Abstract
One of the most challenging issues in the design of phase II/III clinical trials of antimicrobial agents is dose selection. The choice is often based on preclinical data from pharmacokinetic (PK) studies with animals and healthy volunteers but is rarely linked directly to the target organisms except by the MIC, an in vitro measure of antimicrobial activity with many limitations. It is the thesis of this paper that rational dose-selection decisions can be made on the basis of the pharmacodynamics (PDs) of the test agent predicted by a mathematical model which uses four data sets: (i) the distribution of MICs for clinical isolates, (ii) the distribution of the values of the PK parameters for the test drug in the population, (iii) the PD target(s) developed from animal models of infection, and (iv) the protein binding characteristics of the test drug. In performing this study with the new anti-infective agent evernimicin, we collected a large number (n = 4,543) of recent clinical isolates of gram-positive pathogens (Streptococcus pneumoniae, Enterococcus faecalis and Enterococcus faecium, and Staphylococcus aureus) and determined the MICs using E-test methods (AB Biodisk, Stockholm, Sweden) for susceptibility to evernimicin. Population PK data were collected from healthy volunteers (n = 40) and patients with hypoalbuminemia (n = 12), and the data were analyzed by using NPEM III. PD targets were developed with a neutropenic murine thigh infection model with three target pathogens: S. pneumoniae (n = 5), E. faecalis (n = 2), and S. aureus (n = 4). Drug exposure or the ratio of the area under the concentration-time curve/MIC (AUC/MIC) was found to be the best predictor of microbiological efficacy. There were three possible microbiological results: stasis of the initial inoculum at 24 h (107 CFU), log killing (pathogen dependent, ranging from 1 to 3 log10), or 90% maximal killing effect (90% Emax). The levels of protein binding in humans and mice were similar. The PK and PD of 6 and 9 mg of evernimicin per kg of body weight were compared; the population values for the model parameters and population covariance matrix were used to generate five Monte Carlo simulations with 200 subjects each. The fractional probability of attaining the three PD targets was calculated for each dose and for each of the three pathogens. All differences in the fractional probability of attaining the target AUC/MIC in this PD model were significant. For S. pneumoniae, the probability of attaining all three PD targets was high for both doses. For S. aureus and enterococci, there were increasing differences between the 6- and 9-mg/kg evernimicin doses for reaching the 2 log killing (S. aureus), 1 log killing (enterococci), or 90% Emax AUC/MIC targets. This same approach may also be used to set preliminary in vitro MIC breakpoints.
The drug development process traditionally follows the initial “first-in-human” pharmacokinetic (PK) studies with phase II dose-finding studies. Such studies are often relatively small and provide little power to discriminate among doses for adequacy of effect in the clinical setting. There is growing pressure to develop new antimicrobial agents more quickly to meet the need presented by emerging resistance and new infectious diseases with high morbidity and mortality rates (e.g., vancomycin-resistant enterococcal and staphylococcal infections). To meet these emerging infectious disease challenges, it is not always practical to amass clinical databases which have the size necessary to gain optimal statistical precision, and often, there is a need to push forward and conduct much smaller phase II/III studies supported by the results of preclinical and phase I studies. Consequently, doses for large phase II/III clinical treatment trials are often chosen almost empirically. It would be of interest to use preclinical data to identify a dose and schedule which would produce a high likelihood of a successful clinical and/or microbiological outcome.
Over the last decade there has been an explosion in the understanding of the pharmacodynamics (PDs) related to anti-infective drug administration. Prospective (13) and retrospective (8) studies of fluoroquinolone PDs have been published. Other studies have examined the human PDs of aminoglycosides, beta-lactams, and antiviral agents (4, 5, 11, 14). Perhaps even more importantly, in vitro and animal model systems have been developed for the delineation of the PD properties of drugs and have been shown to be quite robust in their predictions of the endpoint most closely linked to outcome (1, 2, 6). Certainly, the hollow-fiber in vitro system as well as mouse and rat models of infection has provided lessons about PDs which have been well validated in clinical trials.
The result of such investigations is that one can identify a small number of factors which influence the clinical and/or microbiological outcome. Some measure of drug exposure (peak concentration, area under the concentration-time curve [AUC], etc.) relative to a measure of potency of the drug for the organism being treated (MIC, minimal bactericidal concentration, etc.), corrected for the amount of protein binding of the drug, can be linked to outcome. One way of examining the possible adequacy of a drug dose and/or schedule is to calculate a target value based on in vitro or animal PD model data (corrected for protein binding) and examine whether the free plasma drug concentrations achieved in phase I/II trials might achieve this target when related to appropriate pathogens by MICs.
However, in the real world there is true between-patient variability in the PK parameters of a drug and there certainly exists a spectrum of sensitivity to any test drug among organisms of clinical interest. Consequently, any method for examination of the adequacy of a fixed-dose regimen needs to explicitly account for both sources of variability (PK and microbiological variabilities). It was the aim of the investigation described here to examine different doses of evernimicin, the first member of a unique class of oligosaccharide antibiotics highly active against gram-positive organisms (including methicillin-resistant Staphylococcus aureus [MRSA] and vancomycin-resistant enterococci) and, by using population simulation by Monte Carlo methods, examine how frequently specific doses of evernimicin would achieve target endpoints derived from animal PD data.
MATERIALS AND METHODS
PD endpoints.
Thigh infections with different pathogens (Streptococcus pneumoniae [n = 5), S. aureus [n = 4], and Enterococcus faecalis and Enterococcus faecium [n = 2]) were established in Swiss-Webster mice. The animals were rendered neutropenic with cyclophosphamide as described previously (9). After a 2-h delay, the infections were treated with different doses and schedules of evernimicin. One strain was evaluated per animal. At hour 24, the animals were humanely killed and the colony counts of pathogens were enumerated as described by Gerber et al. (9).
Different independent variables were evaluated by fitting an inhibitory sigmoid Emax effect model (where Emax is the maximum killing effect) to the colony count data, with each of the dynamic variables serving as the independent variable in the regression. AUC was determined to be the PD variable that had the closest correlation with significant decreases in the colony counts (number of CFU) from the initial inoculum to those observed in specimens after 24 h (O. Vesga and W. A. Craig, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A-32, 1997).
Three different endpoints were calculated from each experiment: (i) a stasis endpoint (that value of the PD variable which resulted in no net change in the number of bacteria beyond the colony count (107 CFU) at the time of inoculation), (ii) a log killing (log drop) endpoint, calculated from the modeled maximal colony count (for the untreated group) at 24 h, and (iii) a 90% Emax value, calculated as the log drop representing 90% of the maximal log drop achievable (Emax). For the second endpoint, the log drop values at 24 h were 3 log10 for S. pneumoniae, 2 log10 for S. aureus, and 1 log10 for the Enterococcus spp., representing a log drop which was achievable within species for all of the experiments whose data were examined. The endpoints were then averaged across strains within species.
Antimicrobial susceptibility: MIC distribution data.
Organisms were collected from recent clinical specimens as part of a large, multicenter susceptibility survey involving 33 different sites in 24 countries. By the E-test method (AB Biodisk, Stockholm, Sweden), the activity of evernimicin was determined against 1,489 S. pneumoniae isolates (including penicillin- and multiple-drug-resistant strains), 1,449 S. aureus isolates (including MRSA and methicillin-susceptible S. aureus strains), and 1,605 enterococcal isolates (including vancomycin-resistant strains of E. faecalis and E. faecium) (R. Hare, personal communication). The E-test was read at the zone of 80% inhibition, which correlates best with standard methods of the National Committee for Clinical Laboratory Standards (10, 12).
Protein binding.
The protein binding of evernimicin was determined by an ultrafiltration (Centrifree) method with human and murine specimens. This methodology was selected as optimal on the basis of the chemical characteristics of evernimicin in vitro.
PKs and Monte Carlo simulation.
Plasma drug concentrations were determined in two studies of evernimicin conducted with 52 volunteers. Data from the first study were from an intravenous infusion rising-multiple-dose study with healthy volunteers (n = 36) receiving multiple doses of evernimicin at a range of doses from 1.0 to 9.0 mg/kg of body weight/day. The plasma sampling schedule for this study was predosing and 0.25, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 4.0, 5.0, 6.0, 8.0, 10.0, 12.0, 16.0, and 24.0 h after dosing, with sampling repeated at steady state. Additional samples were obtained at 36.0, 48.0, 60.0, and 72.0 h as washout samples after administration of the last dose. Data from the second study were obtained from 12 patients with different degrees of hepatic impairment (Childs-Pugh classes A to C), as well as from four healthy subjects. Infusion times for all studies ranged from 0.5 to 1.0 h and were included in the model.
Evernimicin levels in plasma were measured by a validated high-pressure liquid chromatography (HPLC) assay (data on file, Schering-Plough Research Institute). An HPLC method for the determination of SCH27899 levels was validated with human plasma ultrafiltrate over a concentration range of 25 to 2,500 ng/ml with a 100-μl sample volume. The method involved the mixing of human plasma ultrafiltrate with acetonitrile and injection of the mixture onto an HPLC system (Waters Corp., Milford, Mass.). Reversed-phase separation of SCH27899 was achieved on a PRP-1 column (Hamilton Co., Reno, Nev.) with UV detection at 302 nm via a computerized data acquisition system (Waters Corp.). The limit of quantitation was 25 ng/ml. An acceptable 24-h in-process stability of SCH27899 was established. On three separate occasions calibration curves (external standard) were analyzed along with requisite quality control standards. Mean interassay accuracy and precision for all standard curve and quality control samples remained well within established acceptance criteria. Assay performance with human PK samples demonstrated within-day coefficients of variation (CVs) of 6.1% at 0.05 μg/ml and 1.6% at 20.0 μg/ml. Between-day CVs were 7.3 and 1.1%, respectively.
Population PK modeling was performed with the NPEM III package of programs of Schumitzky (15). One-, two-, and three-compartment models with zero-order infusion (0.5 or 1 h, depending on the dose) and first-order elimination and transfer were evaluated (the three-compartment model was run on BigNPEM at the Supercomputer Center, University of California at San Diego). Model discrimination was accomplished with the Akaike information criterion (16). Parameter ranges were established by first running the iterative Bayesian front end of NPEM III. Weighting was as the inverse of the observation variance, with the variance being determined from the data, with the high-level search option of NPEM III estimating the parameters of third-order polynomial. Maximum a posteriori probability (MAP) Bayesian estimation was performed with the population-of-one utility within NPEM III.
The Monte Carlo simulations were run with the ADAPT II package of programs of D'Argenio and Schumitzky (3). The population mean parameter vector and population covariance matrix were embedded in the Subroutine Prior portion of ADAPT II. A population simulation without noise was performed with the simulation module of ADAPT II. In each instance, simulations for 200 subjects were performed. These simulations were repeated a total of five times, for a total of 1,000 simulated subjects per dose.
Statistical analysis.
The fraction of simulated subjects at each dose at each MIC whose AUC/MIC ratio met each of the three PD goals was determined. For each MIC, the fraction of subjects who met the goal was multiplied by the fraction of the distribution of organisms for which the MIC was at that MIC. This was summed over all MICs, which provided an estimate of the overall response of that pathogen to evernimicin at the specified dose. The statistical program Systat for Windows (version 7.0; SPSS, Inc., Chicago, Ill.) was used for all data transformation and statistical testing. Proportions were tested for differences by the Fisher exact test, and differences between means were tested for significance by the t test for independent means. Alpha was set at 0.05.
RESULTS
PD endpoints.
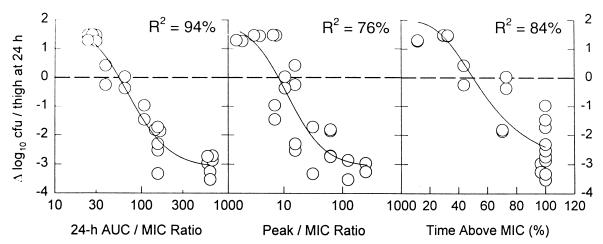
The three endpoints (stasis, log drop, and 90% Emax) for each of the organisms are listed in Table 1. The log drop endpoints differed by species, as not all strains of each species attained the 3-log drop seen for all S. pneumoniae isolates. The log drop for S. aureus was 2 log units (all strains achieved at least a 2-log drop), and the log drop for enterococcal species was 1 log unit (again, all strains achieved at least a 1-log drop). A hierarchy of PD endpoints exists, with the AUC/MIC ratio required to achieve stasis being less than that required for log drop (within species) which, in turn, is less than that required for 90% Emax. A typical exposure-response curve is presented in Fig. 1 for S. pneumoniae. In Fig. 1, the AUC/MIC ratio explains more of the variance than the other independent variables.
TABLE 1.
PD endpoints for evernimicin
| Organism | AUC/MIC ratio
|
||
|---|---|---|---|
| Stasis target | Log drop targeta | 90% Emax target | |
| S. pneumoniae | 115.7 | 239.4 | 1,716.4 |
| S. aureus | 163.4 | 330.1 | 830.8 |
| E. faecalis | 59.6 | 85.4 | 764.4 |
Log drop targets are a 3-log10-unit decline in the number of CFU per milliliter from pretreatment number at the primary infection site for S. pneumoniae, a 2-log10-unit decline for S. aureus, and a 1-log10-unit decline for E. faecalis.
FIG. 1.
Change in number of CFU recovered from mouse thigh at 24 h after initiation of therapy with SCH27899 as a function of the 24-hour AUC/MIC ratio, peak concentration/MIC ratio, and time above the MIC.
MIC distributions.
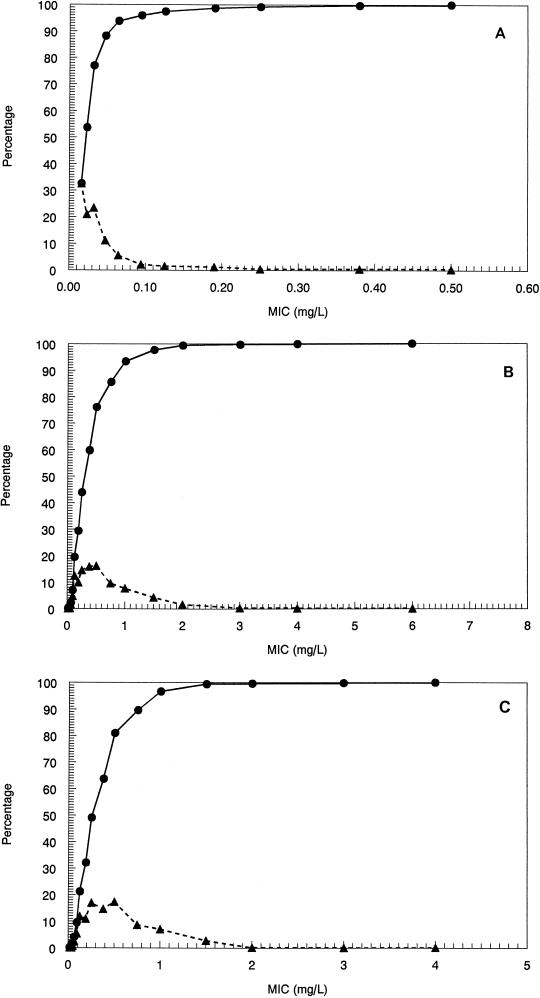
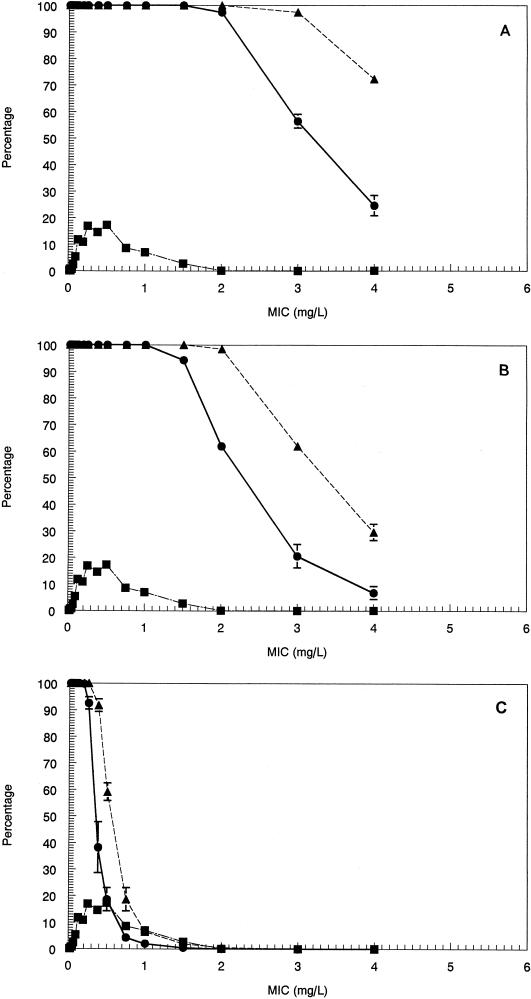
The MICs at which 90% of isolates are inhibited for the pneumococci, staphylococci, and enterococci examined ranged from 0.064 μg/ml for S. pneumoniae to 1.0 μg/ml for MRSA. The MIC distributions are displayed in Fig. 2A to C as cumulative and interval susceptibility plots. For the animal model, MICs were <0.03 μg/ml for the pneumococcal strains. For S. aureus, these were 0.25 or 0.5 μg/ml, and for enterococcal strains, these were also 0.25 or 0.5 μg/ml.
FIG. 2.
Cumulative percentage (●) and interval percentage (▴) of 1,489 strains of S. pneumoniae (A), 1,449 strains of S. aureus (B), and 1,605 strains of enterococcal species (C) sensitive to SCH27899 at the indicated MIC as determined by the E-test.
Protein binding.
The protein binding of evernimicin in humans was determined to be approximately 96.5% at 130 μg/ml. In mouse serum, this value was 96.0%. However, given the reproducibility of the assay and the extensive degree of protein binding, the PD targets were not corrected for protein binding. That is, the targets were for total drug, as determined with mice. As the binding in humans was not significantly different, the targets were not changed.
PK parameters.
A two-compartment model for evernimicin was chosen by Akaike's information criterion. The population mean parameter values and the population covariance matrix are presented in Table 2. The clearance is quite low, but the variability is also reasonably low, given that we have included patients with hepatic dysfunction. Hepatic clearance represents the major clearance pathway for evernimicin, with an increase in clearance being observed for patients with hepatic dysfunction and for patients with low serum albumin concentrations. Renal failure does not alter the clearance for this drug (C. Banfield, S. Pai, S. K. Swan, L. Lambrecht, M. Laughlin, and M. Affrime, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A-50, 1998).
TABLE 2.
Population PK modeling for evernimicin: mean parameter vector and covariance matrixa
| Value | Volume of central compartment (liters) | Plasma clearance (liters/h) | Kcp (h−1) | Kpc (h−1) |
|---|---|---|---|---|
| Mean | 6.36 | 2.46 | 0.302 | 0.089 |
| Covariance matrix | 3.8096 | |||
| 0.0530 | 0.0034 | |||
| 0.0063 | 0.0001 | 0.0003 | ||
| 1.0481 | 0.0120 | 0.0051 | 0.5551 |
Population pharmacokinetic analysis was for 52 subjects, including 4 each with Child-Pugh Class A, B, and C cirrhosis. Overall, 1,689 plasma samples were analyzed.
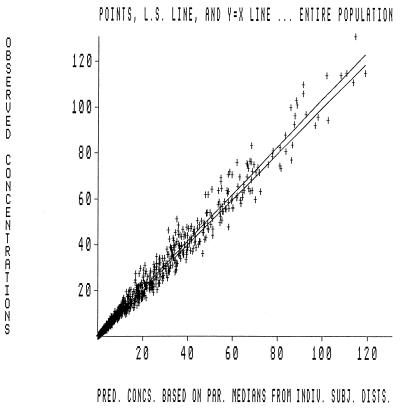
The population PK analysis was quite robust. The MAP Bayesian step allowed construction of an observed concentration versus predicted concentration plot. This is displayed in Fig. 3. The overall r2 was in excess of 0.98, indicating that the fit of the model to the data was excellent. A separate analysis (data not shown) was performed only with data for the healthy subjects. The parameter values and overall r2 were not significantly different (r2 = 0.985), indicating that the data for the patients with hepatic dysfunction were also well fit by the model.
FIG. 3.
Plot of predicted versus observed concentrations for 52 patients contributing 1,689 plasma samples for which evernimicin concentrations were determined. The r2 value was 0.966; P was ≪0.0001. The values on the x axis indicate the predicted concentrations based on the parameter medians for the distribution of individual subjects. The figure shows the individual datum points, l.s. line, and the y equal to x line for the entire population.
Attaining the PD target.
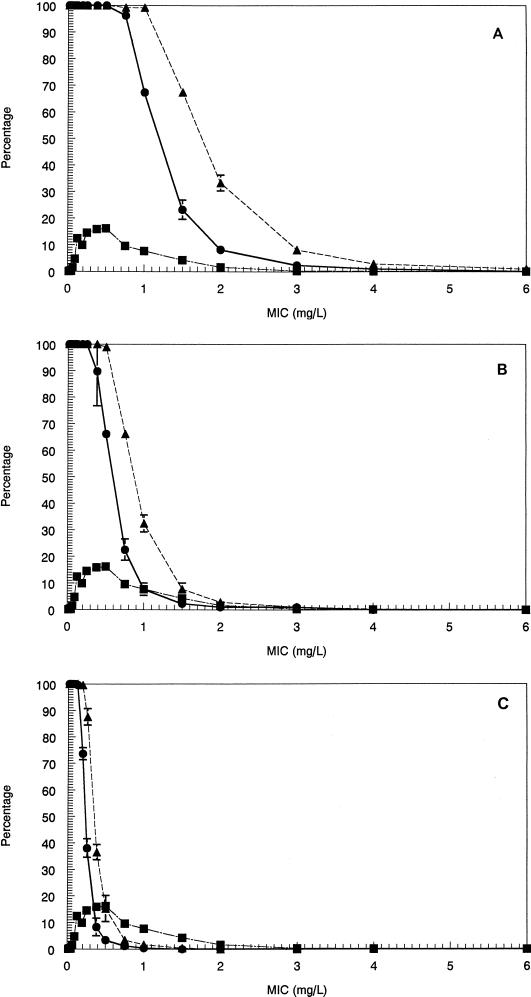
The Monte Carlo simulations allowed calculation of the AUC achieved for each of the simulated subjects for evernimicin doses of 6 and 9 mg/kg/day. The fraction of patients (±standard deviation [SD]) who attained the target for each of the endpoints by MIC is displayed in Fig. 4 to 6.
FIG. 4.
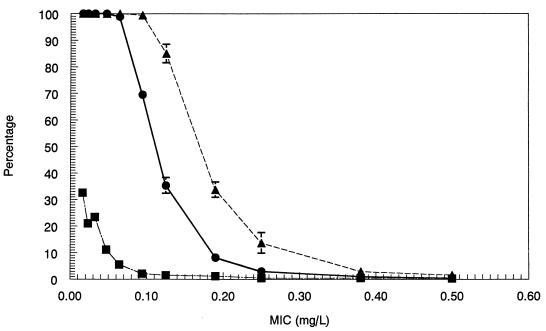
Fractional attainment of the 90% Emax target for S. pneumoniae for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■).
FIG. 6.
(A) The fractional attainment of the stasis target for enterococcal species for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■). (B) Fractional attainment of the 1-log10 CFU drop target for enterococcal species for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■). (C) Fractional attainment of the 90% Emax target for enterococcal species for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■).
These results were then integrated with the MIC distribution data for each organism. In order to properly interpret the likelihood of response by dose, it is necessary examine the full distribution of MICs, as they may be maldistributed and located primarily at one end or the other of the range of MICs. When performing the calculations displayed in Fig. 4 to 6, all observed MICs were used. The data in Fig. 4 to 6 give the expected frequency (±SD) of attaining the therapeutic target at a specific MIC.
The overall estimate of attainment of the therapeutic goal (±SD) for each of the endpoints for each pathogen is displayed in Table 3. Each of the contrasts between the 6- and 9-mg/kg/day doses for each endpoint for each pathogen is statistically significant (P <0.05), although some may not be biologically significant.
TABLE 3.
Fractional attainment of target by evernimicin
| Dose (mg/kg/day) | % Attainment of response
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
S. pneumoniae
|
S. aureus
|
Enterococcal spp.
|
|||||||
| Stasis | Log drop (3 log10 units) | 90% Emax | Stasis | Log drop (2 log10 units) | 90% Emax | Stasis | Log drop (1 log10 unit) | 90% Emax | |
| 6 | 100 ± 0.0 | 99.9 ± 0.009 | 95.87 ± 0.07 | 91.64 ± 0.2 | 71.79 ± 1.89 | 34.25 ± 0.68 | 99.70 ± 0.11 | 99.41 ± 0.02 | 58.14 ± 2.82 |
| 9 | 100 ± 0.0 | 100 ± 0.0 | 97.71 ± 0.02 | 96.83 ± 0.08 | 85.10 ± 0.84 | 50.74 ± 0.84 | 99.93 ± 0.004 | 99.77 ± 0.009 | 74.84 ± 0.59 |
DISCUSSION
The aim of a phase II/III clinical trial of an investigational anti-infective agent is demonstration of the safety and efficacy for a particular indication and/or pathogen(s) directly compared to a standard of care for which a satisfactory safety profile and efficacy in that same setting have been established. The selection of a dose for such a trial is made difficult by the limited nature of the data sets provided by phase I and II studies. This is an especially difficult decision when the test agent is examined for efficacy against new pathogens with a higher prevalence of in vitro resistance to standard comparator drugs than the prevalence observed in the phase II studies or against pathogens for which there is no approved standard of treatment. It is imperative, therefore, that we maximize the information generated from preclinical studies for use in decision support for dose selection and preliminary MIC breakpoints.
Evernimicin is an agent which is highly active in vitro against all clinically relevant gram-positive pathogens. It is very potent, with the MIC for more than 99% of pneumococci being ≤0.25 μg/ml. When the MICs of evernimicin for gram-positive pathogens that are resistant to beta-lactams and macrolides are compared, the MICs for resistant strains are no different from those observed for susceptible strains. The unimodal distribution of the MICs of evernimicin also indicates that no subpopulation of heteroresistant strains has been observed in large, multicenter studies (9a; data on file, Schering-Plough Research Institute).
Evernimicin is highly protein bound. The MICs should be examined relative to the AUC for the free drug or to the AUCs for a target set of drugs, with protein binding built into the calculation, as we have done here.
The PKs of evernimicin are interesting. The volume of the central compartment is small (6.36 ± 1.95 liter), consistent with the highly protein-bound nature of this drug. It should be realized, however, that sufficient concentrations of free drug are reached at extravascular sites in the mouse model and in human clinical subjects that antimicrobial efficacy can be demonstrated. The clearance of evernimicin is low (2.46 liters), with a relatively low coefficient of variation (30.3%; SD, 0.74 liters/h). Particularly when one considers that patients with all levels of hepatic dysfunction were included in the analysis, the PK parameters are relatively consistent with those observed for healthy volunteers. A more detailed discussion of evernimicin PKs is the subject of another publication.
The construction of multiple Monte Carlo simulations is critical, in that it explicitly brings PK variability into the evaluation. Currently, many evaluations of new drugs by use of PKs examine only the mean or median value for the PK parameter values. When one evaluates the median value of clearance (or another PK parameter value[s]), however, 50% of the population has a higher value of clearance, with consequent lower levels of drug exposure and a lower probability of achievement of a stated therapeutic target. Therefore, such evaluations may not accurately predict the microbiological performance of a regimen in large controlled clinical trials.
Monte Carlo simulation also allows calculation of multiple point estimates of the ability of a dose to achieve a therapeutic target based on an animal model of established predictive value. As can be seen by examining the data in Table 3, the increase in dose by 50% (6 to 9 mg/kg/day) allows some improvement in the achievement of the therapeutic goal for the stasis target (0% for pneumococcus, 5.2% for S. aureus, and 0.2% for Enterococcus spp.). However, the incremental improvement is greater for the log drop target (0.014% for pneumococcus with a 3-log drop, 13.3% for S. aureus with a 2-log drop, and 0.36% for Enterococcus spp. with a 1-log drop) and for the 90% Emax target (1.8% for pneumococcus, 16.5% for S. aureus, and 16.7% for Enterococcus spp.). Because an SD is associated with the average goal achievement, it is possible to test the differences between regimens statistically.
Furthermore, one can use the data in Fig. 4 to 6 to highlight other points. First, it is clear that the basis of establishing preliminary MIC breakpoints should, of necessity, also involve the proposed range of drug doses, the site of infection, as well as the distribution of MICs for the pathogens of interest. Second, this approach allows rational consideration of a preclinical breakpoint on the basis of achievement of therapeutic goals derived from nonclinical sources. However, it should be made clear that such breakpoints would be preliminary only until clinical data can be gathered from well-controlled studies and analyzed by the U.S. Food and Drug Administration and the National Committee for Clinical Laboratory Standards. However, once these data are gathered, much the same approach will allow further, clinically based decision supports to be generated and breakpoints arrived at in a rational manner.
While this approach allows rational consideration of breakpoints, it still requires an explicit judgment to be made. At what probability of success (probability of microbiologically adequate therapy) do we consider an MIC to represent susceptibility. This is not a question that can be definitively solved by any mathematical technique. Rather, it is a judgment to be reached by consensus among clinicians and microbiologists. These types of simulations represent decision support rather than decisions themselves.
In the analyses described above, the key issue revolves about the believability of the targets set through the use of the animal system. It is important, therefore, that there be concordance between the results obtained with animal model systems and the results of clinical trials. Perhaps the best example of this concordance can be seen with the fluoroquinolone class of antimicrobials.
In a retrospective analysis of patients receiving ciprofloxacin for hospital-acquired pneumonia, Forrest et al. (8) reported that the AUC/MIC ratio was the pharmacodynamically linked variable. In a prospective, multicenter controlled trial, Preston and colleagues (13) demonstrated that for patients with a variety of community-acquired infections the peak concentration/MIC ratio was the best pharmacodynamically-linked variable. The outcome differences in the trials are explained to a large degree by the findings from the results of a study with an animal model of Drusano et al. (6), which demonstrated that either the peak concentration/MIC ratio or AUC/MIC ratio could be linked to outcome, depending upon whether the peak concentration/MIC ratio significantly exceeded 10/1. The study of Forrest et al. (8) had a median peak concentration/MIC ratio of 12/1, while the study of Preston et al. (13) had a median peak concentration/MIC ratio in excess of 20/1 and more than 80% of patients developed a peak concentration/MIC ratio in excess of 10/1, identified as significant by the data of Blaser et al. (1). Consequently, the clinical results are in perfect concordance with the in vitro and animal model data.
Perhaps more importantly, the actual goals of therapy which were derived from the clinical studies are in concordance with those derived from animal model data. The clearest example is given in the data of both Fantin et al. (7) and Drusano et al. (6). By examining the data published by Fantin et al. (7), one can see that the pefloxacin effect is near maximal at an AUC/MIC ratio of approximately 140/1. Likewise, from the data of Drusano et al. (6), the 90% Emax of the AUC/MIC ratio is approximately 100/1. One can also see this in the data published by Craig (2) for multiple animal models, in which the near-Emax value of the AUC/MIC ratio is approximately 100/1. In the clinical data sets, the study of Forrest et al. (8) identified a breakpoint value of AUC/MIC of 125/1. In the study of Preston et al. (13), the dynamically linked variable for both clinical and microbiological outcome was a peak concentration/MIC ratio with a breakpoint of 12/1. However, if one takes the data from the study of Preston et al. (13) and examines the microbiological outcomes, by use of the AUC/MIC ratio as the independent variable, a breakpoint of 100/1 is obtained. The microbiological outcomes are most appropriate in that the breakpoint described by Forrest et al. (8) was for a microbiological endpoint. Consequently, we can say that there is excellent predictability in this case between different animal model studies and the breakpoints that arise from them and the PD targets derived from data from clinical trials. It should be noted, however, that these clinical trials examined patients who had, in the main, respiratory tract infections with an admixture of skin and skin structure infections. It is likely that different animal models would need to be examined for infections in specialized spaces, such as the central nervous system, prostate, or eye, where absolute drug concentrations and their time profiles differ significantly from those seen in plasma.
Given the information presented above, it is reasonable to set our targets on the basis of the results obtained with animal models. When one examines the data in Table 3, it is apparent that there is not a great deal of difference between doses in achieving the stasis endpoint for S. pneumoniae or Enterococcus spp. However, the difference between doses in achieving the stasis endpoint for S. aureus is more than 5%. When one examines the log drop endpoints, there is little appreciable difference for the first two pathogens, but again, for S. aureus there is an appreciable difference, at an average of 13.3%. For the 90% Emax endpoint, there is a small difference in the fraction of the evaluations which achieve the target for the pneumococcus (1.8%), which markedly increases for both S. aureus and Enterococcus spp. (16.5 and 16.7% differences between doses, respectively).
It may be that different targets derived from in vitro or animal model systems may have different correlates in the clinical arena. As described above, we examined a 90% Emax target and correlated this with the microbiological targets derived from the data from clinical trials. A successful clinical outcome may correlate with stasis or log drop targets derived from in vitro or animal systems.
The curves generated for the fractional probability that evernimicin will attain the AUC/MIC targets required for stasis or log decrease (3 log units and 1 log unit, respectively) in the infectious inoculum of pneumococci and enterococci indicate a high and consistent likelihood of microbiological efficacy for doses of 6 and 9 mg/kg/day. The differences between doses in the probability of success in reaching the PD target AUC for 90% Emax versus stasis or a 1-log drop for enterococci and a 2-log drop for staphylococci may indicate that the activity of the drug is more slowly bactericidal against enterococci and staphylococci. The time-killing kinetics are similar to those observed with vancomycin against susceptible strains of these pathogens.
From this model, it was predicted that higher doses of evernimicin may have acceptably high probabilities of achieving microbiological success. However, how each of these PD targets predicts microbiological efficacy and, subsequently, clinical efficacy is the subject of human clinical studies. It is also necessary to use clinical studies to validate which PD target is most valid for prediction of microbiological and, subsequently, clinical success, as well as for study of the safety and tolerance of all trial doses in patients with active disease. Clinical development of evernimicin was stopped after the completion of phase II/III trials on the basis of data which failed to show sufficient advantage of evernimicin for the treatment of infections caused by vancomycin-susceptible and -resistant gram-positive pathogens compared with the clinical safety and efficacy profiles of approved products. The results of these trials are consistent with the predictions made by these simulations.
In summary, we have delineated a method for using preclinical microbiological and animal model data along with PK data from early phase I studies to set reasonable targets for the drug in terms of exposure in plasma relative to the MIC for the organism and to explicitly factor in both PK and microbiological (MIC) variabilities to evaluate how often such a target is likely to be hit at different doses of drug. This method is also flexible enough to evaluate the impact of altering the schedule of administration. It can also be used to properly compare drugs of different classes (e.g., fluoroquinolones with macrolides or beta-lactams with aminoglycosides), as different targets will be set for each drug class. For evernimicin, it was clear that the 6-mg/kg/day dose provided exposures near the top of the response curve for all the organisms in the distribution and for all targets for the pneumococcus. In addition, the 6-mg/kg/day dose will provide a high likelihood of achieving stasis for all target pathogens. For the enterococci, the 9-mg/kg/day dose will likely be advantageous only when a true maximal effect is required (e.g., for bacteremic patients, the 90% Emax target). For S. aureus, the 9-mg/kg/day dose appears advantageous at the 2-log10-drop and 90% Emax targets. Results from controlled clinical trials appeared to validate these predictions (data on file, Schering-Plough Research Institute).
Appendix
The values displayed in Table 3 provide an estimate of the overall attainment of the microbiological target by the drug at the indicated dose. This estimate takes into account the variability of the drug exposure in the population, as embodied in the Monte Carlo simulation. It also takes into account the variability in the MIC of the drug for clinically appropriate pathogens, as embodied in the measured distribution of MICs for the pathogens (Fig. 2A to C).
The estimates are obtained in a straightforward manner. The example presented in Table A1 is for S. aureus for the 90% Emax endpoint for the 6-mg/kg/day dose. The values for target attainment are from Fig. 5C. One can then sum the data in the final column, giving a point estimate of the expected response rate (hence, the phrase “taking an expectation over the MIC distribution”). In this case, the estimate is 0.342. That is, 34.2% of subjects would be expected to attain the exposure target for evernimicin necessary to achieve 90% of the maximal bacterial killing effect (i.e., an AUC/MIC ratio of 830.8). This assumes that the distribution of the MICs for the organisms encountered in clinical trials are the same as those for the original collection and that the Monte Carlo simulation accurately reflects the drug's PK parameter distribution in patients. Readers should note that the value in Table 3 is 34.25%. The discrepancy comes from rounding errors in this example when only the mean value for target attainment is considered; for calculation of the data presented in Table 3, however, this analysis was performed five times, with the displayed value being the average of the values of these analyses.
TABLE A1.
Example of data used to determine response rates
| MIC (mg/liter) | Fraction of distribution at the indicated MIC | Fractional target attainment (90% Emax) at the MIC | Product of fractions |
|---|---|---|---|
| 0.016 | 0.001 | 1.0 | 0.001 |
| 0.023 | 0.001 | 1.0 | 0.001 |
| 0.032 | 0.003 | 1.0 | 0.003 |
| 0.047 | 0.002 | 1.0 | 0.002 |
| 0.064 | 0.015 | 1.0 | 0.015 |
| 0.094 | 0.048 | 1.0 | 0.048 |
| 0.125 | 0.124 | 0.996 | 0.124 |
| 0.19 | 0.099 | 0.736 | 0.073 |
| 0.25 | 0.145 | 0.38 | 0.055 |
| 0.38 | 0.159 | 0.083 | 0.013 |
| 0.5 | 0.162 | 0.034 | 0.0055 |
| 0.75 | 0.096 | 0.012 | 0.0012 |
| 1.0 | 0.077 | 0.004 | 0.0003 |
| 1.5 | 0.043 | 0.0 | 0 |
| 2.0 | 0.017 | 0.0 | 0 |
| 3.0 | 0.003 | 0.0 | 0 |
| 4.0 | 0.002 | 0.0 | 0 |
| 6.0 | 0.001 | 0.0 | 0 |
FIG. 5.
(A) Fractional attainment of the stasis target for S. aureus for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■). (B) Fractional attainment of the 2-log10 CFU drop target for S. aureus for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■). (C) Fractional attainment of the 90% Emax target for S. aureus for the 6-mg/kg dose (●) and the 9-mg/kg dose (▴). The interval MIC distribution information is included (■).
Other methods can be used. One could fit a distributional model to the MIC distribution for the organism and perform a double Monte Carlo simulation. While not incorrect, this process involves another fitting procedure. The simplest way of displaying the data in this case is with a cumulated frequency plot, with the target cutoff indicated.
REFERENCES
- 1.Blaser J, Stone B B, Groner M C, Zinner S H. Comparative study with enoxacin and netilmicin in a pharmacodynamic model to determine importance of ratio of antibiotic peak concentration to MIC for bactericidal activity and emergence of resistance. Antimicrob Agents Chemother. 1987;31:1054–1060. doi: 10.1128/aac.31.7.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig W A. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis. 1998;26:1–10. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 3.D'Argenio D Z, Schumitzky A. ADAPT II user's guide. Los Angeles: University of Southern California; 1995. [Google Scholar]
- 4.Drusano G L, Aweeka F, Gambertoglio J, Jacobson M, Polis M, Lane H C, Eaton C, Martin-Munley S. Relationship between foscarnet exposure, baseline cytomegalovirus blood culture and the time to progression of cytomegalovirus retinitis in HIV-positive patients. AIDS. 1996;10:1113–1119. [PubMed] [Google Scholar]
- 5.Drusano G L, Bilello J A, Stein D S, Nessly M, Meibohm A, Emini E A, Deutsch P, Condra J, Chodakewitz J, Holder D J. Factors influencing the emergence of resistance to indinavir: role of virologic, immunologic, and pharmacologic variables. J Infect Dis. 1998;178:360–367. doi: 10.1086/515631. [DOI] [PubMed] [Google Scholar]
- 6.Drusano G L, Johnson D E, Rosen M, Standiford H C. Pharmacodynamics of a fluoroquinolone antimicrobial in a neutropenic rat model of Pseudomonas sepsis. Antimicrob Agents Chemother. 1993;37:483–490. doi: 10.1128/aac.37.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fantin B, Leggett J, Ebert S, Craig W A. Correlation between in vitro and in vivo activity of antimicrobial agents against gram-negative bacilli in a murine infection model. Antimicrob Agents Chemother. 1991;35:1413–1422. doi: 10.1128/aac.35.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrest A, Nix D E, Ballow C H, Goss T F, Birmingham M C, Schentag J J. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob Agents Chemother. 1993;37:1073–1081. doi: 10.1128/aac.37.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber A U, Craig W A, Brugger H P, Feller C, Vastola A P, Brandel J. Impact of dosing intervals on activity of gentamicin and ticarcillin against Pseudomonas aeruginosa in granulocytopenic mice. J Infect Dis. 1983;147:910–917. doi: 10.1093/infdis/147.5.910. [DOI] [PubMed] [Google Scholar]
- 9a.Jones, R. N., R. S. Hare, F. J. Sabatelli, and the Ziracin Susceptibility Testing Group. In vitro Gram-positive antimicrobial activity of evernimicin (SCH 27899), a novel oligosaccharide, compared with other antimicrobials: a multicentre international trial. J. Antimicrob. Chemother., in press. [DOI] [PubMed]
- 10.Jones R N, Marshall S A, Erwin M E. Antimicrobial activity and spectrum of SCH27899 (Ziracin) tested against gram-positive species, including recommendations for routine susceptibility testing methods and quality control. Quality Control Study Group. Diagn Microbiol Infect Dis. 1999;34:103–110. doi: 10.1016/s0732-8893(98)00093-5. [DOI] [PubMed] [Google Scholar]
- 11.Kashuba A D, Nafziger A N, Drusano G L, Bertino J S., Jr Optimizing aminoglycoside therapy for nosocomial pneumonia caused by gram-negative bacteria. Antimicrob Agents Chemother. 1999;43:623–629. doi: 10.1128/aac.43.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall S A, Jones R N, Ewin M E. Antimicrobial activity of SCH27899 (Ziracin), a novel everninomycin derivative, tested against Streptococcus spp.: disk diffusion-test method evaluations and quality control guidelines: Quality Control Study Group. Diagn Microbiol Infect Dis. 1999;33:19–25. doi: 10.1016/s0732-8893(98)00105-9. [DOI] [PubMed] [Google Scholar]
- 13.Preston S L, Drusano G L, Berman A L, Fowler C L, Chow A T, Dornseif B, Reichl V, Natarajan J, Corrado M. Pharmacodynamics of levofloxacin: a new paradigm for early clinical trials. JAMA. 1998;279:125–129. doi: 10.1001/jama.279.2.125. [DOI] [PubMed] [Google Scholar]
- 14.Schentag J J, Smith I L, Swanson D J, DeAngelis C, Fracasso J E, Vari A, Vance J W. Role for dual individualization with cefmenoxime. Am J Med. 1984;77:43–50. doi: 10.1016/s0002-9343(84)80074-1. [DOI] [PubMed] [Google Scholar]
- 15.Schumitzky A. Nonparametric EM algorithms for estimating prior distributions. Appl Math Comput. 1991;45:141–157. [Google Scholar]
- 16.Yamaoka K, Nakagawa T, Uno T. Application of Akaike's Information Criterion (AIC) in the evaluation of linear pharmacokinetic equations. J Pharmacokinet Biopharm. 1978;6:165–175. doi: 10.1007/BF01117450. [DOI] [PubMed] [Google Scholar]