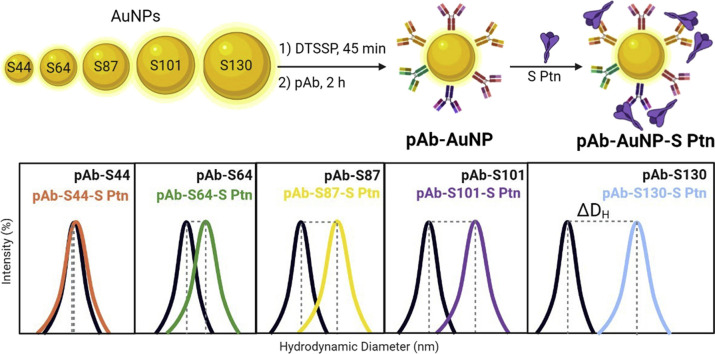
Abstract
Due to the unprecedented and ongoing nature of the coronavirus outbreak, the development of rapid immunoassays to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its highly contagious variants is an important and challenging task. Here, we report the development of polyclonal antibody-functionalized spherical gold nanoparticle biosensors as well as the influence of the nanoparticle sizes on the immunoassay response to detect the SARS-CoV-2 spike protein by dynamic light scattering. By monitoring the increment in the hydrodynamic diameter (ΔDH) by dynamic light scattering measurements in the antigen–antibody interaction, SARS-CoV-2 S-protein can be detected in only 5 min. The larger the nanoparticles, the larger ΔDH in the presence of spike protein. From adsorption isotherm, the calculated binding constant (KD) was 83 nM and the estimated limit of detection was 13 ng/mL (30 pM). The biosensor was stable up to 90 days at 4 °C. Therefore, the biosensor developed in this work could be potentially applied as a fast and sensible immunoassay to detect SARS-CoV-2 infection in patient samples.
Keywords: Immunoassay, Gold nanoparticles size effect, Dynamic light scattering, SARS-CoV-2 spike protein
Graphical abstract
1. Introduction
The development of fast, low-cost, and easy manipulation diagnostic tests for viruses and related diseases has always been of great interest in science and industry. The SARS-CoV-2 outbreak in late 2019 highlighted, even more, this necessity [1,2]. The recent Omicron variant has infected the population all over the globe with an unprecedented transmission scale and submerging health systems [3]. The most common diagnostic tests for viruses can be divided into three major groups: the amplification of specific regions of the viral RNA (molecular), rapid antibody tests, and rapid antigen tests [4]. Molecular detection tests, such as PCR (Polymerase Chain Reaction), are the gold standard due to high specificity and sensibility. However, the PCR test involves several processing steps, long analysis time, high cost, and requires highly trained personnel, which can be a bottleneck in terms of scale and population coverage particularly in poor countries. SARS-CoV-2 antibody rapid tests – also referred to as serology – usually detect the antibodies in 17–19 days after symptom onset in the patients, which limits their use for containing pandemic spread [5]. There are available various antigen rapid diagnostic tests (Ag-RDTs), which can detect the SARS-CoV-2 proteins in 15–20 min visually or using small portable devices [6]. These tests are highly specific; however, they are not sensitive as the PCR. Nevertheless, Ag-RDTs can fast track and identify COVID-19 disease in individuals. Therefore, the development of cheap Ag-RDTs is of great interest [4,7].
Gold nanoparticles (AuNPs) have been widely explored as transducers in biosensors applied for detection of viruses [8], bacteria [9], metals [10,11], DNA [12], chemical warfare agents [13], contaminants [14], and for the early diagnosis of cancer [15,16] and heart disease [17]. The successful use of AuNPs for sensing applications is mainly related to their facile synthesis and functionalization with various molecules, including the biological ones and their unique optical properties from the localized surface plasmon resonance (LSPR) [[18], [19], [20]]. This phenomenon occurs when the radiation forms a dipole in metallic nanoparticles, with a consequent restoring force to compensate it [21]. The result is a coherent collective oscillation of electrons in a specific resonant frequency [22]. The resonance condition only appears when the frequency of the incident radiation matches the frequency of the oscillating electrons [23]. This frequency depends on the nanoparticle composition, size, and shape, which are the parameters commonly used to control their optical properties. However, the dielectric properties of the environment, the presence of ligands at the surface, and the interparticle approximation generally can have more significant effects on the resonance band than the size [21]. The influence of the surrounding medium or ligands is related to their refractive index, whereas the interparticle distance is important due to the coupling of plasmon resonances, which leads to the formation of the so-called ‘hot-spots’, broadening the LSPR band and causing electromagnetic field enhancements [24]. The control of all these parameters is key to the application in diagnostics.
When a substance of interest (analyte) causes the aggregation of AuNPs, it induces a color change, which can be visualized with the ‘naked’ eye to detect its presence. There are also reports of biosensing based on LSPR shift caused by changes in the surrounding refractive index [22,25]. This is only possible because the energy levels of d-d transitions lies in the visible range of the spectrum of AuNPs [26]. The control of color changes is not the only aspect that can be explored from LSPR. This phenomenon also causes an enhancement of the electric fields close to the surface of the particles, making them excellent scatters [22]. Considering the equation which describes Rayleigh scattering, the intensity of light scattered by small particles (<λ/20) is proportional to the size of the particles to the sixth power [27]. However, for particles with LSPR, Rayleigh scattering can be many orders of magnitude larger than for non-plasmonic particles. The increase in size results in an increase of the extinction cross-section, and consequently, intensifies the relative contribution of scattering. For AuNPs with sizes up to 80 nm, the optical extinction is dominated by absorption, but scattering takes place when their sizes are larger than that [[28], [29], [30]].
Dynamic light scattering (DLS) also known as photon correlation spectroscopy or quasi-elastic light scattering is a versatile, sensible, and easy operation technique that gives size distributions of colloidal systems [31,32]. The combination of DLS with plasmonic nanoparticles as detection method has receiving attention since the pioneering work of Qun Huo et al. in 2008 on the quantification of prostate specific antigen for cancer diagnosis [33]. They reported AuNPs-based one-step immunoassays capable to detect aggregation occurring in the presence of prostate specific antigen in a 1 ng/mL concentration. Since then, DLS and plasmonic nanoparticles have been used for the detection of many other species, such as other cancer biomarkers [34,35], DNA [36], glucose [37], acetylcholinesterase inhibitors [38], drugs [39,40], metal ions [31,41,42], and particles in gases [43]. Regarding virus detection, DLS-based biosensors have been less explored. The open literature reports successful detection of influenza (H1N1) [8], hepatitis B [44], and SARS-CoV-2 virus [45]. Regarding SARS-CoV-2 protein detection, various biosensors based on the above mentioned LSPR properties of AuNPs have been recently reported in the open literature, such as colorimetric [46,47], electrochemical [48], electrochemiluminescent [49], opto-microfluidic [50] and so on [51,52]. However, to the best of our knowledge, a different-sized AuNPs study combined with DLS as read-out method for SARS-CoV-2 S-protein detection have not been reported in the open literature.
Herein, the size effect of spherical AuNPs capped with horse polyclonal anti-spike antibody (pAb-AuNPs) on SARS-CoV-2 spike protein (S protein) detection using the DLS technique is reported. The AuNPs were named using their sizes determined using transmission electron microscopy (TEM): S44 (44 ± 4 nm), S64 (64 ± 5 nm), S87 (87 ± 10 nm), S101 (101 ± 10 nm), and S130 (130 ± 13 nm). The biosensor response was evaluated by placing the pAb-AuNPs with S protein and reading the average hydrodynamic diameter shift (ΔDH) caused by the antigen–antibody recognition. The ΔDH was correlated to the concentration of S protein in the medium and the largest nanoparticle pAb-S130 provided the best results. Also, we determine the stability, reproducibility, binding constant, and limit of detection (LOD) of the pAb-S130 biosensor.
2. Experimental procedures
2.1. Materials and methods
Chemicals were purchased from Sigma-Aldrich and used as received: gold (III) chloride hydrate (99.995%), sodium citrate dihydrate (99%), boric acid (99.8%), sodium tetraborate decahydrate, 3,3′-dithiobis (sulfosuccinimidyl propionate) – DTSSP, bovine serum albumin lyophilized powder (BSA). Buffer solutions were filtered with polyethersulfone membrane 0.22 μm pore size (Kasvi) prior to the use. AuNPs, pAb-AuNPs were stocked at 4 °C in Falcon or low-binding microcentrifuge tubes. All glassware was washed with aqua regia and deionized water prior to the use. Horse hyperimmune serum was produced by Instituto Vital Brasil (IVB-Brazil) after animal inoculation with the SARS-CoV-2 S protein [53]. The polyclonal antibody (pAb) was purified using affinity chromatography and 1 mg/mL solutions were stocked at −18 °C. The trimeric S protein of SARS-CoV-2 produced in stable recombinant HEK293 cells as described in Alvim et al. was provided by the Cell Culture Engineering Laboratory – LECC from the Universidade Federal do Rio de Janeiro - UFRJ) [54].
The samples were centrifuged in low binding microcentrifuge tubes using a Mikro 200R (Hettich) centrifuge in two conditions:(i) 9000 rpm (7690 RCF) at 4 °C for 4 min or (ii) 5000 rpm (2370 RCF) at 10 °C for 4 min. All samples were purified at each synthetic step by two centrifugation/washing cycles (borate buffer pH 8.9, 2 mM). For the DLS measurements, a Zetasizer Nano-ZS90 (Malvern) operating at 90° fitted with a 633 nm ‘red’ laser was used. The samples were placed in 50 μL or 1 mL polystyrene cuvettes and analyzed at room temperature (18 °C). Each measurement was carried with 10 runs of 10 s in triplicate to calculate the average and standard deviation. The hydrodynamic diameters (DH) and PDI (polydispersity index) reported in this work are the Z-average obtained from the Malvern Zetasizer 8.01.4906 software. Visible spectra of the dispersions were acquired using a Cary 60 UV–Vis Spectrophotometer (Agilent) at room temperature using 1 mL quartz cuvettes. TEM images were obtained in a Tecnai Spirit microscope operating at 120 kV. For citrate-coated AuNPs samples, one drop of the analyzed dispersions was placed on FCF300–Cu/formvar/carbon film grids and air-dried. AuNPs diameters were determined using ImageJ software (1.53e version) and the histograms of particle-size distributions were plotted for each sample. Attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectra were obtained in the range of 4000–500 cm−1 with a spectral resolution of 4 cm−1 using a Thermo Nicolet iS50 instrument. For ATR-FTIR analysis, pAb-S44, pAb-S64, pAb-S87, pAb-S101, pAb-S130 biosensors with and without BSA the precursors were concentrated by centrifugation and air-dried.
2.2. Citrate-coated gold nanoparticle synthesis
Citrate-coated AuNPs were synthetized using the seeded-growth method with minor modifications [55]. Briefly, to grow Au seeds, 1 mL of 25 mM HAuCl4 aqueous solution was added into a 500 mL round-bottom flask containing 150 mL of 2.2 mM Na3C6H5O7 aqueous solution under stirring. The reaction remained under reflux for 30 min, then cooled to 90 °C, followed by the addition of 2 mL of 60 mM Na3C6H5O7 and 1 mL of 25 mM HAuCl4 solution. Subsequent additions of HAuCl4 (1 mL, 25 mM) solutions were repeated for 2 times in an interval of 30 min, resulting in one growing cycle. After each growing cycle, a dilution process was carried out by extracting 55 mL of the reaction medium, followed by the addition of 53 mL of deionized H2O. Then, a new growing cycle was initiated by adding the same volume of Na3C6H5O7 (2 mL) and HAuCl4 (3 x 1 mL). We conducted this procedure 9 times up to reach the desired particle sizes. Each extracted aliquots were characterized by TEM, visible spectroscopy and DLS. The AuNPs were named using their sizes determined by TEM: S44 (44 ± 4 nm), S64 (64 ± 5 nm), S87 (87 ± 10 nm), S101 (101 ± 10 nm), and S130 (130 ± 13 nm). The method used to calculate the nanoparticle concentration is described in the Supplementary Data.
2.3. General procedure for the synthesis of anti-spike antibody-coated AuNPs
Each gold nanoparticle dispersion (300 μL of S44, S64, S87, S101, and S130) was added to a low-binding microcentrifuge tube followed by the addition of deionized H2O (800 μL). The tube was centrifugated according to the conditions previously described. Then 1.0 mL of the supernatant was carefully removed, followed by the addition of deionized H2O (750 μL) and sodium borate buffer (50 μL, 50 mM, pH 8.9). DTSSP solution (20 μL, 10 mM) was added to the tube, which was shaken for 45 min at 18 °C. The dispersion was centrifugated and washed with 2 mM sodium borate buffer (2x). Variable volumes of pAb anti-S protein were added to the tubes and incubated for 2 h at 18 °C. To purify, the dispersions were washed with 2 mM sodium borate buffer (2x). Then, sodium borate buffer containing 0.25% w/v BSA solution, previously filtered in polyethersulfone membrane, was added to the dispersions, yielding the anti-spike pAb-coated AuNPs. The samples were stored at 4 °C.
2.4. General procedure for the pAb-AuNP immunoassay
The pAb-AuNPs (10 μL), phosphate buffered saline (PBS) solution (80 μL, 150 mM), and variable volumes of S protein (0.65 mg/mL dispersed in PBS and 300 mM of imidazole) in low binding microcentrifuge tubes were incubated for 30 min at 18 °C in an orbital shaker at 300 rpm. The dispersions were transferred to 50 μL polystyrene disposable cuvettes and analyzed by DLS at 18 °C. Control experiments were performed by measuring the hydrodynamic diameters (DH) of the dispersions without the spike protein (S protein = S Ptn) in the same conditions and medium. The interactions between all biosensors and S protein (immunoassay) were represented as the hydrodynamic size difference between the DH of pAb-AuNP-S-protein and the DH of the pAb-AuNP control experiment (ΔDH = DH of pAb-AuNP-S-protein – DH of the pAb-AuNP control experiment). The LOD was calculated from the adsorption isotherm (Fig. 6a) and determined using the first observed point with ΔDH value 3 times above the standard deviation of the immunoassay without S protein [56].
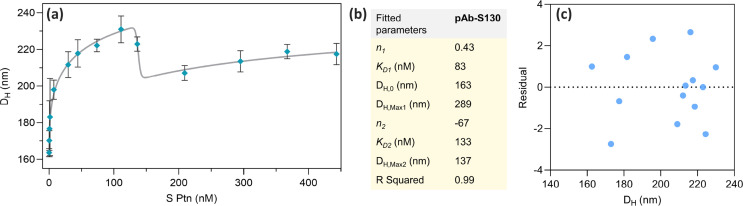
Fig. 6.
(a) S Ptn adsorption isotherm on the pAb-S130 biosensor fitted using Eq. (3); (b) table containing the parameters obtained from Eq. (3); (c) residual plot obtained from the fit of the adsorption isotherm. pAb, polyclonal antibody.
2.5. Model employed to fit the data obtained from the immunoassay
The immunoassay can be described as an analytical method in which the quantification of an analyte is realized through its recognition by an antibody. More specifically, it involves the reversible binding through non-covalent interactions of a receptor (R = IgG) to a ligand (L = S Ptn), originating an immune complex (RL). The affinity of the ligand by the receptor is given by the dissociation constant, K D. This constant can be obtained from the ratio K off/K on at the equilibrium, Eq. (1) [[57], [58], [59]].
| (1) |
The binding affinity can be determined using an adsorption isotherm, in which a variable amount of the ligand is added to a fix concentration of the receptor. The formation of an immune complex can be measured by a diversity of methods and the signal detected for [RL] depends on the ligand concentration in a non-linear manner, usually producing sigmoidal curves. A model commonly used to fit the data is the four-parameter logistic (4-PL), also named the Hill equation, Eq. (2) [60,61].
| (2) |
In Eq. (2), DH is the hydrodynamic diameter (nm) for a given value of x = [S Ptn]. DH,0 is the hydrodynamic diameter when the concentration of S protein is zero (the minimum asymptote). DH,Max is the DH for an infinite concentration of S Ptn, or the maximum asymptote. K D is the dissociation constant, or the concentration that provides the half-response (sometimes referred as EC50), and n is the slope, or the Hill coefficient of cooperativity. When n is > 0, the curve is ascendant and indicates a stimulatory effect; and when n is < 0, the curve is descendent and the effect is inhibitory.
As one-step immunoassays often present the hook effect at high concentration of the analyte, their curves present a bell aspect and are not well-described by a simple logistic curve. To overpass this difficult, we employed an approach that consists the sum of two logistic equations, each one with different n, K D and D H,Max, Eq. (3) [62].
| (3) |
The immunoassay was fitted employing non-linear regression with the software GraphPad Prism version 9.3.1.471.
3. Results and discussions
3.1. Synthesis of AuNPs and pAb-AuNPs and mAb-AuNPs
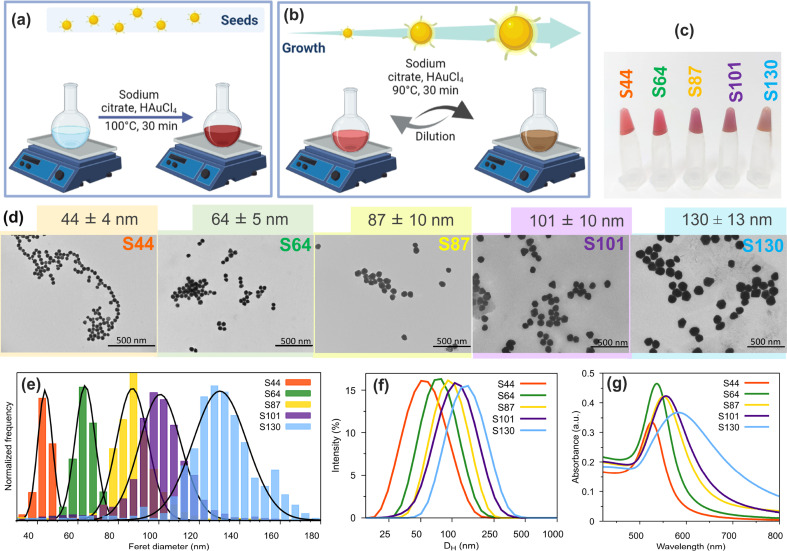
To study the influence of gold nanoparticle sizes (AuNPs) in the biosensor response monitored by DLS, we synthesized spherical citrate-coated AuNPs with diameters ranging from 44 to 130 nm using the seeded-growth method with minor adaptations (Fig. 1 a and b). With this method, monodispersed nanoparticles of various sizes can be synthesized by the kinetic control of the reaction conditions, such as the amount of Au atoms added to the reaction medium [55]. To synthesize the biosensors, we selected five different sizes of the nanoparticles measured by TEM: S44 (44 ± 4 nm), S64 (64 ± 5 nm), S87 (87 ± 10 nm), S101 (101 ± 10 nm), and S130 (130 ± 13 nm). All synthesized AuNPs show monomodal size distributions and spherical shapes, as evidenced by TEM images (Fig. 1d) and DLS analysis (Fig. 1f). The distributions of hydrodynamic diameters (DH) represented by particle volume and number are also monomodal and without aggregates for all citrate-coated AuNPs (Figs. S3a–e, Supplementary Data). The colors of AuNPs dispersions changed from red to purple and the plasmonic resonance peak broadened and shifted from 525 to 583 nm with their growth (Fig. 1c and g).
Fig. 1.
Synthesis of AuNPs: (a), (b) reaction scheme, (c) photography of the AuNP dispersions as synthesized, (d) TEM images (magnification – 46000 × , scale bars 500 nm), (e) size distributions and Gaussian fits (TEM), (f) distribution of hydrodynamic diameters (DH) measured by DLS (dilution in water 1:10), (g) visible spectra (dilution in water 1:10). AuNP, gold nanoparticle; DLS, dynamic light scattering; TEM, transmission electron microscopy.
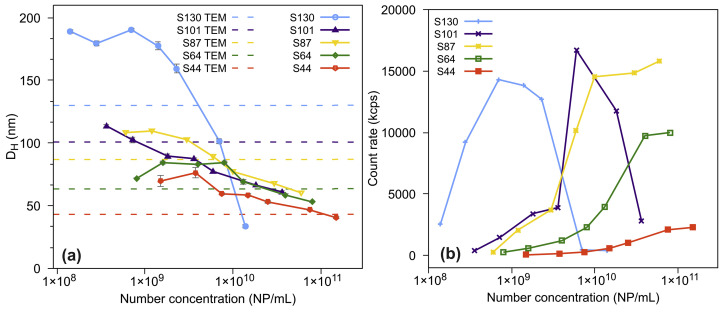
The average hydrodynamic diameters (DH) (Table 1 ) of all citrate-coated AuNPs obtained from DLS show an interesting result. The nanoparticle diameters of S44 and S64 are smaller by TEM than by DLS (DH), as expected. However, for the larger AuNPs, S87, S101, and S130, the hydrodynamic diameters are smaller than those measured by TEM. Zheng et al. associated this effect to multiple scattering mainly observed for nanoparticles larger than 80 nm in concentrated dispersions [63]. To avoid misinterpretation of the average hydrodynamic diameters of the synthesized AuNPs exceeding 80 nm, we performed serial dilutions of the suspensions and the DH was measured by DLS (Fig. 2 a).
Table 1.
Data obtained from citrate-coated AuNPs dispersions and from the biosensors (pAb-AuNPs).
| Size | DHa AuNP/nm | PDIb | λmaxc/nm | [AuNP]/NP /mL | [AuNP]/μg/mL | Sd/nm2/NP | Sd/nm2/mL | DHa DTSSP-AuNP/nm | DHa pAb-AuNP/nm (35 μg/mL) |
|---|---|---|---|---|---|---|---|---|---|
| S44 | 49 ± 12 | 0.22 | 525 | 21 × 1010 | 175 | 6 × 103 | 13 × 1014 | 50 ± 12 | 70 ± 18 |
| S64 | 65 ± 15 | 0.21 | 536 | 8.4 × 1010 | 222 | 13 × 103 | 11 × 1014 | 61 ± 17 | 85 ± 20 |
| S87 | 74 ± 18 | 0.25 | 552 | 3.6 × 1010 | 241 | 24 × 103 | 8.6 × 1014 | 91 ± 22 | 117 ± 28 |
| S101 | 88 ± 22 | 0.26 | 556 | 2.4 × 1010 | 246 | 32 × 103 | 7.7 × 1014 | 95 ± 25 | 109 ± 26 |
| S130 | 121 ± 27 | 0.19 | 583 | 1.1 × 1010 | 251 | 53 × 103 | 5.8 × 1014 | 128 ± 30 | 146 ± 30 |
Hydrodynamic diameters.
Polydispersity index (PDI) of citrate-coated AuNPs.
Plasmonic resonance peak.
Surface area of AuNPs.
Fig. 2.
(a) DH of the serial dilutions of AuNPs measured by DLS; and (b) scattering (DLS) for the serial dilutions of AuNPs (derived count rate). AuNP, gold nanoparticle; DLS, dynamic light scattering.
The results showed that the DH was strongly underestimated for more concentrated dispersions of S101 and S130, reaching sizes smaller than those obtained using TEM. Fig. 2b shows that the scattered light intensity detected for S44–S87 increases with AuNP concentrations. However, for S101 and S130, the scattered light intensity reaches a maximum at 0.6 × 1010 and 0.7 × 109 NP/mL, respectively, and then decreases confirming multiple scattering at higher concentrations. Therefore, for the S101 and S130, we worked at concentrations below 0.6 × 1010 and 0.7 × 109 NP/mL, respectively, to avoid errors in the DH measurements and, consequently, misinterpreted results due to multiple scattering. For practical applications, in the experimental design in which gold aggregate formation measured using DLS will be used for chemical or biological sensing applications, the optimal concentration of the nanoparticles must be investigated in each case [63]. All DLS dataset obtained from the serial dilutions of the AuNP dispersions are shown in (Figs. S4a–c Supplementary Data).
The concentrations of the AuNPs were calculated in micrograms per mL (μg/mL, Au mass) and in nanoparticle per mL (NP/mL) (Table 1). As the particle grows from S44 to S130, the Au concentration (μg/mL) in the dispersions increases and the number of particles per unit of volume decreases. As the diameter increases, the surface area of each NP also increases (nm2/NP). The total surface area should become even larger in the latter cycles of growth. Nonetheless, the number of particles decreases at each dilution cycle because of both volume extraction and HAuCl4 solution addition. The gold added to the dispersion will recover the particle surface instead of forming new nucleus.
The total surface area per volume unit available to be functionalized ranges from 5.8 to 13 × 1014 nm2/mL (Table 1). The nanoparticle surface areas (S) do not significantly change with their sizes; therefore, we decided to prepare the biosensors using the same amount of DTSSP (20 μL of a 10 mM aqueous solution) for all AuNPs.
Citrate-coated AuNPs (S44 to S130) reacted with DTSSP. This molecule is a crosslinker broadly used for covalent anchoring of proteins onto gold surfaces [64]. The molecular structure presents a dithiol group in the central moiety, which is cleaved under basic pH and allows the free sulfur atoms strongly bind onto AuNP surface. The hydrodynamic diameters of DTSSP-AuNPs slightly increase compared to the citrate-coated AuNPs (Table 1) because both molecules (citrate and half DTSSP) show similar estimated sizes (Fig. 3 a). The hydrodynamic diameters of the DTSSP-AuNPs were 10% larger (average) than those observed for the citrate-coated AuNPs.
Fig. 3.
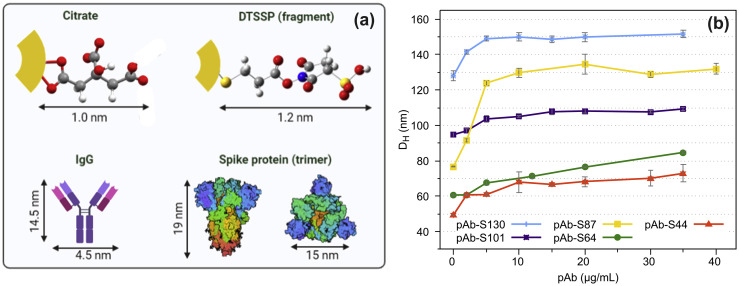
(a) Estimated sizes to citrate molecule [65], a fragment of the crosslinker (DTSSP) molecule, [66], IgG [67] antibody and spike protein (trimer) [68] and (b) DH measured by DLS as function of pAb concentrations. DLS, dynamic light scattering; DTSSP, dithiobis(sulfosuccinimidyl propionate); pAb, polyclonal antibody.
The next step was to bind the polyclonal IgG antibody to DTSSP-AuNPs. The free electron pair of the amino group on IgG can attack the carbonyl group of DTSSP-AuNP forming a stable amide after releasing the N-hydroxysulfosuccinimide leaving group (Fig. S5, Supplementary Data). ATR-FTIR spectra of the pAb-AuNP biosensors show the most common absorption bands of proteins, such as Amide I and II from the peptide groups, CH2 and CH3 vibrations, and C–NH2 stretching from Amide III (Fig. S6, Supplementary Data) [69]. Therefore, the ATR-FTIR spectra confirm that the IgG antibody is anchored on AuNP surface.
The optimal concentration for coating polyclonal IgG antibody (pAb) on DTSSP-AuNPs was investigated by ranging the antibody concentration from 5 to 35 μg/mL and measuring the DH (Fig. 3d). The hydrodynamic diameters of all nanoparticles increased as the concentration of the IgG increased, then they stabilized reaching a plateau when the concentration of IgG was around 10 or 20 μg/mL. The polyclonal IgG antibody concentration employed to prepare the biosensors used in the immunoassay experiments was 35 μg/mL because in this concentration, we could ensure that all nanoparticles were recovered. The biosensors (pAb-S44, pAb-S64, pAb-S87, pAb-S100, pAb-S130) were stored in 0.25% BSA solution prepared in borate buffer (pH 8.9).
3.2. Immunoassay with S protein
To investigate the S protein recognition by the biosensors with different sizes (pAb-S44, pAb-S67, pAb-S87, pAb-S101, pAb-S130), they were (10 μL) incubated with S protein (0.65 mg/mL, 10 μL) in PBS buffer (80 μL) for 30 min, and then their hydrodynamic sizes (DH) were measured using DLS (Fig. 4 a). Control experiments for each biosensor (pAb-S44, pAb-S67, pAb-S87, pAb-S101, pAb-S130) without S protein in PBS buffer and 300 mM imidazole were carried out and the hydrodynamic sizes were measured to calculate the DH shift.
Fig. 4.
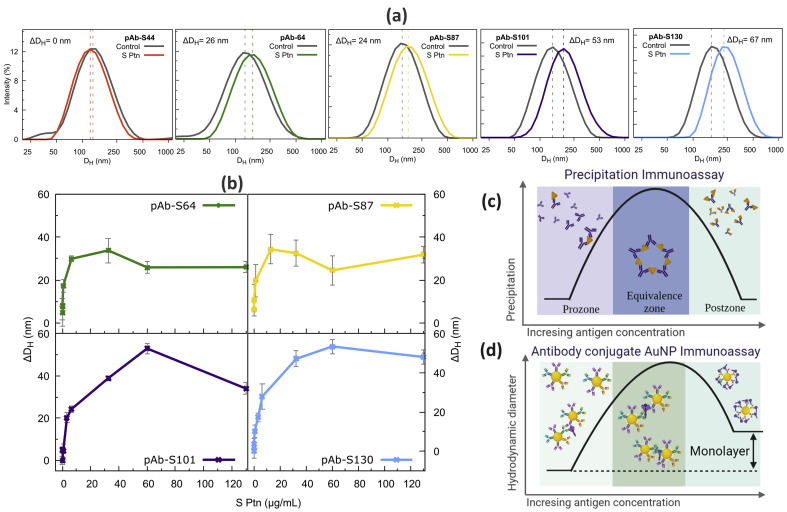
Immunoassay with S Ptn: (a) ΔDH measured in the presence of 65 μg/mL S Ptn for all biosensors; (b) S Ptn adsorption isotherm on the pAb-AuNP biosensors; (c) precipitation immunoassay scheme; and (d) scheme showing the DH variation for antibody conjugate AuNP immunoassay as function of antigen concentration. AuNP, gold nanoparticle; pAb, polyclonal antibody; S Ptn, spike protein.
Fig. 4a shows that the hydrodynamic sizes (DH) of the biosensors increase in the presence of 0.65 mg/mL of S protein confirming its recognition by the pAb-AuNP systems. The pAb-S44 biosensor results were discharged because the DH of its control experiment – the pAb-S44 dispersion in PBS buffer medium without S protein – also increased, resulting in a negligible variation of ΔDH (ΔDH = DH of pAb-S44-S-protein – DH of the pAb-S44 control experiment); therefore, the pAb-S44 biosensor is not stable in the investigated medium (PBS buffer and 300 mM imidazole, control experiment). For the larger nanoparticles, the differences between the DH with S protein and the DH without it (control), ΔDH, are bigger than for the smaller ones. To explain these results, we obtained the adsorption isotherm by measuring the ΔDH for the biosensors as function of S protein concentration, Fig. 4b.
Fig. 4b shows that pAb-S64 to pAb-S130 biosensors exhibit similar behavior with the increase of S protein concentration, which are an abrupt increase in the ΔDH with low S protein concentrations, followed by a maximum in the ΔDH, then the values decline and finally stabilize at high S protein concentrations. Such phenomenon is usually called the high-dose hook effect [70].
To illustrate the hook effect, we can make an analogy with the precipitation immunoassay of polyclonal antibodies and proteins, Fig. 4c. In this type of assay, three distinct zones can be defined: (i) the prozone, with the lowest antigen concentration; (ii) the equivalence zone, where the precipitation takes place; and (iii) the postzone, with high antigen concentrations. In the equivalence zone, there is an optimal concentration of both antigen and antibody – which depends on the nature and the specificity of the interaction between them – where crosslinking occurs, and the aggregates are formed. At very high concentrations of the antigen, all interaction sites of the antibodies are occupied, and agglomeration process is inhibited, therefore, precipitation is avoided. When antibodies are anchored on AuNP surface, many factors can affect the antibody–antigen complex formation. For example, IgG structure has two binding sites in its Fab portion (paratopes) and they sterically influence each other. With the confinement on the nanoparticle surface, the binding sites from neighboring antibodies can influence each other as well. Moreover, the IgG can also adopt different conformations allowing an unpredictable number of paratopes available to interact. Similarly to the precipitation immunoassay, increasing the amount of S protein, an increase in the ΔDH is noted and then the system reaches a maximum in ΔDH (Fig. 4b–d). After that, any extra increment of S protein decreases ΔDH up to reach an approximately constant value. This result can be associated to a postzone-like region in the hook effect, in which there is the formation of S protein monolayer around pAb-AuNPs inhibiting their aggregation. In the postzone-like region – where the S protein monolayer is formed around pAb-AuNPs – the ΔDH is in the range of 30–40 nm for the pAb-S64 to pAb-S130 biosensors, these shifts correspond to ca. twice the size of the S protein (trimer, see Fig. 3a) [68], Figs. 4b–d.
Furthermore, the ΔDH was bigger to pAb-S101 and pAb-S130 than to the smaller biosensors, Fig. 4a. The agglomeration in the equivalence zone of pAb-AuNP immunoassay mainly occurs due to dimer formation, e.g. two pAb-AuNPs and one S protein, thus, the larger the nanoparticles, the bigger the dimers [71]. Also, those larger pAb-AuNP biosensors require higher concentrations of S protein to reach the postzone region – where the saturation of the binding sites takes place – which accounts for the broader equivalence zone observed in Fig. 4b for pAb-S101 and pAb-S130. Therefore, the pAb-S101 and pAb-S130 biosensors show broader antigen quantification regions (dynamic range).
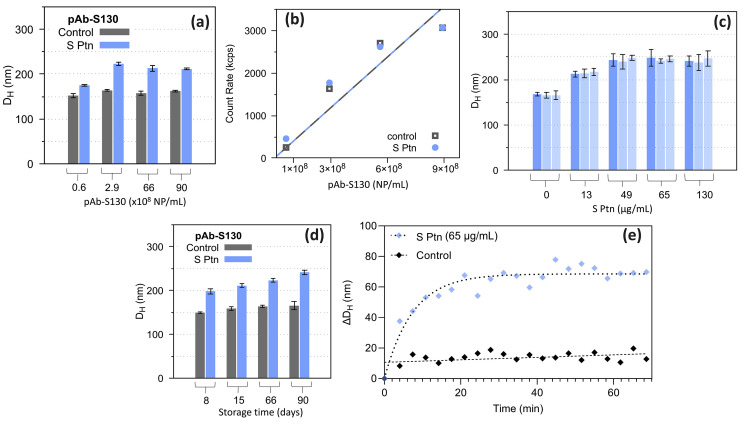
On the light of the mentioned results and considering the higher scattering power of larger AuNPs, we selected the pAb-S130 biosensor for further investigations. As in the case of S130 AuNPs, we also analyzed the optimal concentration range of the pAb-S130 biosensor with S protein (immunoassay) and without it to avoid multiple scattering, which could lead to misinterpretation on the DH [63]. To find the ideal pAb-S130 concentration range for the immunoassay, we kept the S protein concentration constant at 65 μg/mL and we changed the concentration of the pAb-S130 biosensor from 0.6 × 108 to 8.7 × 108 NP/mL and the DH was measured, Fig. 5 a. For the control experiment, DH was measured without S protein in the same conditions. The DH of the control experiment did not significantly change as the pAb-S130 concentration increased, indicating no biosensor aggregation in this concentration range. The count rate of photons for the control experiment and for the immunoassay (Fig. 5b) shows a linear answer from 0.6 to 8.7 × 108 NP/mL evidencing no multiple scattering in this concentration range, therefore, a good working concentration for DLS measurements. For the immunoassay, however, the pAb-S130 concentration of 0.6 × 108 NP/mL resulted in an almost negligible shift in the DH with S protein (Fig. 5a). Therefore, considering the interval investigated, the optimal pAb-S130 biosensor concentration for the immunoassay is in the range from 2.9 × 108 to 8.7 × 108 NP/mL. The pAb-S130 concentration employed in the following experiments were 2.9 × 108 NP/mL. Fig. S7 (Supplementary Data) shows DLS dataset obtained from the serial dilutions of the pAb-S130 biosensor.
Fig. 5.
pAb-S130 immunoassay: (a) DH as function of pAb-S130 concentration without S Ptn (control) and with 65 μg/mL of S Ptn; (b) scattering (count rate) measured for different concentrations of pAb-S130 without S Ptn (control) and with 65 μg/mL of S Ptn; (c) reproducibility experiment; (d) colloidal stability over the time; (e) ΔDH measured versus time for pAb-S130 with 65 μg/mL Ptn S and without S Ptn (control), dashed lines are only eye guides. pAb, polyclonal antibody; S Ptn, spike protein.
Fig. 5c shows the DH reproducibility for pAb-S130 measured in the presence of different concentrations of S protein. All experiments were performed in triplicates for each concentration of S protein and showed good reproducibility. The colloidal stability of pAb-S130 biosensor (stored at 4 °C) regarding DH was monitored over 90 days, Fig. 5d. The results of the control experiment (without S protein) reveal that the DH of the pAb-S130 biosensor remains almost the same. Furthermore, upon the addition of S protein (65 μg/mL) to the stored biosensor, the DH increased showing pAb-S130 remains active for at least 90 days.
We also carried out an experiment to determine the minimum time required to detect DH variation upon the addition of 65 μg/mL of S protein in the pAb-S130 biosensor (Fig. 5e). Control experiment without S protein was also conducted. The results show that after 5 min upon the addition of S protein, a shift of 28 nm in the DH compared to the control experiment (without S-protein) is observed, indicating a fast positive result. After 30 min, the equilibrium was reached and the ΔDH value was ca. 50 nm.
The adsorption isotherm was obtained by measuring the DH vs. S protein concentration in nM for the pAb-S130 biosensor, Fig. 6 a. To fit the data obtained in this experiment, an adapted Hill equation Eq. (3) was used (see Section 2.5 in the Experimental Procedure) [62]. Considering the ascending part of the curve, we observed a K D of 83 nM (Table inserted in Fig. 6b). This result is in the same order of magnitude observed for different IgG attached to AuNP immunoassays reported in the open literature [61]. Also, the K D obtained in this work is in the same order of magnitude of those reported for llama nanobodies that bind SARS-CoV-2 receptor binding domain in the spike trimer [72]. The Hill coefficient was n 1 = 0.4306, indicating a stimulatory effect (n 1 > 0) (Table inserted in Fig. 6b). The maximum DH value occurs at the concentration of 111 nM of S protein. Above this concentration, the DH decreases indicating an inhibitory effect. Nearby 200 nM of S protein, a monolayer formation on the biosensor surface takes place, as we illustrated in Fig. 4d. For S protein concentrations higher than 300 nM, a small increase in the DH values is noted, indicating a multilayer formation tendency. A residual plot (Fig. 6c) shows the selected model satisfactorily fitted the data displayed in Fig. 6a. The LOD was calculated from Fig. 6a using the first observed point with ΔDH value 3 times above the standard deviation of the immunoassay without S protein [56]. The obtained value for ΔDH = 6.6 nm was of 13 ng/mL or 30 pM falls under relevant concentration range [73].
4. Conclusions
In this work, we report the development of an one-step detection method for SARS-CoV-2 S protein employing spherical AuNPs capped with horse pAb using DLS technique as signal read-out. The AuNPs sizes of the biosensors were analyzed and the larger one, pAb-S130, showed the best response. Furthermore, pAb-S130 demonstrated to be very stable and active for S protein detection up to 90 days and reproducible for different antigen concentrations. Time response experiments showed a positive answer within only 5 min and the maximum ΔDH within 20 min. The pAb-S130 biosensor displayed LOD of 13 ng/mL (30 pM). Therefore, due to the affordable, sensitive, user-friendly, robust, and rapid aspect, the proposed method can be considered a potential antigen rapid test.
Credit author statement
The authors of the manuscript gave the following contributions: Carolina B. P. Ligiero, Tamires S. Fernandes, Dayenny L. D'Amato, Francisco V. Gaspar, Paola S. Duarte, Patrícia Bento da Silva, Ricardo B. Azevedo, Glêndara Aparecida de Souza Martins, Bráulio S. Archanjo, Camilla D. Buarque, Giovanna Machado, Ana M. Percebom: these authors were responsible for the experimental work up, characterization and S protein detection studies of the different size biosensors, data curation and first draft writing. Marcelo Abrahão Strauch, Juliana G. Fonseca, Leonardo G. R. Meirelles were responsible for the production and biological purification of the horse polyclonal anti-spike antibody used in this work. Célia M. Ronconi: this author was responsible for conceptualization, formal analysis, project administration, supervision and final writing and review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
This work was financially supported by the Brazilian agencies National Council for Scientific and Technological Development (CNPq grant numbers: 434141/2018-6, 402816/2020-0, 312949/2020-0 (C. M. R. research fellowship), 309160/2020-0 (T S F. fellowship)) and Rio de Janeiro Research Foundation (FAPERJ, Ação Emergencial Covid-19 grant numbers E26/210.194/2020, E26202.157/2020 [Instruction: (C.B.P.L.)], E-26/201.552/2021 (D.L.D.L) and E-26/200.046/2020 (P.S.)). We are grateful to the Material Characterization (http://www.uff.br/lamate/) and Molecular Spectroscopy (http://www.uff.br/lame/) Multiuser Laboratories from Universidade Federal Fluminense (UFF). The authors gratefully acknowledge the donation of trimeric spike protein of SARS-COV-2 by the Cell Culture Engineering Laboratory of COPPE/UFRJ and to Dr. Amilcar Tanuri from Virology Molecular Laboratory from Universidade Federal do Rio de Janeiro (UFRJ) for suggestions. C.B.P.L acknowledge the Brazilian Chemical Society and Royal Society of Chemistry for the prize “Young Researcher of Royal Society of Chemistry – RASBQ 2021”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtchem.2022.100924.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Pokhrel P., Hu C., Mao H. Detecting the coronavirus (COVID-19) ACS Sens. 2020;5:2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Managing Epidemics: Key Facts about Major Deadly Diseases.
- 3.World Health Organization . 14 December 2021. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19.https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-14-december-2021 [Google Scholar]
- 4.Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis - a review of current methods. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(June):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Peeling R.W., Olliaro P.L., Boeras D.I., Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect. Dis. 2021;21(9):e290–e295. doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouzas S. SARS-CoV-2 rapid antigen detection tests. Lancet Infect. Dis. 2021;21(8):1068–1069. doi: 10.1016/S1473-3099(21)00206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Driskell J.D., Jones C.A., Tompkins S.M., Tripp R.A. One-step assay for detecting influenza virus using dynamic light scattering and gold nanoparticles. Analyst. 2011;136(15):3083. doi: 10.1039/c1an15303j. [DOI] [PubMed] [Google Scholar]
- 9.Altintas Z., Akgun M., Kokturk G., Uludag Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018;100:541–548. doi: 10.1016/j.bios.2017.09.046. [DOI] [PubMed] [Google Scholar]
- 10.Sugunan A., Thanachayanont C., Dutta J., Hilborn J.G. Heavy-metal ion sensors using chitosan-capped gold nanoparticles. Sci. Technol. Adv. Mater. 2005;6(3–4):335–340. doi: 10.1016/j.stam.2005.03.007. [DOI] [Google Scholar]
- 11.Sener G., Uzun L., Denizli A. Colorimetric sensor array based on gold nanoparticles and amino acids for identification of toxic metal ions in water. ACS Appl. Mater. Interfaces. 2014;6(21):18395–18400. doi: 10.1021/am5071283. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Song S., Wang L., Pan D., Fan C. A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat. Protoc. 2007;2(11):2888–2895. doi: 10.1038/nprot.2007.419. [DOI] [PubMed] [Google Scholar]
- 13.Yue G., Su S., Li N., Shuai M., Lai X., Astruc D., Zhao P. Gold nanoparticles as sensors in the colorimetric and fluorescence detection of chemical warfare agents. Coord. Chem. Rev. 2016;311:75–84. doi: 10.1016/j.ccr.2015.11.009. [DOI] [Google Scholar]
- 14.Fang C., Dharmarajan R., Megharaj M., Naidu R. Gold nanoparticle-based optical sensors for selected anionic contaminants. TrAC Trends Anal. Chem. 2017;86:143–154. doi: 10.1016/j.trac.2016.10.008. [DOI] [Google Scholar]
- 15.Zheng T., Pierre-Pierre N., Yan X., Huo Q., Almodovar A.J.O., Valerio F., Rivera-Ramirez I., Griffith E., Decker D.D., Chen S., Zhu N. Gold nanoparticle-enabled blood test for early stage cancer detection and risk assessment. ACS Appl. Mater. Interfaces. 2015;7(12):6819–6827. doi: 10.1021/acsami.5b00371. [DOI] [PubMed] [Google Scholar]
- 16.Caputo D., Papi M., Coppola R., Palchetti S., Digiacomo L., Caracciolo G., Pozzi D. A protein corona-enabled blood test for early cancer detection. Nanoscale. 2017;9(1):349–354. doi: 10.1039/C6NR05609A. [DOI] [PubMed] [Google Scholar]
- 17.Pawula M., Altintas Z., Tothill I.E. SPR detection of cardiac troponin T for acute myocardial infarction. Talanta. 2016;146:823–830. doi: 10.1016/j.talanta.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari P.M., Vig K., Dennis V.A., Singh S.R. Functionalized gold nanoparticles and their biomedical applications. Nanomaterials. 2011;1:31–63. doi: 10.3390/nano1010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain P.K., Lee K.S., El-Sayed I.H., El-Sayed M.A. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J. Phys. Chem. B. 2006;110(14):7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 20.Huang X., El-Sayed M.A. Gold nanoparticles: optical properties and implementations in cancer diagnosis and photothermal therapy. J. Adv. Res. 2010;1(1):13–28. doi: 10.1016/j.jare.2010.02.002. [DOI] [Google Scholar]
- 21.Liz-Marzán L.M. Tailoring surface plasmons through the morphology and assembly of metal nanoparticles. Langmuir. 2006;22(1):32–41. doi: 10.1021/la0513353. [DOI] [PubMed] [Google Scholar]
- 22.Mayer K.M., Hafner J.H. Localized surface plasmon resonance sensors. Chem. Rev. 2011;111(6):3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 23.Bodelón G., Costas C., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L.M. Gold nanoparticles for regulation of cell function and behavior. Nano Today. 2017;13:40–60. doi: 10.1016/j.nantod.2016.12.014. [DOI] [Google Scholar]
- 24.Myroshnychenko V., Rodríguez-Fernández J., Pastoriza-Santos I., Funston A.M., Novo C., Mulvaney P., Liz-Marzán L.M., García de Abajo F.J. Modelling the optical response of gold nanoparticles. Chem. Soc. Rev. 2008;37(9):1792–1805. doi: 10.1039/b711486a. [DOI] [PubMed] [Google Scholar]
- 25.Sepúlveda B., Angelomé P.C., Lechuga L.M., Liz-Marzán L.M. LSPR-based nanobiosensors. Nano Today. 2009;4(3):244–251. doi: 10.1016/j.nantod.2009.04.001. [DOI] [Google Scholar]
- 26.Petryayeva E., Krull U.J. Localized surface plasmon resonance: nanostructures, bioassays and biosensing—a review. Anal. Chim. Acta. 2011;706(1):8–24. doi: 10.1016/j.aca.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Bohren G.F., Huffman D.R. Wiley VCH; Weinheim: 2004. Absorption and Scattering of Light by Small Particles. [Google Scholar]
- 28.Yguerabide J., Yguerabide E.E. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications. I. Theory. Anal. Biochem. 1998;262(2):137–156. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 29.Yguerabide J., Yguerabide E.E. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications. II. Experimental characterization. Anal. Biochem. 1998;262(2):157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 30.Anker J.N., Hall W.P., Lyandres O., Shah N.C., Zhao J., Van Duyne R.P. Biosensing with plasmonic nanosensors. Nat. Mater. 2008;7(6):442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Z., Tang A., Huang H., Wang Z. A simple and sensitive sensor for silver ions based on unmodified gold nanoparticles by using dynamic light scattering techniques. Can. J. Chem. 2017;95(12):1267–1272. doi: 10.1139/cjc-2017-0241. [DOI] [Google Scholar]
- 32.Stetefeld J., Mckenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Dai Q., Austin L., Coutts J., Knowles G., Zou J., Chen H., Huo Q. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J. Am. Chem. Soc. 2008;130(9):2780–2782. doi: 10.1021/ja711298b. [DOI] [PubMed] [Google Scholar]
- 34.Li J., Saleem M., Duan Q., Kakuchi T., Chen Y. Aggregation-induced fluorescent response of urea- bearing polyphenyleneethynylenes toward anion sensing. Sci. Technol. Adv. Mater. 2021;22(1):597–606. doi: 10.1080/14686996.2021.1942982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Ma J., Fan Q., Tao Y., Li G. Dynamic light scattering (DLS)-Based immunoassay for ultra-sensitive detection of tumor marker protein. Chem. Commun. 2016;52(50):7850–7853. doi: 10.1039/c6cc02633h. [DOI] [PubMed] [Google Scholar]
- 36.Dai Q., Liu X., Coutts J., Austin L., Huo Q. A one-step highly sensitive method for DNA detection using dynamic light scattering. J. Am. Chem. Soc. 2008;130:8138–8139. doi: 10.1021/ja801947e. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Yang L., Yang X., Wang K., Liu J. Use of mercaptophenylboronic acid functionalized gold nanoparticles in a sensitive and selective dynamic light scattering assay for glucose detection in serum. Analyst. 2013;138:5146–5150. doi: 10.1039/c3an00645j. [DOI] [PubMed] [Google Scholar]
- 38.Alami A. El, Lagarde F., Huo Q., Zheng T., Baitoul M., Daniel P. Acetylcholine and acetylcholinesterase inhibitors detection using gold nanoparticles coupled with dynamic light scattering. Sensors Int. 2020;1:100007. doi: 10.1016/j.sintl.2020.100007. April. [DOI] [Google Scholar]
- 39.Zheng X.T., Goh W.L., Yeow P., Lane D.P., Ghadessy F.J., Tan Y.N. Ultrasensitive dynamic light scattering based nanobiosensor for rapid anticancer drug screening. Sensor. Actuator. B Chem. 2019;279:79–86. doi: 10.1016/j.snb.2018.09.088. May 2018. [DOI] [Google Scholar]
- 40.Levin A.D., Ringaci A., Alenichev M.K., Drozhzhennikova E.B., Shevchenko K.G., Cherkasov V.R., Nikitin M.P., Nikitin P.I. Dynamic light scattering biosensing based on analyte-induced inhibition of nanoparticle aggregation. Anal. Bioanal. Chem. 2020:3423–3431. doi: 10.1007/s00216-020-02605-9. [DOI] [PubMed] [Google Scholar]
- 41.Miao X., Ling L., Shuai X. Ultrasensitive detection of lead (II) with DNAzyme and gold nanoparticles probes by using a dynamic light scattering technique. Chem. Commun. 2011;47:4192–4194. doi: 10.1039/c0cc05344a. [DOI] [PubMed] [Google Scholar]
- 42.Fu C., Ma H., Huang C., Jia N. A simple and dual functional dynamic light scattering (DLS) probe for rapid detection of mercury ions and biothiols. Anal. Methods. 2015;7:7455–7460. doi: 10.1039/c5ay01880c. [DOI] [Google Scholar]
- 43.Chicea D., Leca C., Olaru S., Chicea L.M. An advanced sensor for particles in gases using dynamic light scattering in air as solvent. Sensors. 2021;21:5115. doi: 10.3390/s21155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Li Y., Quan D., Wang J., Zhang Y., Du J., Peng J., Fu Q., Zhou Y., Jia S., Wang Y., Zhan L. Detection of hepatitis B surface antigen by target-induced aggregation monitored by dynamic light scattering. Anal. Biochem. 2012;428(2):119–125. doi: 10.1016/j.ab.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Bento da Silva P., da Silva J.R., Rodrigues M.C., Vieira J.A., de Andrade I.A., Nagata T., Santos A.S., da Silva S.W., da Rocha M.C.O., Báo S.N., Moraes-Vieira P.M., Proença-Modena J., Angelim M.K.C., de Souza G.F., Muraro S.P., de Barros A.L.B., Aparecida de Souza Martins G., Ribeiro-Dias F., Machado G., Fessel M.R., Chudzinski-Tavassi A.M., Ronconi C.M., Goncalves D., Curi R., Oliveira O.N., Jr., Azevedo R.B. Detection of SARS-CoV-2 virus via dynamic light scattering using antibody-gold nanoparticle bioconjugates against viral spike protein. Talanta. 2022;243:123355–123361. doi: 10.1016/j.talanta.2022.123355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lew T.T.S., Aung K.M.M., Ow S.Y., Amrun S.N., Sutarlie L., Ng L.F.P., Su X. Epitope-functionalized gold nanoparticles for rapid and selective detection of SARS-CoV - 2 IgG antibodies. ACS Nano. 2021;15:12286–12297. doi: 10.1021/acsnano.1c04091. [DOI] [PubMed] [Google Scholar]
- 47.Behrouzi K., Lin L. Gold nanoparticle based plasmonic sensing for the detection of SARS-CoV-2 nucleocapsid proteins. Biosens. Bioelectron. 2022;195:113669. doi: 10.1016/j.bios.2021.113669. September 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV - 2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K., Fan Z., Huang Y., Ding Y., Xie M. A strategy combining 3D-DNA walker and CRISPR-cas12a trans-cleavage activity applied to MXene based electrochemiluminescent sensor for SARS-CoV-2 RdRp gene detection. Talanta. 2022;236:122868. doi: 10.1016/j.talanta.2021.122868. September 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funari R., Chu K., Shen A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. September. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Functionalized terahertz plasmonic metasensors: femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens. Bioelectron. 2021;177:112971. doi: 10.1016/j.bios.2021.112971. June 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang L., Ding L., Zhou J., Chen S., Chen F., Zhao C., Xu J., Hu W., Ji J., Xu H., Liu G.L. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021;171:112685. doi: 10.1016/j.bios.2020.112685. September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunha L.E.R., Stolet A.A., Strauch M.A., Pereira V.A.R., Dumard C.H., Gomes A.M.O., Monteiro F.L., Higa L.M., Souza P.N.C., Fonseca J.G., Pontes F.E., Meirelles L.G.R., Albuquerque J.W.M., Sacramento C.Q., Fintelman-Rodrigues N., Lima T.M., Alvim R.G.F., Marsili F.F., Caldeira M.M., Zingali R.B., de Oliveira G.A.P., Souza T.M.L., Silva A.S., Muller R., Rodrigues D., del R.F., Costa L. J. da, Alves A.D.R., Pinto M.A., Oliveira A.C., Guedes H.L.M., Tanuri A., Castilho L.R., Silva J.L. Polyclonal F(Ab’)2 fragments of equine antibodies raised against the spike protein neutralize SARS- CoV-2 variants with high potency. iScience. 2020;24(103315):1–23. doi: 10.1016/j.isci.2021.103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvim R.G.F., Lima T.M., Rodrigues D.A.S., Marsili F.F., Bozza V.B.T., Higa L.M., Monteiro F.L., Leitão I.C., Carvalho R.S., Galliez R.M., Castineiras T.M.P.P., Nobrega A., Travassos L.H., Ferreira O.C., Jr., Tanuri A., Vale A.M., Castilho L.R. An affordable anti-SARS-COV-2 spike protein elisa test for early detection of IgG seroconversion suited for large-scale surveillance studies in low-income countries. medRxiv. 2020;1–19 doi: 10.1101/2020.07.13.20152884. [DOI] [Google Scholar]
- 55.Bast N.G., Comenge J., Puntes V. Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 Nm : size focusing versus ostwald ripening. Langmuir. 2011;27:11098–11105. doi: 10.1021/la201938u. [DOI] [PubMed] [Google Scholar]
- 56.Glass T.R., Ohmura N., Saiki H. Least detectable concentration and dynamic range of three immunoassay systems using the same antibody. Anal. Chem. 2007;79(5):1954–1960. doi: 10.1021/ac061288z. [DOI] [PubMed] [Google Scholar]
- 57.Immunoassay, Diamandis E.P., Christopoulos T.K., editors. Academic Press. 1996. [Google Scholar]
- 58.De Jong L.A.A., Uges D.R.A., Franke J.P., Bischoff R. Receptor-ligand binding assays: technologies and applications. J. Chromatogr. B. 2005;829(1–2):1–25. doi: 10.1016/j.jchromb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Darwish I.A. Immunoassay methods and their applications in pharmaceutical analysis: basic methodology and recent advances. Int. J. Biomed. Sci. 2006;2(3):217–235. [PMC free article] [PubMed] [Google Scholar]
- 60.Gadagkar S.R., Call G.B. Computational tools for fitting the Hill equation to dose-response curves. J. Pharmacol. Toxicol. Methods. 2015;71:68–76. doi: 10.1016/j.vascn.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz G., Ryan N., Rutschke K., Awotunde O., Driskell J.D. Antibodies irreversibly adsorb to gold nanoparticles and resist displacement by common blood proteins. Langmuir. 2019;35(32):10601–10609. doi: 10.1021/acs.langmuir.9b01900. [DOI] [PubMed] [Google Scholar]
- 62.Motulsky H., Christopoulos A. GraphPad Software Inc.; 2003. Fitting Models to Biological Data Using Linear and Nonlinear Regression. A Practical Guide to Curve Fitting. [DOI] [Google Scholar]
- 63.Zheng T., Bott S., Huo Q. Techniques for accurate sizing of gold nanoparticles using dynamic light scattering with particular application to chemical and biological sensing based on aggregate formation. ACS Appl. Mater. Interfaces. 2016;8:21585–21594. doi: 10.1021/acsami.6b06903. [DOI] [PubMed] [Google Scholar]
- 64.Filbrun S.L., Filbrun A.B., Lovato F.L., Oh S.H., Driskell E.A., Driskell J.D. Chemical modification of antibodies enables the formation of stable Antibody-gold nanoparticle conjugates for biosensing. Analyst. 2017;142(23):4456–4467. doi: 10.1039/c7an01496a. [DOI] [PubMed] [Google Scholar]
- 65.Rammohan A., Kaduk J.A. Crystal structures of alkali metal (group 1) citrate salts. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2018;74(2):239–252. doi: 10.1107/S2052520618002330. Citrate size estimated from crystal structure CSD (Cambridge Structural Database) code FETKOH from. [DOI] [PubMed] [Google Scholar]
- 66.DTSSP Geometry Optimized in Vaccum with Gaussian 7.0, DFT/B3LYP(31G+p,D).
- 67.Ruiz G., Tripathi K., Okyem S., Driskell J.D. PH impacts the orientation of antibody adsorbed onto gold nanoparticles. Bioconjugate Chem. 2019;30(4):1182–1191. doi: 10.1021/acs.bioconjchem.9b00123. [DOI] [PubMed] [Google Scholar]
- 68.Wrapp D. Cryo-EM structure of the 2019-NCoV spike in the prefusion conformation. Wang N., Corbett K.S., Goldsmith J.A., Hsieh C., Abiona O., Graham B.S., Mclellan J.S., editors. Sci. 2020;367:1260–1263. doi: 10.1126/science.abb2507. S Ptn (S protein) size estimated from crystal structure from the Protein Data Bank under accession code 6VSB. March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamnev A.A., Dykman L.A., Tarantilis P.A., Polissiou M.G. Spectroimmunochemistry using colloidal gold bioconjugates. Biosci. Rep. 2002;2(5–6):541–547. doi: 10.1023/A:1022077920527. [DOI] [PubMed] [Google Scholar]
- 70.Namburi R.P., Kancherla V., Ponnala A.R. High-dose hook effect. J. Dr. NTR Univ. Heal. Sci. 2014;3(1):5–7. doi: 10.4103/2277-8632.128412. [DOI] [Google Scholar]
- 71.Levin A.D., Filimonov I.S., Alenichev M.K., Goidina T.A. Mathematical modeling of nanosensor systems based on dynamic light scattering. Nanotechnologies Russ. 2018;13:406–413. doi: 10.1134/S1995078018040092. [DOI] [Google Scholar]
- 72.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 73.Albaz A.A., Rafeeq M.M., Sain Z.M., Almutairi W.A., Alamri A.S., Aloufi A.H., Almalki W.H., Tarique M. Nanotechnology-based approaches in the fight against SARS-CoV-2. AIMS Microbiol. 2021;7(4):368–398. doi: 10.3934/MICROBIOL.2021023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.