Abstract
CRISPR base editing techniques tend to edit multiple bases in the targeted region, which is a limitation for precisely reverting disease-associated single-nucleotide polymorphisms (SNPs). We designed an imperfect gRNA (igRNA) editing methodology, which utilized a gRNA with one or more bases that were not complementary to the target locus to direct base editing toward the generation of a single-base edited product. Base editing experiments illustrated that igRNA editing with CBEs greatly increased the single-base editing fraction relative to normal gRNA editing with increased editing efficiencies. Similar results were obtained with an adenine base editor (ABE). At loci such as DNMT3B, NSD1, PSMB2, VIATA hs267 and ANO5, near-perfect single-base editing was achieved. Normally an igRNA with good single-base editing efficiency could be selected from a set of a few igRNAs, with a simple protocol. As a proof-of-concept, igRNAs were used in the research to construct cell lines of disease-associated SNP causing primary hyperoxaluria construction research. This work provides a simple strategy to achieve single-base base editing with both ABEs and CBEs and overcomes a key obstacle that limits the use of base editors in treating SNP-associated diseases or creating disease-associated SNP-harboring cell lines and animal models.
INTRODUCTION
Base editors were initially developed for precise cytosine (C) to thymine (T) editing (cytosine base editor, CBE) without DNA double-strand breaks and the use of an editing template (1,2) and were then expanded to adenine (A) to guanine (G) editing (adenine base editor, ABE) (3), cytosine (C) to guanine (G) editing (glycosylase base editor, GBE), and, recently, cytosine (C) to adenine (A) editing in bacteria (4,5). These techniques represent a breakthrough for precise base conversion in the chromosomes of various species (6–9).
Normally, multiple bases are edited instead of a single-base, causing unwanted base conversions (bystander editing) if multiple Cs or As exist within or near the editing window. Specific single-base editing is one of the most desired properties for the application of base editing techniques. Within the editing window, editing at a position other than the target position, that is, at nearby C or A nucleotides, may lead to undesirable effects in most cases. For example, a large proportion of single-gene genetic diseases are caused by individual mutations, known as single-nucleotide polymorphisms (SNPs). When a base editor is employed to correct such SNPs, only the target nucleotide is expected to be edited, and any other nucleotides within the same editing window should be avoided except synonymous mutations. Unfortunately, most base editors, including CBEs and ABEs, have editing windows with multiple target nucleotides.
By studying the interaction between the Cas9/gRNA complex and DNA, it was determined that in the base editing process, a DNA R-loop is formed after the Cas9 protein binds its target (1,10). The formation of an R-loop provides a single-stranded DNA substrate for the deaminase of the base editor (11). Since, theoretically, most nucleotides within the R-loop may be accessed by the deaminase, multiple targeted nucleotides are normally edited in the editing process. Thus, an editing window is formed and defined as protospacer positions that support a certain fraction (typically 50%) or higher of the average peak editing efficiency (12). Since the R-loop, which is the molecular basis of the editing window of base editors, is formed by binding of the Cas protein, the Cas domain of base editors is considered to be one of the main determinants of its editing window (12,13).
Currently, all base editors based on natural Cas have a multiple-nucleotide editing window. For example, SaCas9 typically supports a broader editing window of protospacer positions 3–12 for CBEs and 4–12 for ABEs (14), while SpCas9 editors have editing windows of positions 4–8 for CBEs and 4–7 for ABEs. In addition, the activity of the fused deaminase and the fusion linker of the base editor also affect the editing window (14,15). To increase the location specificity and narrow the editing window, researchers have developed various base editors by engineering their constituted domains (16). For example, modification of the base editor linker and deaminases was reported for narrowing the editing window of some CBEs (15,17); mutations in the deaminase could reduce the size of the editing window (14,18) of CBEs; and using an alternative deaminase that requires a specific motif could also narrow the editing window of CBEs (19,20). However, no universal base editors with a single-nucleotide editing window have been reported. In addition, no modified ABE with a single-base window has been constructed to date, although approximately half of the known disease-associated SNPs need ABEs for correction (3).
In this work, we analyzed the distribution of various types of editing products generated by CBEs and found that base editors had a strong preference to edit multiple cytosine bases together. It was reported that an RNA bubble hairpin was added to gRNAs to decrease the off-target effect, proving that gRNA engineering has the potential to change the base editing performance (21). Thus, we attempted to design and test gRNAs to develop a simple and universal strategy for convenient specific single-base editing with both ABEs and CBEs.
MATERIALS AND METHODS
Strains and culture conditions
Escherichia coli DH5α was used as a cloning host and grown at 37°C in lysogeny broth (LB, 1% (w/v) tryptone, 0.5% (w/v) yeast extract and 1% (w/v) NaCl. Ampicillin (100 mg/l) was added to the medium when appropriate.
Plasmid construction
HEK293T and HeLa cells gRNA expression plasmids were assembled with the Golden Gate method with the N20 sequence embedded in the primers, and RNF2 sgRNA expression plasmids were used as the template (1). All the DNA templates were PCR amplified with Phusion DNA polymerase (NEB, USA). PCR products were gel purified, digested with the restriction enzyme DpnI (NEB, USA), and assembled with the Golden Gate assembly method.
Cell culture and transfection
HeLa cells and HEK293T cells (from ATCC) were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37°C under 5% CO2. Cells were seeded in 24-well plates (Corning, USA). Approximately 24 h after seeding, cells were transfected at ∼40% confluency with Lipofectamine 2000 (Life Technologies, Invitrogen, USA) according to the manufacturer's protocols. Then, 600 ng of CBE or ABE plasmids and 300 ng of sgRNA-expressing plasmids were transfected with 50 μl of DMEM containing 1.8 μl of Lipofectamine 2000. Twenty-four hours after transfection, 5 μg/ml puromycin (Merck, USA) was added to the medium besides the groups without puromycin. In addition, 120 h after transfection, genomic DNA was extracted from the cells using QuickExtract DNA Extraction Solution (Epicentre, USA). Puromycin was also added to the medium at twenty-four hours in the experiment groups with different treatment times, and genomic DNA was extracted from the edited cells at 24, 48, 72, 96, 120 and 144 h after transfection, respectively. Ultimately, the target genomic regions (200–300 bp) of interest were amplified by PCR for high-throughput DNA sequencing.
High-throughput DNA sequencing of genomic DNA samples and data analysis
Next-generation sequencing library preparations were constructed following the manufacturer's protocol (VAHTS Universal DNA Library Prep Kit for Illumina), as described previously (5). Then, libraries with different indexes were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer's instructions (Illumina, San Diego, CA, USA). Sequencing was carried out using a 2 × 150 paired-end configuration; image analysis and base calling were conducted by HiSeq Control Software (HCS) + RTA 2.7 (Illumina) on a HiSeq instrument. For paired-end sequencing results, read 1 and read 2 were merged to generate a complete sequence according to their overlapping regions, and a file in FASTA (fa) format was generated. Data were split according to their barcodes. The merged sequences were aligned to the reference sequence by using BWA (version 0.7.12) software. Examined target sites that mapped with ∼100 000 independent reads were selected, and obvious base substitutions were observed at only the targeted base editing sites. Base substitution frequencies were calculated by dividing the base substitution read number by the total read number.
Statistical analysis
The experimental data were presented as mean ± s.d. of n = 3 independent biological replicates, and the significant differences were conducted using two-tailed Student's t-tests.
RESULTS
Design of an imperfect guide RNA (igRNA) for single-base editing with a CBE
To understand the actual distribution of various types of editing products generated by CBEs, nine loci were edited using the BE4max (22) editor, and the PCR products amplified from edited loci were subjected to deep sequencing. By analyzing the results illustrated in Supplementary Figure S1A, we found that Cs within the editing window are preference to co-edited at most loci. For example, at the RNF2 locus, there were four major editing products, including conversion products with Ts at position C6, positions C3 and C6, positions C6 and C12, and positions C3, C6 and C12. The editing efficiencies of each type ranged from 1% to 41%, with a total editing efficiency of 54.48 ± 0.74%, and single C6-to-T conversion was only 5.02 ± 0.08%. Similar editing results were also observed from the editing of most loci (Supplementary Figure S1A).
Based on these results, we propose that in the process of base editing, after one target base is converted, the gRNA of the base editor could still bind to its complementary sequence to convert other target bases within the editing window. Such a binding pattern favors editing of multiple but not one base. However, if we start with igRNA, after one base is converted, the igRNA would have more non-complementary bases to the target locus and might lose its ability to guide the base editor complex to the modified locus while keeping its ability to bind with the original DNA sequence. This results in the ending of the editing process with a modified locus, but not the original sequence. Thus, the whole editing process may cause increased single-base editing conversion as shown in Supplementary Figure S1B.
CBE-mediated single-base editing with igRNA
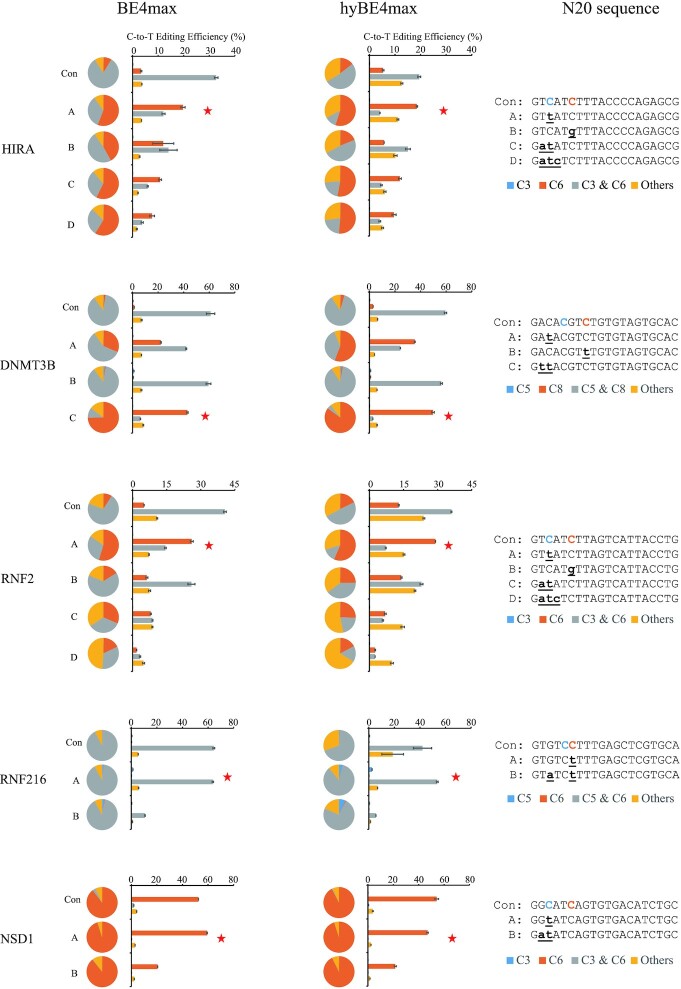
To test the above hypothesis, base editing experiments were carried out using BE4max and hyBE4max (23), an editor increased activity and expanded the editing window by inserting a non-sequence-specific single-stranded DNA-binding domain from Rad51 protein between Cas9 nickase and the deaminase in BE4max, with both normal gRNAs and igRNAs. To achieve optimal single-base editing efficiency and specificity, multiple igRNAs were employed to edit each locus. igRNAs were designed with one, two, or three noncomplementary bases with the target DNA sequences. The editing results in HEK293T cells were obtained by deep sequencing (Figure 1, Supplementary Table S1, and Supplementary Table S2). At the HIRA locus, HIRA-A igRNA with one noncomplementary base achieved the highest single-base editing efficiency. The editing efficiencies for single C6-to-T conversion were improved from 3.34 ± 0.29% to 19.72 ± 0.81% and 5.35 ± 0.34% to 18.57 ± 0.26% with BE4max and hyBE4max, respectively. The fraction of single C6-to-T conversion among all the editing types also improved from 8.40 ± 0.66% to 55.75 ± 0.62% and 14.22 ± 0.39% to 54.90 ± 0.50% with BE4max and hyBE4max, respectively. At the DNMT3B and RNF2 loci, similar improvements were achieved by utilizing igRNAs. At RNF216 locus, the editing efficiency of single C5 in the context of dual Cs (CC) is low with igRNAs, but is significantly higher than the control. Even if the single-base editing efficiency at the NSD1 locus was already high with normal gRNA, NSD1-A igRNA could still improve the C6-to-T editing efficiencies and fractions from 52.49 ± 0.06% to 59.34 ± 0.18% and 89.58 ± 0.27% to 95.06 ± 0.11% with BE4max, respectively. With hyBE4max at the NSD1 locus, NSD1-A igRNA reduced the C6-to-T editing efficiency from 54.19 ± 1.44% to 47.03 ± 0.60%, but the fraction of C6-to-T was improved from 92.35 ± 0.43% to 94.94 ± 0.16%.
Figure 1.
Editing efficiencies and editing product fractions of cytosine base editing with igRNA in HEK293T cells. The base editing efficiencies and fractions of editing products were obtained from five loci using gRNAs and igRNAs with the BE4max and hyBE4max editors. Con, the control gRNAs that exactly matched the target sequence. Capital A to D, different igRNAs. The red asterisk represents the igRNA with the best performance. The bars represent mean values, and the error bars represent the s.d. of three independent biological replicates. The pie charts represent the fractions of various types of editing products.
To verify the generalizability of igRNAs in CBE, base editing experiments have also been tested in HeLa cells under the same conditions (Supplementary Figure S2, Supplementary Table S3 and Supplementary Table S4). Similar to the results in HEK293T cells, we found that igRNAs can also increase the editing probability at one preferred protospacer position compared to other positions. Four more loci were tested using gRNAs and igRNAs with the BE4max editors in HEK293T cells (Supplementary Figure S3, Supplementary Table S5). At 4 tested loci, igRNAs can also improve the efficiencies of single-base editing. Especially at the EMX1-SITE1 locus, EMX1-SITE1-B igRNA performed a great effect that improved, the C7-to-T editing efficiencies and fractions from 1.12 ± 0.07% to 50.83 ± 0.33% and 1.44 ± 0.08% to 74.78 ± 0.31%, respectively. Nevertheless, we found igRNAs might perform an ordinary effect on dual Cs. Although it partly decreased the portion of bystanders or other products, igRNA yet couldn’t increase the absolute editing efficiency of target C.
For the design of igRNAs, we conclude from the results for the 9 tested loci that effective igRNAs for the CBE normally contained one or two noncomplementary bases, while the editing efficiencies of the igRNA containing three noncomplementary bases were greatly reduced.
ABE-mediated single-base editing with igRNA
Since half of the total reported disease-associated SNPs can be corrected by ABEs, this single-base conversion technique is extremely important for the potential treatment of human genetic diseases (24). Further modification of the deaminase TadA could be difficult, most likely because it is already highly evolved; currently, there are no reported ABEs with single-base editing windows (12). This could be a major obstacle preventing the development of genetic therapies based on ABEs.
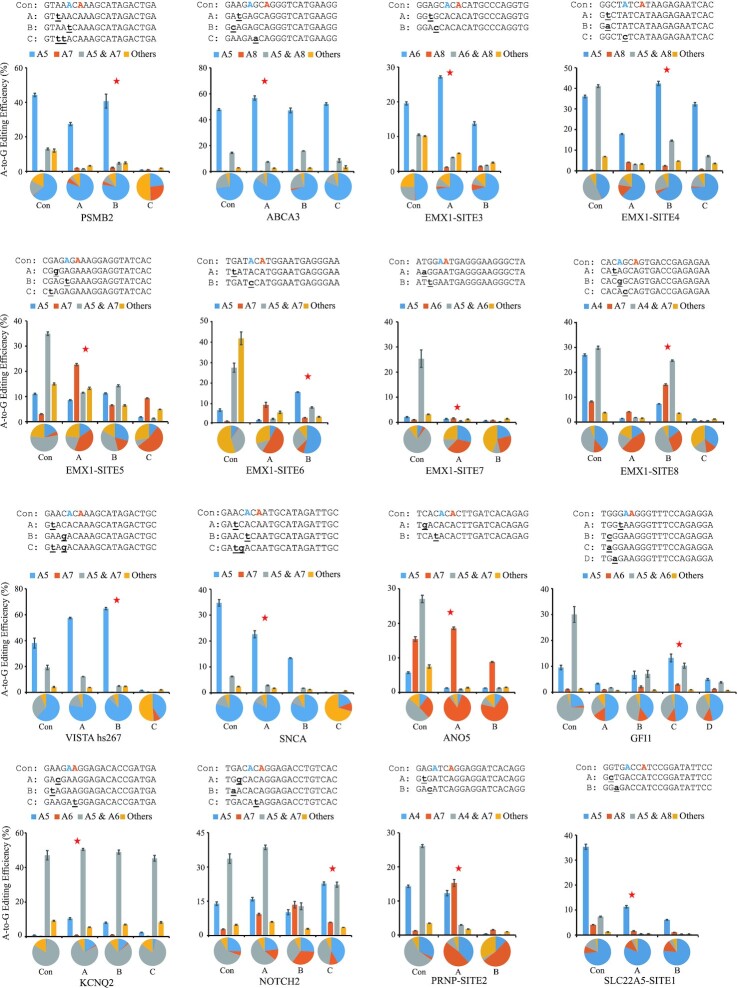
To minimize the editing window of ABEs, igRNAs for the NG-ABEmax (13) editor were designed to perform base editing experiments both in HEK293T and HeLa cells, the editing results were obtained by deep sequencing. Optimal igRNAs were found to have greatly narrowed the editing window of NG-ABEmax to mainly one base, and the fractions of edited products containing single A-to-G conversion were improved at most tested loci in HEK293T cells (Figure 2, Supplementary Table S6 and Supplementary Table S7). Optimal single-base editing was achieved with the igRNAs PSMB2-B, ABCA3-A, EMX1-SITE3-A, EMX1-SITE4-B, EMX1-SITE5-A, EMX1-SITE6-B, EMX1-SITE7-A, EMX1-SITE8-B, VISTA hs267-B, SNCA-A, ANO5-A, GFI1-C, KCNQ2-A, NOTCH2-C, PRNP-SITE2-A and SLC22A5-SITE1-A. At these tested loci, the fractions of single A-to-G conversion were improved from 63.49 ± 1.74% to 77.40 ± 0.21%, 73.33 ± 0.32% to 84.86 ± 0.45%, 48.20 ± 0.31% to 72.37 ± 0.40%, 42.70 ± 0.27% to 66.12 ± 0.52%, 4.77 ± 0.21% to 40.46 ± 0.50%, 8.24 ± 0.80% to 53.83 ± 1.06%, 3.26 ± 0.09% to 33.61 ± 0.23%, 12.00 ± 0.09% to 29.73 ± 0.70%, 61.85 ± 0.55% to 87.04 ± 0.41%, 79.73 ± 0.41% to 82.63 ± 0.37%, 27.75 ± 0.03% to 84.23 ± 0.98%, 1.39 ± 0.18% to 15.50 ± 0.54%, 25.36 ± 0.72% to 41.91 ± 0.70%, 22.76 ± 0.15% to 48.25 ± 0.81%, 2.81 ± 0.17% to 47.33 ± 0.51% and 73.56 ± 0.33% to 81.50 ± 0.81% at A5 of PSMB2, A5 of ABCA3, A6 of EMX1-SITE3, A5 of EMX1-SITE4, A7 of EMX1-SITE5, A5 of EMX1-SITE6, A6 of EMX1-SITE7, A7 of EMX1-SITE8, A5 of VISTA hs267, A5 of SNCA, A7 of ANO5, A5 of KCNQ2, A5 of NOTCH2, A5 of GFI1, A7 of PRNP-SITE2 and A5 of SLC22A5-SITE1, respectively. The corresponding A-to-G editing efficiencies changed from 44.22 ± 0.97% to 40.66 ± 4.04%, 47.77 ± 0.68% to 56.72 ± 1.64%, 19.54 ± 0.44% to 27.15 ± 0.29%, 36.06 ± 0.63% to 42.36 ± 1.07%, 3.05 ± 0.19% to 22.67 ± 0.47%, 6.18 ± 0.58% to 14.95 ± 0.14%, 1.04 ± 0.16% to 1.61 ± 0.21%, 8.28 ± 0.20% to 15.04 ± 0.36%, 38.10 ± 3.82% to 64.71 ± 0.85%, 34.72 ± 1.27% to 22.61 ± 1.36%, 15.46 ± 0.66% to 18.56 ± 0.35%, 0.79 ± 0.11% to 10.41 ± 0.50%, 13.93 ± 0.76% to 22.74 ± 0.69%, 9.57 ± 0.87% to 13.24 ± 1.49%, 1.27 ± 0.07% to 15.27 ± 1.02% and 35.30 ± 1.04% to 11.32 ± 0.49%. Surprisingly, increased editing efficiency was observed at 13 out of the 16 tested loci. Single-base editing with the consecutive dual As (AA) with igRNAs is similar to consecutive dual Cs (CC). Similar editing results are found in HeLa cells with igRNA (Supplementary Figure S4, Supplementary Table S8 and Supplementary Table S9). In addition, igRNA can also reduce bystander editing without puromycin selection (Supplementary Figure S5A, and Supplementary Table S10). Compare to the experiments with puromycin, the editing efficiency was lower, probably due to the lower fractions of cells with plasmids.
Figure 2.
Editing efficiencies and editing product fractions of adenine base editing with igRNA in HEK293T cells. Base editing efficiencies and fractions of editing products obtained from 16 loci by the NG-ABEmax editor using gRNAs and igRNAs. Con, the control gRNAs that exactly matched the target sequence. Capital A to D, different igRNAs. The red asterisk represents the igRNA with the best performance. The bars represent mean values, and the error bars represent the s.d. of three independent biological replicates. The pie charts represent the fractions of various types of editing products.
The single-base conversion fractions with igRNA were improved at tested loci, demonstrating successful ABE-mediated single-base editing. Once again, surprisingly good results were obtained, revealing that the optimal igRNAs had both improved editing specificity and efficiency relative to their parent gRNA for the ABE at most loci, as observed with the CBE. In contrast to CBEs, ABEs have a more rigid requirement for complementary base numbers between gRNA and target DNA, and all igRNAs with two noncomplementary bases were found to have greatly reduced editing efficiencies. Therefore, the rule for designing ABE igRNAs is to make igRNAs with only one noncomplementary site.
Controllable single-base editing strategy with the SpRY-editors and igRNA
Based on our experiment, it is not possible to edit one position dominantly by using an igRNA, and to edit another position dominantly by using another igRNA at one locus. One set of igRNA derived from one gRNA can only increase the single-base editing of the dominant position. So that it is difficult to achieve controllable single-base editing at one locus, especially at the loci containing consecutive dual Cs or dual As.
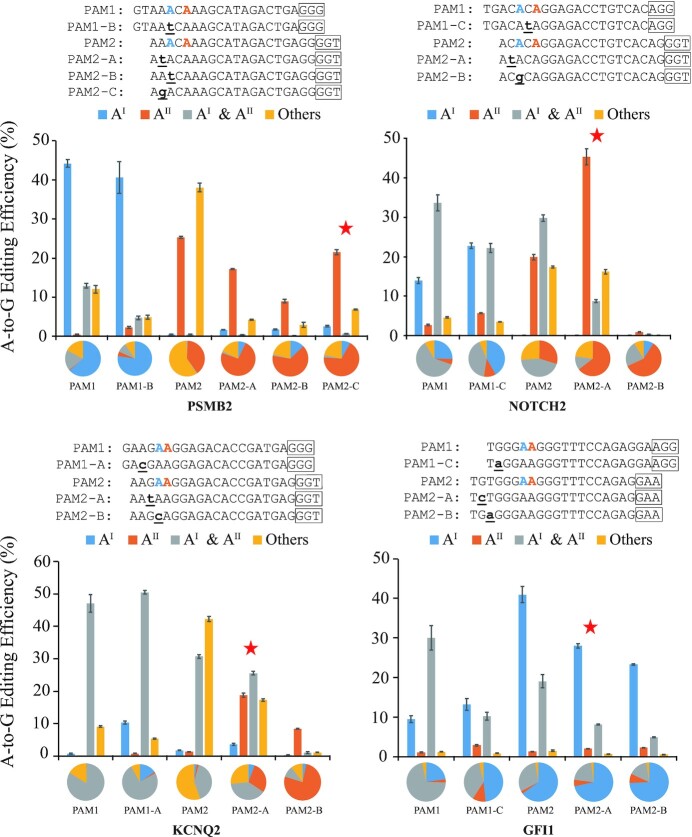
Here, we provide a strategy to use SpRY-editors (near-PAMless editors) (25) combined with igRNA to achieve controllable single-base editing and single-base editing with dual consecutive bases. Theoretically, variation of the PAM position changes the major editing substrate position, and igRNA reduces the bystander editing. The SpRY-ABEmax editor, consisting of an engineering cas9 variant and adenine deaminases, can recognize almost all PAMs (25). With this editor, the gRNA for the protospacers of PSMB2, NOTCH2, KCNQ2 and GFI1 were redesigned in two sets designated as PAM1 and PAM2, targeting different bases, AI and AII respectively within one locus (Figure 3 and Supplementary Table S11).
Figure 3.
The base editing results of controllable single-base editing with SpRY-editor and igRNA. Base editing efficiencies and fractions of editing products were obtained from 4 loci by the NG-ABEmax and SpRY-ABEmax editors using gRNAs or igRNAs. AI represents blue A, AII represents orange A. PAM1, the control gRNAs for NG-ABEmax editor exactly matched the target sequence. PAM2, the gRNAs for SpRY-ABEmax editor also exactly matched the target sequence. Capital A to C, different igRNAs. The frames represent PAMs for different loci. The red asterisk represents the igRNA with the best performance. The bars represent mean values, and the error bars represent the s.d. of three independent biological replicates. The pie charts represent the fractions of various types of editing products.
Experiment results shown that moving of PAM position could change the dominant editing position, that the editing efficiencies and product fractions of AI at PSMB2 and NOTCH2 loci were sharply decreased with AII increased (Figure 3 and Supplementary Table S11). At these tested loci, the fractions of single A-to-G conversion of AI were changed from 63.49 ± 1.74% to 0.77 ± 0.04%, 25.36 ± 0.72% to 0.23 ± 0.01%, 1.39 ± 0.18% to 2.36 ± 0.10% and 22.76 ± 0.15% to 64.99 ± 1.27% at PSMB2, NOTCH2, KCNQ2 and GFI1, respectively. The corresponding A-to-G editing efficiencies changed from 44.22 ± 0.97% to 0.49 ± 0.02%, 13.93 ± 0.76% to 0.16 ± 0.01%, 0.79 ± 0.11% to 1.80 ± 0.10% and 9.57 ± 0.87% to 40.95 ± 2.02%, respectively. In contrast, the fractions of single A-to-G conversion of AII were changed from 0.68 ± 0.08% to 39.32 ± 0.46%, 4.95 ± 0.14% to 29.59 ± 0.41%, 0.22 ± 0.03% to 1.86 ± 0.09% and 2.70 ± 0.10% to 2.25 ± 0.14% at PSMB2, NOTCH2, KCNQ2 and GFI1, respectively. The corresponding A-to-G editing efficiencies changed from 0.47 ± 0.05% to 25.30 ± 0.25%, 2.72 ± 0.18% to 19.89 ± 0.68%, 0.13 ± 0.01% to 1.41 ± 0.05% and 1.13 ± 0.10% to 1.42 ± 0.01%. Obviously, the editing preferability between AI and AII could be exchanged by moving the PAM position.
In the following step, we designed igRNAs derived from PAM1 gRNA or PAM2 to gRNA to reduce the bystanders as in previous experiments, from a mall set of igRNAs, optimal single-base editing was achieved with the igRNAs PSMB2-PAM2-C, NOTCH2- PAM2-A, KCNQ2- PAM2-A and GFI1-PAM2-A (Figure 3 and Supplementary Table S11). At three out of four tested loci, the fractions of single A-to-G conversion of AII was increased from 39.32 ± 0.46% to 67.99 ± 0.42%, 29.59 ± 0.41% to 64.28 ± 0.27%, and 1.86 ± 0.09% to 28.77 ± 0.23% at PSMB2, NOTCH2, and KCNQ2, respectively. The corresponding A-to-G editing efficiencies changed from 25.30 ± 0.25% to 21.51 ± 0.66%, 19.89 ± 0.68% to 45.23 ± 2.03% and 1.41 ± 0.05% to 18.82 ± 0.67%. These results demonstrated the successful controllable single-base editing. At GFI1 loci, it was observed that the editing preferability between AI and AII was not exchanged, but the proportion of single-A of the dual As was increased with the igRNAs and SpRY-ABEmax combined strategy. This strategy was also applied to CBE at the RNF216 locus, the results show very low editing efficiency for both Cs, probably because of PAM or protospacer changing. Thus, combined igRNAs with the PAM-less editors, such as SpRY-ABEmax, controllable single-base editing at any position might be achieved at some genomic loci.
Research for construction of disease-associated SNPs cell lines with igRNA
Most SNPs are located at genomic loci surrounded by other bases editable by current base editors (bystanders). The low single-base editing specificity makes it difficult to obtain cleanly edited cells, which is the major obstacle preventing the application of base editing to efficiently create model cell lines or animal models with disease-associated SNPs. This problem is even more severe in the development of efficient and safe genetic therapies by onsite SNP correction with base editors. To demonstrate the technical advancement of igRNA for the construction of cell lines with SNPs, a locus containing disease-associated SNPs among multiple editable bases in the editing window was selected for editing with igRNAs.
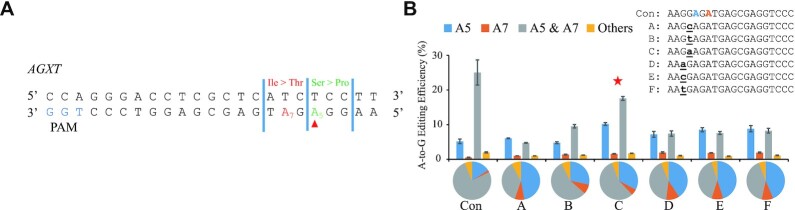
The 661T > C SNP (T-to-C conversion SNP at position 661, A > G in the complementary chain) in the AGXT gene causes the Ser221Pro missense mutation and leads to primary hyperoxaluria, which is a rare condition characterized by the recurrent kidney and bladder stones (26,27). As shown in Figure 4A, A5-to-G conversion at the target site causes a Ser-to-Pro amino acid exchange (indicated in green); however, the presence of A7 or other As around A5 causes bystander editing and undesired nonsynonymous amino acid exchange (indicated in red). To construct cell lines bearing a clean 661T > C disease-associated SNP, six igRNAs with one noncomplementary base either at different sites or with different base types were designed. Among these igRNAs, AGXT-C gave the best A5-to-G single-base editing performance. The single-base editing fraction was improved from 15.89 ± 0.27% to 32.78 ± 0.33%, and the editing efficiency was improved from 7.8 ± 0.98% to 15.30 ± 0.61% relative to the parent gRNA, while the A5 and A7 double-base editing fraction dropped from 76.22 ± 0.67% to 56.43 ± 0.45% (Figure 4B, Supplementary Table S6 and Supplementary Table S7). The increased editing efficiency indicated that the number of correctly edited cells increased 2-fold with igRNA, and the proportion of edited cells with A5-to-G single-base conversion was also increased 2-fold by igRNA. The results demonstrated the great capacity of igRNA techniques for creating model cell lines and, more importantly, for correction of disease-associated SNPs to treat human diseases.
Figure 4.
Cell lines bearing the disease-associated SNP AGXT 661T > C construction research with igRNA single-base editing. (A) The target sequence in the AGXT gene. The PAM sequence is shown in blue. The desired A5-to-G conversion, which causes a Ser-to-Pro amino acid exchange, is indicated in green. Base conversion of the bystander A7 is indicated in red and causes undesired nonsynonymous amino acid changes. (B) Six igRNAs with one noncomplementary base either at different sites or with different base types were employed to accomplish 661T > C conversion in the AGXT gene, which causes primary hyperoxaluria. Con, the control gRNAs. A∼F, different igRNAs. The red asterisk represents the igRNA with the best performance. The bars represent mean values, and the error bars represent the s.d. of three independent biological replicates. The pie charts represent the fractions of various types of editing products.
DISCUSSION
In this work, we found that Cs within the editing window are preference to co-edited in most loci (Supplementary Figure S1A), and designed an imperfect gRNA (igRNA) editing methodology for convenient specific single-base editing with both ABEs and CBEs. For genome editing applications, especially genetic therapies, the single-base editing fraction parameter could be even more important than the editing efficiency, since bystander editing with target SNP correction might cause unknown problems. The fraction of single-base editing with igRNA was improved by 5.64-, 38.50-, 5.13-, 1.58-, 0.06-, 50.93-, 0.57-, 6.48- and 3.31-fold relative to that with normal gRNA at the nine loci tested with BE4max, demonstrating a tremendous improvement (Supplementary Table S1, and Supplementary Table S5). In above loci, igRNAs were able to increase the absolute single-base editing efficiency, which was a surprisingly good result that we did not expect. We considered that mismatched igRNAs are inferior to normal perfectly matched gRNAs and certainly reduce the editing efficiency. In our systematic experiment of various igRNAs derived from one gRNA, the data suggested that it was quite possible that one or more igRNAs might have a superior performance relative to their parent gRNA at most loci. Nevertheless, it is actually difficult to achieve single-base editing with the consecutive dual Cs or dual As with igRNAs, we come up with a strategy to use SpRY-editors (near-PAMless editors) combined with igRNA, this strategy provides a possible way to solve single-base editing with consecutive dual Cs or dual As. We thus suggest that researchers attempt or adopt the igRNA strategy with their projects in cellular research or molecular therapy development to further improve both the efficiency and specificity of base editing.
To obtain some support for our hypothesis that BEs (ABEs or CBEs) firstly bind on-target and convert one nucleotide before converting other bases, we have measured the editing outcome at different time points (e.g. 24, 48, 72, 96, 120, 144 h) in one base editing process using NG-ABEmax (Supplementary Figure S5B, and Supplementary Table S12). We found with the original gRNA, the editing efficiency of single A7 of the target locus was much higher than bystander editing at the 24 h time point, which gradually increased over time. However, the major bystander editing, A5 and A7 dual editing, increased faster than the single A7 editing, which surpassed A7 at around 60 h and became the dominant bystander. Based on changed the editing results along with the time period, we might be able to propose the hypothesis, that one base located at the best position of the R-loop might be converted first, after which the base editor might still bind to the target locus and convert to other bases. Such a binding pattern favors editing of multiple but not one base. However, as illustrated in Supplementary Figure S5B, when an imperfect guide RNA (igRNA) that already has one or more bases not complementary to the target locus was used, at 24 h, both A7 editing and bystander editing were lowered, however, as the editing proceeded, the A7 editing continued to increase but bystander editing had almost no change. This result might support the hypothesis, that if we start with igRNA, after one base is converted, the igRNA would have more non-complementary bases to the target locus and might lose its ability to guide the base editor complex to the modified locus while keeping its ability binding with original DNA sequence. This results in the ending of the editing process with a modified locus, but not the original sequence. Thus, the whole editing process may cause increased single-base editing conversion as shown in Supplementary Figure S1B.
Currently, off-target effects and bystander editing are two major obstacles to be overcome for better application of base editing techniques. While our work resolved the bystander editing issue to some extent, it did not tackle the off-target effect problem. The off-target (28–31) include the effect of base editing is contributed by two factors: deaminase and Cas9. Since the igRNA techniques change only the gRNA part and still use the established base editors, the off-target effect due to the deaminase should be the same. The nonspecific binding of Cas9 is mainly observed at genomic loci containing protospacer sequences similar to those of the gRNA. To estimate Cas9-induced off-target effects, researchers normally sequence and analyze multiple potential off-target sites, which are genomic loci with the most similar protospacer. We have run all of the parent gRNAs with its according igRNAs of the different target sites with Cas-OFFinder (up to 3 mismatches, no bulges) (32). Based on the results, there were differences in the number of off-target sites between the gRNAs and igRNAs of each locus, the numbers of predicted off-target sites were listed in Supplementary Table S13 and Supplementary Table S14. The data revealed that for any given igRNA, the number of off-target sites could be either more or less relative to that of the original gRNA. For example, the number of off-target sites of PRNP-site1 original gRNA was 25, and that of the two igRNA, igRNA A and B, were 34 and 4 respectively. In general, the off-target status of igRNAs is comparable with their parental gRNA. When using igRNAs, we could select one having the same or fewer potential off-target sites as the original gRNA to maintain the same off-target potential.
According to the research of the article, we generalize a relatively simple guideline for igRNA design. First, several igRNAs are designed from the parent gRNA with mismatches up to 2 positions spanning approximately protospacer positions 2–6 (where position 1 is the first nucleotide of the protospacer and the PAM is at positions 21–23) with CBEs, and only 1 position spans approximately protospacer positions 2–5 with ABEs. Here, the nucleotide type of mismatches could be random. Choosing the same or fewer potential Cas9 dependent off-target sites as the original gRNA. Second, editing experiments were performed with the set of igRNAs to select the best one. From the experiment we have done, we found an improved single-base editing result could be obtained from four or fewer igRNAs at most loci. Of course, for a single important edit locus, such as a disease-associated mutation, the researcher can also design a larger library of igRNAs based on the capacity to screen for the best igRNA. We think this is a generally usable rule for igRNA implementation.
However, if conditions allowed, a more systematic approach might be employed to construct a mathematical model helping design igRNA for each target. We found that different base type changes, such as G-to-A, G-to-C, G-to-T, at the same site also showed different editing results (Figure 4B). Furthermore, the number of mismatched nucleotides had also different effects at various loci. An ideal approach would be generating a systemically mismatched igRNA library, which would contain (420–1) gRNAs even only mismatches one N20 position, to cover the variations at one locus. To generate enough data, editing experiment with this number of igRNAs needs to be carried out at a large number of loci. Then, artificial intelligence could be employed to analyze the large data to form a mathematical model to help design igRNAs. Apparently, such a completely systematic approach requires a great researching capacity and investment. And whether a functional model could be constructed considering the very diverse data set. Hopefully, with the decrease of cost for high-throughput DNA synthesis and DNA sequencing, along with more powerful computers, this systematic approach could be realized.
This work describes the innovation of a simple strategy to be able to conveniently obtain single-base editing with both ABEs and CBEs. The igRNA technique achieves more controllable and specific genomic manipulation and overcomes the most significant obstacle of base editors for applications in genetic therapies and for the creation of disease-associated models.
DATA AVAILABILITY
There is no restriction on data associated with this study. High-throughput sequencing data have been deposited in the NCBI database (accession code PRJNA741886 and PRJNA807106).
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: X.Z., C.B. and J.L. designed the research, analyzed data and wrote the manuscript. D.Z. and G.J. designed the research, performed experiments, analysed data and wrote the manuscript. X.C., S.L. and J.W. performed experiments, analysed data. Z.Z., S.P., Z.D. and Y.M. designed the research.
Contributor Information
Dongdong Zhao, College of Life Science, Tianjin Normal University, Tianjin, China; Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China.
Guo Jiang, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; School of Life Sciences, Guangxi Normal University, Guilin, China.
Ju Li, College of Life Science, Tianjin Normal University, Tianjin, China.
Xuxu Chen, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; School of Life Sciences, Guangxi Normal University, Guilin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
Siwei Li, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
Jie Wang, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China; School of Biological Engineering, Dalian Polytechnic University, Dalian, China.
Zuping Zhou, School of Life Sciences, Guangxi Normal University, Guilin, China; Guangxi Universities Key Laboratory of Stem cell and Biopharmaceutical Technology, Guangxi Normal University, Guilin, China.
Shiming Pu, School of Life Sciences, Guangxi Normal University, Guilin, China; Guangxi Universities Key Laboratory of Stem cell and Biopharmaceutical Technology, Guangxi Normal University, Guilin, China.
Zhubo Dai, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
Yanhe Ma, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
Changhao Bi, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
Xueli Zhang, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China; Key Laboratory of Systems Microbial Biotechnology, Chinese Academy of Sciences, Tianjin, China; National Technology Innovation Center of Synthetic Biology, Tianjin, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFA0903700]; National Natural Science Foundation of China [31861143019, 31770105, 32171449, 32001041, 81972700, 61827819]; Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project [TSBICIP-KJGG-017]; Youth Innovation Promotion Association CAS [2022177] and Tianjin Natural Science Foundation [20JCYBJC00310]. Funding for open access charge: Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-017).
Conflict of interest statement. A provisional patent has been submitted in part entailing the reported approach.
REFERENCES
- 1. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y.et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016; 353:aaf8729. [DOI] [PubMed] [Google Scholar]
- 3. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kurt I.C., Zhou R., Iyer S., Garcia S.P., Miller B.R., Langner L.M., Grünewald J., Joung J.K.. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021; 39:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao D., Li J., Li S., Xin X., Hu M., Price M.A., Rosser S.J., Bi C., Zhang X.. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021; 39:35–40. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y., Qin W., Lu X., Xu J., Huang H., Bai H., Li S., Lin S.. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 2017; 8:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banno S., Nishida K., Arazoe T., Mitsunobu H., Kondo A.. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 2018; 3:423–429. [DOI] [PubMed] [Google Scholar]
- 8. Li C., Zong Y., Wang Y., Jin S., Zhang D., Song Q., Zhang R., Gao C.. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018; 19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suh S., Choi E.H., Leinonen H., Foik A.T., Newby G.A., Yeh W.H., Dong Z., Kiser P.D., Lyon D.C., Liu D.R.et al.. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 2021; 5:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lapinaite A., Knott G.J., Palumbo C.M., Lin-Shiao E., Richter M.F., Zhao K.T., Beal P.A., Liu D.R., Doudna J.A.. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science. 2020; 369:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R.. Direct observation of R-loop formation by single RNA-guided cas9 and cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anzalone A.V., Koblan L.W., Liu D.R.. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020; 38:824–844. [DOI] [PubMed] [Google Scholar]
- 13. Huang T.P., Zhao K.T., Miller S.M., Gaudelli N.M., Oakes B.L., Fellmann C., Savage D.F., Liu D.R.. Circularly permuted and PAM-modified cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 2019; 37:626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R.. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017; 35:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan J., Zhang F., Karcher D., Bock R.. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 2019; 10:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong Y.K., Lee S., Hwang G.H., Hong S.A., Park S.E., Kim J.S., Woo J.S., Bae S.. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 2021; 39:1426–1433. [DOI] [PubMed] [Google Scholar]
- 17. Tan J., Zhang F., Karcher D., Bock R.. Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat. Commun. 2020; 11:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin S., Fei H., Zhu Z., Luo Y., Liu J., Gao S., Zhang F., Chen Y.H., Wang Y., Gao C.. Rationally designed APOBEC3B cytosine base editors with improved specificity. Mol. Cell. 2020; 79:728–740. [DOI] [PubMed] [Google Scholar]
- 19. Gehrke J.M., Cervantes O., Clement M.K., Wu Y., Zeng J., Bauer D.E., Pinello L., Joung J.K.. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018; 36:977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee S., Ding N., Sun Y., Yuan T., Li J., Yuan Q., Liu L., Yang J., Wang Q., Kolomeisky A.B.et al.. Single C-to-T substitution using engineered APOBEC3G-nCas9 base editors with minimum genome- and transcriptome-wide off-target effects. Sci. Adv. 2020; 6:eaba1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Z., Wang Y., Liu Q., Qiu Y., Zhong Z., Li K., Li W., Deng Z., Sun Y.. Improving the precision of base editing by bubble hairpin single guide RNA. Mbio. 2021; 12:e00342-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., Chen L., Zhu B., Wang L., Chen C., Hong M., Huang Y., Li H., Han H., Cai B.et al.. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 2020; 22:740–750. [DOI] [PubMed] [Google Scholar]
- 24. Rees H.A., Liu D.R.. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018; 19:770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P.. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020; 368:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cochat P., Hulton S.A., Acquaviva C., Danpure C.J., Daudon M., De Marchi M., Fargue S., Groothoff J., Harambat J., Hoppe B.et al.. Primary hyperoxaluria type 1: indications for screening and guidance for diagnosis and treatment. Nephrol., Dial. Transplant. 2012; 27:1729–1736. [DOI] [PubMed] [Google Scholar]
- 27. Li Y., Zheng R., Xu G., Huang Y., Li Y., Li D., Geng H.. Generation and characterization of a novel rat model of primary hyperoxaluria type 1 with a nonsense mutation in alanine-glyoxylate aminotransferase gene. Am. J. Physiol. Renal Physiol. 2021; 320:F475–f484. [DOI] [PubMed] [Google Scholar]
- 28. Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017; 35:475–480. [DOI] [PubMed] [Google Scholar]
- 29. Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H.. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019; 364:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K.. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019; 569:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R.et al.. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019; 571:275–278. [DOI] [PubMed] [Google Scholar]
- 32. Bae S., Park J., Kim J.S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There is no restriction on data associated with this study. High-throughput sequencing data have been deposited in the NCBI database (accession code PRJNA741886 and PRJNA807106).