, Yao YC, Yu HY, Han QW, Yu B, Liu FX, et al. 2010. The existence of multipotent stem cells with epithelial-mesenchymal transition features in the human liver bud. Int J Biochem Cell Biol. 42(12):2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindall AF, Londono-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. 2013. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 73:2368–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoratou E, Campbell H, Ventham NT, Kolarich D, Pucic-Bakovic M, Zoldos V, Fernandes D, Pemberton IK, Rudan I, Kennedy NA, et al. 2014. The role of glycosylation in IBD. Nat Rev Gastroenterol Hepatol. 11:588–600. [DOI] [PubMed] [Google Scholar]
- Torres-Pinedo R, Mahmood A. 1984. Postnatal changes in biosynthesis of microvillus membrane glycans of rat small intestine: I. Evidence of a developmental shift from terminal sialylation to fucosylation. Biochem Biophys Res Commun. 125:546–553. [DOI] [PubMed] [Google Scholar]
- Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, Guttman DS, Becker AB, Mandhane PJ, Turvey SE, et al. 2017. Exposure to household furry pets influences the gut microbiota of infant at 3–4 months following various birth scenarios. Microbiome. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkonen KH, Wadstrom T, Moran AP. 1997. Identification of the N-acetylneuraminyllactose-specific laminin-binding protein of Helicobacter pylori. Infect Immun. 65:916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bodegom M, Homberg JR, Henckens M. 2017. Modulation of the hypothalamic-pituitary-adrenal axis by early life stress exposure. Front Cell Neurosci. 11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen WJ, Laman JD, t Hart BA. 2017. Modulation of multiple sclerosis and its animal model experimental autoimmune encephalomyelitis by food and gut microbiota. Front Immunol. 8:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertino-Bell A, Ren J, Black JD, Lau JT. 1994. Developmental regulation of beta-galactoside alpha 2,6-sialyltransferase in small intestine epithelium. Dev Biol. 165:126–136. [DOI] [PubMed] [Google Scholar]
- Vimr ER. 2013. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013:816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 68:132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Smith TJ, Lau JT. 1990. Transcriptional regulation of the liver beta-galactoside alpha 2,6-sialyltransferase by glucocorticoids. J Biol Chem. 265:17849–17853. [PubMed] [Google Scholar]
- Wang X, Vertino A, Eddy RL, Byers MG, Jani-Sait SN, Shows TB, Lau JT. 1993. Chromosome mapping and organization of the human beta-galactoside alpha 2,6-sialyltransferase gene. Differential and cell-type specific usage of upstream exon sequences in B-lymphoblastoid cells. J Biol Chem. 268:4355–4361. [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science. 334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tian J, Wang S. 2018. Insight into non-pathogenic Th17 cells in autoimmune diseases. Front Immunol. 9:1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuensch SA, Huang RY, Ewing J, Liang X, Lau JT. 2000. Murine B cell differentiation is accompanied by programmed expression of multiple novel beta-galactoside alpha2, 6-sialyltransferase mRNA forms. Glycobiology. 10:67–75. [DOI] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, Littman DR. 2018. C-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 554:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Shun CT, Chien CT, Wang TH. 2008. Effective prevention and treatment of Helicobacter pylori infection using a combination of catechins and sialic acid in AGS cells and BALB/c mice. J Nutr. 138:2084–2090. [DOI] [PubMed] [Google Scholar]
- Yang JC, Yang HC, Shun CT, Wang TH, Chien CT, Kao JY. 2013. Catechins and sialic acid attenuate Helicobacter pylori-triggered epithelial Caspase-1 activity and eradicate Helicobacter pylori infection. Evid Based Complement Alternat Med. 2013:248585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. 2012. Human gut microbiome viewed across age and geography. Nature. 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KY, Yeh M, Holt PR. 1991a. Thyroxine and cortisone cooperate to modulate postnatal intestinal enzyme differentiation in the rat. Am J Phys. 260:G371–G378. [DOI] [PubMed] [Google Scholar]
- Yeh KY, Yeh M, Montgomery RK, Grand RJ, Holt PR. 1991b. Cortisone and thyroxine modulate intestinal lactase and sucrase mRNA levels and activities in the suckling rat. Biochem Biophys Res Commun. 180:174–180. [DOI] [PubMed] [Google Scholar]
- Zagon IS, Rahn KA, Bonneau RH, Turel AP, McLaughlin PJ. 2010. Opioid growth factor suppresses expression of experimental autoimmune encephalomyelitis. Brain Res. 1310:154–161. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. 2015. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 21:895–905. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lu J, Xu Z, Zou X, Sun X, Xu Y, Shan A, Lu J, Yan X, Cui Y, et al. 2017. Differential expression of ST6GAL1 in the tumor progression of colorectal cancer. Biochem Biophys Res Commun. 486:1090–1096. [DOI] [PubMed] [Google Scholar]
- Zheng W, Ma Y, Zhao A, He T, Lyu N, Pan Z, Mao G, Liu Y, Li J, Wang P, et al. 2019. Compositional and functional differences in human gut microbiome with respect to equol production and its association with blood lipid level: a cross-sectional study. Gut Pathog. 11:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietak M, Kovatcheva-Datchary P, Markiewicz LH, Stahlman M, Kozak LP, Backhed F. 2016. Altered microbiota contributes to reduced diet-induced obesity upon cold exposure. Cell Metab. 23:1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.