Abstract
The tumour necrosis factor (TNF) is the most potent inducer of cell death amongst cytokines. It is crucial for processes including homeostasis, the development of the immune system and fighting infections. However, high levels of TNF due to genetic disorders or persistent infections can contribute to autoinflammatory and autoimmune diseases or life-threatening conditions like sepsis. These diseases generally display increased levels of cell death, which, downstream of the TNF receptor, can either be caspase-dependent (apoptosis) or caspase-independent (necroptosis). Significant efforts have been invested in unravelling and manipulating signalling mechanisms regulating these two different types of cell death. Here I discuss how modern proteomic approaches like phosphoproteomics and secretomics provide a novel perspective on this central cytokine and its effect on inflammation and cell survival.
Keywords: apoptosis, necroptosis, post-translational modification, proteomics, tumor necrosis factors
Introduction
The pro-inflammatory cytokine TNF is a master regulator of the immune system. It is produced by a range of immune cells of the adaptive and innate immune system [1]. TNF enhances antigen-specific responses and is important for the resolution phase of the adaptive immune response [2]. Investigations into TNF expression in innate immune cells revealed its up-regulation during various pathogenic infections. TNF stimulation, in turn, leads to the up-regulation of hundreds of target genes important to fight infections by recruiting other immune cells and amplifying the immune response. Sustained high levels of TNF in persistent infections or autoimmune disorders can have detrimental consequences [3]. This also applies to other proinflammatory cytokines like Interleukin-1β (IL-1b) and Interleukin-6 (IL-6).
However, in contrast with these cytokines, TNF potently induces cell death, which greatly contributes to its pro-inflammatory nature [4]. While cell death of infected cells can prevent further proliferation and spread of the pathogen, persistent cell death can damage the host by exacerbating inflammation. Socrates saying that ‘Death may be the greatest of all human blessings' may not always hold true on the cellular level. Intensive research in the past decades have uncovered a range of different ways a cell can die in response to infections and cytokines with different inflammatory consequences [5]. These various cell death pathways have been developed by the host to fight infection, and over years of co-evolution, they have also been harnessed and manipulated to the pathogen's advantage [6]. TNF has been found to directly induce two types of cell death, which can either be caspase-dependent called apoptosis or caspase-independent called necroptosis [4]. To manipulate TNF signalling for therapeutic purposes, its signalling mechanisms have to be studied in detail. Proteomics offers a global, unbiased and discovery-driven approach that is ideally suited to this purpose. In this perspective, I use TNF signalling as a paradigm case to highlight the power of that technology in (immune) signalling. For illustration I focus on two recent global and extensive studies of our group [7,8].
TNF signalling and cell death
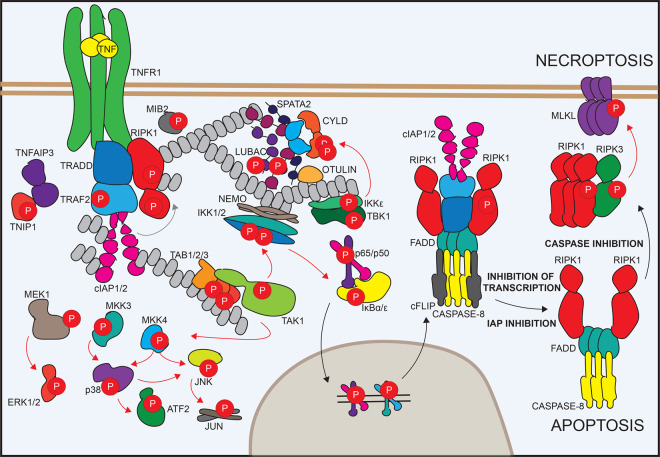
The binding of TNF to its TNF receptor induces the recruitment of several cytosolic proteins via mutual domains (Figure 1) [9]. The Fas-associated protein with death domain (FADD), the tumor necrosis factor receptor type 1-associated death domain (TRADD), and receptor-interacting serine/threonine-protein kinase 1 (RIPK1) interact via their death domains with the TNF receptor and each other [10]. Thereby they build a stable signalling platform that allows other proteins to bind, including TNF receptor associated factor 2 (TRAF2) and the E3 ligases inhibitor of apoptosis proteins (IAPs) and the linear ubiquitin assembly complex (LUBAC) complex. Ubiquitination of the complex builds additional recruitment platforms for the kinases transforming growth factor-β-activated kinase 1 (TAK1), IκB kinase (IKKs) and mitogen-activated protein kinase (MAPKs), which upon association can become activated. Intact activation of the nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells (NF-κB) and MAPK signalling results in the up-regulation of many TNF target genes, including cytokines, chemokines, and also pro-survival proteins such as the cellular FADD-like IL-1β-converting enzyme inhibitory protein (cFLIP), which inhibits caspase activity [11,12]. Therefore, intact TNF signalling does not lead to cell death. Perturbation to this system, including infections, or upon genetic aberrations — which lead to the inhibition, down-regulation, or chronic activation of members of the TNF signalling complex — result in a balance shift from the formation of complex I (pro-survival) to complex II (apoptosis-inducing) with the increased binding and activation of caspase-8/caspase-10 (in human) [3,13,14]. When cFLIP levels are no longer sufficient to keep these caspases in check, they become active and degrade their substrates resulting in apoptosis [15]. Apoptosis is an ‘immunological silent’ cell death, with features including cellular disintegration, blebbing, shrinkage and nuclear fragmentation, with minimal inflammatory stimulation of the environment [4]. Additionally, apoptotic corpses are quickly removed by phagocytes [16]. In several cell types, inhibiting caspases does not block cell death upon TNF stimulation but leads to an alternative, lytic form of cell death called necroptosis [17]. This involves the autophosphorylation of RIPK1, which leads to the activation of the receptor-interacting serine/threonine-protein kinase 3 (RIPK3) and phosphorylation of the mixed-lineage kinase domain-like pseudokinase (MLKL) [18–20]. Active MLKL oligomerises, translocates to the membrane and induces plasma membrane rupture, leading to the uncontrolled release of cellular content, which is thought to drive a strong inflammatory signal [21–26].
Figure 1. Scheme of TNF signalling including proteins involved in TNF-induced apoptosis and necroptosis.
Signalling is primarily mediated through PTMs like phosphorylation and ubiquitylation. Hence, most proteins involved in this signalling are found to be phosphorylated.
Investigation of TNF signalling using mass spectrometry
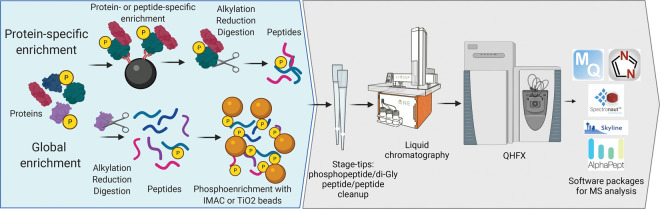
In the last decade, seminal studies investigating TNF signalling used elegant mouse models lacking one or more members of the TNF signalling pathway to analyse their role in homeostasis or upon inflammatory challenges in vivo [27–30]. The identification of TNF complex members has primarily occurred through yeast two-hybrid screens and mass spectrometry (MS) [20,31–33]. For example, the central mediator of extrinsic apoptosis caspase-8 was identified in 1996 by Muzio et al. [31] using mass spectrometry, which was then in its infancy. The enormous leap of development and accessibility of this technology enabled the identification and characterisation of many further TNF signalling members [34–37]. Alongside the development of MS technology, genetic screening presented itself as a helpful tool to identify functional members of signalling cascades. MS, however, is the only technology to unbiasedly determine interaction partners in endogenous systems [38]. Just recently several novel members of the TNF signalling complex including the spermatogenesis associated 2 protein (SPATA2), mind bomb 2 (MIB2), the TANK-binding kinase 1 (TBK1) and the IκB kinase epsilon (IKKε) were identified using classical pulldown approaches in combination with MS [34,35,37,39]. MS is also becoming more readily utilised in structural studies and has proven exceptionally successful in combination with various pulldown- and enrichment strategies to unravel enzyme-substrate relationships (Figure 2). It is primarily used in the signalling field to investigate post-translational modifications like phosphorylation, ubiquitylation and less studied PTMs including acetylation, methylation, citrullination and other modifications like protein cleavage [7,35,40]. An enrichment strategy for caspase substrates recently identified the NEDD4-binding protein 1 (N4BP1) as a target of caspase-8 and suppressor of the cytokine response upon lipopolysaccharide (LPS) or TNF stimulation [41]. Also poly(ADP-ribosyl)ation (PARylation) of complex II has been presented as regulator of TNF-induced death, which was discovered through the identification of tankyrase-1 (TNKS1) as a novel interaction partner of complex II via MS (Lin et al. 2021, Biorxiv). Enrichment strategies for these less studied PTMs like PARylation still require optimizations, but offer exciting avenues to unravel novel signalling aspects. MS not only distinguishes between all these different types of modifications, but also reports their precise localisation on the respective substrates and their abundance. A combination of diverse enrichment and/or analysis strategies could provide further insights into the interplay between different PTMs in the same biological context. So far, MS-analyses of TNF signalling was primarily focused on phosphorylation and ubiquitylation due to their pronounced and crucial role. These PTMs were either identified through specific enrichment of the proteins of interest or global phospho- and diGly- enrichments (Figure 2) [7,35,36,42–48]. The subsequent functional analyses and generation of site-specific antibodies validated these findings. A range of studies using MS revealed ubiquitylation of most TNF complex members including the NF-κB essential modulator (NEMO), RIPK1, RIPK3, TRAF2, TRADD primarily mediated by LUBAC and cIAPs, which have also shown to autoubiquitylate [36,49,50]. Further E3 ligases like MIB2, the pellino E3 ubiquitin protein ligase 1 (PELI1) and the TNF-induced protein 3 (TNFAIP3/A20) were also found to regulate TNF signalling, counteracted by DUBs like the OTU deubiquitinase with linear linkage specificity (OTULIN), the cylindromatosis (CYLD) and the ubiquitin specific peptidase 22 (USP22) [51,52]. A study reported that ubiquitylation on four positions of MLKL, which were identified by MS, regulate MLKL-induced cell death upon stimulation and restrain activated MLKL at basal conditions, while another study demonstrated that ubiquitylation of MLKL on a different lysine induces higher-order oligomerisation and activation of MLKL [53,54]. There is a constant development of improved diGly enrichment- and analysis strategies and of new tools that investigate ubiquitin linkages. Additionally, more specific inhibitors targeting E3 ligases and other TNF-signalling members are presented. Together, this will not only provide more information on ubiquitylated proteins and ubiquitin chain types but also offer a starting point for the development of the proteolysis targeting chimera (PROTAC)-based therapies targeting TNF signalling [42,55,56].
Figure 2. Scheme of proteomic workflows mostly used to investigate signalling changes including post-translational modifications e.g. phosphorylation.
Phosphoproteomics — signalling during cell death
Global PTM enrichment strategies provide a unique, unbiased perspective on the signalling events changing upon specific stimulations. Combined with precise alterations to the system in question, it can provide a snapshot of modified proteins and information about kinetics, enzyme-substrate relationships and localisations of the modified proteins. Recently, we have used this approach on a large scale to elucidate the TNF signalling pathway and signalling events dominating during TNF-induced cell death [7]. A detailed time course experiment revealed different clusters of phosphorylations based on their temporal induction. While we could distinguish early from late events occurring downstream of the TNF signalling receptor, we could not directly discern kinase-substrate relationships due to their rapid on-off kinetics and simultaneous occurrence. In vitro kinase assays are frequently used to assign kinases to selected substrates. They are, however, not feasible for large-scale studies [57]. Systematic pulldowns of kinases are also rarely successful in providing a comprehensive and reliable list of substrates due to the mostly transient interactions of kinases with their substrates. For most large-scale approaches like phosphoproteome studies, it is very effective to inhibit the activity or delete various kinases known to orchestrate signalling. While this doesn't prove a direct kinase-substrate relationship, it allows the assignment of regulated phosphorylation events to upstream kinases. Final kinase-substrate relationships of selected candidates can then be determined by in vitro kinase assays or by investigating kinase-substrate interactions. Kinase motif analyses can also be of help to pinpoint direct substrates. An earlier study investigated phosphorylation events dependent on RIPK3 by using RIPK3 deficient L929 cells and presented RIPK3 dependent phosphorylations. However, no information on the significance of the phosphorylation changes was reported [45]. Another study presented a deeper phosphoproteome with phosphorylation events downstream of IKK2 using an IKK2 inhibitor upon TNF stimulation [46]. We inhibited a range of kinases driving NF-κB and MAPK signalling, which are known to play a role in TNF signalling and could indeed determine downstream phosphorylation events. Cellular fractionation allowed us to localise these TNF regulated phosphorylation in the nucleus, cytosol and membrane. Combined, this information contributes to a better understanding of the TNF landscape. Another way to determine signalling events important in maintaining intact signalling is to observe regulatory response upon disturbing this balance. One phosphoproteome study investigated TNF signalling upon IAP inhibition, an apoptosis inducing stimulation [44]. This study looked, however, at a very early time point, when caspases are not activated yet. When we triggered TNF-induced apoptosis and necroptosis and looked at phosphorylation changes, we detected evidence of caspase activity specifically during apoptosis based on the activation of the DNA repair pathways. We also observed prominent regulation of transcriptional cyclin-dependent kinases (CDK) phosphorylation upon apoptosis. Zhong et al. [45] showed an increased CDK12 phosphorylation upon TNF treatment in wild-type and RIPK3 knockdown cells, but with no significance reported. With further experiments we revealed a crucial function for CDKs in TNF-induced transcription, affecting levels of target genes like cytokines and pro-survival mediators like cFLIP. Pan-CDK inhibition reduced the induction of all cFLIP isoforms. CDK12/13 inhibition also strongly attenuated up-regulation of cFLIPL. Consequently, inhibition of transcriptional CDKs induced synergistic cell death upon TNF stimulation. Hence, our unbiased, global phosphoproteomic approach revealed the organisation of phosphorylations upon TNF stimulation in time and space and highlighted novel potential drug targets that influence the decision between survival and death, inflammation and homeostasis.
The release of proteins during apoptosis and necroptosis
Our phosphoproteome analysis elucidated cell death type-specific and common signalling events, which can be manipulated to inhibit or enhance cell death. However, information on the inflammatory potential of apoptosis and necroptosis was limited. During apoptosis, the cell membrane integrity remains intact until the later stages of secondary necrosis [58]. Due to the early activation of caspases, many potentially inflammatory components of the cell are cleaved and deactivated, e.g. the damage-associated molecular patterns (DAMPs) like DNA and proteins [59]. In vivo, fast removal of apoptotic cells by phagocytes, also known as efferocytosis, prevents the uncontrolled spilling of its content into the environment [16,60]. Therefore, apoptosis is thought to be less inflammatory. In contrast with apoptosis, necroptosis leads to the immediate release of the cellular content and is therefore believed to be highly inflammatory [59,61]. To investigate the inflammatory potential of both modes of cell death in an unbiased way, we analysed the proteins released by apoptotic and necroptotic cells along a time course using mass spectrometry [8]. The supernatants of necroptotic cells showed an early release of a mixture of most intracellular proteins, which supports its presentation as inflammatory cell death. However, when investigating the release of cytokines, we detected a moderate increase in cytokine release by necroptotic cells, in contrast with TNF only treated or apoptotic cells. This is most likely due to the fast kinetics of necroptosis induction in vitro. The supernatant analysis also uncovered the activation of specific processes during cell death. We detected extracellular domains of receptors in the supernatants of apoptotic and necroptotic cells. Inhibition of disintegrin and metalloproteinase (ADAMs) demonstrated the activation of these metalloproteases during both cell death types, which leads to the cleavage of surface proteins. This can shut down signalling, detach dying cells from tissue and generate decoy receptors.
Furthermore, we specifically detected an increased release of luminal lysosomal proteins in the supernatant and membrane-bound lysosomal proteins in the extracellular vesicles of necroptotic cells. This indicated the activation of lysosomal exocytosis, a process activated due to membrane perturbation and calcium influx to prevent membrane rupture. Hence, the analysis of supernatants gave indications of the inflammatory potential of cell death types and led to the discovery of processes activated during cell death. In our study, we used a monocytic cell line. Further investigations of primary cells or dying cells in tissue upon more physiological conditions would provide an invaluable addition to the existing data.
Prospects and challenges
Many studies have already applied various proteomic techniques to successfully investigate different aspects of TNF signalling and TNF-induced cell death. In this way, a range of regulators that present potential therapeutic targets have been identified. In my opinion, we are only at the beginning of exploiting the potential of proteomics in other signalling pathways, especially in primary tissue. So far, the amount of material required for PTM studies has severely limited such exciting projects. However, the increased sensitivity of mass spectrometers and improved protocols that allow efficient sample preparation will mitigate this problem. A perennial challenge in proteomics is identifying key functional events within these large datasets, especially of PTMs, that may contain hundreds to thousands of regulated events. I suggest three approaches could help with this: Firstly, the computational approach, illustrated recently by the development of a method that assigns scores to all phosphorylation events of human proteins reported in Phosphositeplus based on their predicted functionality [62]. The authors considered more than 59 features, including structure, conservation of amino acids and previous experimental results. This yielded a promising overall score that appears to rank the functionally most important phosphorylation events highly. Secondly, the development of an improved stratification of proteins and PTMs by improving the current assignments to enrichment terms and enrichment analysis strategies and by thorough and systematic investigations of enzyme-substrate relationships in diverse biological contexts. While the former can largely be realised with already existing data, the latter requires extensive and well thought out setups involving genetic or pharmacological inhibition in different cell types, with various stimulations at different time points. Thirdly, the combination of proteomic experiments with large, project-specific genetic screens, which help narrow down on proteins and modifications of interest for further follow-up studies.
Besides the incredible technical development on the MS hardware side in the past years, the growing number of software solutions for analysing mass spectrometry data drives the field's expansion, development, and importance. It is highly likely that in the future, proteomics will provide even more insight into signalling pathways and complex biological processes like cell death in diverse biological samples ranging from the single cell to any tissue of interest.
Perspectives
Importance of the field. TNF is a prominent and clinically relevant cytokine and a shining example of the potential of proteomic applications to uncover signalling mechanisms. The detailed understanding of signalling pathways and their effects on global cellular processes is a prerequisite for successfully developing therapies.
A summary of the current thinking. These proteomic approaches can also be applied to unravel other signalling pathways, including other cell death types like pyroptosis, ferroptosis or the process of dead cell removal.
Future directions. The continuous technical improvement will allow the application of proteomics to investigate various aspects of signalling in vivo with spatial information and unprecedented sensitivity.
Acknowledgements
Thanks to Dr. Che Stafford and Prof. Matthias Mann for editing the manuscript. M.C.T was supported by the Max Planck Society for the Advancement of Science and a Marie Sklodowska-Curie Actions Individual Fellowship (746329). Figure 2 was generated with Biorender.com.
Abbreviations
- ADAM
A disintegrin and metalloproteinase
- CDKs
cyclin-dependent kinases
- cFLIP
cellular FADD-like IL-1β-converting enzyme inhibitory protein
- CYLD
cylindromatosis
- DAMPs
damage-associated molecular pattern
- FADD
Fas-associated protein with death domain
- IAP
inhibitor of apoptosis protein
- IKK
IκB kinase
- IKKε
IκB kinase epsilon
- IL
interleukin
- LPS
lipopolysaccharide
- LUBAC
linear ubiquitin assembly complex
- MAPK
mitogen-activated protein kinase
- MIB2
mind bomb 2
- MLKL
mixed-lineage kinase like domain-like
- MS
mass spectrometry
- N4BP1
NEDD4-binding protein 1
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells
- OTULIN
OTU deubiquitinase with linear linkage specificity
- PARylation
Poly(ADP-ribosyl)ation
- PELI1
Pellino E3 ubiquitin protein ligase 1
- PROTAC
proteolysis targeting chimera
- PTMs
post-translational modifications
- RIPK1
receptor-interacting serine/threonine-protein kinase 1
- RIPK3
receptor-interacting serine/threonine-protein kinase 3
- SPATA2
spermatogenesis associated 2
- TAK1
transforming growth factor-β-activated kinase 1
- TBK1
TANK-binding kinase 1
- TNF
tumour necrosis factor
- TNFAIP3
TNF-induced protein 3
- TNKS1
tankyrase-1
- TRADD
tumour necrosis factor receptor type 1-associated death domain
- TRAF2
TNF receptor associated factor 2
- USP22
ubiquitin specific peptidase 22
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Open Access Statement
Open access for this article was enabled by the participation of Max Planck Digital Library in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with Max Planck Digital Library.
Author Contributions
M.C.T. wrote the manuscript.
References
- 1.Vassalli, P. (1992) The pathophysiology of tumor necrosis factors. Annu. Rev. Immunol. 10, 411–452 10.1146/annurev.iy.10.040192.002211 [DOI] [PubMed] [Google Scholar]
- 2.Kassiotis, G. and Kollias, G. (2001) Uncoupling the proinflammatory from the immunosuppressive properties of tumor necrosis factor (TNF) at the p55 TNF receptor level: implications for pathogenesis and therapy of autoimmune demyelination. J. Exp. Med. 193, 427–434 10.1084/jem.193.4.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster, J.D. and Vucic, D. (2020) The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front. Cell Dev. Biol. 8, 365 10.3389/fcell.2020.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertheloot, D., Latz, E. and Franklin, B.S. (2021) Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell. Mol. Immunol. 18, 1106–1121 10.1038/s41423-020-00630-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi, L., Vitale, I., Aaronson, S.A., Abrams, J.M., Adam, D., Agostinis, P.et al. (2018) Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 25, 486–541 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman, M.M. and McFadden, G. (2006) Modulation of tumor necrosis factor by microbial pathogens. PLoS Pathog. 2, e4 10.1371/journal.ppat.0020004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanzer, M.C., Bludau, I., Stafford, C.A., Hornung, V. and Mann, M. (2021) Phosphoproteome profiling uncovers a key role for CDKs in TNF signaling. Nat. Commun. 12, 6053 10.1038/s41467-021-26289-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanzer, M.C., Frauenstein, A., Stafford, C.A., Phulphagar, K., Mann, M. and Meissner, F. (2020) Quantitative and dynamic catalogs of proteins released during apoptotic and necroptotic cell death. Cell Rep. 30, 1260–1270 e5 10.1016/j.celrep.2019.12.079 [DOI] [PubMed] [Google Scholar]
- 9.Silke, J. (2011) The regulation of TNF signaling: what a tangled web we weave. Curr. Opin. Immunol. 23, 620–626 10.1016/j.coi.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 10.Micheau, O. and Tschopp, J. (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 10.1016/S0092-8674(03)00521-X [DOI] [PubMed] [Google Scholar]
- 11.Zhou, A., Scoggin, S., Gaynor, R.B. and Williams, N.S. (2003) Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22, 2054–2064 10.1038/sj.onc.1206262 [DOI] [PubMed] [Google Scholar]
- 12.Micheau, O., Lens, S., Gaide, O., Alevizopoulos, K. and Tschopp, J. (2001) NF-kappaB signals induce the expression of c-FLIP. Mol. Cell. Biol. 21, 5299–5305 10.1128/MCB.21.16.5299-5305.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalaoui, N., Boyden, S.E., Oda, H., Wood, G.M., Stone, D.L., Chau, D.et al. (2020) Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 10.1038/s41586-019-1828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao, P., Sun, J., Wu, Z., Wang, S., Wang, J., Li, W.et al. (2020) A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114 10.1038/s41586-019-1830-y [DOI] [PubMed] [Google Scholar]
- 15.Tsuchiya, Y., Nakabayashi, O. and Nakano, H. (2015) FLIP the switch: regulation of apoptosis and necroptosis by cFLIP. Int. J. Mol. Sci. 16, 30321–30341 10.3390/ijms161226232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doran, A.C., Yurdagul, Jr, A. and Tabas, I. (2020) Efferocytosis in health and disease. Nat. Rev. Immunol. 20, 254–267 10.1038/s41577-019-0240-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. and Kroemer, G. (2010) Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 11, 700–714 10.1038/nrm2970 [DOI] [PubMed] [Google Scholar]
- 18.Hildebrand, J.M., Kauppi, M., Majewski, I.J., Liu, Z., Cox, A.J., Miyake, S.et al. (2020) A missense mutation in the MLKL brace region promotes lethal neonatal inflammation and hematopoietic dysfunction. Nat. Commun. 11, 3150 10.1038/s41467-020-16819-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, J.M., Czabotar, P.E., Hildebrand, J.M., Lucet, I.S., Zhang, J.G., Alvarez-Diaz, S.et al. (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 20.Sun, L., Wang, H., Wang, Z., He, S., Chen, S., Liao, D.et al. (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 21.Hildebrand, J.M., Tanzer, M.C., Lucet, I.S., Young, S.N., Spall, S.K., Sharma, P.et al. (2014) Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl Acad. Sci. U.S.A. 111, 15072–15077 10.1073/pnas.1408987111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanzer, M.C., Tripaydonis, A., Webb, A.I., Young, S.N., Varghese, L.N., Hall, C.et al. (2015) Necroptosis signaling is tuned by phosphorylation of MLKL residues outside the pseudokinase domain activation loop. Biochem. J. 471, 255–265 10.1042/BJ20150678 [DOI] [PubMed] [Google Scholar]
- 23.Garnish, S.E., Meng, Y., Koide, A., Sandow, J.J., Denbaum, E., Jacobsen, A.V.et al. (2021) Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis. Nat. Commun. 12, 2211 10.1038/s41467-021-22400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samson, A.L., Zhang, Y., Geoghegan, N.D., Gavin, X.J., Davies, K.A., Mlodzianoski, M.J.et al. (2020) MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 11, 3151 10.1038/s41467-020-16887-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, Y., Davies, K.A., Fitzgibbon, C., Young, S.N., Garnish, S.E., Horne, C.R.et al. (2021) Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis. Nat. Commun. 12, 6783 10.1038/s41467-021-27032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrie, E.J., Sandow, J.J., Jacobsen, A.V., Smith, B.J., Griffin, M.D.W., Lucet, I.S.et al. (2018) Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat. Commun. 9, 2422 10.1038/s41467-018-04714-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton, K., Wickliffe, K.E., Maltzman, A., Dugger, D.L., Reja, R., Zhang, Y.et al. (2019) Activity of caspase-8 determines plasticity between cell death pathways. Nature 575, 679–682 10.1038/s41586-019-1752-8 [DOI] [PubMed] [Google Scholar]
- 28.Rickard, J.A., O'Donnell, J.A., Evans, J.M., Lalaoui, N., Poh, A.R., Rogers, T.et al. (2014) RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 157, 1175–1188 10.1016/j.cell.2014.04.019 [DOI] [PubMed] [Google Scholar]
- 29.Dannappel, M., Vlantis, K., Kumari, S., Polykratis, A., Kim, C., Wachsmuth, L.et al. (2014) RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 513, 90–94 10.1038/nature13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dillon, C.P., Weinlich, R., Rodriguez, D.A., Cripps, J.G., Quarato, G., Gurung, P.et al. (2014) RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202 10.1016/j.cell.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muzio, M., Chinnaiyan, A.M., Kischkel, F.C., O'Rourke, K., Shevchenko, A., Ni, J.et al. (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell 85, 817–827 10.1016/S0092-8674(00)81266-0 [DOI] [PubMed] [Google Scholar]
- 32.Stanger, B.Z., Leder, P., Lee, T.H., Kim, E. and Seed, B. (1995) RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 81, 513–523 10.1016/0092-8674(95)90072-1 [DOI] [PubMed] [Google Scholar]
- 33.Hsu, H., Xiong, J. and Goeddel, D.V. (1995) The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 81, 495–504 10.1016/0092-8674(95)90070-5 [DOI] [PubMed] [Google Scholar]
- 34.Feltham, R., Jamal, K., Tenev, T., Liccardi, G., Jaco, I., Domingues, C.M.et al. (2018) Mind bomb regulates cell death during TNF signaling by suppressing RIPK1's cytotoxic potential. Cell Rep. 23, 470–484 10.1016/j.celrep.2018.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, S.A., Satpathy, S., Beli, P. and Choudhary, C. (2016) SPATA2 links CYLD to the TNF-alpha receptor signaling complex and modulates the receptor signaling outcomes. EMBO J. 35, 1868–1884 10.15252/embj.201694300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerlach, B., Cordier, S.M., Schmukle, A.C., Emmerich, C.H., Rieser, E., Haas, T.L.et al. (2011) Linear ubiquitination prevents inflammation and regulates immune signaling. Nature 471, 591–596 10.1038/nature09816 [DOI] [PubMed] [Google Scholar]
- 37.Lafont, E., Draber, P., Rieser, E., Reichert, M., Kupka, S., de Miguel, D.et al. (2018) TBK1 and IKKepsilon prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol. 20, 1389–1399 10.1038/s41556-018-0229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bludau, I. and Aebersold, R. (2020) Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat. Rev. Mol. Cell Biol. 21, 327–340 10.1038/s41580-020-0231-2 [DOI] [PubMed] [Google Scholar]
- 39.Elliott, P.R., Leske, D., Hrdinka, M., Bagola, K., Fiil, B.K., McLaughlin, S.H.et al. (2016) SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol. Cell 63, 990–1005 10.1016/j.molcel.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frauenstein, A., Ebner, S., Hansen, F.M., Sinha, A., Phulphagar, K., Swatek, K.et al. (2021) Identification of covalent modifications regulating immune signaling complex composition and phenotype. Mol. Syst. Biol. 17, e10125 10.15252/msb.202010125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gitlin, A.D., Heger, K., Schubert, A.F., Reja, R., Yan, D., Pham, V.C.et al. (2020) Integration of innate immune signaling by caspase-8 cleavage of N4BP1. Nature 587, 275–280 10.1038/s41586-020-2796-5 [DOI] [PubMed] [Google Scholar]
- 42.Hansen, F.M., Tanzer, M.C., Bruning, F., Bludau, I., Stafford, C., Schulman, B.A.et al. (2021) Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology. Nat. Commun. 12, 254 10.1038/s41467-020-20509-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dondelinger, Y., Delanghe, T., Priem, D., Wynosky-Dolfi, M.A., Sorobetea, D., Rojas-Rivera, D.et al. (2019) Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 10, 1729 10.1038/s41467-019-09690-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohideen, F., Paulo, J.A., Ordureau, A., Gygi, S.P. and Harper, J.W. (2017) Quantitative phospho-proteomic analysis of TNFalpha/NFkappaB signaling reveals a role for RIPK1 phosphorylation in suppressing necrotic cell death. Mol. Cell. Proteomics 16, 1200–1216 10.1074/mcp.M117.068189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, C.Q., Li, Y., Yang, D., Zhang, N., Xu, X., Wu, Y.et al. (2014) Quantitative phosphoproteomic analysis of RIP3-dependent protein phosphorylation in the course of TNF-induced necroptosis. Proteomics 14, 713–724 10.1002/pmic.201300326 [DOI] [PubMed] [Google Scholar]
- 46.Krishnan, R.K., Nolte, H., Sun, T., Kaur, H., Sreenivasan, K., Looso, M.et al. (2015) Quantitative analysis of the TNF-alpha-induced phosphoproteome reveals AEG-1/MTDH/LYRIC as an IKKbeta substrate. Nat. Commun. 6, 6658 10.1038/ncomms7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welz, B., Bikker, R., Junemann, J., Christmann, M., Neumann, K., Weber, M.et al. (2019) Proteome and phosphoproteome analysis in TNF long term-exposed primary human monocytes. Int. J. Mol. Sci. 20, 1241 10.3390/ijms20051241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cantin, G.T., Venable, J.D., Cociorva, D. and Yates, III, J.R. (2006) Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J. Proteome Res. 5, 127–134 10.1021/pr050270m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda, F., Deribe, Y.L., Skanland, S.S., Stieglitz, B., Grabbe, C., Franz-Wachtel, M.et al. (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471, 637–641 10.1038/nature09814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertrand, M.J., Milutinovic, S., Dickson, K.M., Ho, W.C., Boudreault, A., Durkin, J.et al. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700 10.1016/j.molcel.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 51.Roberts, J.Z., Crawford, N. and Longley, D.B. (2021) The role of ubiquitination in apoptosis and necroptosis. Cell Death Differ. 10.1038/s41418-021-00922-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roedig, J., Kowald, L., Juretschke, T., Karlowitz, R., Ahangarian Abhari, B., Roedig, H.et al. (2021) USP22 controls necroptosis by regulating receptor-interacting protein kinase 3 ubiquitination. EMBO Rep. 22, e50163 10.15252/embr.202050163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, Z., Dagley, L.F., Shield-Artin, K., Young, S.N., Bankovacki, A., Wang, X.et al. (2021) Oligomerization-driven MLKL ubiquitylation antagonizes necroptosis. EMBO J. 40, e103718 10.15252/embj.2019103718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia, L.R., Tenev, T., Newman, R., Haich, R.O., Liccardi, G., John, S.W.et al. (2021) Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance. Nat. Commun. 12, 3364 10.1038/s41467-021-23474-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton, K., Matsumoto, M.L., Wertz, I.E.. Kirkpatrick, D.S., Lill, J.R., Tan, J.et al. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 10.1016/j.cell.2008.07.039 [DOI] [PubMed] [Google Scholar]
- 56.Hu, Z. and Crews, C.M. (2021) Recent developments in PROTAC-mediated protein degradation: from bench to clinic. Chembiochem 23, e202100270 10.1002/cbic.202100270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson, S.A. and Hunter, T. (2005) Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 10.1038/nmeth731 [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., Chen, X., Gueydan, C. and Han, J. (2018) Plasma membrane changes during programmed cell deaths. Cell Res. 28, 9–21 10.1038/cr.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidovich, P., Kearney, C.J. and Martin, S.J. (2014) Inflammatory outcomes of apoptosis, necrosis and necroptosis. Biol. Chem. 395, 1163–1171 10.1515/hsz-2014-0164 [DOI] [PubMed] [Google Scholar]
- 60.Boada-Romero, E., Martinez, J., Heckmann, B.L. and Green, D.R. (2020) The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell Biol. 21, 398–414 10.1038/s41580-020-0232-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaczmarek, A., Vandenabeele, P. and Krysko, D.V. (2013) Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 38, 209–223 10.1016/j.immuni.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 62.Ochoa, D., Jarnuczak, A.F., Vieitez, C., Gehre, M., Soucheray, M., Mateus, A., et al. (2020) The functional landscape of the human phosphoproteome. Nat. Biotechnol. 38, 365–373 10.1038/s41587-019-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]