Abstract
The COVID-19 pandemic continues to be a public health threat with emerging variants of SARS-CoV-2. Nirmatrelvir (PF-07321332) is a reversible, covalent inhibitor targeting the main protease (Mpro) of SARS-CoV-2 and the active protease inhibitor in PAXLOVID (nirmatrelvir tablets and ritonavir tablets). However, the efficacy of nirmatrelvir is underdetermined against evolving SARS-CoV-2 variants. Here, we evaluated the in vitro catalytic activity and potency of nirmatrelvir against the Mpro of prevalent variants of concern (VOCs) or variants of interest (VOIs): Alpha (α, B.1.1.7), Beta (β, B.1.351), Delta (δ, B1.617.2), Gamma (γ, P.1), Lambda (λ, B.1.1.1.37/C37), Omicron (ο, B.1.1.529), as well as the original Washington or wildtype strain. These VOCs/VOIs carry prevalent mutations at varying frequencies in the Mpro specifically for α, β, γ (K90R), λ (G15S), and ο (P132H). In vitro biochemical enzymatic assay characterization of the enzyme kinetics of the mutant Mpros demonstrates that they are catalytically comparable to wildtype. We found that nirmatrelvir has similar potency against each mutant Mpro including P132H that is observed in the Omicron variant with a Ki of 0.635 nM as compared to a Ki of 0.933 nM for wildtype. The molecular basis for these observations were provided by solution-phase structural dynamics and structural determination of nirmatrelvir bound to the ο, λ, and β Mpro at 1.63 to 2.09 Å resolution. These in vitro data suggest that PAXLOVID has the potential to maintain plasma concentrations of nirmatrelvir many-fold times higher than the amount required to stop the SARS-CoV-2 VOC/VOI, including Omicron, from replicating in cells.
Keywords: PF-07321332, nirmatrelvir, Mpro, Ki, VOC, crystal structure, Omicron, Beta, Lambda, SARS-CoV-2
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; VOC, variant of concern; VOI, variant of interest
New viral infectious diseases are emerging and have caused major public health crises in recent years. Reported examples in the last 20 years include severe acute respiratory syndrome coronavirus (SARS-CoV), H1N1 influenza, the Middle East respiratory syndrome coronavirus, Ebola virus disease, and Zika virus (1). The world continues to grapple with a global pandemic caused by a novel coronavirus, SARS-CoV-2 that was initially reported to the World Health Organization (WHO) on December 31, 2019 (2). Later, the WHO designated this virus as the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) owing to its similarity with the previous SARS-CoV (https://apps.who.int/iris/handle/10665/330760). Then at the end of January 2020, the WHO declared this viral outbreak as a public health emergency of international concern (3) and subsequently characterized it as a pandemic.
Genetic lineages of SARS-CoV-2 have been emerging and circulating around the world since the beginning of the COVID-19 pandemic. Like all viruses, SARS-CoV-2 is constantly changing through mutation and each virus with a unique sequence is considered a new variant. The WHO as well as other public health organizations monitor all variants that cause COVID-19 for increased risk to global public health and classify as variants being monitored, variants of interest (VOIs), variants of concern (VOCs), and variants of high consequence. As of February 3, 2022, the WHO has designated five VOCs: B.1.1.7 (Alpha, α), B.1.351 (Beta, β), P.1 (Gamma, γ), B.1.617.2 (Delta, δ), and B.1.1.529 (Omicron, ο), while B.1.1.1.37 (Lambda, λ) and B.1.621 (Mu, μ) have been designated as ‘VOIs’ (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
SARS-CoV-2 is a highly infectious beta coronavirus that can be life-threatening in serious cases. While effective COVID-19 vaccines have been developed, for individuals who have yet to be vaccinated or cannot be vaccinated, such as due to pre-existing medical conditions, therapeutics will likely be needed to effectively combat coronavirus disease 2019 (COVID-19) (4).
SARS-CoV-2 main protease (Mpro, also referred to as 3CL protease) is a cysteine protease that is critical for the processing of the two polyproteins (pp1a and pp1ab) encoded by the SARS-CoV-2 genome. The protease cleaves these polyproteins into shorter, nonstructural proteins that are essential for viral replication (4). Owing to this key role in viral replication, small-molecule inhibitors of SARS-CoV-2 Mpro represent attractive therapeutics for the treatment of COVID-19. We have previously reported on the discovery and antiviral efficacy of nirmatrelvir (PF-07321332), an orally bioavailable SARS-CoV-2 Mpro inhibitor with in vitro pan-human coronavirus antiviral activity with excellent off-target selectivity and in vivo safety profiles (4). Nirmatrelvir has demonstrated oral activity in a mouse-adapted SARS-CoV-2 model and has achieved oral plasma concentrations that exceed the in vitro antiviral cell potency, in a phase I clinical trial in healthy subjects (4). Here we report on the catalytic activity of frequently observed Mpro mutations in SARS-CoV-2 VOC/VOI, in vitro efficacy of nirmatrelvir against these mutant Mpros, the solution-phase structural dynamics, and structure of nirmatrelvir bound to the Mpros from three VOCs, β, λ, and ο.
Results
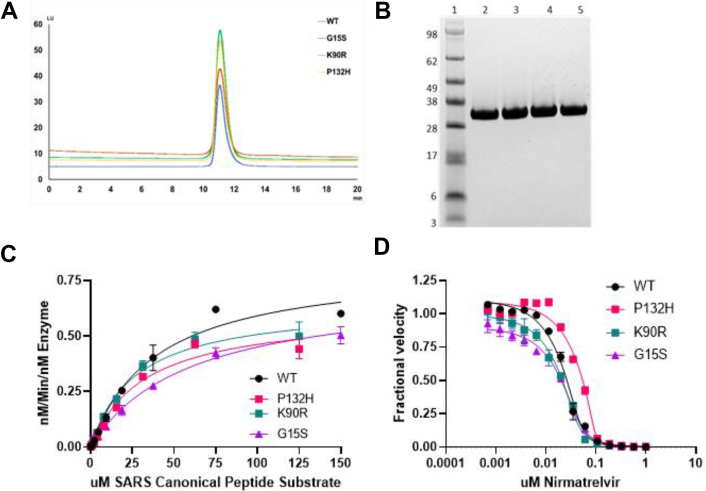
Full-length wildtype Mpro from the original Washington variant (USA-WA1/2020) and VOC/VOI SARS-CoV-2 Mpro were expressed and purified to near homogeneity as demonstrated by a singular protein peak with a confirmed intact mass of 33.8 kDa for a fully authentic form of each protein (Fig. 1A). A final and size-exclusion chromatography step showed the wildtype, K90R, G15S, and P132H Mpro proteins to be nearly 100% pure by Western blot analysis (Fig. 1B). An established Mpro fluorescence resonance energy transfer (FRET)–based cleavage assay was used to determine enzyme catalytic activity by monitoring initial velocities of the proteolytic activities at varying substrate (SARS canonical peptide) concentrations (4, 5, 6) (Fig. 1C). The turnover number (kcat) and Michaelis constants (Km) were determined for the wildtype, K90R, G15S, and P132H Mpro proteins (Table 1). The catalytic efficiencies (kcat/Km) of the K90R (28,255 S−1 M−1), G15S (16,483 S−1 M−1), and P132H (20,800 S−1 M−1) are similar to that of wildtype Mpro (31,500 S−1 M−1). These data suggest that the K90R, G15S, and P132H Mpro variants exhibit comparable enzymatic activities to wildtype Mpro. Next, we evaluated the ability of nirmatrelvir to inhibit wildtype and K90R, G15S and P132H Mpro enzymatic activities (Fig. 1D). Nirmatrelvir potently inhibited wildtype (mean Ki of 0.93 nM) and the mutated enzymes containing the K90R (Ki 1.05 nM), G15S (Ki 4.07 nM), and P132H (Ki 0.64 nM) Mpro (Table 2). A comparison of the VOC/VOI enzyme potency to that of wildtype Mpro by t test using the log of individual Ki values with one tail and unequal variance determined that the Ki values are not statistically different for the K90R and P132H Mpro proteins. There was a statistically significant shift in potency for G15S Mpro mutant, frequently observed in the Lambda variant relative to wildtype (p < 0.0005).
Figure 1.
Purification and characterization of SARS-Cov-2 MproP132H.A, analytical size-exclusion profile using using Zenix SEC-300, 4.6 × 150 mM column B, SDS-PAGE analysis: Lane 1, Marker; Lane 2, SARS-Cov-2 Mpro wildtype; Lane 3, SARS-Cov-2 Mpro G15S; Lane 4, SARS-Cov-2 Mpro K90R; Lane 5, SARS-Cov-2 Mpro P132H. C, enzyme kinetics of Mpro variants: The rate of cleavage of the FRET peptide substrate in the presence of indicated Mpro is monitored by increase in fluorescence over time with the fluorescent signal being converted to nM substrate cleaved by the use of a standard curve generated from cleaved substrate. The data were then normalized to the amount of enzyme used in the experiment. D, enzyme inhibition of Mpro: Mpro activity is monitored in the presence of increasing concentrations of nirmatrelvir with Ki values calculated using the Morrison equation to fit the data.
Table 1.
Enzymatic catalytic constants for SARS-CoV-2 Mpro enzyme
| SARS-CoV-2 | Predominant enzyme mutation | Km (μM) | Kcat S−1 | kcat/Km S−1 M−1 | n |
|---|---|---|---|---|---|
| USA-WA1 | Wildtype | 26.4 ± 9.8 | 0.76 ± 0.41 | 31,500 ± 18,600 | 14 |
| Lambda, λ B.1.1.1.37 |
G15S | 67.7 ± 18.6 | 1.18 ± 0.6 | 16,500 ± 5400 | 4 |
| Alpha, α B1.1.7 Beta, β B.1.351 Gamma, γ P.1 |
K90R | 35.8 ± 17.3 | 0.9 ± 0.48 | 28,300 ± 12,600 | 8 |
| Omicron, ο B.1.1.529 |
P132H | 25.2 ± 11.7 | 0.45 ± 0.14 | 20,800 ± 9350 | 4 |
Table 2.
In vitro potency of nirmatrelvir for inhibiting the SARS-CoV-2 mutant Mpro activity in a FRET assay
| Mutant enzyme | Ki (nM) geomean | Ki (nM) lower 95% CI | Ki (nM) upper 95% | n | p-valuea to wildtype Ki |
|---|---|---|---|---|---|
| G15S | 4.07 | 2.62 | 6.32 | 4 | 0.0002 |
| K90R | 1.05 | 0.19 | 5.82 | 5 | 0.371 |
| P132H | 0.635 | 0.179 | 2.25 | 4b | 0.074 |
| Wildtype | 0.933 | 0.471 | 1.85 | 9b | NA |
p-value calculated as a t test statistic for log Ki values compared to wildtype.
The n value represents the number of Ki values used to determine the geomean and CI which is lower than the experiment count due to censoring, i.e., experimental values that are < are excluded from geomean calculation.
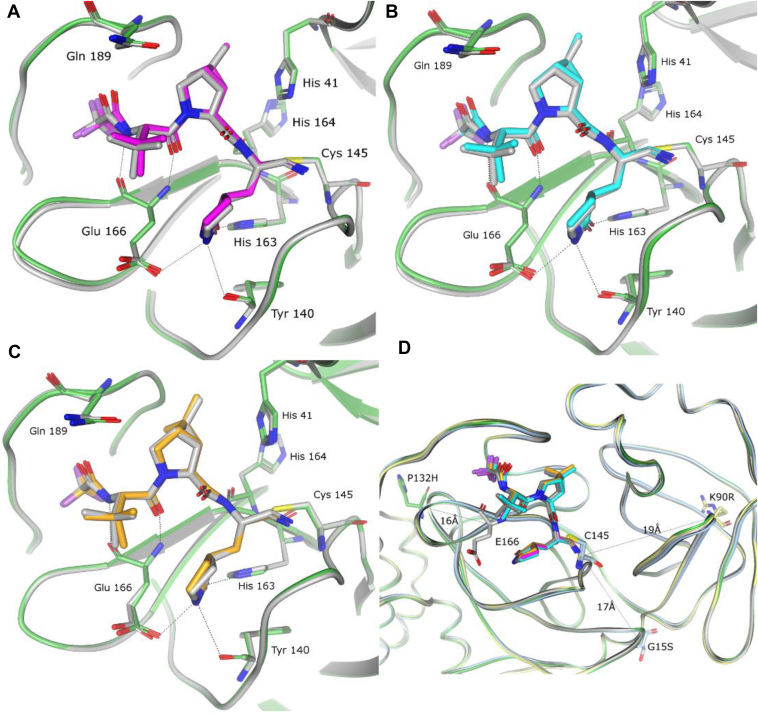
The crystal structures of nirmatrelvir bound to the three VOCs were determined to 2.09-Å (K90R), 1.68-Å (G15S), and 1.63-Å (P132H) resolution (Fig. 2). Superposition of each of these mutant structures with the wildtype Mpro, the structure of which was reported in detail previously (4, 7), shows that the binding mode of nirmatrelvir is unperturbed by the mutations, with the ligand maintaining the protein interactions observed in the wildtype Mpro, as shown in Figure 2. Indeed, these mutations are distal to the PF-07321332-binding pocket, with Pro 132 located approximately 16 Å (Cα-Pro132 to Cα-Glu166) from the binding pocket, while K90R and G15S reside 19 Å and 17 Å, respectively (Cα-R90/S15 to Cα-Cys145) from the PF-07321332-binding pocket (Fig. 2D). The crystal structures also show that the mutations do not give rise to any signification changes of the protein around the binding pocket or the site of the mutation (Fig. 2D)
Figure 2.
Structural characterization of nirmatrelvir bound to SARS-Cov-2 MproVOC.A–C, superposition of the x-ray crystal structures of nirmatrelvir bound to wildtype SARS-CoV-2 Mpro (gray) and (A) SARS-Cov-2 Mpro P132H (in magenta and green), (B) G15S (in cyan and green), and (C) K90R (in orange and green). D, ribbon figure of the three VOC crystal structures superposed on the WT, highlighting the location and distance of the mutant residues (in stick representation) in relation for the nirmatrelvir-binding site. Colors are maintained from A–C. VOC, variant of concern.
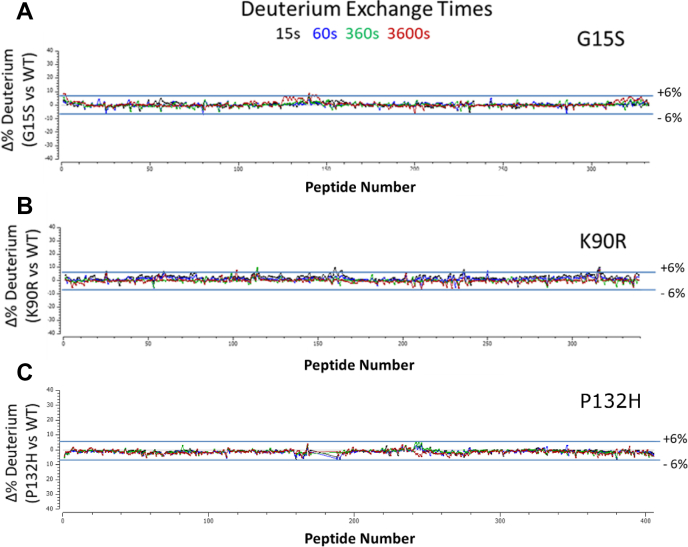
To provide an extended SARS-Cov-2 Mpro structural assessment, the solution-phase structural dynamics of K90R, G15S, and P132H along with the wildtype SARS-Cov-2 Mpro (10 μM, 25 mM Tris pH = 7.2, 150 mM NaCl) were individually profiled using HDX-MS. A residual plot (Fig. 3) comparing wildtype and mutant deuterium uptake profiles (HD Examiner 3.3.0 software, Sierra Analytics) revealed no significance differences (±6% deuterium) in the backbone dynamics of K90R, G15S, or P132H from wildtype SARS-Cov-2 Mpro. These results also suggest that the mutations alone did not produce a significant shift from wildtype solution-phase backbone conformational dynamics.
Figure 3.
Residual deuterium exchange plots indicate no significant differential uptake between wildtype and SARS-CoV-2 Mpromutants.A, G15S. B, K90R. C, P132H.
Discussion
We evaluated the catalytic activity and potency of nirmatrelvir against the Mpro mutations observed in the prevalent VOCs: Alpha (α, B.1.1.7), Beta (β, B.1.351), and Gamma (γ, P.1), which harbor a K90R mutation; Lambda (λ, B.1.1.1.37/C.37) with a G15S mutation; and Omicron (ο, B.1.1.529) with a P132H mutation. The K90R, G15S, and P132H Mpro mutations exhibited comparable enzymatic properties as compared to wildtype Mpro as evidenced by our determined catalytic efficiencies (kcat/Km). We demonstrate that nirmatrelvir has a comparable potency against these mutant Mpros relative to wildtype (Mpro from the original Washington strain [USA-WA1/2020]). Furthermore, deuterium exchange profiles of K90R, G15S, and P132H showed equivalent uptake to wildtype Mpro and revealed no detectable differences in backbone dynamics. In combination with static high-resolution crystal structures, HDX-MS exchange from the ensemble of solution-phase conformations provides evidence of equivalent solution-phase backbone dynamics for K90R, G15S, and P132H to wildtype and suggests no evidence for influence from crystal lattice packing in the crystal structures. Indeed, the x-ray crystal structures of nirmatrelvir bound to K90R, G15S, and P132H Mpro variants shows that not only is the binding mode of nirmatrelvir indistinguishable to the wildtype Mpro structure but also the conformation of the protein is not altered by the mutations, in agreement with the solution-phase data. These observations are consistent with recently reported biochemical characterization with similar kcat/Km and nirmatrelvir potency values (8, 9). Nirmatrelvir also appears to retain its in vitro antiviral efficacy against Omicron relative to wildtype (10, 11, 12, 13).
The emergence of naturally occurring SARS-CoV-2 variants exemplifies its ability to mutate and signify the continued potential for this pandemic to be problematic. It is important to continue monitoring emerging VOCs to quickly understand potential challenges to the efficacy of current and future antiviral therapies. More broadly, the availability of naturally occurring mutations could provide opportunities to more deeply understand additional aspects of protease structure–function and fitness. Studies described here demonstrate the in vitro inhibitory activity of nirmatrelvir against the Alpha, Beta, Gamma, Lambda, and Omicron variants of Mpro and indicate the structural basis for retention of in vitro potency against these mutant proteins. They also inform the methods for assessing activity against subsequent variants possessing mutations in the Mpro protein.
Experimental procedures
Protein production and purification
Briefly, the wildtype SARS-Cov-2 Mpro construct was designed based on Su et al. (14). An additional N-terminal PreScission protease cleavage site was inserted between GST and the self-cleavage site. Site-directed mutagenesis was performed to make each of the variants (K90R, G15S, or P132H). The resulting plasmid was then transformed into BL21 (DE3) cells for protein expression. One liter of LB media was inoculated with 30 ml of overnight culture and grown at 37 °C until an A600 of 0.6 was reached. The culture was induced using a final concentration of 0.2 mM IPTG and harvested 1 h postinduction. Cells were lysed in 20 mM Tris pH 8.0 buffer containing 500 mM NaCl, 10% glycerol, 0.2 mM Tris (2-carboxyethyl) phosphine with a microfluidizer, and the mixture was clarified by centrifugation at 15,000g. The resulting supernatant was purified by a Ni-affinity column using a step gradient, followed by C-terminal His-tag cleavage with PreScission protease, and a secondary Ni-affinity purification to remove noncleaved Mpro and PreScission protease.
Enzyme characterization and kinetics
The enzymatic activity of the main protease Mpro of SARS-CoV-2 wildtype and variants was monitored using a continuous FRET assay (4, 5, 6). The SARS-CoV-2 Mpro assay measures the activity of full-length SARS-CoV-2 Mpro protease to cleave a synthetic fluorogenic substrate peptide with the following sequence DABCYL-KTSAVLQ-SGFRKME-EDANS modeled on a consensus peptide. The fluorescence of the cleaved EDANS peptide (excitation 340 nm/emission 490 nm) is measured using a fluorescence intensity protocol on a PHERAstar FSX microplate reader (BMG Labtech). The assay reaction buffer contained 20 mM Tris–HCl (pH 7.3), 100 nM NaCl, 1 mM EDTA, 5 mM Tris (2-carboxyethyl) phosphine. For enzyme kinetic studies, 30 to 60 nM SARS-CoV-2 Mpro mutant protease was added to low-volume 384-well plates, and enzyme reactions were initiated with the addition of 30 μM peptide substrate. Enzyme kinetic constants were calculated from initial rates with the RFU/s slope converted to μM/s activity from a standard curve of cleaved substrate. For enzyme inhibition studies, 30 to 60 nM SARS-CoV-2 Mpro mutant protease was added to low-volume 384-well plates containing compounds spotted into assay plates using an Echo 555 acoustic liquid handler in a direct dilution scheme that results in a 1 μM top dose with 1.75-fold dilutions over 14 points. Enzyme and compounds were allowed to preincubate for 20 min before addition of substrate to 30 μM final concentration to initiate the reaction. Enzyme activity was calculated from the RFU/s slope collected over 15 min and expressed as percent activity based on control wells containing no compound representing 100% activity and wells containing no enzyme representing 0% activity. Ki values were fit to the Morrison equation with the enzyme concentration parameter allowed to float, the Km parameter fixed to the substrate Km determined for each mutant, and the substrate concentration parameter fixed to 30 μM.
Crystallization and structure determination
Apo crystals of SARS-CoV-2 Mpro mutants were obtained via vapor diffusion in sitting drops using MRC-Maxi (Swissci) plates where protein, at 7.30 mg/ml, was mixed 1:1 with well solution containing 20 to 24% w/v polyethylene glycol (PEG) 3350 and 0.12 to 0.21 M sodium sulfate. Plates were incubated at 21 °C, and crystals grew in under 24 h. PF-07321332 (1 mM final concentration) was added directly to the crystallization drops and allowed to soak into the apo crystals for 3 h at 30 °C. Soaked crystals were then passed through a cryoprotectant consisting of well buffer containing 20% ethylene glycol and flash cooled in liquid nitrogen in preparation for data collection.
X-ray diffraction data were collected at −173 °C at IMCA-CAT 17-ID beamline of the Advanced Photon Source (APS) at Argonne National Labs, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science using the Eiger 2 × 9M detector (Dectris). Use of the IMCA-CAT beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. The structures were determined by difference Fourier and refined using the anisotropically scaled data as described previously for wildtype SARS-CoV-2 Mpro in complex with PF-07321332 (4). Diffraction data processing and model refinement statistics for each of the mutant Mpro are given in Table 3.
Table 3.
Data statistics for X-ray diffraction data
| Mutant | P132H | G15S | K90R |
| PDB entry ID | 7TLL | 7U28 | 7U29 |
| Wavelength (Å) | 1.0 | 1.0 | 1.0 |
| Resolution | 112.84–1.63 | 112.33–1.68 | 110.22–2.09 |
| Space group | P21 | P21 | P21 |
| Unit cell dimensions [Å] | a = 45.4, b = 53.8, c = 115.5 | a = 45.6, b = 53.8, c = 114.3 | a = 45.5, b = 55.8, c = 114.2 |
| Unit cell dimensions [o] | α = γ =90.0, β = 102.4 | α = γ =90.0, β = 100.8 | α = γ =90.0, β = 105.2 |
| Total number of reflectionsa | 178,785 (6869) | 156,159 (6075) | 95,920 (4735) |
| Unique reflectionsa | 53,153 (2659) | 46,261 (2314) | 28,383 (1419) |
| Multiplicitya | 3.4 (2.6) | 3.4 (2.6) | 3.4 (3.3) |
| Completeness (%), sphericala | 78.0 (18.9) | 74.2 (23.9) | 85.7 (35.2) |
| Completeness (%), ellipsoidala | 92.8 (53.2) | 81.0 (46.4) | 87.4 (40.6) |
| Mean I/σ(I)a | 11.5 (1.4) | 10.5 (1.5) | 7.9 (1.6) |
| Rmergeb | 0.056 (0.607) | 0.064 (0.595) | 0.117 (0.789) |
| Rpimc | 0.035 (0.453) | 0.041 (0.454) | 0.075 (0.499) |
| CC1/2d | 0.999 (0.635) | 0.998 (0.601) | 0.996 (0.583) |
| Refinement statistics | |||
| Reflections used | 53,153 | 46,261 | 28,383 |
| Reflections used for Rfree | 2620 | 2222 | 1502 |
| Rcryste | 0.211 | 0.208 | 0.204 |
| Rfree | 0.250 | 0.248 | 0.266 |
| Ramachandran plot | |||
| Favored regions (%) | 98.2 | 98.0 | 97.5 |
| Allowed regions (%) | 1.5 | 1.5 | 2.2 |
| Outlier regions (%) | 0.3 | 0.5 | 0.3 |
Rfree is the same as Rcryst, but for 5% of the data randomly omitted from refinement (15).
Numbers in parentheses refer to the highest resolution shell.
(16).
CC1/2 = xxx as defined by Karplus and Diederichs (17).
, where Fo and Fc are the observed and calculated structure factors, respectively.
Hydrogen deuterium exchange
Native protein was deuterium exchanged at four time points (15 s, 1 min, 6 min, and 1 h) and subsequently digested with two protease columns (Protease XIII/Pepsin [NovaBioAssays] and Nepenthesin-1 [AffiPro]) to generate deuterated peptides for LC-MS analysis. A total of 405 WT peptides were identified (Table 4) on the Fusion Lumos OrbiTrap mass spectrometer (mass tolerance < 5 ppm) using Proteome Discoverer 2.2.0 software (Thermo Fisher Scientific).
Table 4.
SARS-CoV-2 deuterium exchange summary (wildtype, P132H, G15S, K90R Mpro)
| Dataset | CoV-2_WT (control) | Cov-2_P132H | Cov-2_G15S | Cov-2_K90R |
|---|---|---|---|---|
| HDX reaction details | 4C | 4C | 4C | 4C |
| HDX time course (s) | 15, 60, 360, 3600 | 15, 60, 360, 3600 | 15, 60, 360, 3600 | 15, 60, 360, 3600 |
| # of peptides | 405 | 387 | 326 | 339 |
| Sequence coverage | 100.00% | 100.00% | 99.02% | 99.02% |
| Average peptide length/Redundancy | 17.24/22.82 | 17.29/21.86 | 16.33/17.39 | 16.65/18.44 |
| Replicates | 4 | 4 | 4 | 4 |
| Repeatability (avg. stddev of #D) | 0.1123 | 0.1200 | 0.1777 | 0.1412 |
| Significant differences in HDX (delta HDX > X D - 95% CI) | n/a | 0.3123 D | 0.373 D | 0.3657 D |
Data availability
RCSB Protein Data Bank Accession code: The coordinates and structure factors have been deposited with the RCSB Protein Data Bank under accession codes: 7U29 (K90R), 7U28 (G15S), and 7TLL (P132H).
Conflict of interest
All authors are employees of Pfizer, Inc and may hold or own shares of stock.
Acknowledgments
We thank Mark Noe, Gretchen Dean, Annaliesa Anderson, and Charlotte Allerton for critical reading of the manuscript and leadership.
Author contributions
S. E. G., S. N., B. B., Y. Z., and C. M. S. conceptualization; S. E. G., S. N., O. P., R. F., W. L., B. B., K. F., and J. N. data curation; S. E. G., S. N., O. P., R. F., W. L., B. B., K. F., and J. N. formal analysis; S. E. G., S. N., O. P., R. F., W. L., B. B., K. F., and J. N. methodology; S. E. G., T. C., Y. Z., A. E. S., and C. M. S. project administration; S. E. G., S. N., B. B., Y. Z., A. E. S., and C. M. S. validation; S. E. G., S. N., O. P., B. B., Y. Z., A. E. S., and C. M. S. visualization; S. E. G., B. B., and C. M. S. writing–original draft; S. E. G., S. N., O. P., R. F., W. L., B. B., T. C., Y. Z., A. E. S., and C. M. S. writing–review and editing; B. B., K. F., J. N., T. C., A. E. S., and C. M. S. resources; T. C., A. E. S., and C. M. S. supervision.
Edited by Wolfgang Peti
References
- 1.Majumder J., Minko T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021;23:14. doi: 10.1208/s12248-020-00532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 5.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., et al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman R., Kania R., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Murray B.W., et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Lin C., Zhou X., Zhong F., Zeng P., Yang Y., Zhang Y., Yu B., Fan X., McCormick P.J., Fu R., Fu Y., Jiang H., Zhang J. Structural basis of main proteases of coronavirus bound to drug candidate PF-07321332. bioRxiv. 2021 doi: 10.1101/2021.11.05.467529. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ullrich S., Ekanayake K.B., Otting G., Nitsche C. Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg. Med. Chem. Lett. 2022;62:128629. doi: 10.1016/j.bmcl.2022.128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sacco M.D., Hu Y., Gongora M.V., Meilleur F., Kemp M.T., Zhang X., Wang J., Chen Y. The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. Cell Res. 2022;32:498–500. doi: 10.1038/s41422-022-00640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res. 2022;198:105252. doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rai D.K., Yurgelonis I., McMonagle P.M., Rothan H.A., Hao L., Gribenko A., Titova E., Kreiswirth B., White K.M., Zhu Y., Anderson A.S., Cardin R.D. Nirmatrelvir, an orally active Mpro inhibitor, is a potent inhibitor of SARS-CoV-2 variants of concern. bioRxiv. 2022 doi: 10.1101/2021.01.17.476644. [preprint] [DOI] [Google Scholar]
- 12.Rosales R., McGovern B.L., Rodriguez M.L., Rai D.K., Cardin R.D., Anderson A.S., Sordillo E.M., Bakel H.V., Simon V., García-Sastre A., White K.M. Nirmatrelvir, Molnupiravir, and Remdesivir maintain potent in vitro activity against the SARS-CoV-2 Omicron variant. bioRxiv. 2022 doi: 10.1101/2021.01.17.476685. [preprint] [DOI] [Google Scholar]
- 13.Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vries A.C., Rottier R.J., Bruno M.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32:322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brünger A. In: Carter C.W. Jr., Sweet R.M., editors. vol. 277. Academic Press; Cambridge, MA: 1997. (Methods in Enzymology). chap. 19. [Google Scholar]
- 16.Weiss M., Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 1997;30:203–205. [Google Scholar]
- 17.Karplus P., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RCSB Protein Data Bank Accession code: The coordinates and structure factors have been deposited with the RCSB Protein Data Bank under accession codes: 7U29 (K90R), 7U28 (G15S), and 7TLL (P132H).