Abstract
The Cepheid Xpert® Xpress SARS-CoV-2 assay is 1 of the several real-time reverse transcription polymerase chain reaction (RT-PCR) assays that received Emergency Use Authorization from the United States Food and Drug Administration (FDA) for detection of SARS-CoV-2. Here we report 4 SARS-CoV-2 samples that were reported as presumptive positives on the Cepheid platform while reported as positives on alternative RT-PCR platforms. Whole genome sequencing indicated that the samples were Delta variants and had point mutations in the N gene which potentially interfered with SARS-CoV-2 detection. Two types of point mutations were found in these samples in the US CDC 2019-nCoV Real time PCR N2 Probe region: C29203T and C29200T. C29203T is a novel point mutation, and C29200T has not been previously reported in the Delta variants. This underlines the fact that mutations in the real-time RT-PCR assay target region could hinder accurate detection of SARS-CoV-2.
Keywords: Cepheid Xpert® Xpress SARS-CoV-2 assay, presumptive positive, false negative, C29203T, C29200T
1. Introduction
Several polymerase chain reactions (PCR) based coronavirus diseases-19 (COVID-19) testing platforms to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were developed and are available in the market under emergency use authorization (EUA). These platforms have been widely deployed across the world, including the United States (US), for confirming cases of COVID-19. Prompt detection of COVID-19 cases using these testing platforms helps with patient isolation, treatment of infection, and blocking further transmission.
Those primer and/or probes for the PCR assays for these testing platforms are primarily targeted to genomic regions of the nucleoprotein (N), spike protein (S), envelope gene (E), and open reading frame (ORF1ab) of the SARS-CoV-2 virus [1]. However, as the virus genomes change continuously, mutations arise in the test targeting region that could impact testing results. For example, spike protein mutations lead to S-gene target failure (SGTF) for ThermoFisher TaqPath™ COVID-19 testing platforms that use the S protein region. While there are adverse effects on testing performance as a result of SGTF, it has been a useful tool for screening out Delta and Omicron variants [2], [3], [4], [5].
The Cepheid Xpert® Xpress SARS-CoV-2 (Xpert) test on the GeneXpert system targeted to the N and E genes has been affected by mutations in the N gene [6]. Currently, 3 independent point mutations in the N gene (C29200T, C29200A, and C29197T) that affect the Xpert assay have been reported [7], [8], [9], [10]. In the Xpert assay, tested samples are considered positive if either (1) both the N2 and E targets are detected, or (2) only the N2 target is detected. In the case where only the E target is detected, the tested samples are considered presumptive positive. Therefore, failure in N gene detection is likely to generate presumptive positive results which will only be resolved by alternative test platforms [11]. Considering that the accurate detection of SARS-CoV-2 is crucial in the battle against the COVID-19 pandemic, vigilant monitoring for RT-PCR testing platforms is necessary.
Here we report 2 point mutations on the N2 probe binding region which resulted in presumptive positive results on Xpert assay. Per routine clinical care, Central Texas Veterans Health Care System (CTVHCS) performed SARS-CoV-2 testing of patients’ nasopharyngeal swabs collected in viral transport media (VTM) using the Xpert assay. Out of all specimens collected from May 2021 to early November 2021, we found 4 cases of SARS-CoV-2 positives that were initially classified as presumptive positive from the Xpert test then later confirmed as positive by 2 alternative testing platforms, BD MAX™ SARS-CoV-2 assay (BD, Franklin Lakes, NJ) and GenMarks ePlex respiratory pathogen panel 2(RP2) (GenMark Diagnostics, Inc, Carlsbad, CA). The initial presumptive positive results were caused by N gene detection failure by the Xpert tests. Subsequent whole genome sequencing (WGS) found 2 independent point mutations, known C29200T and novel C29203T, in the N2 probe binding regions. SARS-CoV-2 incessantly mutates and therefore continuous mutation analysis on samples with strange SARS-CoV-2 PCR test results is always necessary for monitoring the performance of both FDA-approved tests and laboratory-developed tests.
2. Materials and methods
2.1. SARS-CoV-2 detection
Nasopharyngeal swab samples were collected from patients in VTM at CTVHCS from May through November 2021. The RT-PCR based SARS-CoV-2 FDA EUA approved Cepheid Xpert® Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA), BD MAX™ SARS-CoV-2 assay (BD, Franklin Lakes, NJ), and GenMarks, ePlex respiratory pathogen panel 2(RP2) (GenMark Diagnostics, Inc, Carlsbad, CA) were used to detect SARS-CoV-2 in samples. Samples were processed as per in house written protocols according to the manufacturer's specifications. Briefly, 750 μL VTM for BD MAX, 300 μL for Xpert, and 200 μL for ePlex were used as input, respectively, for individual testing cartridges. All 3 systems are fully automated for downstream real-time RT-PCR analysis. The BD MAX™ system targets both the N1 and N2 regions of the N gene and the human RNase P gene. The ePlex respiratory pathogen panel detects SARS-CoV-2 and 17 other common respiratory pathogens from the VTM specimen used for the BD Max and Xpert assays [12].
According to our standard laboratory protocol, presumptive positive samples from the Xpert assay were subject to repeat testing using the same platform. If test results persisted as presumptive positive, the samples were tested using an alternative platform such as BD Max, ePlex or Cobas 6800.
2.2. Whole genome sequencing
RNA extraction for whole genome sequencing from collected samples was performed using QIAmp Viral RNA Kit (Qiagen) according to the manufacture's protocol. Briefly, 140 μL samples were used as initial inputs and a final elution volume was 35 μL. Libraries for sequencing were prepped using both the COVIDseq Test (Illumina) and the Swift normalase amplicon SARS-CoV-2 panels (SNAP) library prep kit (Swift biosciences) according to the manufacturer's protocol. Briefly, 8.5 μL of extracted RNA was used for 25 μL cDNA synthesis, and 10 μL of cDNA was used for each library prep kit. For library prep using SNAP, optional normalase I and normalase II treatment was applied to all samples. Synthetic SARS-CoV-2 RNA (Twist bioscience) was used as control. Sequencing of prepared libraries was performed using the Illumina NextSeq 550 system according to manufacturer's protocol. Briefly, final concentration, 1.4 pM libraries with 1% phiX control were loaded to 500/550 Mid Output Kit (Illumina) and paired-end reads (2 × 150 bp) were selected.
2.3. Bioinformatics & data analysis
The FASTQ files generated from the NextSeq Local Run manager were uploaded to Illumina BaseSpace Sequence Hub and analyzed with Illumina SARS-CoV-2 NGS Data Toolkit, DRAGEN COVID Lineage App. FASTQ files from both library preps were combined by uploading files under the same Biosample in BaseSpace. Consensus FASTA files generated against reference SARS-Cov-2 sequence (NC_045512) were uploaded for variant analysis and detection of mutation at Pangolin lineage (https://pangolin.cog-uk.io) and Nextclade (https://clades.nextstrain.org). For detailed sequence analysis, NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used. Mutation frequency of C29200T and C29203T was searched on the GISAID database using primer checker (https://www.epicov.org/epi3/frontend#407fad) with input of fwd TTACAAACATTG GCCGCAA, rev GCGCGACATTCCGAAGAC, and prb ACAATTTGCCCCCAGCGCTTCAG.
2.4. Data availability
The sequence has been deposited at GISAID under the accession numbers of EPI_ISL_7235551, EPI_ISL_7235560, EPI_ISL_7235555, EPI_ISL_7235547. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
3. Results
3.1. Detection of Cepheid Xpert® Xpress SARS-CoV-2 assay presumptive positive samples
Our laboratory used the Xpert assay from May 5, 2021 to November 1, 2021 until it was replaced by Xpert® Xpress CoV-2/Flu/RSV plus (Xpert plus). In this time period, we tested 6448 samples using the Cepheid Xpert® Xpress system. Four samples out of 637 positive samples had resulted as presumptive positive on th eCepheid Xpert® Xpress SARS-CoV-2 assay. For all 4 presumptive positive samples, N2 was undetected while E markers were positive. Re-test of the 4 samples in Xpert reproduced the initial presumptive positive results. These 4 samples had low CT values for the E target in Xpert assay and were repeatedly reported as positive in 2 alternative platforms. ePlex identified the samples as SARS-CoV-2 positive and the BD Max test detected N1/N2 with low Ct values (high viral load) (Table 1 ).
Table 1.
Results of multiple assays and WGS of Cepheid Xpert® Xpress SARS-CoV-2 presumptive positive samples.
| Sample ID | N gene mutation | Collection date | Xpert E/N2Ct | BD Max N1/N2 | ePlex | PANGO lineage | NextClade | Genome coverage | Coverage depth |
|---|---|---|---|---|---|---|---|---|---|
| PV2_93 | C29200T | 8/24/21 | 21/0 | 18/19 | + | AY. 24 | 21A (Delta) | 99.78% | 21846 |
| PV2_94 | C29200T | 8/29/21 | 15/0 | 11/13 | + | B.1.617. 2 | 21A (Delta) | 99.78% | 5601 |
| PV2_39 | C29203T | 7/23/21 | 14/0 | 11/13 | + | AY. 3 | 21A (Delta) | 99.78% | 17117 |
| PV2_95 | C29203T | 9/2/21 | 17/0 | 15/16 | + | AY. 3 | 21A (Delta) | 99.78% | 6155 |
3.2. Detection of mutation in the N region of SARS-CoV-2
To assess the failure of detection in the N2 region using the Xpert assay, WGS was performed. Sequencing analysis generated sequencing coverage of 99.78% for all samples, and sequencing depth ranged between 5601X and 21846X (Table 1). The samples were identified as Delta (B.1.617.2) and Delta sub lineage (AY.3 and AY.24) based on PANGO lineage [13] and 21A based on NextClade (Table 1). Based on the current database (https://outbreak.info/situation-reports), there are 4 known mutations in the N region in Delta (B.1.617.2) or Delta sublineages AY.3 or AY.24: A28461G (D63G), G28881T (R203M), G28916T (G215C), G29402T (D377Y). Those mutations, however, do not affect the SARS-CoV-2 detection by the Xpert assay.
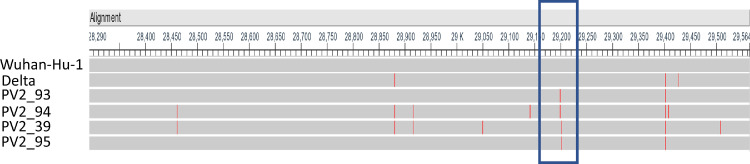
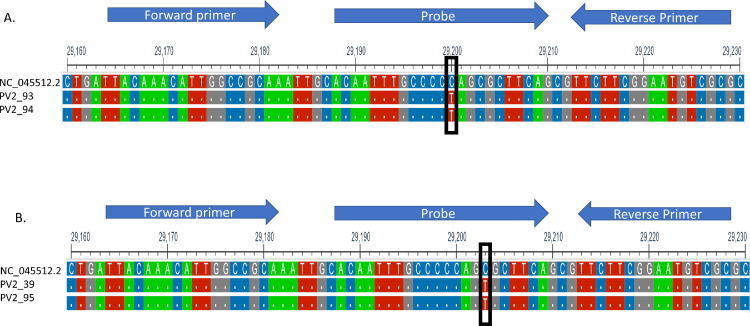
Since the entire N gene encoding region of all of the presumptive positive samples were sequenced with 100% sequence coverage and high read depth, we were able to detect several mutations were detected in the N gene of the 4 samples. G29402T and C29200T were present in PV2-93. G29402T and C29203T were present in PV2-95 (Fig. 1 ). Since G29402T is a common mutation in Delta variants, and most Delta variants samples were detected by Xpert assay, these 2 point mutations, C29200T and C29203T, were suspected to be the mutations that led to N gene detection failure in Xpert assay. PV2-94 and PV2-39 also contain C29200T and C29203T each (Fig. 1). Other mutations in PV2-94 and PV2-39 were either present in only 1 or 2 presumptive positive samples or common mutations with Delta variants (G29402T) (Fig. 1). In addition, these mutations lie within the N2 Probe binding region of US CDC 2019-nCoV Real time PCR primer and probe sequence for assay [9]. The N2 probe is in nucleotide sequence position 29188 ∼29210 of the Wuhan SARS CoV2 reference genome with sequence ACAATTTGCCCCCAGCGCTTCAG (Fig. 2 ). One mutation C to T transition at position 29200 (Fig. 2A) was observed in 2 samples which lineage B.1.617.2 and AY.24. The other C to T transition at position 29203 (Fig. 2B) was observed in the other 2 samples for which lineage was identified as AY.3. Those mutations were synonymous mutations of amino acid sequence P and S, respectively. Because both mutations reside in the probe binding region, we believe these mutations may hinder probe annealing and lead to detection failure of N2.
Fig. 1.
Sequence alignment of the N region of presumptive positive samples. Sequence was compared with SARS-CoV-2 reference genome (Wuhan-Hu-1, NC-045512) and control Delta. The red bar indicates the position of mutation in the sequence. The blue box is the 2019-nCoV_N2 primer and probe region. (Color version of figure is available online.)
Fig. 2.
Sequence alignment of 2 independent point mutations with C to T mutation at position 29200 (box) (A) and a C to T mutation at position 29203 (box) (B) of Wuhan-1 SARS-CoV2-reference sequence (NC_045512.2). Arrows indicate US CDC 2019 nCoV_N2 target primer and probe binding regions.
3.3. Frequency of the C29200T and C29203T
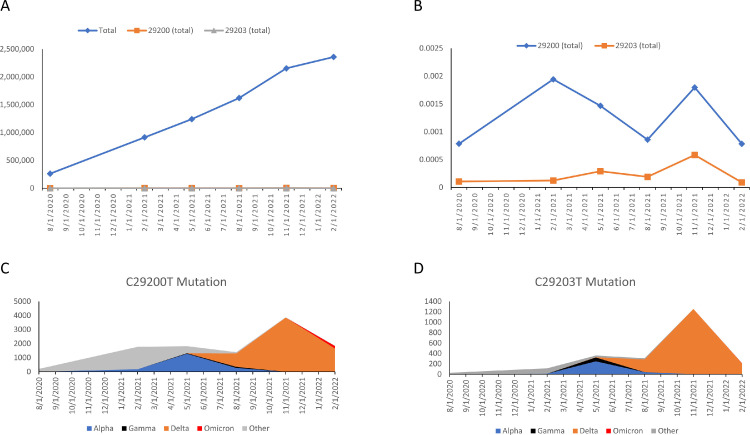
To investigate the frequency of these mutations, the GISAID database with a total of 8,550,976 viruses sequences from January 2020 to February 2022 was analyzed. The frequency of C29200T was between 0.078% and 0.194%. The frequency of C29203T was between 0.009% and 0.058% (Fig. 3 A, B). The C29200T and C29203T mutations were found in most of the prevailing variants including Delta and Omicron (Fig. 3C, D).
Fig. 3.
Mutation frequency of C29200T and C29203T. (A) Total number of virus sequence uploaded to GISAID from January 21, 2020 to February 18, 2022 compared to number of C29200T and C29203T mutations in the same period. (B) Proportion of C29200T and C29203T mutation. (C) Distribution of C29200T mutation across the variants. (D) Distribution of C29203T mutation across the variants.
4. Discussion
The rapid mutation rate of SARS-CoV-2 provides challenges for conventional molecular testing design [14]. Previously, mutations seen in the Alpha variant interfered with diagnosis of SARS-CoV-2 cases when using the ThermoFisher TaqPath COVID combo kit [4] which led to an FDA warning [15]. The Cepheid Xpert® Xpress SARS-CoV-2 (Xpert®) test targets N2 and E genes, but since the E genes is the common marker for Sarbecoronavorus, its sole detection does not confirm SARS-CoV-2 [16]. Therefore, any mutations in primer and/or probe targeted regions in the N domain resulted in presumptive positive results in the Xpert test, and as a result, repeating the same assay or using an alternative testing platform for those samples is required. The new Xpert plus assay, which replaced the Xpert assay, improved the test sensitivity by adding the RdRp target. All 4 presumptive positive samples were identified later as positive by the plus version of the Xpert assay, possibly due to positive detection of RdRp (proprietary information of Cepheid). However, the data from individual target genes are not accessible in the new Xpert plus assay, therefore N gene detection for C29200T and C29203T mutations in the Xpert plus assay is not obtainable.
Our laboratory utilizes multiple testing platforms to resolve false positive or negative cases [16]. WGS helps overcome the challenges faced by conventional molecular detection systems since the extent to which mutations affect the overall result is lower for WGS than PCR testing. With WGS, we found 2 independent point mutations in the N gene residing in the N2 probe binding region that potentially affect the detection of SARS-CoV-2. One mutation of C to T transition at position 29200 has been reported [7,10] but not in the Delta variant. The other mutation of C to T transition at position 29203 has not been reported yet in SARS-CoV-2, including in Delta variants.
The frequency of the C29200T and C29203T mutations were low and only sporadically found in various lineages. Therefore, the C29200T and C29203T mutations are likely to be spontaneously occurring independent of evolution. However, both mutations, C29200T and C29203T, could result in false negatives by target failure.
Presumptive positives in the Xpert assay could be generated by low viral load in the samples. However, these 4 samples contain high viral load which is indicated by low CT values for the E target in th eXpert assay. As the BD Max test detected the 4 samples as positive with low Ct values of the N1/N2 target, certain tests may tolerate the single nucleotide mismatch in the probe region, whereas the Xpert very likely could not tolerate the same single nucleotide mismatch even with high viral load samples.
In conclusion, 2 independent point mutations in the N2 probe binding region were found in the circulating Delta variant of SARS-CoV-2. The mutations interfere with certain SARS-CoV-2 assays that subsequently generate false negative test results. The data supports the need of targeting multiple regions for reliable SARS-CoV-2 detection as mutations emerged in this target region.
Authors’ Contributions
Hosoon Choi: Conceptualization, Formal analysis, Investigation, Methodology, Writing - original draft, Writing - review & editing. Munok Hwang: Formal analysis, Investigation, Methodology, Visualization, Software, Writing - original draft, Writing - review & editing. Janell Lukey: Investigation, Data curation, Methodology, Writing - review & editing. Chetan Jinadatha: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing. Dhammika Navarathna: Conceptualization, Project administration, Investigation, Methodology, Writing - original draft, Writing - review & editing. All authors have read and approved the final manuscript.
Acknowledgments
The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veteran Affairs or the funding agency We acknowledge the CTVHCS clinical microbiology lab staff members; Mr. Shawn Sharp and Ms. Linda Wiley, Ms. Ivy Englett and Ms. Ma Rowena San Juan for clinical samples collection. We also appreciate John David Coppin and Thanuri Navarathna for proofreading the article.
Funding
This work was supported by the VA SeqCURE grant which in turn received funding from the American Rescue Plan Act funds (Grant N/A) with additional support from Central Texas Veterans Healthcare System (Temple, TX).
Conflicts of Interest
All authors report no conflicts of interest.
References
- 1.Benda A, Zerajic L, Ankita A, Cleary E, Park Y, Pandey S. COVID-19 testing and diagnostics: a review of commercialized technologies for cost, convenience and quality of tests. Sensors (Basel) 2021;21:6581. doi: 10.3390/s21196581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backer JA, Eggink D, Andeweg SP, Veldhuijzen IK, van Maarseveen N, Vermaas K, et al. Shorter serial intervals in SARS-CoV-2 cases with Omicron BA.1 variant compared with Delta variant, the Netherlands, 13 to 26 December 2021. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.6.2200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown KA, Gubbay J, Hopkins J, Patel S, Buchan SA, Daneman N, et al. S-Gene target failure as a marker of variant B.1.1.7 among SARS-CoV-2 isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA. 2021;325:2115–2116. doi: 10.1001/jama.2021.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.FDA Center for Devices and Radiological Health . 2020. Risk of inaccurate results with thermo fisher scientific taqpath COVID-19 combo kit - letter to clinical laboratory staff and health care providers.https://www.fda.gov/medical-devices/letters-health-care-providers/risk-inaccurate-results-thermo-fisher-scientific-taqpath-covid-19-combo-kit-letter-clinical Available at: [Google Scholar]
- 5.Rosato AE, Msiha E, Weng B, Mesisca M, Gnass R, Gnass S, et al. Rapid detection of the widely circulating B.1.617.2 (Delta) SARS-CoV-2 variant. Pathology. 2022 doi: 10.1016/j.pathol.2022.01.001. S0031-3025(22)00035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.FDA Center for Devices and Radiological Health . FDA; 2021. SARS-CoV-2 viral mutations: impact on COVID-19 tests.https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests Available at: [Google Scholar]
- 7.Hasan MR, Sundararaju S, Manickam C, Mirza F, Al-Hail H, Lorenz S, et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J Clin Microbiol. 2021;59:e03220–e03278. doi: 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leelawong M, Mitchell SL, Fowler RC, Gonzalez E, Hughes S, Griffith MP, et al. SARS-CoV-2 N gene mutations impact detection by clinical molecular diagnostics: reports in two cities in the United States. Diagn Microbiol Infect Dis. 2021;101 doi: 10.1016/j.diagmicrobio.2021.115468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller S, Lee T, Merritt A, Pryce T, Levy A, Speers D. Single-point mutations in the N gene of SARS-CoV-2 adversely impact detection by a commercial dual target diagnostic assay. Microbiol Spectr. 2021;9 doi: 10.1128/Spectrum.01494-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cepheid . 2021. Xpert® Xpress SARS-CoV-2.https://www.fda.gov/media/136314/download Available at: [Google Scholar]
- 12.GenMark Diagnostics . 2021. ePlex® Respiratory Pathogen Panel 2.https://www.fda.gov/media/142905/download Available at: [Google Scholar]
- 13.Rambaut A, Holmes EC, Á O'Toole, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bano I, Sharif M, Alam S. Genetic drift in the genome of SARS COV-2 and its global health concern. J Med Virol. 2022;94:88–98. doi: 10.1002/jmv.27337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FDA Center for Devices and Radiological Health . 2021. Genetic Variants of SARS-CoV-2 May Lead to False Negative Results with Molecular Tests for Detection of SARS-CoV-2 - Letter to Clinical Laboratory Staff and Health Care Providers.https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2 Available at: [Google Scholar]
- 16.Navarathna DH, Sharp S, Lukey J, Arenas M, Villas H, Wiley L, et al. Understanding false positives and the detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and BD MAX SARS-CoV-2 assays. Diagn Microbiol Infect Dis. 2021;100 doi: 10.1016/j.diagmicrobio.2021.115334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence has been deposited at GISAID under the accession numbers of EPI_ISL_7235551, EPI_ISL_7235560, EPI_ISL_7235555, EPI_ISL_7235547. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.