Abstract
Programmed cell death has long been characterised as a key player in the development of human disease. Necroptosis is a lytic form of programmed cell death that is universally mediated by the effector protein mixed lineage kinase domain-like (MLKL), a pseudokinase. MLKL's activating kinase, receptor interacting protein kinase 3 (RIPK3), is itself activated within context specific scaffolds of receptor interacting protein kinase 1 (RIPK1), Z-DNA Binding Protein-1 (ZBP1) or TIR domain-containing adaptor inducing interferon-β (TRIF). These core necroptosis modulating proteins have been comprehensively revealed as potent drivers and suppressors of disease in inbred mouse strains. However, their roles in human disease within the ‘real world’ of diverse genetic backgrounds, natural infection and environmental challenges remains less well understood. Over 20 unique disease-associated human germline gene variants in this core necroptotic machinery have been reported in the literature and human clinico-genetics databases like ClinVar to date. In this review, we provide an overview of these human gene variants, with an emphasis on those encoding MLKL. These experiments of nature have the potential to not only enrich our understanding of the basic biology of necroptosis, but offer important population level insights into which clinical indications stand to benefit most from necroptosis-targeted drugs.
Keywords: loss of function gene variant, missense gene variant, MLKL, necroptosis, pathogenic mutation
Introduction
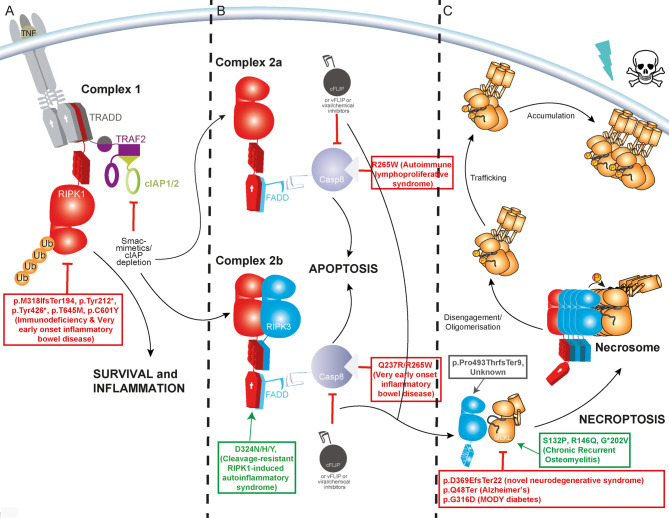
Programmed cell death takes many forms and is crucial to every aspect of normal animal development and homeostasis. The most well studied programmed cell death pathway is the caspase-dependent apoptosis. Apoptosis can be initiated by a range of intrinsic and extrinsic signals, and is commonly regarded as a ‘clean death’ characterised by the caspase-mediated disassembly of cells into highly phagocytosable, membrane enclosed bundles [1,2]. Unlike apoptosis, necroptosis is a caspase-independent form of programmed cell death that is characterised by the release of highly inflammatory cytokines, intracellular proteins, and nucleic acids into the extracellular space [3–6]. In turn, necroptosis is itself induced by inflammatory cytokines and danger- or pathogen-associated molecular patterns via their cognate transmembrane receptors or intracellular pattern recognition proteins like nucleic acid sensor Z-DNA Binding Protein-1. Of these various initiating stimuli, the most well studied route to necroptosis is downstream of tumour necrosis factor receptor 1 (TNFR1). This signal triggers the eventual formation of the RIPK1 and RIPK3-containing necrosome prior to terminal MLKL activation and cell death (Figure 1) [7–9].
Figure 1. TNF-induced necroptosis occurs when upstream pro-survival and pro-apoptotic pathways are inhibited.
(A) The binding of TNF to TNFR1 stimulates downstream nuclear factor-κB activation and other pro-survival, proinflammatory signals. (B) When cIAP1/2 activity is low, signalling is diverted to the formation of a death-induced signalling complex termed Complex II (a/b). This physically distinct complex is variably composed of TRADD, FADD, RIPK1, RIPK3 and the apoptosis initiator Caspase-8. (C) Necroptosis is activated when cellular conditions curb Caspase-8 activity and favour the assembly of RIPK1 and RIPK3, via their RIP Homotypic Interaction Motif (RHIM) domains, into a high molecular mass complex termed the necrosome. Here, RIPK3 is activated by autophosphorylation and MLKL is recruited and phosphorylated by RIPK3. MLKL dissociates from RIPK3 and oligomerised MLKL is trafficked to biological membrane [84,85]. The precise molecular events that lead to lytic permeabilization of the cell are still a matter of contention [86]. Sites of some selected published human germline mutations shown to be associated with human disease due to gain (red) or loss/reduction in function (green) are shown.
Importantly, several additional, highly context-dependent facets to MLKL function have been reported (recently reviewed by Weir et al. [3]). These include roles in potassium channel mediated promotion of inflammasome activation [10], endocytic and exocytic modulation of cytokine release and membrane repair [11–14], myelin sheath remodelling [15], neutrophil extracellular trap formation [16,17] and even the direct suppression of intracellular bacteria [18,19].
Mlkl gene knock-out and knock-in mutant mice have revealed necroptosis as a driver or protective factor in murine disease spanning every bodily system [20]. Simultaneously, massively parallel sequencing technologies have increased the efficiency and reduced the cost of human genotyping. This has led to the rapid accumulation of human genomic data and even paired phenotypic data in large open access databases like gnomAD [21] and the Global Biobank Engine [22]. Rare mutations in human MLKL have revealed neurological and metabolic pathologies where the absence of MLKL function plays a driving role [23–25], whilst rare and de novo patient mutations in MLKL signal modulators like CASP8 and RIPK1 [26], and common polymorphisms in MLKL [27] have provided insight into pathologies where excessive necroptotic signalling is an important contributor to inflammatory disease (Figure 2A). Together these studies of MLKL function in humans reveal some similarities, but also many key differences to observations made in inbred laboratory mouse strains housed under specific pathogen-free conditions [20].
Figure 2. Frequency of gene variation and pathogenic gene variants in MLKL and upstream signalling modulators.
(A) Total unique germline and de novo variants reported in ClinVar [52] as ‘pathogenic’, ‘likely pathogenic’, ‘associated’ or ‘risk factor. Somatic gene variants, contiguous copy-number variants, and gene variants not accompanied by a human disease condition were excluded from these counts. It is important to note that some patient mutations reported in the scientific literature may not have been submitted or updated by authors to the ClinVar database at the time of writing and thus are not included here. Disease causing variants in more upstream MLKL signalling modulators including death receptors, pattern recognition receptors, interferons and the NF-κB pathway are not presented here but are described in recent reviews [87–90]. (B) Summed global Minor Allele Frequency (MAF) of Missense and Loss of Function (LOF) variants as annotated by gnomAD at time of writing, (n = >280 000 alleles sequenced) [54]. Missense and LOF alleles flagged as ‘low confidence’ or ‘variant quality/annotation dubious’, alleles unique to non-canonical transcripts (with exception of CFLAR, where both long and short forms were included) or alleles with MAFs > 0.5 were excluded. (C) The top 10 missense variants used in calculating summed missense allele frequency in (B) and their global allele frequency plotted according to position in protein. *TICAM1 pPro367dup MAF = 0.3.
MLKL truncation/deletion mutations in human neurodegeneration
To date, only four individuals that are homozygous for a predicted loss of function (LOF) MLKL gene variant have been reported in the gnomAD database, a carefully curated collection of whole genome or exome data for over 140 000 human adults of diverse ancestry [21]. In 2020, Faergman et al. reported homozygosity of one of these MLKL variants (p.Asp369GlufsTer22, rs561839347) in two brothers diagnosed with progressive neurodegenerative spectrum disorder [24,28]. MLKL was not detectable by western blot in tissue derived from these patients, and patient-derived fibroblasts did not undergo programmed cell death when exposed to an exogenous necroptotic stimulus in culture. These data support gnomAD's ‘loss of function’ classification of p.Asp369GlufsTer22, making this the first peer-reviewed, clinical description of humans that are homozygous for an MLKL LOF gene variant. The brothers presented with an asymmetric lower limb weakness at the ages of 19 and 30, respectively, which developed into a novel progressive neurodegenerative disorder characterised by paresis, ataxia and dysarthria. Magnetic resonance imaging performed at 28 and 31 years, respectively, after onset of symptoms revealed severe global cerebral, cerebellar, cortical cerebellar fibers tract and brainstem atrophy and small, discrete periventricular T2-hyperintense white matter lesions. All pathology was limited to the nervous system and the patients did not exhibit any increased susceptibility to infections. It is important to note that this MLKL mutation also segregated with a homozygous in-frame deletion of one amino acid in the adjacent FA2H gene (fatty acid 2-hydroxylase), and a maternally inherited missense amino acid substitution in the X-linked gene AP1S2 (vesicle sorting and transport) in these patients. While FA2H variant function was not impaired when examined in vitro and the AP1S2 variant is classified as ‘benign’ based on ACMG guidelines [29], the influence of these and other genetic/environmental modifiers cannot be fully excluded from the severe phenotype exhibited by these individuals.
Another MLKL LOF gene variant (p.Gln48Ter, rs763812068) was found to be >20 fold enriched in a cohort of Hong Kong Chinese patients suffering from the neurodegenerative disease Alzheimer's, relative to a large ancestry-matched population [23]. This MLKL LOF variant is specific to populations of Southern Chinese descent, was only present in heterozygotes, and was not linked to any predicted damaging mutations in the adjacent FA2H gene. Wang et al. reported that shRNA-mediated knock down of MLKL in APPswe-293 cells led to an increased ratio of amyloid beta proteins Aβ42 to Aβ40. Further study of the precise role of MLKL in modulating levels of these proteins in neuronal cells remains an important area of future investigation.
Homozygous and heterozygous Mlkl gene knock-out (MlklKO/KO) mice do not appear to exhibit the spontaneous, progressive neurodegeneration observed in the patients described by Faergman et al. [24] and Wang et al. [23], implicating the contribution of additional genetic and/or environmental modifiers or key-interspecies differences in MLKL function. MLKL protein is present at low to undetectable levels in the healthy adult mouse brain and spinal cord [30–32], and was reported to be dispensable to the onset and severity of mouse autoimmune lateral sclerosis (ALS) [32]. There are reports however that MLKL is induced following injury to the mouse cerebral cortex and spinal cord [33,34]. MlklKO/KO mice also exhibit a reduced propensity for demyelination in several other independent experimental mouse disease models. This manifests as improved outcomes in mouse models of Parkinson's disease [35] and multiple sclerosis [36], and poorer outcomes in models of physical nerve injury [15]. These mouse data, combined with the wide steady state distribution of MLKL protein throughout the human brain [37] supports the existence of important roles for MLKL in cell types essential for neurological function in humans. The collective global frequency of MLKL LOF alleles is 0.0007, (equivalent to 1–2 in every 1000 individuals being heterozygous carriers) (Figure 2B). The identification and clinical description of additional human individuals that are homozygous or compound heterozygous for MLKL LOF alleles will shed further light on whether the absence of MLKL alone is sufficient to give rise to neurodegenerative disease, or if other genetic or environmental events are indeed at play.
An MLKL missense hypomorph in Maturity Onset Diabetes of the Young (MODY)
Recently, a heterozygous MLKL missense variant p.G316D (rs375490660) was reported to be associated with Maturity Onset Diabetes of the Young (MODY) in a Palestinian family [25]. This variant segregates with a known MODY-associated LOF gene variant in the gene encoding the pancreatic transcription factor PDX1, and other predicted deleterious variants in the genes ERN2 (endoplasmic reticulum stress), NIPAL4 (Mg2+ transporter activity) and SPTBN4 (actin binding). When compared with exogenously produced MLKLWT, protein levels of exogenously produced MLKL p.G316D are significantly reduced. RIPK3-phosphorylated (Ser358) MLKL p.G316D are basically undetectable despite normal upstream signalling. Exogenous expression of MLKL p.G316D was also unable to reconstitute necroptotic cell death in two lineage-distinct CRISPR-Cas9 MLKLKO/KO human cell lines and did not appear to influence the function of endogenously expressed MLKLWT in MLKL wild-type cells (simulating the heterozygous state of patients). While the stability and function of endogenously expressed MLKL p.G316D were not verified in patient-derived cells in its endogenous heterozygous context, normal levels of MLKL p.G316D mRNA in patient derived cells indicates that this missense mutation does not lead to reduced mRNA stability [25]. For this reason, we have chosen to classify this missense variant for the purpose of this review as a ‘hypomorph’ as opposed to a full LOF variant, as the phospho-Ser358-independent functions of MLKL p.G316D are yet to be investigated.
With a global minor allele frequency of 1.062 × 10−5, MLKL p.G316D homozygotes have not been recorded in the gnomAD database. How a monoallelic reduction in necroptotic cell death contributes to the aetiology of diabetes in this family remains a matter of investigation. MlklKO/KO mice do not display any of the hallmarks of diabetes when fed a standard laboratory mouse chow diet, and actually demonstrate improved glucose and insulin tolerance relative to MlklWt/Wt controls when placed on a high fat diet for more than 12 weeks [38,39]. Investigating whether Mlkl gene knockout or mutation is a genetic modifier in mouse models of spontaneous diabetes (e.g. Pdx1Wt/KO mice [40]) could provide a useful route for the mechanistic dissection of these human findings.
Importantly, such studies can also prompt examination of potential risk posed by other human MLKL LOF and missense variants to the incidence of MODY. With no ‘gold standard’ criterion or threshold for diagnosing MODY in human patients, broadening gene panels for precision genetic testing will inform clinical diagnosis and management [41].
Common MLKL missense variants in inflammatory disease, historically important hypermorphs?
The biological consequences of spontaneous and rationally designed MLKL gain of function or ‘activating’ mutations have been described both in vitro and in vivo in murine models [30,42,43]. Human MLKL however is less amenable to rationally designed modifications that confer RIPK3 independent activity [43,44]. In 2020, a random, chemically induced germline missense mutation in mouse Mlkl was shown to result in systemic lethal neonatal inflammation in homozygotes and hematopoietic dysfunction in heterozygotes [27]. This mutation, Mlkl p.D139V, confers constitutive cell death activity to MLKL, removing the requirement of upstream necroptotic signalling and RIPK3-mediated phosphorylation. The human equivalent, MLKL p.D140V, has been observed only once, and only in heterozygous form in humans in the gnomAD database (rs1330913532). Interestingly, three very closely situated MLKL missense gene variants are amongst the most prevalent MLKL single nucleotide polymorphisms (SNPs) present in humans. These SNPs, MLKL p.Ser132Pro (rs35589326, global MAF — 0.0138), p.Arg146Gln (rs34515646, global MAF — 0.0152) and p.Gly*202Val (rs144526386, global MAF — 0.0123, *non canonical transcript)- have not been previously associated with human disease in any previous genome-wide- or phenome-wide-association studies (GWAS or PheWAS) individually. They were however shown to occur in trans at 10–12 times the expected frequency in a cohort of patients suffering from chronic recurrent multifocal osteomyelitis (CRMO) [27]. While this association has not been independently replicated in a second CRMO patient cohort to date, this finding does fit nicely with mouse and human studies that implicate ‘unchecked’ necroptosis in the progression of inflammatory diseases [20,26].
An analysis of the cellular function of the protein products of these common human MLKL SNPs relative to wild-type MLKL is yet to be reported in the literature. However, the hypermorphic nature of the closely situated mouse mutation Mlkl p.D139V does raise important questions as to the evolutionary significance of such high allele frequencies. An estimated 8% of the global population are heterozygous carriers of MLKL p.S132P, p.R146Q or p.G*202V based on gnomAD data, and 167 are homozygotes. Many reported hypermorphic variants in immune-related genes that confer a survival advantage to specific pathogens have achieved high allele frequency in certain populations [45,46]. Many such SNPs are also associated with an increased relative risk of autoinflammatory diseases like inflammatory bowel disease and systemic lupus erythematosus [46]. The identification of such evolutionary triads (pathogen - gene hypermorph — inflammatory disease) has the potential to blaze important trails into the development of novel anti-microbials and immunomodulatory therapies alike.
By this same evolutionary logic, MLKL's implication in diseases like diabetes and Alzheimer's also warrants the investigation of any historical survival advantage conferred by MLKL gene variants or expression Quantitative Trait Loci (eQTL) with regards to nutrient acquisition and storage. An intriguing example of this was described recently for MLKL's upstream activator RIPK1. Human gene variants that increase RIPK1 gene expression were found to be associated with obesity [47]. Nutrition and metabolism serve alongside pathogens as major evolutionary drivers of human genetic variation, and are core determinants of the common disorders (e.g. cardiovascular disease), that contribute substantially to disease burden in the modern world [48].
MLKL gene variants as modifiers of complex polygenic human traits and diseases
To date, MLKL gene variants have only been weakly associated with human traits or changes in disease risk in conventional GWAS. The highest 'P' value reported for an MLKL variant in a published GWAS was for a non-coding SNP rs2057805 (global MAF 0.2489) located in an upstream regulatory region in adolescent idiopathic scoliosis [49]. This association reached genome wide significance with a Bonferroni-adjusted P-value of 3 × 10−9. Other notable associations of MLKL SNPs with quantitative human traits that are significant or approaching genome wide significance include one with comparative height at age 10 (P = 5.26 × 10−7) [50] and another with mean reticulocyte volume, P = 1 × 10−8 (Global Biobank Engine, Stanford, CA (URL: http://gbe.stanford.edu accessed September 2021)) [22]. Interestingly, three non-coding SNPs in MLKL's obligate activating kinase RIPK3 are each very strongly associated with standing height in humans, P values <4 × 10−23 [22]. While the small ‘beta’ log odds ratios for each of these MLKL and RIPK3 SNPs described indicate that they account for only very low proportions of the phenotypic variance in these quantitative traits, they nonetheless highlight the potential of such unbiased approaches for uncovering unexpected gene-function links [51].
Disease causing gene variants identified in important upstream regulators of MLKL
When considering the pathophysiological impacts of necroptosis, we must not only consider genetic variation in MLKL itself, but also variation in genes that can inappropriately unleash, amplify, or even dampen the cellular activity of MLKL. Based on observations of important MLKL regulatory genes gleaned from genetically modified mice [20], we have plotted for the purposes of this review the number of disease associated human mutations reported in ClinVar [52], as accessed in September 2021 (Figure 2A). This tally is strongly dominated by RIPK1, TNFAIP3 (encoding A20) and CASP8, where several unique LOF or missense mutations are characterised by autoinflammatory and lymphoproliferative syndromes [53]. It is important to note however, that the identification of pathogenic gene variants is heavily biased towards those that associated with severe or atypical disease. A higher number of pathogenic RIPK1, TNFAIP3 and CASP8 mutations in ClinVar does not equate to a larger contribution to the heritable variation in disease risk, when considering all disease burden in the human population. In fact, it is likely quite the opposite. Examining the mutational spectrum of ∼141 000 individuals catalogued by the gnomAD database permits a simple visualisation of the high evolutionary constraint on LOF variation in RIPK1, TNFAIP3 and CASP8 [54] (Figure 2B). MLKL and ZBP1 each exhibit a higher summed frequency of predicted LOF alleles than RIPK1, TNFAIP3 and CASP8 combined, and both are dotted with a larger repertoire of more common (>MAF 0.001) missense gene variants (Figure 2C). The higher missense and LOF variation tolerance profiles of MLKL, RIPK3 and ZBP1 indicate that gene variants are less likely to result in a loss of reproductive fitness (e.g. severe disease in children and young adults) that would cause them to be subject to purifying selection over time [55]. By this same logic however, the higher levels of standing protein-coding variation in MLKL, RIPK3 and ZBP1 genes also increases the statistical likelihood that these genes can act as low effect size modifiers of polygenic traits in humans (e.g. disease risk, infection response, height or weight) [56].
MLKL and the varied manifestations of inborn errors in CASP8
A full decade before MLKL's role in cell death was discovered, inherited deficiency of Caspase 8 was reported to mediate autoimmune lymphoproliferative syndrome (ALPS). Chun et al. [57] reported homozygosity of a CASP8 variant (p.R248W) in two siblings that presented with lymphadenopathy and splenomegaly. Caspase-8 p.R248W exhibited reduced stability and resulted in an enzymatically inactive protein, with peripheral blood lymphocytes defective in CD95-induced apoptosis. Caspase-8 activity is an important determinant of programmed cell death downstream of death receptor and inflammasome activation [58]. The absence or inhibition of Caspase-8 or c-FLIPL activity facilitates the stabilisation of RIPK1 and RIPK3 [59–62] (both shown to be cleaved by Caspase-8 and/or Caspase-8-c-FLIPL heterodimers). Amongst other signalling events, this also promotes the formation of the necrosome and MLKL-mediated cell death. Mice deficient in Caspase 8 die at embryonic day 10.5 but Casp8WT/KO heterozygotes are phenotypically normal [63]. Casp8KO/KO embryonic lethality does not occur on a MlklKO/KO background, but Casp8KO/KO, MlklKO/KOmice do go on to develop progressive lymphadenopathy [64]. In light of this later observation, dysregulated MLKL function and necroptotic cell death is likely to be more pertinent to the many examples of human CASP8 variant -borne disease that do not manifest as ALPS.
Caspase-8 deficiency manifests as very early-onset Inflammatory Bowel Disease (VEO-IBD) in some individuals. Lehle et al. [65] reported three unrelated patients with homozygous CASP8 mutations (p.Q237R and p.R265W (equivalent position to canonical isoform R248W)) that presented with diarrheal, perianal disease, failure to thrive and discontinuous severe proctocolitis, as well as increased susceptibility to bacterial and viral infections. Primary patient cells and cells engineered to express CASP8 p.Q237R exogenously exhibited increased IL-1β release in response to lipopolysaccharide (LPS) priming, that could be blocked by NLRP3-mediated inflammasome inhibitor MCC950 and the MLKL inhibitor necrosulfonamide. Furthermore, CASP8 p.Q237R, but not p.R265W, was also shown to predispose engineered HT-29 colon carcinoma cells to increased MLKL oligomerisation and death when exposed to a necroptotic stimulus [65]. The important role of MLKL in this disease scenario is further supported by recent observations that MLKL fully mediates the ileitis and colitis observed in mice with intestinal epithelial cell-specific deficiency of Caspase-8 [66]. Chun et al.’s [65] and Lehle et al.’s [67] clinical descriptions of distinct disease manifestation in individuals harbouring CASP8 p.R248W/R265W also highlights that unchecked necroptosis may be variably expressed as a phenotype of the same CASP8 mutation, depending on the individual . The genetic or environmental determinants of this disparate role of MLKL remains an important line of future investigation into the potential toxicity of Caspase-8 targeted drugs.
Choose your battle: MLKL and disease associated with inborn errors of RIPK1
Given the important position of RIPK1 at the nexus of several cellular signalling pathways, MLKL and necroptosis account for a significant portion, but not all of, the downstream effects born of genetically encoded RIPK1 dysfunction. In 2018, Cuchet-Lourenco et al. [68] reported four individuals that were homozygous for three unique RIPK1 LOF nucleotide deletions, and a number of additional patients with homozygous RIPK1 LOF alleles were reported soon after [69]. Patients presented with early-onset inflammatory bowel disease, lymphopenia and arthritis of varying severity. One patient also suffered from growth restriction, severe motor delay and mild intellectual disability [70]. An increased susceptibility to viral, bacterial and fungal infection was also reported [53,71]. Patient fibroblasts isolated from one of these patients were shown to be more susceptible to cell death in the presence of TNF and polyI:C. These cells were protected from death by the MLKL inhibitor necrosulfonamide, strongly implicating necroptosis [68]. Similarly to Casp8, Ripk1 knockout in murine models is homozygous lethal, with mice dying during the immediate post-natal period due to systemic inflammation and cell death in multiple tissues. This inflammation is ameliorated in a MlklKO/KO background, but mouse pup survival is only extended by a few days [72]. The severe phenotype of RIPK1 KO/KO mice is only bypassed when both necroptosis and extrinsic apoptosis are removed from the equation by compound genetic crosses [72–74]. Homozygous RIPK1 LOF in humans can lead to severe disease, but is not incompatible with life as it is in mice. Is this difference intrinsic to inter-species differences in RIPK1 itself, or can it be explained by key species-specific differences in the regulation of downstream signal effectors like MLKL?
Compounding these questions are disease causing missense RIPK1 gene variants that prevent RIPK1 cleavage by caspases. Heterozygosity of such ‘cleavage resistant’ alleles is sufficient to induce a severe periodic inflammatory syndrome in humans characterised by fever and lymphadenopathy [75–77]. Homozygosity reduces the lifespan of mouse embryos to E10.5, well shorter than that observed for Ripk1KO/KO mice and for reasons that cannot be remedied by the deletion of Mlkl [76,78]. The hyperinflammatory phenotype of heterozygous adult mice is similarly unaltered by the genetic deletion of Mlkl (N. Lalaoui, unpublished).
RIPK3 variants in human health and disease
To date, there have been no clinical descriptions of humans that are homozygous for RIPK3 LOF gene variants. One individual that is homozygous for a predicted LOF RIPK3 allele was recorded as part of the ‘human knockout project’, though details of this individual's phenotype were not provided [79]. The allele in question, RIPK3 p.Pro493ThrfsTer9 (rs531266348), contains a frame shift insertion that may lead to a short C terminal truncation as opposed to a full LOF. As such, its classification as ‘Loss of Function’ is classified as low confidence in the gnomAD database, as is the only other potential LOF variant found in homozygous form; RIPK3 p.Arg422Ter (rs146886719). Similar to the MlklKO/KO mouse, Ripk3KO/KO mice do not show any overt signs of spontaneous neurodegenerative disease or diabetes under steady state conditions, even when aged beyond 12 months [80,81]. Interestingly, Ripk3 KO/KO mice do show evidence of disturbed bone architecture and increased osteoclasts, supporting the reported association of the RIPK3 cis-eQTL rs3212240 with lower estimated bone mineral density in a human cohort [82]. Given our discussion of gene variation and evolutionary fitness, it would be remiss not to mention reports that MlklKO/KO and Ripk3KO/KO mice show a significantly reduced propensity for reproductive organ aging [81,83]. Necroptosis deficient mice sire more pups at advanced age than their wild-type counterparts. This advantage is offset by the reduced fitness of the pups born to older fathers, but certainly warrants consideration of the influence of necroptosis gene variation and necroptosis drugs in human fertility going forward [81].
Conclusion
This review gives an overview of the current landscape of MLKL's role in heritable disease. Two distinct, de novo MLKL LOF variants have been shown to segregate with a severe familial neurodegenerative disorder and familial maturity onset diabetes of the young (MODY). While MLKL LOF alleles are more prevalent in the population relative to other key genes involved in programmed cell death, they are dwarfed in frequency (>100-fold less frequent) by a series of common non-conservative protein modifying mutations that cluster at key functional domains of MLKL. The identification and clinical description of additional families and individuals carrying homozygous and heterozygous MLKL LOF gene variants is not unlikely given the growing uptake of whole genome sequencing for clinical diagnosis. While rare, these individuals will reveal new and important insights into the role of MLKL and disease aetiology and the basic molecular mechanism of MLKL's varied cellular functions. They will help to focus and fine-tune the study of the trait- and disease- modifying potential of the MLKL protein coding variants carried by >10% of the global population, unlocking new clinical indications and contraindications for drugs that target necroptosis and its important upstream regulators.
Perspectives
Naturally occurring human gene variants are important tools for the study of necroptosis at the molecular and pathophysiological level.
Rare and de novo necroptosis gene variants have been implicated in autoinflammatory disease, neurodegeneration and metabolic disease.
More than 10% of individuals carry protein-coding single nucleotide polymorphisms in MLKL. Studying their role in human health and disease will uncover new clinical indications and contraindications for drugs that target necroptosis and its important upstream regulators.
Acknowledgements
We wish to thank Michael S. Hildebrand for his careful reading of this manuscript and helpful suggestions and James M. Murphy for generously providing the graphical elements used in Figure 1. Due to length restrictions, we have cited recent long-form reviews in place of some relevant primary sources.
Abbreviations
- ALPS
autoimmune lymphoproliferative syndrome
- CRMO
chronic recurrent multifocal osteomyelitis
- GWAS
Genome Wide Association Studies
- LOF
loss of function
- MLKL
mixed lineage kinase domain-like
- MODY
Maturity Onset Diabetes of the Young
- RIPK1
receptor interacting protein kinase 1
- SNPs
single nucleotide polymorphisms
- TNFR1
tumour necrosis factor receptor 1
- ZBP1
Z-DNA Binding Protein-1
Competing Interests
S.E.G. and J.M.H. contribute to a project developing necroptosis inhibitors in collaboration with Anaxis Pty Ltd.
Funding
J.M.H. is the recipient of an Australian NHMRC Career Development Fellowship 1142669 and Ideas Grant 2011584. S.E.G. is the recipient of The Wendy Dowsett Scholarship and an Australian Government Research Training Program Stipend Scholarship. We also thanks the Victorian Government Operational Infrastructure Support scheme.
Author Contributions
S.E.G. and J.M.H. performed analyses of publicly available human gene variant frequencies, conceived, constructed figures for and wrote this manuscript.
References
- 1.Kolb, J.P., Oguin, III, T.H., Oberst, A. and Martinez, J. (2017) Programmed cell death and inflammation: winter is coming. Trends Immunol. 38, 705–718 10.1016/j.it.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boada-Romero, E., Martinez, J., Heckmann, B.L. and Green, D.R. (2020) The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell Biol. 21, 398–414 10.1038/s41580-020-0232-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weir, A., Hughes, S., Rashidi, M., Hildebrand, J.M. and Vince, J.E. (2021) Necroptotic movers and shakers: cell types, inflammatory drivers and diseases. Curr. Opin. Immunol. 68, 83–97 10.1016/j.coi.2020.09.008 [DOI] [PubMed] [Google Scholar]
- 4.Tanzer, M.C., Frauenstein, A., Stafford, C.A., Phulphagar, K., Mann, M. and Meissner, F. (2020) Quantitative and dynamic catalogs of proteins released during apoptotic and necroptotic cell death. Cell Rep. 30, 1260–1270.e5 10.1016/j.celrep.2019.12.079 [DOI] [PubMed] [Google Scholar]
- 5.Shlomovitz, I., Erlich, Z., Arad, G., Edry-Botzer, L., Zargarian, S., Cohen, H.et al. (2021) Proteomic analysis of necroptotic extracellular vesicles. Cell Death Dis. 12, 1059 10.1038/s41419-021-04317-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murai, S., Yamaguchi, Y., Shirasaki, Y., Yamagishi, M., Shindo, R., Hildebrand, J.M.et al. (2019) A FRET biosensor for necroptosis uncovers two different modes of the release of DAMPs. Nat. Commun. 9, 4457 10.1038/s41467-018-06985-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson, A.L., Garnish, S.E., Hildebrand, J.M. and Murphy, J.M. (2021) Location, location, location: a compartmentalized view of TNF-induced necroptotic signaling. Sci. Signal. 14, eabc6178 10.1126/scisignal.abc6178 [DOI] [PubMed] [Google Scholar]
- 8.Sun, L., Wang, H., Wang, Z., He, S., Chen, S., Liao, D.et al. (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 9.Zhao, J., Jitkaew, S., Cai, Z., Choksi, S., Li, Q., Luo, J.et al. (2012) Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl Acad. Sci. U.S.A. 109, 5322–5327 10.1073/pnas.1200012109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conos, S.A., Chen, K.W., De Nardo, D., Hara, H., Whitehead, L., Nunez, G.et al. (2017) Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc. Natl Acad. Sci. U.S.A. 114, E961–E9E9 10.1073/pnas.1613305114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, Y.N., Guy, C., Olauson, H., Becker, J.U., Yang, M., Fitzgerald, P.et al. (2017) ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and Its consequences. Cell 169, 286–300.e16 10.1016/j.cell.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon, S., Kovalenko, A., Bogdanov, K. and Wallach, D. (2017) MLKL, the protein that mediates necroptosis, also regulates endosomal trafficking and extracellular vesicle generation. Immunity 47, 51–65.e7 10.1016/j.immuni.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 13.Zargarian, S., Shlomovitz, I., Erlich, Z., Hourizadeh, A., Ofir-Birin, Y., Croker, B.A.et al. (2017) Phosphatidylserine externalization, “necroptotic bodies” release, and phagocytosis during necroptosis. PLoS Biol. 15, e2002711 10.1371/journal.pbio.2002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, W., Guo, J., Gao, B., Zhang, W., Ling, L., Xu, T.et al. (2019) Flotillin-mediated endocytosis and ALIX-syntenin-1-mediated exocytosis protect the cell membrane from damage caused by necroptosis. Sci. Signal. 12, eaaw3423 10.1126/scisignal.aaw3423 [DOI] [PubMed] [Google Scholar]
- 15.Ying, Z., Pan, C., Shao, T., Liu, L., Li, L., Guo, D.et al. (2018) Mixed lineage kinase domain-like protein MLKL breaks down myelin following nerve injury. Mol. Cell 72, 457–468.e5 10.1016/j.molcel.2018.09.011 [DOI] [PubMed] [Google Scholar]
- 16.D'Cruz, A.A., Speir, M., Bliss-Moreau, M., Dietrich, S., Wang, S., Chen, A.A.et al. (2018) The pseudokinase MLKL activates PAD4-dependent NET formation in necroptotic neutrophils. Sci. Signal. 11, eaao1716 10.1126/scisignal.aao1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakazawa, D., Desai, J., Steiger, S., Muller, S., Devarapu, S.K., Mulay, S.R.et al. (2018) Activated platelets induce MLKL-driven neutrophil necroptosis and release of neutrophil extracellular traps in venous thrombosis. Cell Death Discov. 4, 6 10.1038/s41420-018-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sai, K., Parsons, C., House, J.S., Kathariou, S. and Ninomiya-Tsuji, J. (2019) Necroptosis mediators RIPK3 and MLKL suppress intracellular Listeria replication independently of host cell killing. J. Cell Biol. 218, 1994–2005 10.1083/jcb.201810014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon, S., Bogdanov, K. and Wallach, D. (2022) Site-specific ubiquitination of MLKL targets it to endosomes and targets Listeria and Yersinia to the lysosomes. Cell Death Differ. 10.1038/s41418-021-00924-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tovey Crutchfield, E.C., Garnish, S.E. and Hildebrand, J.M. (2021) The role of the key effector of necroptotic cell death, MLKL, in mouse models of disease. Biomolecules 11, 803 10.3390/biom11060803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T.et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInnes, G., Tanigawa, Y., DeBoever, C., Lavertu, A., Olivieri, J.E., Aguirre, M.et al. (2019) Global Biobank Engine: enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics 35, 2495–2497 10.1093/bioinformatics/bty999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, B., Bao, S., Zhang, Z., Zhou, X., Wang, J., Fan, Y.et al. (2018) A rare variant in MLKL confers susceptibility to ApoE varepsilon4-negative Alzheimer's disease in Hong Kong Chinese population. Neurobiol. Aging 68, 160.e1–160.e7 10.1016/j.neurobiolaging.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 24.Faergeman, S.L., Evans, H., Attfield, K.E., Desel, C., Kuttikkatte, S.B., Sommerlund, M.et al. (2020) A novel neurodegenerative spectrum disorder in patients with MLKL deficiency. Cell Death Dis. 11, 303 10.1038/s41419-020-2494-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildebrand, J.M., Lo, B., Tomei, S., Mattei, V., Young, S.N., Fitzgibbon, C.et al. (2021) A family harboring an MLKL loss of function variant implicates impaired necroptosis in diabetes. Cell Death Dis. 12, 345 10.1038/s41419-021-03636-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speir, M., Djajawi, T.M., Conos, S.A., Tye, H. and Lawlor, K.E. (2021) Targeting RIP kinases in chronic inflammatory disease. Biomolecules 11, 646 10.3390/biom11050646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hildebrand, J.M., Kauppi, M., Majewski, I.J., Liu, Z., Cox, A.J., Miyake, S.et al. (2020) A missense mutation in the MLKL brace region promotes lethal neonatal inflammation and hematopoietic dysfunction. Nat. Commun. 11, 3150 10.1038/s41467-020-16819-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor, J.C., Martin, H.C., Lise, S., Broxholme, J., Cazier, J.B., Rimmer, A.et al. (2015) Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet. 47, 717–726 10.1038/ng.3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J.et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405–424 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, J.M., Czabotar, P.E., Hildebrand, J.M., Lucet, I.S., Zhang, J.G., Alvarez-Diaz, S.et al. (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 31.Wu, J., Huang, Z., Ren, J., Zhang, Z., He, P., Li, Y.et al. (2013) Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 23, 994–1006 10.1038/cr.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, T., Perera, N.D., Chiam, M.D.F., Cuic, B., Wanniarachchillage, N., Tomas, D.et al. (2020) Necroptosis is dispensable for motor neuron degeneration in a mouse model of ALS. Cell Death Differ. 27, 1728–1739 10.1038/s41418-019-0457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, J., Zhao, Y., Zhang, L., Fan, H., Qi, C., Zhang, K.et al. (2018) RIPK3/MLKL-Mediated neuronal necroptosis modulates the M1/M2 polarization of microglia/Macrophages in the ischemic cortex. Cereb Cortex 28, 2622–2635 10.1093/cercor/bhy089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan, H., Zhang, K., Shan, L., Kuang, F., Chen, K., Zhu, K.et al. (2016) Reactive astrocytes undergo M1 microglia/macrohpages-induced necroptosis in spinal cord injury. Mol. Neurodegener. 11, 14 10.1186/s13024-016-0081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, Q.S., Chen, P., Wang, W.X., Lin, C.C., Zhou, Y., Yu, L.H.et al. (2020) RIP1/RIP3/MLKL mediates dopaminergic neuron necroptosis in a mouse model of Parkinson disease. Lab. Invest. 100, 503–511 10.1038/s41374-019-0319-5 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, S., Su, Y., Ying, Z., Guo, D., Pan, C., Guo, J.et al. (2019) RIP1 kinase inhibitor halts the progression of an immune-induced demyelination disease at the stage of monocyte elevation. Proc. Natl Acad. Sci. U.S.A. 116, 5675–5680 10.1073/pnas.1819917116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlen, M., Fagerberg, L., Hallstrom, B.M., Lindskog, C., Oksvold, P., Mardinoglu, A.et al. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 38.Saeed, W.K., Jun, D.W., Jang, K., Oh, J.H., Chae, Y.J., Lee, J.S.et al. (2019) Decrease in fat de novo synthesis and chemokine ligand expression in non-alcoholic fatty liver disease caused by inhibition of mixed lineage kinase domain-like pseudokinase. J. Gastroenterol. Hepatol. 34, 2206–2218 10.1111/jgh.14740 [DOI] [PubMed] [Google Scholar]
- 39.Xu, H., Du, X., Liu, G., Huang, S., Du, W., Zou, S.et al. (2019) The pseudokinase MLKL regulates hepatic insulin sensitivity independently of inflammation. Mol. Metab. 23, 14–23 10.1016/j.molmet.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, J.D., Ahmed, N.T., Luciani, D.S., Han, Z., Tran, H., Fujita, J.et al. (2003) Increased islet apoptosis in Pdx1+/- mice. J. Clin. Invest. 111, 1147–1160 10.1172/JCI200316537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hattersley, A.T. and Patel, K.A. (2017) Precision diabetes: learning from monogenic diabetes. Diabetologia 60, 769–777 10.1007/s00125-017-4226-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hildebrand, J.M., Tanzer, M.C., Lucet, I.S., Young, S.N., Spall, S.K., Sharma, P.et al. (2014) Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl Acad. Sci. U.S.A. 111, 15072–15077 10.1073/pnas.1408987111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Z., Dagley, L.F., Shield-Artin, K., Young, S.N., Bankovacki, A., Wang, X.et al. (2021) Oligomerisation-driven MLKL ubiquitylation antagonises necroptosis. EMBO J. 40, e103718 10.15252/embj.2019103718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrie, E.J., Sandow, J.J., Jacobsen, A.V., Smith, B.J., Griffin, M.D.W., Lucet, I.S.et al. (2018) Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat. Commun. 9, 2422 10.1038/s41467-018-04714-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werren, E.A., Garcia, O. and Bigham, A.W. (2021) Identifying adaptive alleles in the human genome: from selection mapping to functional validation. Hum. Genet. 140, 241–276 10.1007/s00439-020-02206-7 [DOI] [PubMed] [Google Scholar]
- 46.Ramos, P.S., Shedlock, A.M. and Langefeld, C.D. (2015) Genetics of autoimmune diseases: insights from population genetics. J. Hum. Genet. 60, 657–664 10.1038/jhg.2015.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karunakaran, D., Turner, A.W., Duchez, A.C., Soubeyrand, S., Rasheed, A., Smyth, D.et al. (2020) RIPK1 gene variants associate with obesity in humans and can be therapeutically silenced to reduce obesity in mice. Nat. Metab. 2, 1113–1125 10.1038/s42255-020-00279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsson, E.K., Kwiatkowski, D.P. and Sabeti, P.C. (2014) Natural selection and infectious disease in human populations. Nat. Rev. Genet. 15, 379–393 10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, J., Zhou, Y., Liu, S., Song, X., Yang, X.Z., Fan, Y.et al. (2018) The coexistence of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) at a locus can result in distorted calculations of the significance in associating SNPs to disease. Hum. Genet. 137, 553–567 10.1007/s00439-018-1910-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, K., Stringer, S., Frei, O., Umicevic Mirkov, M., de Leeuw, C., Polderman, T.J.C.et al. (2019) A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 10.1038/s41588-019-0481-0 [DOI] [PubMed] [Google Scholar]
- 51.Crouch, D.J.M. and Bodmer, W.F. (2020) Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl Acad. Sci. U.S.A. 117, 18924–18933 10.1073/pnas.2005634117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landrum, M.J., Lee, J.M., Benson, M., Brown, G.R., Chao, C., Chitipiralla, S.et al. (2018) Clinvar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D10D7 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, J., Jin, T., Aksentijevich, I. and Zhou, Q. (2021) RIPK1-Associated inborn errors of innate immunity. Front. Immunol. 12, 676946 10.3389/fimmu.2021.676946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alfoldi, J., Wang, Q.et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silk, M., Petrovski, S. and Ascher, D.B. (2019) MTR-Viewer: identifying regions within genes under purifying selection. Nucleic Acids Res. 47, W121–W1W6 10.1093/nar/gkz457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lappalainen, T. and MacArthur, D.G. (2021) From variant to function in human disease genetics. Science 373, 1464–1468 10.1126/science.abi8207 [DOI] [PubMed] [Google Scholar]
- 57.Chun, H.J., Zheng, L., Ahmad, M., Wang, J., Speirs, C.K., Siegel, R.M.et al. (2002) Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 419, 395–399 10.1038/nature01063 [DOI] [PubMed] [Google Scholar]
- 58.Schwarzer, R., Laurien, L. and Pasparakis, M. (2020) New insights into the regulation of apoptosis, necroptosis, and pyroptosis by receptor interacting protein kinase 1 and caspase-8. Curr. Opin. Cell Biol. 63, 186–193 10.1016/j.ceb.2020.02.004 [DOI] [PubMed] [Google Scholar]
- 59.Oberst, A., Dillon, C.P., Weinlich, R., McCormick, L.L., Fitzgerald, P., Pop, C.et al. (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471, 363–367 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin, Y., Devin, A., Rodriguez, Y. and Liu, Z.G. (1999) Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13, 2514–2526 10.1101/gad.13.19.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng, S., Yang, Y., Mei, Y., Ma, L., Zhu, D.E., Hoti, N.et al. (2007) Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 19, 2056–2067 10.1016/j.cellsig.2007.05.016 [DOI] [PubMed] [Google Scholar]
- 62.Shindo, R., Ohmuraya, M., Komazawa-Sakon, S., Miyake, S., Deguchi, Y., Yamazaki, S.et al. (2019) Necroptosis of intestinal epithelial cells induces type 3 innate lymphoid cell-dependent lethal ileitis. iScience 15, 536–551 10.1016/j.isci.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varfolomeev, E.E., Schuchmann, M., Luria, V., Chiannilkulchai, N., Beckmann, J.S., Mett, I.L.et al. (1998) Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9, 267–276 10.1016/S1074-7613(00)80609-3 [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Diaz, S., Dillon, C.P., Lalaoui, N., Tanzer, M.C., Rodriguez, D.A., Lin, A.et al. (2016) The pseudokinase MLKL and the kinase RIPK3 have distinct roles in autoimmune disease caused by loss of death-receptor-Induced apoptosis. Immunity 45, 513–526 10.1016/j.immuni.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehle, A.S., Farin, H.F., Marquardt, B., Michels, B.E., Magg, T., Li, Y.et al. (2019) Intestinal inflammation and dysregulated immunity in patients with inherited caspase-8 deficiency. Gastroenterology 156, 275–278 10.1053/j.gastro.2018.09.041 [DOI] [PubMed] [Google Scholar]
- 66.Schwarzer, R., Jiao, H., Wachsmuth, L., Tresch, A. and Pasparakis, M. (2020) FADD and caspase-8 regulate Gut homeostasis and inflammation by controlling MLKL- and GSDMD-mediated death of intestinal epithelial cells. Immunity 52, 978–993 e6 10.1016/j.immuni.2020.04.002 [DOI] [PubMed] [Google Scholar]
- 67.Dietrich, M.H., Chan, L.J., Adair, A., Keremane, S., Pymm, P., Lo, A.W.et al. (2021) Nanobody generation and structural characterization of plasmodium falciparum 6-cysteine protein Pf12p. Biochem. J. 478, 579–595 10.1042/BCJ20200415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cuchet-Lourenco, D., Eletto, D., Wu, C., Plagnol, V., Papapietro, O., Curtis, J.et al. (2018) Biallelic RIPK1 mutations in humans cause severe immunodeficiency, arthritis, and intestinal inflammation. Science 361, 810–813 10.1126/science.aar2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li, Y., Fuhrer, M., Bahrami, E., Socha, P., Klaudel-Dreszler, M., Bouzidi, A.et al. (2019) Human RIPK1 deficiency causes combined immunodeficiency and inflammatory bowel diseases. Proc. Natl Acad. Sci. U.S.A. 116, 970–975 10.1073/pnas.1813582116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uchiyama, Y., Kim, C.A., Pastorino, A.C., Ceroni, J., Lima, P.P., de Barros Dorna, M.et al. (2019) Primary immunodeficiency with chronic enteropathy and developmental delay in a boy arising from a novel homozygous RIPK1 variant. J. Hum. Genet. 64, 955–960 10.1038/s10038-019-0631-3 [DOI] [PubMed] [Google Scholar]
- 71.Liu, L. and Lalaoui, N. (2021) 25 years of research put RIPK1 in the clinic. Semin. Cell Dev. Biol. 109, 86–95 10.1016/j.semcdb.2020.08.007 [DOI] [PubMed] [Google Scholar]
- 72.Rickard, J.A., O'Donnell, J.A., Evans, J.M., Lalaoui, N., Poh, A.R., Rogers, T.et al. (2014) RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 157, 1175–1188 10.1016/j.cell.2014.04.019 [DOI] [PubMed] [Google Scholar]
- 73.Dillon, C.P., Weinlich, R., Rodriguez, D.A., Cripps, J.G., Quarato, G., Gurung, P.et al. (2014) RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202 10.1016/j.cell.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaiser, W.J., Daley-Bauer, L.P., Thapa, R.J., Mandal, P., Berger, S.B., Huang, C.et al. (2014) RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc. Natl Acad. Sci. U.S.A. 111, 7753–7758 10.1073/pnas.1401857111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, X., Dowling, J.P. and Zhang, J. (2019) RIPK1 can mediate apoptosis in addition to necroptosis during embryonic development. Cell Death Dis. 10, 245 10.1038/s41419-019-1490-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lalaoui, N., Boyden, S.E., Oda, H., Wood, G.M., Stone, D.L., Chau, D.et al. (2020) Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 10.1038/s41586-019-1828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tao, P., Sun, J., Wu, Z., Wang, S., Wang, J., Li, W.et al. (2020) A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114 10.1038/s41586-019-1830-y [DOI] [PubMed] [Google Scholar]
- 78.Newton, K., Wickliffe, K.E., Dugger, D.L., Maltzman, A., Roose-Girma, M., Dohse, M.et al. (2019) Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431 10.1038/s41586-019-1548-x [DOI] [PubMed] [Google Scholar]
- 79.Saleheen, D., Natarajan, P., Armean, I.M., Zhao, W., Rasheed, A., Khetarpal, S.A.et al. (2017) Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544, 235–239 10.1038/nature22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newton, K., Sun, X. and Dixit, V.M. (2004) Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and toll-like receptors 2 and 4. Mol. Cell. Biol. 24, 1464–1469 10.1128/MCB.24.4.1464-1469.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li, D., Meng, L., Xu, T., Su, Y., Liu, X., Zhang, Z.et al. (2017) RIPK1-RIPK3-MLKL-dependent necrosis promotes the aging of mouse male reproductive system. elife 6, e27692 10.7554/eLife.27692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mullin, B.H., Tickner, J., Zhu, K., Kenny, J., Mullin, S., Brown, S.J.et al. (2020) Characterisation of genetic regulatory effects for osteoporosis risk variants in human osteoclasts. Genome Biol. 21, 80 10.1186/s13059-020-01997-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li, D., Ai, Y., Guo, J., Dong, B., Li, L., Cai, G.et al. (2020) Casein kinase 1G2 suppresses necroptosis-promoted testis aging by inhibiting receptor-interacting kinase 3. eLife 9, e61564 10.7554/eLife.61564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garnish, S.E., Meng, Y., Koide, A., Sandow, J.J., Denbaum, E., Jacobsen, A.V.et al. (2021) Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis. Nat. Commun. 12, 2211 10.1038/s41467-021-22400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samson, A.L., Zhang, Y., Geoghegan, N.D., Gavin, X.J., Davies, K.A., Mlodzianoski, M.J.et al. (2020) MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 11, 3151 10.1038/s41467-020-16887-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petrie, E.J., Hildebrand, J.M. and Murphy, J.M. (2017) Insane in the membrane: a structural perspective of MLKL function in necroptosis. Immunol. Cell Biol. 95, 152–159 10.1038/icb.2016.125 [DOI] [PubMed] [Google Scholar]
- 87.Steiner, A., Harapas, C.R., Masters, S.L. and Davidson, S. (2018) An update on autoinflammatory diseases: relopathies. Curr. Rheumatol. Rep. 20, 39 10.1007/s11926-018-0749-x [DOI] [PubMed] [Google Scholar]
- 88.Di Donato, G., d'Angelo, D.M., Breda, L. and Chiarelli, F. (2021) Monogenic autoinflammatory diseases: state of the Art and future perspectives. Int. J. Mol. Sci. 22, 6360 10.3390/ijms22126360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duncan, C.J.A., Randall, R.E. and Hambleton, S. (2021) Genetic lesions of type I interferon signalling in human antiviral immunity. Trends Genet. 37, 46–58 10.1016/j.tig.2020.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Notarangelo, L.D., Bacchetta, R., Casanova, J.L. and Su, H.C. (2020) Human inborn errors of immunity: an expanding universe. Sci. Immunol. 5, eabb1662 10.1126/sciimmunol.abb1662 [DOI] [PMC free article] [PubMed] [Google Scholar]