Abstract
In recent years, protein kinases have been one of the most pursued drug targets. These determined efforts have resulted in ever increasing numbers of small-molecule kinase inhibitors reaching to the market, offering novel treatment options for patients with distinct diseases. One essential component related to the activation and normal functionality of a protein kinase is the regulatory spine (R-spine). The R-spine is formed of four conserved residues named as RS1–RS4. One of these residues, RS3, located in the C-terminal part of αC-helix, is usually accessible for the inhibitors from the ATP-binding cavity as its side chain is lining the hydrophobic back pocket in many protein kinases. Although the role of RS3 has been well acknowledged in protein kinase function, this residue has not been actively considered in inhibitor design, even though many small-molecule kinase inhibitors display interactions to this residue. In this minireview, we will cover the current knowledge of RS3, its relationship with the gatekeeper, and the role of RS3 in kinase inhibitor interactions. Finally, we comment on the future perspectives how this residue could be utilized in the kinase inhibitor design.
Keywords: kinase inhibitors, point mutations, protein kinases, protein–ligand interaction, regulatory-spine
Introduction
Protein kinases are dynamic proteins that regulate a multitude of cellular signalling processes. They control the activity of their downstream targets mainly by phosphorylation, and their own activity is also usually controlled in the same manner. The human kinome comprises more than 500 protein kinases [1], and nearly 500 proteins contain a typical kinase domain [2]. Still, the biological role of many protein kinases is largely unknown, and there are ongoing efforts aiming to characterize these poorly understood protein kinases [3]. Although protein kinases display high similarity in their kinase domain, there is a higher level diversity in their structures; while some kinases consist (almost) solely of the kinase domain (e.g. MAPK14, GSK3B), other are larger multidomain proteins (e.g. LRRK2 [4]). The structure and function of the protein kinase domain is well-established (Figure 1A). For a comprehensive view of structural history of protein kinases, the reader is recommend a recent review by Taylor et al. [5].
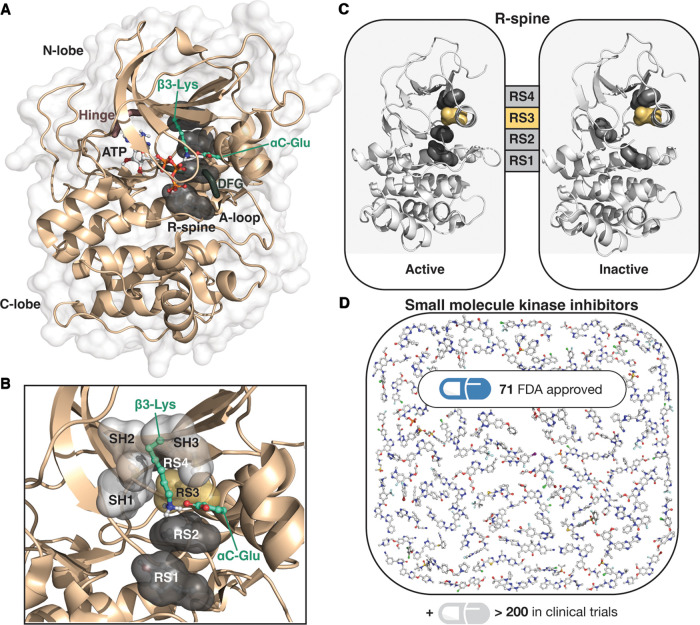
Figure 1. Protein kinase domains are important drug targets.
(A) A typical structure of a protein kinase domain. ATP-binding cleft is located between the N- and C-lobes of the kinase. In the figure, structure of cAMP-dependent protein kinase catalytic subunit alpha is depicted (PDB ID: 4wb5 [20]; inhibitory peptide is hidden). R-spine residues are illustrated with black surface. ATP, β3-Lys and αC-Glu are shown with stick model. (B) Shell residues SH1–SH3 (grey surface) are located next to the R-spine RS3 and RS4 residues. Shell residue SH2 (gatekeeper) is located close to RS3 (yellow surface). (C) The R-spine of a protein kinase is assembled in active conformation and disassembled in inactive conformation. In the figure, active and inactive configurations are illustrated with BRAF (PDB IDs: 4e26 [21] and 1uwh [22]). (D) Several small-molecule kinase inhibitors are already in clinical use, and dozens are in clinical trials.
In the protein kinase domain, one of the key dynamic elements in regulating protein kinase function is the hydrophobic regulatory-spine (R-spine), which was discovered already 15 years ago in 2006 [6]. The R-spine consists of four residues, named RS1–RS4, which connects the two lobes of the kinase domain (Figure 1A,B). Two of these residues, RS1 and RS2, belong the C-lobe. RS1 is His (sometimes Tyr) residue from the HRD (or YRD) motif [7]. RS2 is Phe (or Leu) from the DFG-motif, which is part of the activation loop of a protein kinase. The other two R-spine residues belong to the N-lobe of the protein kinase. RS4 is a residue from the β4-strand, which is less conserved but frequently Leu can be found in this position. Finally, RS3 is located four residues C-terminal from the αC-helix Glu that forms a salt-bridge to the Lys of β3-sheet. RS3 is usually (not always) accessible from the ATP binding site as its side chain lines the active site cleft. Overall, αC-helix, where RS3 is located, has a central role in the kinase activation process [8]. In the catalytically active form of a protein kinase, R-spine is assembled and in the inactive state it is disassembled (Figure 1C). In the active state the location of RS2 as part of the assembled R-spine results in an open and extended conformation of the activation loop (A-loop), while a closed A-loop configuration is preferred in the inactive state. Notably, additional stabilization of the R-spine, such as via in-frame insertions or RS3 mutations [9,10], may result in increased catalytic activity of the protein kinase.
Next to the R-spine in the N-lobe are located three conserved residues, named as Shell (SH) residues (Figure 1C) [9]. These residues, which are usually hydrophobic, have a role in supporting R-spine and are therefore important for kinase activity. One of these residues, SH2, is found close to RS3. This SH2 residue is more commonly known as the gatekeeper residue, which is named due to its role in controlling access to the hydrophobic pocket [11]. This shell residue participates in regulating R-spine dynamics, and gatekeeper mutations may stabilize the R-spine promoting the kinase activation [12].
In recent years, ever increasing efforts have been conducted by the pharmaceutical industry to target protein kinases [13]. These efforts have resulted in numerous small-molecule kinase inhibitors, totalling now over 70 FDA approved small-molecule inhibitors (Figure 1D). According to the Protein Kinase Inhibitors in Clinical Trials database (PKIDB) [14,15], approximately 300 small-molecule kinase inhibitors are either in clinical trials or already approved. Comprehensive reviews of the kinase inhibitor drug discovery and kinase inhibitor development are available [16–18]. Currently, oncology is dominating indication for the kinase inhibitors, but there is potential also in other therapeutic areas such as autoimmune and inflammatory diseases, and degenerative disorders [19].
Here, we review the characteristics of RS3 and its relationship with the neighbouring gatekeeper (SH2) based on the publicly available structural data. We also have a look at RS3 interactions to small-molecule kinase inhibitors, including the approved drugs. Finally, we end the review with available mutational data of RS3.
RS3 in the human kinome
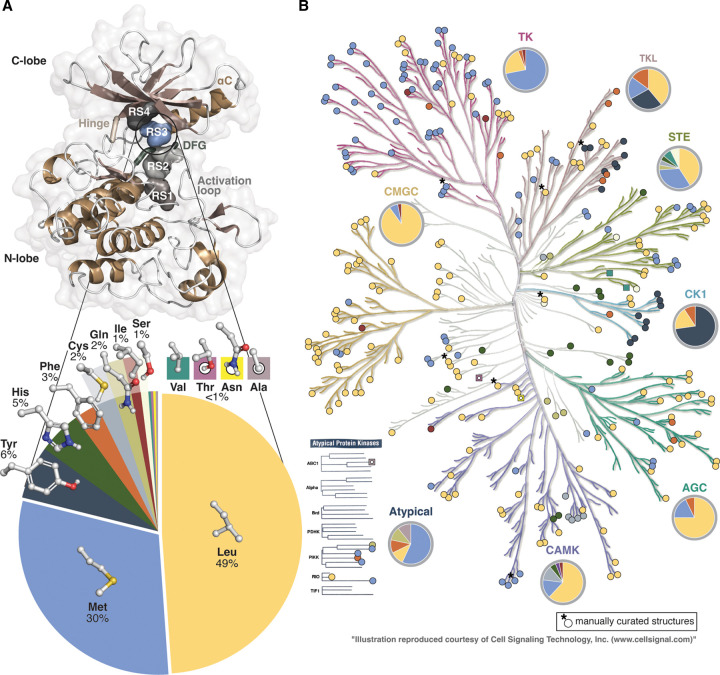
A majority of the human protein kinases with publicly disclosed structures display a nonpolar aliphatic RS3 residue (Figure 2). Nearly half of these kinases exhibit Leu in RS3, and almost a third have Met (Figure 2A). In the overall human proteome Leu is also the most abundant residue (9.97%), while Met has the second lowest frequency (2.13%) of all amino acids [23]. Following the abundant Leu and Met in RS3, next preferred are aromatic residues. Tyr, His and Phe appear with the frequencies of 3–6%. Cys and Gln exist in 2%, Ile and Ser in 1%. Even more rare residues that are observed in this location are Val, Thr, Asn and Ala. The charged residues, Asp, Glu, Lys and Arg, as well as structurally more unique Trp, Gly or Pro are not present in the analysed set of human protein kinases in RS3. Based on the sequence alignment of a larger set of eucaryotic protein kinases these residues have been suggested to exist as RS3 residues, although rarely [7]. Regardless of the high existence of hydrophobic residues in RS3, there exists no clear trend related to the hydrophobicity ranking of the residues and their observed frequencies [24,25].
Figure 2. RS3 in human protein kinases with available structures.
(A) Occurrence of RS3 residues in human protein kinases with publicly available structural data (289 kinases). The location of RS3 is highlighted in IGFR1 kinase domain (PDB ID: 3qqu [28]). The shown frequencies are rounded up to the nearest %, for residues with <1% frequency, percentage is not shown. (B) RS3 residue distribution in human kinome. Colours of the residue types are as in A. Eight structures with KLIFS annotation errors that were manually curated (RS3 was properly assigned) are indicated with an asterisk. Data in A and B consist of human protein kinases with publicly available structures with (with lipid kinases excluded). Human kinome tree illustration was made with the help of KinMap [29].
Protein kinases of different groups and families display distinct preferences for RS3 residues (Figure 2B). The majority of the kinases (72%) belonging to the TK group display Met RS3. More than a fifth (22%) of this group present Leu in this position, including protein kinases belonging to JakA family (JAK1–3; TYK2) and Trk family (NTRK1–3 also known as TRKA–C). Four protein kinases have either Ile (ALK; ERBB3 (ErbB3)) or a Phe (PTK7 (CCK4); LMTK3 (LMR3)). In EGFR family Met is preferred in RS3, except ErbB3 has Ile. Interestingly, ErbB3 has been identified to display considerably lower kinase activity [26,27]. However, ErbB3 displays also other unique characteristics that differ from other EGFRs (for instance, instead of αC-Glu ErbB3 has a His in this location). In the PDGFR family, KIT displays Leu instead of Met that is observed in other family members (FLT, PDGFRA, CSF1R (FMS)).
In contrast with the TKs, in the CMGC group Leu is clearly dominating RS3 (90%). Only three protein kinases have Met in RS3 (MAPK8 (JNK1); MAPK10(JNK3); GSK3B), and MAPK6 (Erk3) displays Ile in this position.
The TKLs prefer quite diversely Leu, Met, Tyr and Phe in their RS3. Members of the STKR family prefer an aromatic residue in this position: Tyr is observed in ACVR1 (ALK2), ACVR2A (ACTR2), ACVRL1 (ALK1), BMBR1B, BMBR2 and TGFBR1 (TGFbR1); Phe in ACVR2B (ACTR2B) and TGFBR2 (TGFbR2). Aromatic Phe is also present in two MLK family members: MAP3K9 (MLK1) and MAP3K21 (MLK4), while other kinases in this family display Leu (ILK; MAP3K12 (DLK); MAP3K20 (ZAK); MAP3K7 (TAK1); TNNI3K (HH498)).
Kinases of the STE group display mainly Leu and Met in RS3. However, also other RS3 residues are observed in this group, including His (STE11) and Ser (STRADA (STLK5); MAP2K6), Gln (MAP3K8 (COT)), Cys (MAP3K14 (NIK)) and Val (MAP2K4; MAP2K7). From these, MAP3K14 (NIK) and MAP3K8 (COT) belong to the STE-Unique family. Three members of the STE7 family display rare RS3 residues. These kinases are MAP2K6 (Ser), MAP2K4 also known as MKK4 (Val) and MAP2K7 also known as MKK7 (Val). Interestingly, unique autoinhibited conformational states have been reported for these kinases state [30–32]; with MKK4 this state may be related to its dimer form [33].
In the CK1 group, Tyr is dominating in RS3 (73%). VRK3 displays an aromatic Phe in this position. TTBK family kinases (TTBK1, TTBK2) are more diverse in this group with their aliphatic Leu in RS3.
The majority of the AGC and CAMK group kinases exhibit Leu in RS3, which is followed by Met with lower frequencies. Two kinases with an aromatic RS3 (Phe) are observed in AGC group, in PKN2 and PRKCI (PKCi). In CAMK group, kinases of the CAMK2 family (CAMK2A, CAMK2B, CAMK2D, CAMK2G) display Cys in RS3, as well as CASK. His is observed in MAPKAP family (MAPKAP2, MAPKAP3). TRIB1 (Trb1) represents Ile in RS3; however, in this pseudokinase the neighbouring Tyr may actually occupy the canonical RS3 position [34,35].
The protein kinases that are not belonging to any specific group display also family specific preferences. For instance, protein kinases of WEE family and PLK family exhibit His in RS3 (located above and below CK1 group in the kinome tree). Of the Atypical kinases, COQ8A that is also known as ADCK3 [36], is the only structure in the dataset that displays Ala in RS3.
Polar RS3 are rare
Not only hydrophobic RS3 residues exist, but also polar residues are observed in this position. AURKA is an example of a widely studied kinase that has a polar RS3 (Gln) is. It was disclosed by Levinson et al. that this polar residue has a specific role in AURKA activation via a water-network [37]. Similarly, AURKB and AURKB have also Gln as their RS3. In addition, Gln is observed in MAP3K8 (COT, TPL2) and ATM. MAP3K8 controls inflammation [38] and ATR DNA damage responses [39].
In the available data, Ser is observed in three kinases. While in STRADA (STLK5, STE20) this residue appears unreachable from the binding cleft (PDB ID: 3gni [40], 2wtk [41]), in Haspin Ser is accessible (participates in water coordination next to αC-Glu (PDB ID: 4ouc [42]). RS3 Ser may also be accessible in MAP2K6 when it is not in its autoinhibited state (PDB ID: 3fme). In the autoinhibited state its neighbouring Met appears to take the regular RS3 position (PDB ID: 3vn9 [32]).
Two unique polar RS3 residues are present in the data. Asn is observed in the RS3 of CHK1, while all the other kinases belonging to the same CAMKL family have either a lipophilic Leu or Met in this position. CHK1 inhibition could be useful in the treatment of KRAS driven pancreatic ductal adenocarcinoma [43]. Thr is observed in ULK4, while other members of the ULK family (ULK1–ULK3) have Leu in the respective position. ULK4 is a pseudokinase and has an unusual structural characteristic in its αC-helix: it exhibits Trp residue in the location of αC-Glu. This Trp appears to participate in its R-spine formation [44].
RS3 relationship with gatekeeper
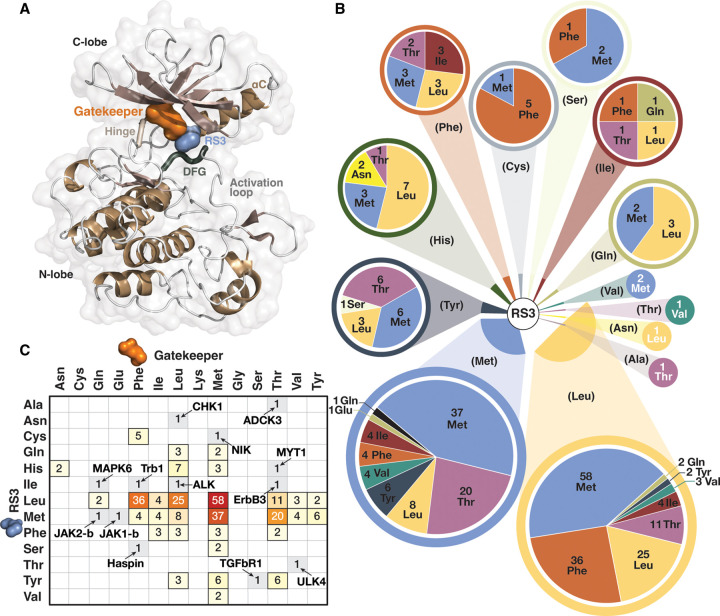
The access towards the hydrophobic pocket (and towards RS3) is controlled by gatekeeper, also known as SH2 residue. This residue may also influence R-spine dynamics and it can be found in close contact to RS3 (Figure 3A). Generally, protein kinases prefer Met, Leu, Phe and Thr gatekeepers (Figure 3B,C). In the available structures, Met is the most abundant gatekeeper (40%), followed by Leu (18%), Phe (16%) and Thr (15%). Less frequent gatekeepers — but presented in more than eight kinase domains — are Ile 4%, Tyr 3%, Val 3%.
Figure 3. Gatekeeper and RS3.
(A) Gatekeeper is located near RS3, and in some cases these residues are in contact. In the figure, gatekeeper of the protein kinase IGF1R (PDB ID: 3qqu [28]) is shown with orange surface and RS3 with blue surface. (B) Distribution of gatekeeper residues according to their RS3 residue (in parenthesis) represented in pie charts. (C) Correlation matrix of RS3 and gatekeeper residues. The unique RS3-gatekeeper combination containing kinases are labelled.
Kinases with different RS3 residues display also distinct gatekeeper preferences (Figure 3B,C). With Met in RS3, less Phe gatekeepers are observed in comparison with when RS3 is Leu. Bulky aromatic RS3 residues (Tyr, His, Phe) do not exist in combination with aromatic gatekeepers. The kinase domains with Cys in RS3 appear to prefer Phe gatekeepers. Polar Gln and Asn display either Leu or Met as their gatekeepers. While Thr is rare RS3 residue, as a gatekeeper it is common with Leu, Met and Tyr in RS3. In addition to Thr, other polar gatekeepers do exist. Asn gatekeeper is observed only in combination with His RS3 in WEE1 (Wee1) (PDB ID: 3biz [45]) and WEE2 (Wee1B) (PDB ID: 5vdk [46]). Gln gatekeeper is observed in four kinases, including MAPK1 (Erk2) (PDB ID: 5hnv [47]), MAPK3 (Erk1) (PDB ID: 4qtb [48]), MAPK6 (Erk3) (PDB ID: 7aqb [49]), and pseudokinase domain JAK2-b (PDB ID: 4fvq [50]). Pseudokinase domain Jak1-b has a unique polar Glu as its gatekeeper (PDB ID: 4l00 [51]). Overall, thirteen protein kinases in the analysed dataset display unique RS3-gatekeeper combinations (Figure 3C).
RS3 and small-molecule kinase inhibitors
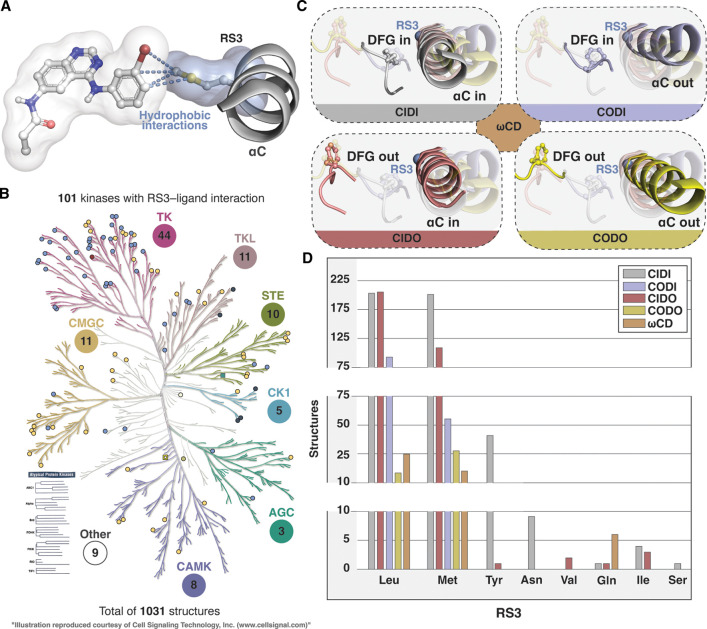
We searched the KLIFS database [52] and complemented our search using Protein–Ligand Database (PLDB) tool of Maestro (Schrödinger LLC) to map out all the existing protein kinase–ligand complexes that have contacts between RS3 and the ligand (Figure 4). In KLIFS, RS3 is named as residue #28 (αC-Glu is #24) [53]. Over 100 protein kinases have structures where RS3–ligand interactions are observed (Figure 4B). In total, more than 1000 structures with RS3–ligand interactions are available.
Figure 4. Publicly available structures that display RS3–ligand contacts.
(A) An example of a structure displaying hydrophobic contacts to RS3 (PDB ID: 4lqm [54]) (B) Structures with RS3–ligand interactions. Results of the KLIFS search, which was complemented with Protein–Ligand database (PLDB) (Schrödinger LLC) that identified 197 additional structures. (C) Conformation classification of protein kinases based on DFG and αC-helix conformations as disclosed by Ung et al.[55]. The ambiguous conformations are named as ωCD, which may represent the transition conformations between the four states. Location of RS3 Cα-atom is illustrated with a blue sphere. (D) Number of structures with RS3–ligand contacts and divided by RS3 residue type and KinaMetrix [56] defined kinase conformations. For 11 structures the conformation was undefined and those were excluded.
RS3–ligand interactions are observed with kinases containing Leu, Met, Tyr, Asn, Val, Gln, Ile or Ser in their RS3. No direct RS3–ligand contacts were observed for kinases with Ala, Cys, Phe, His and Thr. Polar interactions to RS3 are extremely rare. H-bond interaction occurs between inhibitor and RS3 only in 12 structures, including AURKA (Gln) (PDB ID: 4uzd [57]); CHK1 (Asn) (PDB IDs: 4fsq, 4fst, 4ftk, 4ftl, 4ftm, 4ftn); Haspin (Ser) (PDB ID: 6z5a); VRK1 (Tyr) (PDB IDs: 6btw, 6cfm, 6cnx, 6npn). Hydrophobic interactions are abundant, and hydrophobic contacts to Leu and Met appear in 551 and 410 structures, respectively. This is not surprising, based on their high frequency in RS3 (79%) among the available structures. Interactions to Tyr appear in 41 structures, while other RS3 residues with interactions are represented each with 10 or less structures.
Interactions to RS3 appear independent on the kinase conformation (Figure 4C,D) [55]. Based on the KinaMetrix [56], ‘αC-helix in' conformations are dominating in the structures. CIDI is the most populated with 459 structures and CIDO appears in 323 structures. CODI and CODO structures with αC-helix out configuration exist in 148 and 42 structures, respectively. The ambiguous ωCD occurs in 47 structures. Leu and Met RS3–ligand interaction structures display all configurations, albeit less structures of CODI (16%), CODO (4%) and ωCD (4%) conformations exists. With less structural information containing RS3 residues the conformational representation does not cover all configurations. Nevertheless, all conformations are present in the structures with RS3–ligand interactions. As there exists distinct protein kinase conformation classifications, we also analysed the conformational distribution of RS3 contact structures with Kincore [58]. Table 1 shows the conformational distribution of these structures assigned with the Kincore. Overall, majority of the available compounds with the RS3 interactions exist in DFGin and DFGout spatial classes, covering different conformational classes (dihedrals).
Table 1. RS3–ligand contact structures with Kincore defined conformations.
| Spatial Label | Dihedral label | Annotation | Leu1 | Met1 | Tyr | Asn | Val | Gln | Ile | Ser |
|---|---|---|---|---|---|---|---|---|---|---|
| DFGin | BLAminus | Active | 71 | 96 | 39 | 9 | - | - | - | 1 |
| BLBplus | SRC-inactive | 120 | 50 | - | - | - | - | - | - | |
| ABAminus | Active-like | 21 | 21 | 1 | - | - | - | 3 | - | |
| BLBminus | 36 | 33 | - | - | - | - | - | - | ||
| BLAplus | FGFR-inactive | 23 | 39 | - | - | - | - | - | - | |
| BLBtrans | CDK-inactive | - | 7 | - | - | - | - | - | - | |
| None | 38 | 12 | - | - | - | 2 | 1 | - | ||
| DFGinter | BABtrans | - | - | - | - | - | - | - | - | |
| None | AURKA-inactive | 13 | 6 | - | - | - | 3 | - | - | |
| DFGout | BBAminus | Type-2 binding | 151 | 101 | 1 | - | 1 | 1 | 2 | - |
| None | DFGout-like | 59 | 32 | - | - | 1 | 2 | 1 | - | |
| None | None | 13 | 4 | - | - | - | 2 | - | - | |
| Total | 545 | 401 | 41 | 9 | 2 | 10 | 7 | 1 |
Conformation definitions were unavailable in Kincore for six (Leu) and nine (Met) individual structures included in Figure 4D.
Approved small-molecule kinase inhibitors and RS3
A publicly available structure exists for 49 out of the 71 FDA-approved small-molecule kinase inhibitors. From these, interaction to RS3 is displayed by 26 inhibitors (55%). These structures include targets with Met (Figure 5) and Leu RS3 residues (Figure 6). Inhibitors which exhibit RS3 interactions represent all types of kinase inhibitors that bind to the ATP-binding cleft. Of note, inhibitors of different type may engage RS3 site in a different manner. For instance, type II inhibitors, which bind the kinase in its inactive conformation, reach beyond the RS3 on the αC-helix side, and thereby can interact with the side chain to its ‘side' or ‘head' or both. Conversely, type I inhibitors, which bind to the active conformation of the kinase, interact mainly with the head of the RS3 side chain.
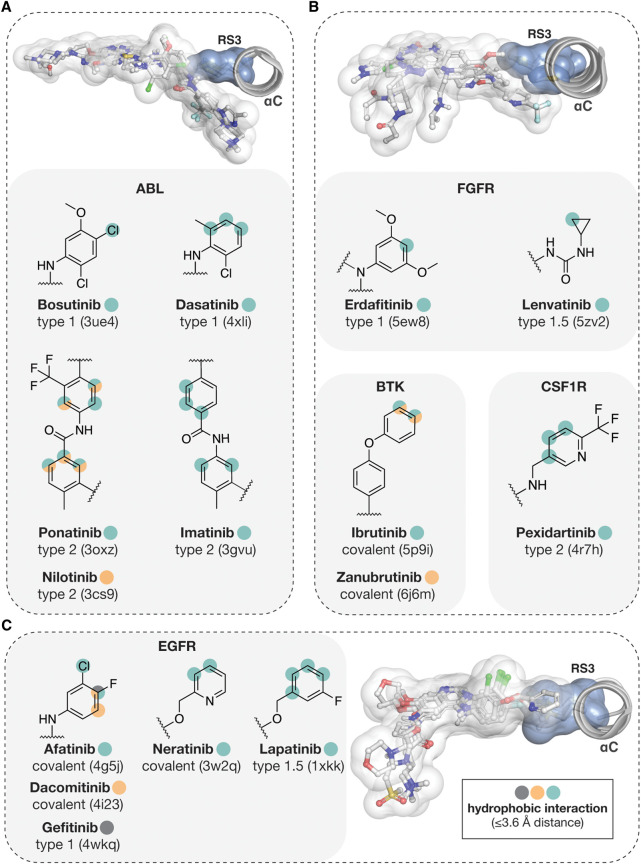
Figure 5. FDA approved small-molecule kinase inhibitors with Met RS3 contacts.
(A) ABL inhibitors. (B) FGFR, BTK and CSF1R inhibitors. (C) EGFR inhibitors. In 3D images, structures of the superimposed kinases are shown with their bound inhibitors. Full kinase inhibitor structures are represented with transparent white surface and stick models. RS3 Met is shown with transparent blue surface and sticks. The RS3 interacting atoms of the inhibitors are highlighted in 2D-structures of the structural moieties that are located near the RS3 residue. Hydrophobic interaction was defined based on a 3.6 Å (or shorter) distance between two hydrophobic atoms.
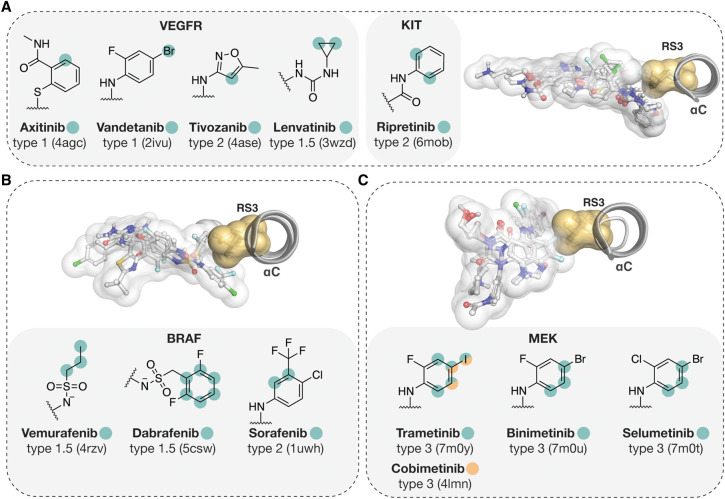
Figure 6. FDA approved small-molecule kinase inhibitors with Leu RS3 contacts.
(A) VEGFR and KIT inhibitors. (B) BRAF inhibitors. (C) MEK inhibitors. In 3D images, structures of the superimposed kinases are shown with their bound inhibitors. Full kinase inhibitor structures are represented with transparent white surface and stick models. RS3 Leu is shown with transparent yellow surface and sticks. The RS3 interacting atoms of the inhibitors are highlighted in 2D-structures of the structural moieties that are located near the RS3 residue. Hydrophobic interaction was defined based on a 3.6 Å (or shorter) distance between two hydrophobic atoms.
In occupying the region next to RS3, mainly six membered aromatic ring containing structural moieties are preferred (Figures 5, 6). Correspondingly, these aromatic rings participate in RS3 interactions in most cases. Tivozanib is the only inhibitor that presents a five-membered heteroaromatic ring in this location. Furthermore, aromatic ring attached halogens, Cl with RS3 Met (bosutinib; afatinib) and Br or I with RS3 Leu (vandetanib; trametinib, cobimetinib), display contacts to RS3. Vemurafenib and lenvatinib are the only exceptions that utilize nonaromatic moieties to interact with RS3. In addition to VEGFRs (Leu), lenvatinib also binds and inhibits FGFR-1 (Met). To both, Leu or Met, lenvatinib is in contact from its cyclopropyl urea moiety (PDB IDs: 3wzd [59], 5zv2 [60]). Vemurafenib displays contacts to RS3 Leu from aliphatic carbons of its propylsulfonamide group (PDB ID: 4rzv [61]).
Some of these drugs display interactions with other kinases RS3 that are distinct from their main target. Bosutinib displays hydrophobic contacts to RS3 with MST3 (Leu) (PDB ID: 4qmn [62]), as well as dasatinib to Leu with MAPK14 (p38a) (PDB ID: 3lfa). Ponatinib displays hydrophobic interactions to Leu with RIPK2 (PDB ID: 4c8b [63]), KIT (PDB ID: 4u0i [64]) and BRAF (PDB ID: 6p3d [65]). Nilotinib to Leu with MAPK11 (p38b) (PDB ID: 3gp0). Imatinib displays contact to Leu when bound to KIT (PDB ID: 1t46 [66]) and MAPK14 (p38a) (PDB ID: 3hec [67]). Also, Pexidartinib exhibits contact to RS3 Leu with KIT (PDB ID: 7khg [68]). For inhibitors which main targets display Leu in RS3, apart from lenvatinib no Met RS3 interaction containing structures are available.
RS3 interaction is kinase dependent. Approved drugs with interactions to RS3 do not necessarily exhibit contacts to RS3 with other kinases that they bind to. For instance, gefitinib has been co-crystallized with GAK (Met), where it displays no contact to RS3 (PDB ID: 5y7z [69]). Ibrutinib has been also co-crystallized with MAP2K7 that has Val in RS3, but it does not display any contacts to this residue in this complex (PDB ID: 6yg2 [70]).
Mutations in RS3 exist rarely
According to the Catalogue of Somatic Mutations In Cancer (COSMIC; v.95) database [71], no clear tendency for mutations in RS3 exists. In total 82 kinases display at least one mutation (missense or silent) (Table 2). Only with ALK, several mutations at this location appear in the data. These mutations include, I1171N, I1171T and I1171S. For BRAF, L505H mutation is found in eight samples. Perhaps the low number of observed RS3 mutations is not surprising, due to the crucial role of this residue in the kinase function. In comparison, RS2 mutations are also rare, with BRAF F595L (13 samples in COSMIC v.95) being the most frequent in the analysed kinases. Meanwhile, RS2 flanking residues are common oncogenic drivers; for instance, BRAF V600E is found in 52 733 samples and EGFR L858R is present in 10 642 samples. Mutations at αC-helix may activate the kinase via destabilizing the kinase inactive conformation [72], but they are mainly found in other locations on the αC-helix than on RS3 [73].
Table 2. Missense mutations in RS3 (COSMIC v.95).
| Kinase | RS3 | RS3-contact1 | Mutation | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | -2 | Total | ||||
| TK | ||||||||||||||||||||
| 1 | ABL1 | M290 | + | 1 | 1 | |||||||||||||||
| 2 | ALK | I1171 | + | 18 | 11 | 5 | 2 | 36 | ||||||||||||
| 3 | DDR2 | M629 | + | 2 | 2 | |||||||||||||||
| 4 | EGFR | M766 | + | 1 | 1 | 1 | 1 | 4 | ||||||||||||
| 5 | FGFR2 | M538 | + | 1 | 1 | 1 | 3 | |||||||||||||
| 6 | FLT1 | L882 | + | 1 | 1 | |||||||||||||||
| 7 | JAK2 | L902 | + | 1 | 1 | |||||||||||||||
| 8 | TYK2 | L951 | + | 1 | 1 | |||||||||||||||
| 9 | KDR | L889 | + | 1 | 1 | 2 | ||||||||||||||
| 10 | KIT | L644 | + | 1 | 1 | |||||||||||||||
| 11 | PDGFRa | M648 | + | 1 | 1 | |||||||||||||||
| 12 | LCK | M292 | + | 4 | 4 | |||||||||||||||
| 13 | SYK | M424 | + | 1 | 1 | |||||||||||||||
| 14 | AXL | M589 | - | 2 | 1 | 2 | 5 | |||||||||||||
| 15 | EphA3 | M674 | - | 2 | 2 | |||||||||||||||
| 16 | EphA7 | M686 | - | 1 | 1 | |||||||||||||||
| 17 | EphB3 | M686 | - | 1 | 1 | |||||||||||||||
| 18 | ErbB3 | I744 | - | 2 | 2 | |||||||||||||||
| 19 | ITK | M410 | - | 1 | 1 | |||||||||||||||
| 20 | JAK3 | L875 | - | 2 | 1 | 5 | 8 | |||||||||||||
| 21 | ROR2 | M526 | - | 1 | 1 | |||||||||||||||
| 22 | ROS1 | M2001 | - | 1 | 1 | |||||||||||||||
| 23 | ZAP70 | M390 | - | 1 | 1 | |||||||||||||||
| TKL | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 24 | MAP3K7 | L81 | + | 1 | 1 | |||||||||||||||
| 25 | BRAF | L505 | + | 8 | 8 | |||||||||||||||
| 26 | ACVR2B | F234 | - | 4 | 4 | |||||||||||||||
| 27 | MAP3K9 | F195 | - | 3 | 3 | |||||||||||||||
| 28 | RAF1 | L397 | - | 1 | 1 | |||||||||||||||
| 29 | HH498 | L513 | - | 2 | 1 | 1 | 4 | |||||||||||||
| STE | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 30 | PAK1 | M319 | + | 1 | 1 | 2 | ||||||||||||||
| 31 | PAK4 | M370 | + | 1 | 1 | |||||||||||||||
| 32 | STK10 | L85 | + | 1 | 1 | |||||||||||||||
| 33 | SLK | L83 | + | 1 | 1 | 2 | ||||||||||||||
| 34 | MAP2K1 | L118 | + | 1 | 2 | 3 | ||||||||||||||
| 35 | MAP2K6 | S103 | - | 2 | 2 | |||||||||||||||
| 36 | MAP3K14 | C444 | - | 1 | 1 | |||||||||||||||
| 37 | OXSR1 | M67 | - | 1 | 1 | |||||||||||||||
| 38 | PAK5 | M498 | - | 1 | 1 | |||||||||||||||
| 39 | TNIK | L73 | - | 1 | 1 | |||||||||||||||
| AGC | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 40 | AKT2 | L204 | + | 1 | 1 | |||||||||||||||
| 41 | ROCK1 | M128 | + | 1 | 1 | |||||||||||||||
| 42 | AKT1 | L202 | - | 2 | 2 | |||||||||||||||
| 43 | CDC42BPB | L128 | - | 1 | 1 | |||||||||||||||
| 44 | DMPK1 | L123 | - | 1 | 1 | 2 | ||||||||||||||
| 45 | GPRK5 | L238 | - | 1 | 1 | |||||||||||||||
| 46 | MASTL | L85 | - | 1 | 1 | |||||||||||||||
| 47 | PRKCh | L407 | - | 1 | 1 | |||||||||||||||
| 48 | SGK1 | L150 | - | 2 | 1 | 1 | 2 | 6 | ||||||||||||
| CAMK | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 49 | CHEK1 | N59 | + | 1 | 1 | |||||||||||||||
| 50 | DRAK2 | L84 | + | 1 | 1 | |||||||||||||||
| 51 | PIM1 | L93 | + | 1 | 3 | 4 | ||||||||||||||
| 52 | RPS6KA3 | L467 | + | 1 | 1 | 2 | ||||||||||||||
| 53 | CAMK1d | L73 | - | 1 | 1 | |||||||||||||||
| 54 | CAMK4 | L93 | - | 3 | 1 | 3 | 7 | |||||||||||||
| 55 | MARK2 | M104 | - | 1 | 1 | |||||||||||||||
| 56 | MNK1 | L98 | - | 2 | 2 | |||||||||||||||
| 57 | MNK2 | L133 | - | 1 | 1 | |||||||||||||||
| 58 | SgK085 | M155 | - | 2 | 2 | |||||||||||||||
| 59 | PHKg2 | L81 | - | 1 | 1 | 2 | ||||||||||||||
| 60 | AMPKa2 | L68 | - | 1 | 1 | |||||||||||||||
| 61 | RPS6KA2 | L113 | - | 2 | 2 | |||||||||||||||
| 62 | SNRK | M67 | - | 1 | 1 | |||||||||||||||
| 63 | STK17B | L84 | - | 1 | 1 | |||||||||||||||
| CDK | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 64 | GSK3B | M101 | + | 1 | 1 | |||||||||||||||
| 65 | JNK3 | M115 | + | 1 | 1 | |||||||||||||||
| 66 | Erk5 | L106 | + | 2 | 2 | |||||||||||||||
| 67 | CDK4 | L60 | - | 1 | 3 | 4 | ||||||||||||||
| 68 | CDK6 | L65 | - | 2 | 2 | |||||||||||||||
| 69 | CK2a1 | L85 | - | 1 | 1 | |||||||||||||||
| 70 | DYRK1A | L207 | - | 1 | 1 | 2 | ||||||||||||||
| 71 | HIPK2 | L247 | - | 1 | 1 | |||||||||||||||
| 72 | JNK1 | M77 | - | 1 | 1 | |||||||||||||||
| Atypical | ||||||||||||||||||||
| N | Q | M | D | T | S | V | I | R | F | K | H | W | P | L | - | |||||
| 73 | ATM | Q2729 | - | 1 | 2 | 3 | ||||||||||||||
| 74 | ATR | M2339 | - | 1 | 1 | |||||||||||||||
| 75 | ADCK3 | A415 | - | 4 | 4 | |||||||||||||||
| Other | ||||||||||||||||||||
| N | M | D | T | S | V | I | R | F | K | H | W | P | L | - | ||||||
| 76 | TTK | L575 | + | 1 | 1 | 2 | ||||||||||||||
| 77 | Haspin | S539 | - | 1 | 1 | |||||||||||||||
| 78 | NEK7 | H86 | - | 1 | 1 | |||||||||||||||
| 79 | TBK1 | L59 | - | 2 | 2 | |||||||||||||||
| 80 | TLK2 | H518 | - | 2 | 2 | |||||||||||||||
| 81 | ULK1 | L67 | - | 1 | 1 | |||||||||||||||
| 82 | WEE2 | H263 | - | 3 | 3 | |||||||||||||||
Hydrophobic interaction between ligand and RS3: +at least one structure with RS3–ligand contact available; - no contacts observed in available structures;
Silent mutation.
In the literature, only few cases of studies including RS3 mutations have been reported. With BRAF, a secondary mutation in RS3 (L505H) induces resistance to vemurafenib [74]. Also, in another study resistance to dabrafenib or vemurafenib was demonstrated with BRAF L505H mutation [10]. Moreover, alectinib resistance was reported with I1171S and I1171N mutations in ALK [75]. The I1171N mutation was demonstrated to increase autophosphorylation level of ALK in vitro [76]. I1171T has been identified to induce crizotinib resistance [77]. Interestingly, these two inhibitors, crizotinib and alectinib do not display direct contacts to RS3. EGFR M766T mutation was reported to induce resistance to gefitinib and erlotinib [78].
Conclusions
The function of the R-spine and the role of RS3 is quite conserved with typical protein kinases. Couple of residues are dominating RS3 in the available protein kinase domain structures. Nevertheless, also unique RS3 residues are observed, and in combination with gatekeeper even more kinase specific profiles for these residues are observed. Obviously, even with identical RS3–gatekeeper combinations the 3D-environment within this region can be quite different between two kinases. Kinase specific angles and absolute positions of these residues may provide important opportunities for selective targeting. Obviously, one must carefully consider this case-by-case, as RS3 is not targetable in all kinases. With pseudokinases [79–81], which compared with regular protein kinases can vary more in their structure in this region, the role of RS3 and its targeting would require further research.
The general understanding of RS3–ligand interactions are quite limited, even though numerous structures that contain these mainly hydrophobic interactions are available. Currently, no studies investigating specific effect of RS3 on ligand binding affinity exist that directly compare a set of ligands with selected mutations of this residue. Further research is needed to disclose the influence of RS3 residue for ligand binding and should be also extended to the cases where no direct contact between the residue and inhibitor exists. Of note, even with hydrophobic interactions (in the case of hydrophobic RS3) this should not be overlooked as these interactions may be crucial for the inhibitor binding [82]. For example, non-canonical interactions play a detrimental role in binding affinity of the ultra-potent small-molecule biotin [83]. There may be good possibilities available to optimize RS3-specific interactions, for instance, with enhanced interactions with the sulfur atom of Met [84].
Infrequency of mutations in RS3 may indicate a defiance against plausible point mutation in this position that could cause drug resistance [85]. Perhaps the somewhat buried location of RS3 in the quite rigid αC-helix position that offers a limited flexibility, renders the mutations in this location (at least in most cases) incompetent to drive kinase activation. This motivates further to optimize protein–ligand interactions for RS3. However, the data at hand may not necessarily cover potential drug therapy induced mutations in cancer patients. We believe that in near future, with accumulation of this data, this information will be more accessible, and a better estimate can be provided.
The full data presented in this review are freely available at https://doi.org/10.5281/zenodo.5796550
Perspectives
The role of the conserved R-spine and RS3 residue in protein kinase function is well established.
Protein kinases display diversity in their RS3 residue and in its surroundings. Many small-molecule protein kinase inhibitors, including approved drugs, display contacts to RS3.
Considering the RS3 residue more carefully in the design of small-molecule kinase inhibitors may offer important advantage for the inhibitor binding and selectivity.
Abbreviations
- ATP
adenosine triphosphate
- PLDB
Protein–Ligand Database
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research is financially supported by Academy of Finland GeneCellNano Flagship funding 337120 and T.P. acknowledges funding by Orion Research Foundation sr.
Author Contributions
Both authors wrote the manuscript and made the figures. The original concept was conceived by T.P. T.P. directed the project.
References
- 1.Manning, G., Whyte, D.B., Martinez, R., Hunter, T. and Sudarsanam, S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 2.Modi, V. and Dunbrack, R.L. (2019) A structurally-validated multiple sequence alignment of 497 human protein kinase domains. Sci. Rep. 9, 19790 10.1038/s41598-019-56499-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berginski, M.E., Moret, N., Liu, C., Goldfarb, D., Sorger, P.K. and Gomez, S.M. (2021) The dark kinase knowledgebase: an online compendium of knowledge and experimental results of understudied kinases. Nucleic Acids Res. 49, D529–D535 10.1093/nar/gkaa853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myasnikov, A., Zhu, H., Hixson, P., Xie, B., Yu, K., Pitre, A.et al. (2021) Structural analysis of the full-length human LRRK2. Cell 184, 3519–3527.e10 10.1016/j.cell.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor, S.S., Wu, J., Bruystens, J.G.H., Rio, J.C.D., Lu, T.-W., Kornev, A.P.et al. (2021) From structure to the dynamic regulation of a molecular switch: a journey over 3 decades. J. Biol. Chem. 296, 100746 10.1016/j.jbc.2021.100746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornev, A.P., Haste, N.M., Taylor, S.S. and Eyck, L.F.T. (2006) Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl Acad. Sci. U.S.A. 103, 17783–17788 10.1073/pnas.0607656103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meharena, H.S., Chang, P., Keshwani, M.M., Oruganty, K., Nene, A.K., Kannan, N.et al. (2013) Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol. 11, e1001680 10.1371/journal.pbio.1001680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor, S.S., Shaw, A.S., Kannan, N. and Kornev, A.P. (2015) Integration of signaling in the kinome: architecture and regulation of the αC helix. Biochim. Biophys. Acta Proteins Proteomics 1854, 1567–1574 10.1016/j.bbapap.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, J., Ahuja, L.G., Meharena, H.S., Kannan, N., Kornev, A.P., Taylor, S.S.et al. (2015) Kinase regulation by hydrophobic spine assembly in cancer. Mol. Cell. Biol. 35, 264–276 10.1128/MCB.00943-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap, J., Deepak, R.N.V.K., Tian, Z., Ng, W.H., Goh, K.C., Foo, A.et al. (2021) The stability of R-spine defines RAF inhibitor resistance: a comprehensive analysis of oncogenic BRAF mutants with in-frame insertion of αC-β4 loop. Sci. Adv. 7, eabg0390 10.1126/sciadv.abg0390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, Y., Shah, K., Yang, F., Witucki, L. and Shokat, K.M. (1998) A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg. Med. Chem. 6, 1219–1226 10.1016/S0968-0896(98)00099-6 [DOI] [PubMed] [Google Scholar]
- 12.Nussinov, R., Zhang, M., Maloney, R., Tsai, C.-J., Yavuz, B.R., Tuncbag, N.et al. (2021) Mechanism of activation and the rewired network: new drug design concepts. Med. Res. Rev. 42, 770–799 10.1002/med.21863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih, H.-P., Zhang, X. and Aronov, A.M. (2018) Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat. Rev. Drug Discov. 17, 19–33 10.1038/nrd.2017.194 [DOI] [PubMed] [Google Scholar]
- 14.Bournez, C., Carles, F., Peyrat, G., Aci-Sèche, S., Bourg, S., Meyer, C.et al. (2020) Comparative assessment of protein kinase inhibitors in public databases and in PKIDB. Molecules 25, 3226 10.3390/molecules25143226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carles, F., Bourg, S., Meyer, C. and Bonnet, P. (2018) PKIDB: a curated, annotated and updated database of protein kinase inhibitors in clinical trials. Molecules 23, 908 10.3390/molecules23040908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayala-Aguilera, C.C., Valero, T., Lorente-Macías, Á., Baillache, D.J., Croke, S. and Unciti-Broceta, A. (2021) Small molecule kinase inhibitor drugs (1995–2021): medical indication, pharmacology, and synthesis. J. Med. Chem. 65, 1047–1131 10.1021/acs.jmedchem.1c00963 [DOI] [PubMed] [Google Scholar]
- 17.Cohen, P., Cross, D. and Jänne, P.A. (2021) Kinase drug discovery 20 years after imatinib: progress and future directions. Nat. Rev. Drug Discov. 20, 551–569 10.1038/s41573-021-00195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attwood, M.M., Fabbro, D., Sokolov, A.V., Knapp, S. and Schiöth, H.B. (2021) Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov. 20, 839–861 10.1038/s41573-021-00252-y [DOI] [PubMed] [Google Scholar]
- 19.Ferguson, F.M. and Gray, N.S. (2018) Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 10.1038/nrd.2018.21 [DOI] [PubMed] [Google Scholar]
- 20.Cheung, J., Ginter, C., Cassidy, M., Franklin, M.C., Rudolph, M.J., Robine, N.et al. (2015) Structural insights into mis-regulation of protein kinase A in human tumors. Proc. Natl Acad. Sci. U.S.A. 112, 1374–1379 10.1073/pnas.1424206112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin, J., Xie, P., Ventocilla, C., Zhou, G., Vultur, A., Chen, Q.et al. (2012) Identification of a novel family of BRAFV600E inhibitors. J. Med. Chem. 55, 5220–5230 10.1021/jm3004416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan, P.T.C., Garnett, M.J., Roe, S.M., Lee, S., Niculescu-Duvaz, D., Good, V.M.et al. (2004) Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 10.1016/S0092-8674(04)00215-6 [DOI] [PubMed] [Google Scholar]
- 23.Kozlowski, L.P. (2017) Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 45, D1112–D1116 10.1093/nar/gkw978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas, K.M., DeVido, D.R. and Dorsey, J.G. (2003) Evaluation of methods for measuring amino acid hydrophobicities and interactions. J. Chromatogr. A 1000, 637–655 10.1016/S0021-9673(03)00182-1 [DOI] [PubMed] [Google Scholar]
- 25.Simm, S., Einloft, J., Mirus, O. and Schleiff, E. (2016) 50 years of amino acid hydrophobicity scales: revisiting the capacity for peptide classification. Biol. Res. 49, 31 10.1186/s40659-016-0092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roskoski, R. (2019) Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol. Res. 139, 395–411 10.1016/j.phrs.2018.11.014 [DOI] [PubMed] [Google Scholar]
- 27.Shi, F., Telesco, S.E., Liu, Y., Radhakrishnan, R. and Lemmon, M.A. (2010) Erbb3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl Acad. Sci. U.S.A. 107, 7692–7697 10.1073/pnas.1002753107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchanan, J.L., Newcomb, J.R., Carney, D.P., Chaffee, S.C., Chai, L., Cupples, R.et al. (2011) Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors. Bioorg. Med. Chem. Lett. 21, 2394–2399 10.1016/j.bmcl.2011.02.075 [DOI] [PubMed] [Google Scholar]
- 29.Eid, S., Turk, S., Volkamer, A., Rippmann, F. and Fulle, S. (2017) Kinmap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics 18, 16 10.1186/s12859-016-1433-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, T., Kinoshita, T., Kirii, Y., Yokota, K., Hamada, K. and Tada, T. (2010) Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state. Biochem. Biophys. Res. Commun. 400, 369–373 10.1016/j.bbrc.2010.08.071 [DOI] [PubMed] [Google Scholar]
- 31.Sogabe, Y., Hashimoto, T., Matsumoto, T., Kirii, Y., Sawa, M. and Kinoshita, T. (2016) A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7. Biochem. Biophys. Res. Commun. 473, 476–481 10.1016/j.bbrc.2016.03.036 [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto, T., Kinoshita, T., Matsuzaka, H., Nakai, R., Kirii, Y., Yokota, K.et al. (2012) Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state. J. Biochem. 151, 541–549 10.1093/jb/mvs023 [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko, E., Poso, A. and Pantsar, T. (2020) The autoinhibited state of MKK4: phosphorylation, putative dimerization and R134W mutant studied by molecular dynamics simulations. Comput. Struct. Biotechnol. J. 18, 2687–2698 10.1016/j.csbj.2020.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, J.M., Nakatani, Y., Jamieson, S.A., Dai, W., Lucet, I.S. and Mace, P.D. (2015) Molecular mechanism of CCAAT-enhancer binding protein recruitment by the TRIB1 pseudokinase. Structure 23, 2111–2121 10.1016/j.str.2015.08.017 [DOI] [PubMed] [Google Scholar]
- 35.Jamieson, S.A., Ruan, Z., Burgess, A.E., Curry, J.R., McMillan, H.D., Brewster, J.L.et al. (2018) Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci Signal. 11, eaau0597 10.1126/scisignal.aau0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asquith, C.R.M., Murray, N.H. and Pagliarini, D.J. (2019) ADCK3/COQ8A: the choice target of the UbiB protein kinase-like family. Nat. Rev. Drug Discov. 18, 815–815 10.1038/d41573-019-00158-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cyphers, S., Ruff, E.F., Behr, J.M., Chodera, J.D. and Levinson, N.M. (2017) A water-mediated allosteric network governs activation of aurora kinase A. Nat. Chem. Biol. 13, 402–408 10.1038/nchembio.2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, D., Matsumoto, M.L., McKenzie, B.S. and Zarrin, A.A. (2018) TPL2 kinase action and control of inflammation. Pharmacol. Res. 129, 188–193 10.1016/j.phrs.2017.11.031 [DOI] [PubMed] [Google Scholar]
- 39.Lee, J.-H. and Paull, T.T. (2021) Cellular functions of the protein kinase ATM and their relevance to human disease. Nat. Rev. Mol. Cell Biol. 22, 796–814 10.1038/s41580-021-00394-2 [DOI] [PubMed] [Google Scholar]
- 40.Zeqiraj, E., Filippi, B.M., Goldie, S., Navratilova, I., Boudeau, J., Deak, M.et al. (2009) ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7, e1000126 10.1371/journal.pbio.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeqiraj, E., Filippi, B.M., Deak, M., Alessi, D.R. and van Aalten, D.M.F. (2009) Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326, 1707–1711 10.1126/science.1178377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiolica, A., de Medina-Redondo, M., Schoof, E.M., Chaikuad, A., Villa, F., Gatti, M.et al. (2014) Modulation of the chromatin phosphoproteome by the haspin protein kinase*. Mol. Cell. Proteomics 13, 1724–1740 10.1074/mcp.M113.034819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klomp, J.E., Lee, Y.S., Goodwin, C.M., Papke, B., Klomp, J.A., Waters, A.M.et al. (2021) CHK1 protects oncogenic KRAS-expressing cells from DNA damage and is a target for pancreatic cancer treatment. Cell Rep. 37, 110060 10.1016/j.celrep.2021.110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preuss, F., Chatterjee, D., Mathea, S., Shrestha, S., St-Germain, J., Saha, M.et al. (2020) Nucleotide binding, evolutionary insights, and interaction partners of the pseudokinase Unc-51-like kinase 4. Structure 28, 1184–1196.e6 10.1016/j.str.2020.07.016 [DOI] [PubMed] [Google Scholar]
- 45.Smaill, J.B., Lee, H.H., Palmer, B.D., Thompson, A.M., Squire, C.J., Baker, E.N.et al. (2008) Synthesis and structure-activity relationships of soluble 8-substituted 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of the Wee1 and Chk1 checkpoint kinases. Bioorg. Med. Chem. Lett. 18, 929–933 10.1016/j.bmcl.2007.12.046 [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J.-Y., Cuellar, R.A., Berndt, N., Lee, H.E., Olesen, S.H., Martin, M.P.et al. (2017) Structural basis of wee kinases functionality and inactivation by diverse small molecule inhibitors. J. Med. Chem. 60, 7863–7875 10.1021/acs.jmedchem.7b00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, R.A., Bethel, P., Cook, C., Davies, E., Debreczeni, J.E., Fairley, G.et al. (2017) Structure-guided discovery of potent and selective inhibitors of ERK1/2 from a modestly active and promiscuous chemical start point. J. Med. Chem. 60, 3438–3450 10.1021/acs.jmedchem.7b00267 [DOI] [PubMed] [Google Scholar]
- 48.Chaikuad, A., Tacconi, E.M.C., Zimmer, J., Liang, Y., Gray, N.S., Tarsounas, M.et al. (2014) A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 10, 853–860 10.1038/nchembio.1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schröder, M., Filippakopoulos, P., Schwalm, M.P., Ferrer, C.A., Drewry, D.H., Knapp, S.et al. (2020) Crystal structure and inhibitor identifications reveal targeting opportunity for the atypical MAPK kinase ERK3. Int. J. Mol. Sci. 21, 7953 10.3390/ijms21217953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bandaranayake, R.M., Ungureanu, D., Shan, Y., Shaw, D.E., Silvennoinen, O. and Hubbard, S.R. (2012) Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol. 19, 754–759 10.1038/nsmb.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toms, A.V., Deshpande, A., McNally, R., Jeong, Y., Rogers, J.M., Kim, C.U.et al. (2013) Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat. Struct. Mol. Biol. 20, 1221–1223 10.1038/nsmb.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanev, G.K., de Graaf, C., Westerman, B.A., de Esch, I.J.P. and Kooistra, A.J. (2021) KLIFS: an overhaul after the first 5 years of supporting kinase research. Nucleic Acids Res. 49, D562–D569 10.1093/nar/gkaa895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Linden, O.P.J., Kooistra, A.J., Leurs, R., de Esch, I.J.P. and de Graaf, C. (2014) KLIFS: a knowledge-based structural database To navigate kinase–ligand interaction space. J. Med. Chem. 57, 249–277 10.1021/jm400378w [DOI] [PubMed] [Google Scholar]
- 54.Yasuda, H., Park, E., Yun, C.-H., Sng, N.J., Lucena-Araujo, A.R., Yeo, W.-L.et al. (2013) Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci. Transl. Med. 5, 216ra177 10.1126/scitranslmed.3007205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ung, P.M.-U., Rahman, R. and Schlessinger, A. (2018) Redefining the protein kinase conformational space with machine learning. Cell Chem. Biol. 25, 916–924.e2 10.1016/j.chembiol.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman, R., Ung, P.M.-U. and Schlessinger, A. (2019) Kinametrix: a web resource to investigate kinase conformations and inhibitor space. Nucleic Acids Res. 47, D361–D366 10.1093/nar/gky916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carry, J.-C., Clerc, F., Minoux, H., Schio, L., Mauger, J., Nair, A.et al. (2015) SAR156497, an exquisitely selective inhibitor of aurora kinases. J. Med. Chem. 58, 362–375 10.1021/jm501326k [DOI] [PubMed] [Google Scholar]
- 58.Modi, V. and Dunbrack, R.L. (2022) Kincore: a web resource for structural classification of protein kinases and their inhibitors. Nucleic Acids Res. 50, D654–D664 10.1093/nar/gkab920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamoto, K., Ikemori-Kawada, M., Jestel, A., von König, K., Funahashi, Y., Matsushima, T.et al. (2015) Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization. ACS Med. Chem. Lett. 6, 89–94 10.1021/ml500394m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuki, M., Hoshi, T., Yamamoto, Y., Ikemori-Kawada, M., Minoshima, Y., Funahashi, Y.et al. (2018) Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models. Cancer Med. 7, 2641–2653 10.1002/cam4.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karoulia, Z., Wu, Y., Ahmed, T.A., Xin, Q., Bollard, J., Krepler, C.et al. (2016) An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell 30, 485–498 10.1016/j.ccell.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olesen, S.H., Zhu, J.-Y., Martin, M.P. and Schönbrunn, E. (2016) Discovery of diverse small-molecule inhibitors of mammalian Sterile20-like kinase3 (MST3). ChemMedChem 11, 1137–1144 10.1002/cmdc.201600115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Canning, P., Ruan, Q., Schwerd, T., Hrdinka, M., Maki, J.L., Saleh, D.et al. (2015) Inflammatory signaling by NOD-RIPK2 Is inhibited by clinically relevant type II kinase inhibitors. Chem. Biol. 22, 1174–1184 10.1016/j.chembiol.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garner, A.P., Gozgit, J.M., Anjum, R., Vodala, S., Schrock, A., Zhou, T.et al. (2014) Ponatinib inhibits polyclonal drug-resistant KIT oncoproteins and shows therapeutic potential in heavily pretreated gastrointestinal stromal tumor (GIST) patients. Clin. Cancer Res. 20, 5745–5755 10.1158/1078-0432.CCR-14-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cotto-Rios, X.M., Agianian, B., Gitego, N., Zacharioudakis, E., Giricz, O., Wu, Y.et al. (2020) Inhibitors of BRAF dimers using an allosteric site. Nat. Commun. 11, 4370 10.1038/s41467-020-18123-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mol, C.D., Dougan, D.R., Schneider, T.R., Skene, R.J., Kraus, M.L., Scheibe, D.N.et al. (2004) Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 279, 31655–31663 10.1074/jbc.M403319200 [DOI] [PubMed] [Google Scholar]
- 67.Namboodiri, H.V., Bukhtiyarova, M., Ramcharan, J., Karpusas, M., Lee, Y. and Springman, E.B. (2010) Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases. Biochemistry 49, 3611–3618 10.1021/bi100070r [DOI] [PubMed] [Google Scholar]
- 68.Wagner, A.J., Severson, P.L., Shields, A.F., Patnaik, A., Chugh, R., Tinoco, G.et al. (2021) Association of combination of conformation-specific KIT inhibitors with clinical benefit in patients with refractory gastrointestinal stromal tumors: a phase 1b/2a nonrandomized clinical trial. JAMA Oncol. 7, 1343–1350 10.1001/jamaoncol.2021.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohbayashi, N., Murayama, K., Kato-Murayama, M., Kukimoto-Niino, M., Uejima, T., Matsuda, T.et al. (2018) Structural basis for the inhibition of cyclin G-associated kinase by gefitinib. ChemistryOpen 7, 713–719 10.1002/open.201800177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schröder, M., Tan, L., Wang, J., Liang, Y., Gray, N.S., Knapp, S.et al. (2020) Catalytic domain plasticity of MKK7 reveals structural mechanisms of allosteric activation and diverse targeting opportunities. Cell Chem. Biol. 27, 1285–1295.e4 10.1016/j.chembiol.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 71.Tate, J.G., Bamford, S., Jubb, H.C., Sondka, Z., Beare, D.M., Bindal, N.et al. (2019) COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barouch-Bentov, R. and Sauer, K. (2011) Mechanisms of drug resistance in kinases. Expert Opin. Investig. Drugs. 20, 153–208 10.1517/13543784.2011.546344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim, P., Li, H., Wang, J. and Zhao, Z. (2021) Landscape of drug-resistance mutations in kinase regulatory hotspots. Brief Bioinform. 22, bbaa108 10.1093/bib/bbaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoogstraat, M., Hooijdonk, C.G.G., Ubink, I., Besselink, N.J.M., Pieterse, M., Veldhuis, W.et al. (2015) Detailed imaging and genetic analysis reveal a secondary BRAFL505H resistance mutation and extensive intrapatient heterogeneity in metastatic BRAF mutant melanoma patients treated with vemurafenib. Pigment Cell Melanoma Res. 28, 318–323 10.1111/pcmr.12347 [DOI] [PubMed] [Google Scholar]
- 75.Ou, S.-H.I., Klempner, S.J., Greenbowe, J.R., Azada, M., Schrock, A.B., Ali, S.M.et al. (2014) Identification of a novel HIP1-ALK fusion variant in non-small-cell lung cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J. Thorac. Oncol. 9, 1821–1825 10.1097/JTO.0000000000000368 [DOI] [PubMed] [Google Scholar]
- 76.Bresler, S.C., Weiser, D.A., Huwe, P.J., Park, J.H., Krytska, K., Ryles, H.et al. (2014) ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 26, 682–694 10.1016/j.ccell.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friboulet, L., Li, N., Katayama, R., Lee, C.C., Gainor, J.F., Crystal, A.S.et al. (2014) The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 4, 662–673 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruan, Z., Katiyar, S. and Kannan, N. (2017) Computational and experimental characterization of patient derived mutations reveal an unusual mode of regulatory spine assembly and drug sensitivity in EGFR kinase. Biochemistry 56, 22–32 10.1021/acs.biochem.6b00572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mace, P.D. and Murphy, J.M. (2021) There's more to death than life: noncatalytic functions in kinase and pseudokinase signaling. J. Biol. Chem. 296, 100705 10.1016/j.jbc.2021.100705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shrestha, S., Byrne, D.P., Harris, J.A., Kannan, N. and Eyers, P.A. (2020) Cataloguing the dead: breathing new life into pseudokinase research. FEBS J. 287, 4150–4169 10.1111/febs.15246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kung, J.E. and Jura, N. (2019) Prospects for pharmacological targeting of pseudokinases. Nat. Rev. Drug Discov. 18, 501–526 10.1038/s41573-019-0018-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bissantz, C., Kuhn, B. and Stahl, M. (2010) A medicinal chemist's guide to molecular interactions. J. Med. Chem. 53, 5061–5084 10.1021/jm100112j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McConnell, D.B. (2021) Biotin's lessons in drug design. J. Med. Chem. 64, 16319–16327 10.1021/acs.jmedchem.1c00975 [DOI] [PubMed] [Google Scholar]
- 84.Beno, B.R., Yeung, K.-S., Bartberger, M.D., Pennington, L.D. and Meanwell, N.A. (2015) A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem. 58, 4383–4438 10.1021/jm501853m [DOI] [PubMed] [Google Scholar]
- 85.Ward, R.A., Fawell, S., Floc'h, N., Flemington, V., McKerrecher, D. and Smith, P.D. (2021) Challenges and opportunities in cancer drug resistance. Chem. Rev. 121, 3297–3351 10.1021/acs.chemrev.0c00383 [DOI] [PubMed] [Google Scholar]