Summary
Two doses of Pfizer/BioNTech BNT162b2 mRNA vaccine elicit robust severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-neutralizing antibodies with frequent adverse events. Here, by applying a high-dimensional immune profiling on 92 vaccinees, we identify six vaccine-induced immune dynamics that correlate with the amounts of neutralizing antibodies, the severity of adverse events, or both. The early dynamics of natural killer (NK)/monocyte subsets (CD16+ NK cells, CD56high NK cells, and non-classical monocytes), dendritic cell (DC) subsets (DC3s and CD11c− Axl+ Siglec-6+ [AS]-DCs), and NKT-like cells are revealed as the distinct cell correlates for neutralizing-antibody titers, severity of adverse events, and both, respectively. The cell correlates for neutralizing antibodies or adverse events are consistently associated with elevation of interferon gamma (IFN-γ)-inducible chemokines, but the chemokine receptors CCR2 and CXCR3 are expressed in distinct manners between the two correlates: vaccine-induced expression on the neutralizing-antibody correlate and constitutive expression on the adverse-event correlate. The finding may guide vaccine strategies that balance immunogenicity and reactogenicity.
Keywords: mRNA vaccine, SARS-CoV-2, neutralizing antibodies, adverse events, cell dynamics, immune correlates, innate immunity
Graphical abstract
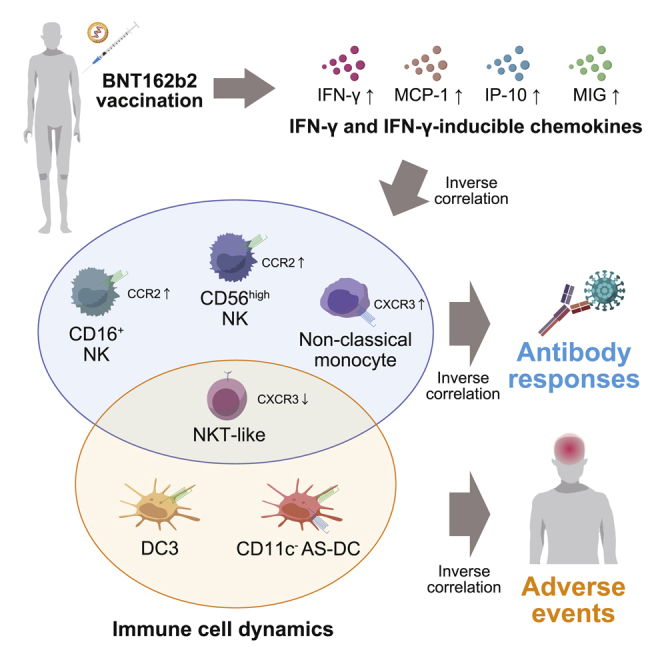
Highlights
-
•
Immune profiling of mRNA vaccinees reveals dramatic immunological changes
-
•
The dynamics of NK/monocyte subsets correlate with neutralizing-antibody response
-
•
The dynamics of dendritic cell subsets correlate with severity of adverse events
-
•
IFN-γ-inducible chemokines, but not the receptors, are related to these correlates
Takano et al. identify cellular correlates for neutralizing-antibody titers and systemic adverse events following SARS-CoV-2 mRNA vaccination. The involvement of IFN-γ-mediated pathways is proposed. The findings give insights into balancing the immunogenicity and reactogenicity of mRNA vaccine.
Introduction
The Pfizer/BioNTech BNT162b2 mRNA vaccine in a two-dose setting induces robust neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with 95% efficacy in preventing coronavirus 2019 (COVID-19).1 However, the immunogenicity produced by the mRNA vaccine is frequently associated with adverse events (AEs), including both local and systemic symptoms. The high incidence of AEs is likely caused by the pro-inflammatory nature of mRNA vaccines; however, the mechanism underlying the high immunogenicity and reactogenicity are limited owing to the lack of sufficient information on the early immune responses following vaccination. To this end, it is crucial to characterize the early immune cell dynamics that are initiated soon after vaccination and trigger subsequent immune responses behind the immunogenicity and reactogenicity.
In previous studies, a systems vaccinology approach based on transcriptional profiling was applied to dissect the early immune responses following administration of the influenza inactivated,2,3 live-attenuated yellow fever,4 live-attenuated tularemia,5 and SARS-CoV-2 mRNA vaccines.6 Influenza vaccination of individuals with pre-existing immunity triggers a rapid reduction of natural killer (NK) cell numbers in peripheral blood; however, it fails to reduce the numbers of NK cells in elderly individuals with compromised antibody responses.2 Alternatively, Arunachalam et al. revealed, following the second dose of Pfizer/BioNTech BNT162b2 mRNA vaccine, an expansion of heterogeneous cell clusters, comprising CD14+ monocytes, cDC2, and other unknown cell subsets. Moreover, the monocyte-related gene signatures were found to be associated with neutralizing antibody titers 3 weeks after the secondary vaccination.6 Thus, the early immune cell signatures associated with neutralizing antibody responses are elucidated by transcriptional profiling, while the immune cell dynamics underlying the AEs remain unexplored. Elucidation of these aspects has the potential to provide key insights for developing vaccine strategies to elicit strong neutralizing-antibody responses while minimizing the associated reactogenicity.
In this study, we sought to elucidate the early immune cell dynamics associated with eliciting neutralizing antibodies and post-vaccination symptoms in mRNA vaccinees. To this end, healthcare workers without prior history of SARS-CoV-2 infection were enrolled, and the frequencies of 18 immune cell subsets were quantitated in peripheral blood using high-dimensional flow cytometric analysis in the pre- and post-vaccination state during primary and booster vaccinations. Based on our analysis, we then classified early immune events into three groups: correlates for neutralizing antibodies, correlates for AEs, and correlates for both. Identification of these immune correlates advances our mechanistic understanding regarding the immunogenicity and reactogenicity of the Pfizer/BioNTech BNT162b2 mRNA vaccine.
Results
Immune profiling of BNT162b2 vaccination reveals dramatic immunological changes after secondary vaccination
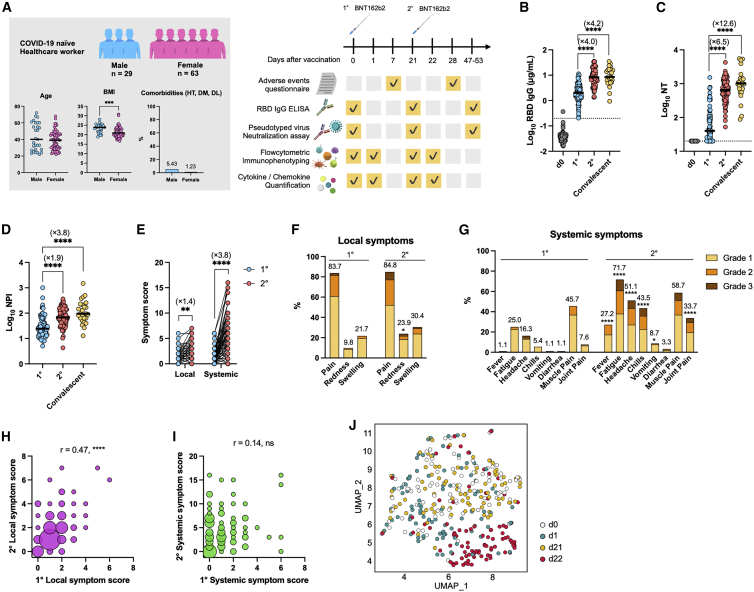
A total of 92 healthcare workers who received the Pfizer/BioNTech BNT162b2 mRNA vaccine were enrolled in this study. The median age was 39 years (interquartile range [IQR]: 28–50), ranging from 22 to 72 years, and 29/92 vaccinees (32%) were male. No significant difference was detected in median age between males (n = 29, median: 40, IQR: 28–60) and females (n = 63, median: 39, IQR: 29–46). Males (n = 29, median: 23.9, IQR: 21.6–24.7) had a significantly higher body mass index (BMI) than females (n = 58, median: 20.9, IQR: 19.4–23.4). The prevalence of comorbidities (hypertension, diabetes mellitus, and dyslipidemia) was higher in males (5.43%, n = 29) than females (1.23%, n = 63) (Figure 1A). None of the participants had prior histories of COVID-19 diagnosis or anti-nucleocapsid antibodies in pre- or post-vaccinated plasma during the study period, supporting that SARS-CoV-2 immunity observed in this study was activated by the vaccination, and not by the infection.
Figure 1.
Two doses of BNT162b2 vaccine elicits robust antibody responses and adverse events with distinct immune cell dynamics
(A) Cohort demographics are summarized: age, body mass index (BMI), and comorbidities (left). HT, hypertension; DM, diabetes mellitus; DL, dyslipidemia. A cohort of healthcare workers with two BNT162b2 doses (n = 92) was evaluated for antibody responses, incidence of adverse events, dynamics of immune cells among PBMCs, and cytokine/chemokine production (right).
(B–D) Antibody responses elicited by vaccination. RBD IgG titer (B), pseudotyped virus-neutralizing-antibody (NT) titers (C), and neutralization potency index (NPI) (D) were assessed; vaccinees, n = 92; convalescent, n = 25. Bars represent the median, and each dot represents the data for an individual participant in (B)–(D). RBD IgG threshold titer for seropositive is shown as a dotted line in (B). Detection limit of peudotyped virus NT titer is shown as a dotted line in (C).
(E) Total symptom-severity scores for local and systemic symptoms were assessed. Circles connected with a line represent data from the same individual.
(F and G) Frequencies and severity of local (F) and systemic (G) symptoms are shown; n = 92.
(H and I) Correlations between primary and secondary symptom-severity scores of local (H) or systemic (I) symptoms. The sizes of circles indicate the number of individuals. n = 92.
(J) UMAP of 18 immune cell frequencies was performed; n = 92. Each dot represents the data for an individual participant and is colored by time point.
Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Mann-Whitney test in (A); Kruskal-Wallis test in (B)–(D); Wilcoxon test in (E); Fisher’s exact test in (F) and (G); and Spearman’s rank-order coefficient test in (H) and (I).
See also Figures S1–S3 and Tables S1 and S2.
AEs, including local and systemic symptoms, within 1 week after vaccination were evaluated as symptom scores based on a self-reported questionnaire (Figure 1A; Table S1). The same criteria used for the clinical trial of the BNT162b2 mRNA vaccine was applied in this study.1 Immunoglobulin G (IgG) against spike protein receptor-binding domain (RBD) and neutralizing (NT) antibodies in plasma were quantified at the indicated time points (Figure 1A). The vaccinees who received the first dose were seropositive for RBD IgG (100%) and NT titers (74%) at 3 weeks post-vaccination, and the secondary vaccination further boosted the magnitudes of antibody titers to the levels in convalescent plasma from those who had recovered from severe COVID-19 (Figures 1B and 1C).7 A more dramatic increase in NT titers relative to RBD IgG titers resulted in a significant elevation of the NT potency index (NPI; NT activity per RBD IgG) (Figure 1D), an antibody parameter correlating with affinity maturation.7
Consistent with previous studies,1,8, 9, 10, 11, 12, 13 both the local and systemic symptom scores increased 1.4- and 3.8-fold after secondary vaccination, respectively, relative to those after primary vaccination (Figure 1E). Increased scores of AEs resulted from higher incidence and elevated grades of systemic symptoms, including fever, fatigue, headache, and chills after the secondary vaccination (Figures 1F and 1G). However, the frequencies of AEs in this study were slightly elevated compared with those in the original trial.1 For example, a higher incidence of fever (28% versus 16%) and fatigue (72% versus 59%), possibly reflecting a younger median age (39 versus 52 on average) and a higher female ratio (68% versus 49%), were observed in this study. Those who had severe local symptoms following primary vaccination tended to suffer from local symptoms again after secondary vaccination (Figure 1H), whereas no significant correlation was noted for systemic symptoms between primary and secondary vaccination (Figure 1I). The dissociation between the primary and secondary responses and exaggeration of systemic symptoms during the secondary response suggests the possible involvement of immune memory, which includes not only the responses by memory T and B lymphocytes14, 15, 16, 17, 18 but also epigenetic remodeling (so-called trained immunity) in myeloid cells.3,6
To elucidate the early vaccine-induced immune responses, peripheral blood, collected on day 1 after primary and secondary vaccinations, was subjected to high-dimensional flow cytometric phenotyping, which quantifies 18 immune cell subsets at high resolution (Figure S1). The eighteen cell subsets included three myeloid-derived suppressor cell (MDSC) subsets (early-stage MDSCs [e-MDSCs], monocytic MDSCs [M-MDSCs] and polymorphonuclear MDSCs [PMN-MDSCs]), B cells, CD4+ T cells, CD8+ T cells, two NK cell subsets (CD16+ NK cells and CD56high NK cells), NKT-like cells, which are known as a subset of T cells that serve as a bridge between innate and adaptive immunity,19 three monocyte subsets (classical monocytes, non-classical monocytes, and intermediate monocytes), and six dendritic cell (DC) subsets (conventional DC1s [cDC1s], cDC2s, DC3s, CD11c+ Axl+ Siglec-6+ DCs [AS-DCs], CD11c- AS-DCs, and plasmacytoid DCs [pDCs]) (Table S2). The frequency of each cell subset was quantified as a percentage of CD45+ cells (Figure S2), and then the fold increases or decreases from pre- to post-vaccination were calculated and defined as cell dynamics hereafter. Uniform manifold approximation and projection (UMAP) was applied to visualize the trajectory of vaccine-induced dynamics of 18 cell types following primary and secondary vaccinations (Figure 1J). Pre-vaccination samples (days 0 and 21) were merged with each other and revealed the baseline landscape based on 18 cell-type frequencies in the UMAP space. Of note, post-vaccination samples following primary (day 1) and secondary (day 22) vaccinations showed distinct distributions from the pre-vaccination states. The samples shifted slightly to the lower left in the primary responses, but those from the same donors shifted away from the pre-vaccination space to the lower right space in the secondary responses. The distinct trajectories and clustering obtained using UMAP between the primary and secondary vaccinations are likely independent of age, gender, and BMI (Figure S3), as these parameters failed to define the clusters revealed after vaccination. Thus, the UMAP results provided a visualization of the profound and distinct dynamics of immune cells during secondary responses.
Early dynamics of NK and monocyte subsets link to NT antibody response
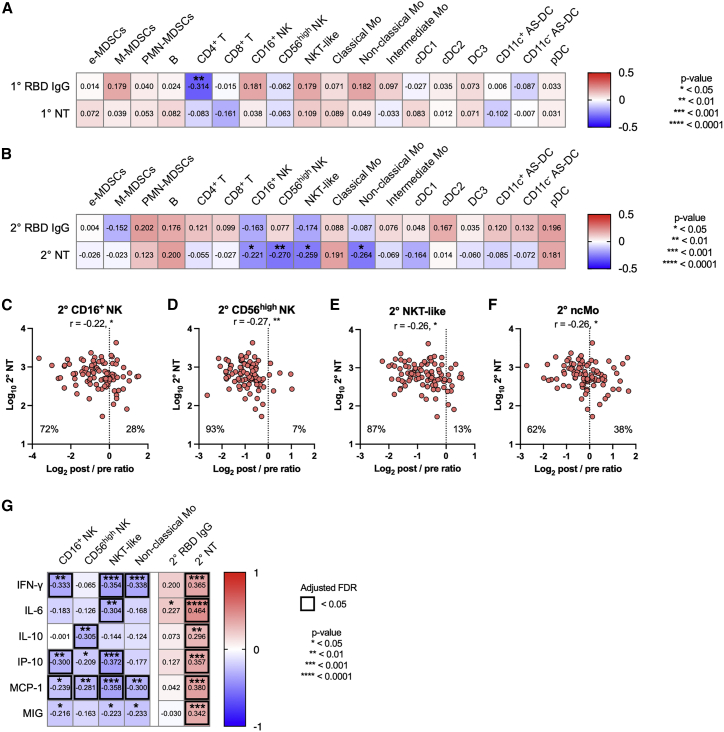
The possible links between these immune cell dynamics and antibody responses were evaluated during the primary (Figure 2A) and secondary (Figure 2B) responses. The dynamics of CD4+ T cells after primary vaccination were inversely correlated with primary RBD IgG titers (Figure 2A); however, this correlation was not extended to functional NT titers (Figure 2A). Following secondary vaccination, negative correlations were detected between NT titers and the dynamics of four cell populations (CD16+ NK cells, CD56high NK cells, NKT-like cells, and non-classical monocytes) (Figures 2B–2F), indicating that vaccinees with a more profound reduction in these cells had higher NT titers. It should be noted that the frequency of NK and NKT-like cells decreased in >70% of the vaccinees in response to secondary vaccination, likely reflecting the rapid recruitment of these cell subsets into inflammatory sites, as previously observed.20,21
Figure 2.
Early dynamics of NK, NKT, and monocyte subsets link to neutralizing-antibody response
(A) Heatmap representation of Spearman correlation matrix between cell dynamics (d1–d0 post/pre ratios) and antibody responses after primary vaccination. n = 92.
(B) Heatmap representation of Spearman correlation matrix between cell dynamics (d22–d21 post/pre ratios) and antibody responses after secondary vaccination. n = 92.
(C–F) Correlation of NT titer (d47–51) and cell dynamics (d22–d21 post/pre ratios). (C) CD16+ NK cells. (D) CD56high NK cells. (E) NKT-like cells. (F) Non-classical monocytes. n = 92. Each dot represents the data for an individual participant in (C)–(F).
(G) Heatmap representation of Spearman correlation matrix between cell dynamics (d22–d21 post/pre ratios)/antibody responses and cytokines/chemokines. n = 92.
Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Spearman’s rank-order coefficient test. Spearman’s r values are indicated in each cell in (A), (B), and (G) and above the plots in (C)–(F).
See also Figure S4.
Monocytes are identified as a correlate for antibody responses following SARS-CoV-2 mRNA vaccines.6 In our study, the vaccinees with a more profound reduction in non-classical monocytes mounted higher amounts of NT (Figure 2F). Likewise, those who had reduced numbers of NK subsets following vaccination produced greater amounts of NT (Figures 2C and 2D), resembling previous findings in influenza vaccinees.2 Overall, the cellular subsets identified by our immune-profiling approach partially overlapped with those from previous studies using transcriptional profiling, suggesting the convergence of monocyte and NK cell subsets during early vaccine-induced responses irrespective of vaccine modality.
To assess the possible regulatory cytokines and chemokines in the identified cell dynamics, we quantified the concentrations of multiple cytokines and chemokines in the plasma (Figure S4A), and then the post/pre ratios were calculated and defined as cytokine dynamics. The amounts of interferon gamma (IFN-γ), interleukin-6 (IL-6), IL-10, IFN-γ-inducible protein 10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), and monokine induced by IFN-γ (MIG) increased following secondary vaccination (Figure S4A) and were inversely correlated with the dynamics of NK and NKT-like cells, known producers of IFN-γ (Figure 2G). Furthermore, the dynamics of these cytokines and chemokines following secondary vaccination positively correlated with NT titers (Figure 2G). Together, these data suggest that rapid migration of these cell subsets into inflammatory sites enhances the systemic production of IFN-γ and its inducible chemokines and that the NK-IFN-γ pathway correlates with the subsequent magnitude of antibody responses,22 which is consistent with previous results using systems vaccinology approaches6 and systemic cytokine/chemokine analysis.23
Correlations between AEs and NT antibodies
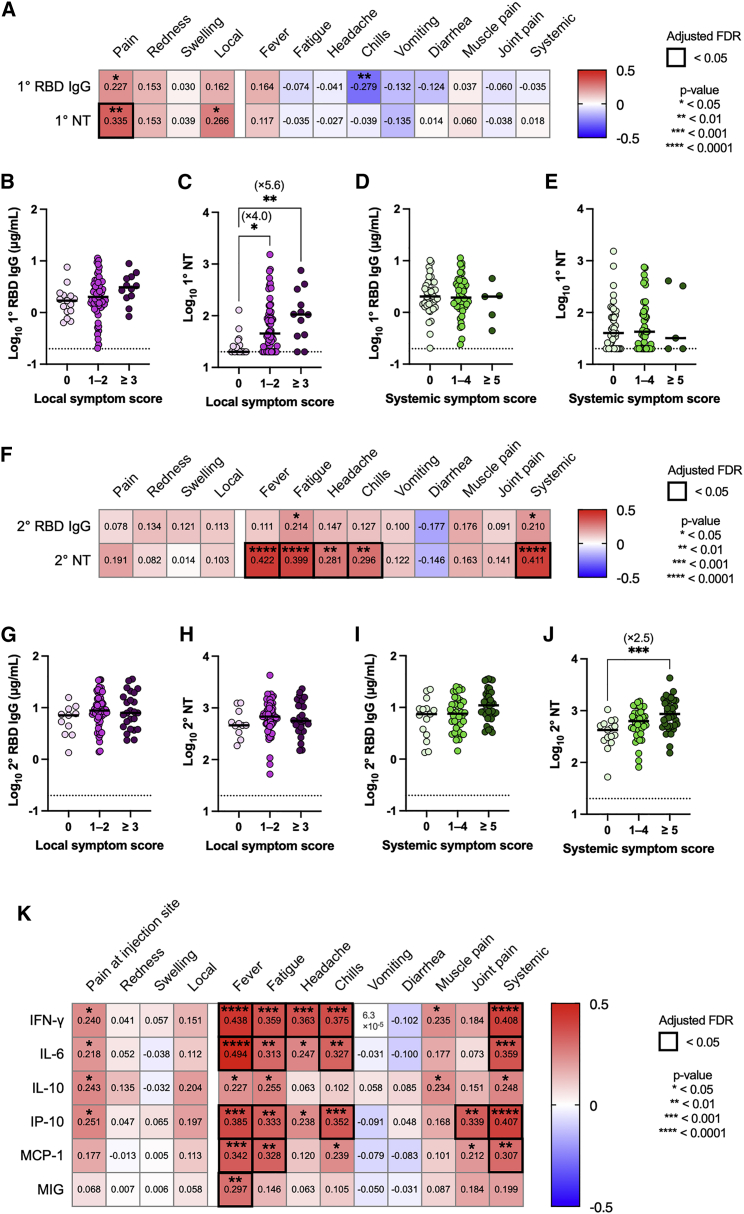
Considering that correlations between antibody titers (RBD IgG) and AEs remain controversial,8, 9, 10, 11, 12,24 we quantified the functional NT titers in our cohort. We found a significant correlation between pain at the injection site and RBD IgG/NT titers (Figure 3A), while the sum of local symptoms was linked to the NT titers only (Figure 3A). The vaccinees were then classified into three groups based on their symptom scores, and antibody titers were compared among the groups with different symptom severity (Figures 3B–3E). Upon primary vaccination, NT titers, not RBD IgG, were >5-fold higher in the vaccinees with severe local symptoms (>3) compared to those without symptoms (Figures 3B and 3C). However, antibody titers following primary vaccination were not correlated with the severity of systemic symptoms (Figures 3D and 3E), suggesting that primary antibody responses are largely governed by local immune responses. No significant correlations between the vaccine-elicited cytokines/chemokines (IL-6, IL-8, IL-10, and IP-10) and the antibody titers or the symptom-severity scores following the primary vaccination suggested the dissociation of these systemic events with local immune responses (Figure S4B).
Figure 3.
Adverse events correlate with neutralizing antibodies
(A) Heatmap representation of Spearman correlation matrix between symptom severity scores and antibody responses (primary vaccination). n = 92.
(B and C) RBD IgG (B) and NT titers (C) were evaluated based on the local symptom severity scores after primary vaccination. 0, n = 15; 1–2, n = 65; ≥3, n = 12.
(D and E) RBD IgG (D) and NT titers (E) were evaluated based on the systemic symptom severity scores after primary vaccination. 0, n = 40; 1–4, n = 47; ≥5, n = 5.
(F) Heatmap representations of Spearman correlation matrix between symptom severity scores and antibody responses (secondary vaccination). n = 92.
(G and H) RBD IgG (G) and NT titers (H) were evaluated based on the local symptom severity scores after secondary vaccination. 0, n = 11; 1–2, n = 56; ≥3, n = 25.
(I and J) RBD IgG (I) and NT titers (J) were evaluated based on the systemic symptom severity scores after secondary vaccination. 0, n = 15; 1–4, n = 39; ≥5, n = 38.
(K) Heatmap representations of Spearman correlation matrix between symptom severity scores and cytokine dynamics (d22–d21 post/pre ratios). n = 92. Data are shown as the median and points indicate each participant in (B–E). Statistical significance is indicated as follows; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Spearman’s rank-order coefficient test in (A, F, and K); Kruskal-Wallis test in (B–E, G–J). Spearman’s r values are indicated in each cell in (A, F, and K).
In contrast to the primary immune responses, the NT titers following secondary vaccination correlated with multiple systemic symptoms but not with local symptoms (Figures 3F–3J). A comparative analysis revealed a 2.5-fold increase in NT titers in those who suffered from severe systemic symptoms, while RBD IgG titers remained unchanged (Figures 3I and 3J). These results revealed the dose- and symptom-dependent correlations between NT titers and symptoms, highlighting the links between local symptoms and primary NT titers as well as those between systemic symptoms and secondary NT titers. In analogy to the NT titers, the vaccine-elicited IFN-γ, IL-6, IP-10, and MCP-1 positively correlated with the severity of systemic symptoms following secondary vaccination (Figures 2G and 3K).
Early dynamics of DC subsets link to AEs
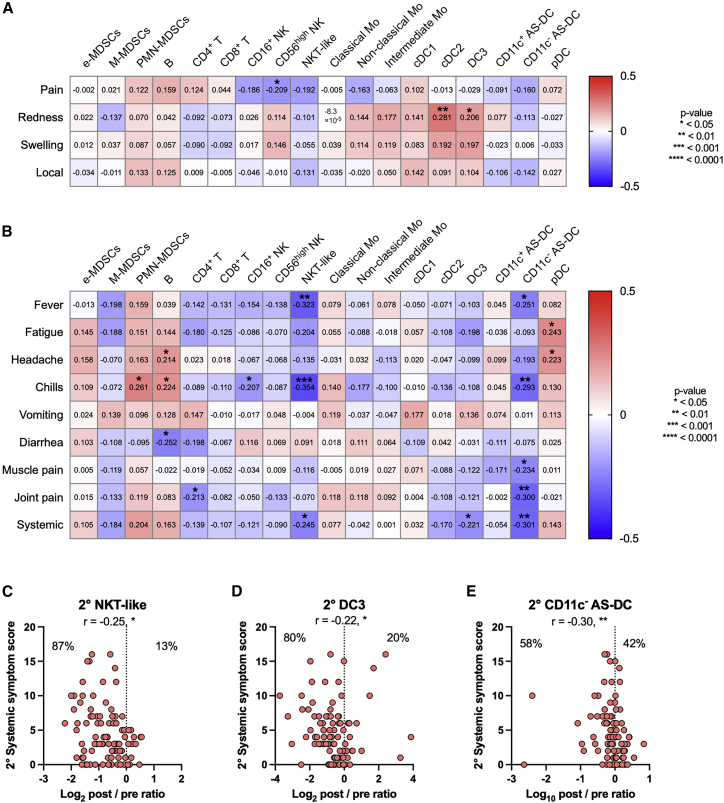
Next, we attempted to identify the cell dynamics that are more tightly linked to AEs than to antibody responses, as such information is important for attenuating AEs without compromising the immunogenicity of vaccine. To identify such immune events, correlation analyses were performed between the cell dynamics and local/systemic symptoms during the primary (Figure S5) and secondary responses (Figures 4A and 4B). No significant correlations were observed between the sum of symptoms and cellular dynamics during the primary response (Figure S5); however, three cellular populations correlated with the sum of systemic, not local, symptoms during the secondary responses (Figures 4A and 4B). Among them, the dynamics of NKT-like cells after secondary vaccination were also found to negatively correlate with the severity of systemic symptoms (Figure 4C); meanwhile, the dynamics of CD11c− AS-DCs and DC3s after secondary vaccination were negatively correlated with the severity of systemic symptoms, not with antibody (Ab) responses (Figures 2B, 4D, and 4E).
Figure 4.
Early dynamics of NKT and DC subsets link to adverse events
(A) Heatmap representations of Spearman correlation matrix between cell dynamics (d22–d21 post/pre ratios) and local symptom severity scores (secondary vaccination). n = 92.
(B) Heatmap representations of Spearman correlation matrix between cell dynamics (d22–d21 post/pre ratios) and systemic symptom severity scores. n = 92.
(C–E) Correlation of systemic symptom scores and cell dynamics (d22–d21 post/pre ratios). (C) NKT-like cells. (D) DC3s. (E) CD11c− AS-DCs. n = 92. Statistical significance is indicated as follows; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Spearman’s rank-order coefficient test. Spearman’s r values are indicated in each cell in (A and B), and above the plots in (C–E). See also Figure S5.
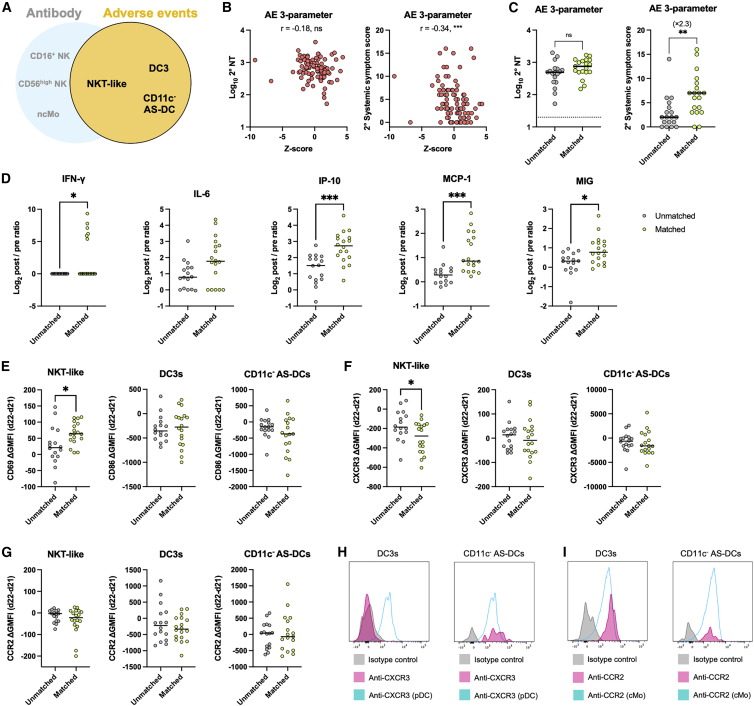
We evaluated the discriminative value of the identified cellular dynamics for systemic symptoms and antibody titers following secondary vaccination. First, three cell populations (NKT-like cells, DC3, and CD11c− AS-DCs) were selected (Figure 5A) and combined into a Z score to visualize the correlation with NT titers and systemic-symptom scores (Figure 5B). The combined parameter showed a stronger correlation with systemic-symptom scores than individual parameters (Figure 5B; r = −0.34 versus r = −0.30 to −0.22 in Figures 4C–4E); however, it failed to correlate with NT titers. Next, to assess the utility of these three parameters for identifying a subgroup of vaccinees with severe reactogenicity, the vaccinees were separated into matched groups that were positive (i.e., post/pre ratios were below the median of the cohort) for all three parameters and unmatched groups that were negative (i.e., post/pre ratios were above the median of the cohort) for three parameters. There was no significant difference in gender distribution between the groups (matched group: male, n = 4; female, n = 15, and unmatched group: male, n = 7; female, n = 11, p = 0.2953, Fisher’s exact test). Classification by the three parameters slightly, but not significantly, increased the NT titers in the matched group, whereas the matched group led to a significant 2.3-fold increase in systemic symptoms (Figure 5C).
Figure 5.
The combination of NKT and DC subset dynamics classify a subgroup of vaccinees with severe adverse events
(A) Scheme of the 3-parameter model related to adverse events (AEs).
(B) Z score calculated based on three parameters (i.e., the post/pre ratios of NKT-like cells, DC3s, and CD11c- AS-DCs). Correlation of Z score and NT titers at days 47–51 and systemic-symptom-severity scores. n = 92.
(C) Participants were stratified into matched and unmatched groups for the 3-parameter model related to AEs. NT titers and systemic-symptom-severity scores compared between matched and unmatched groups. Matched group, n = 19; unmatched group, n = 18.
(D) Cytokine dynamics (d22–d21 post/pre ratios) are compared between AE-related matched and unmatched groups. Matched group, n = 18; unmatched group, n = 16 (AE 3-parameter).
(E–G) Differences in expression levels of CD69 or CD86 (E), CXCR3 (F), and CCR2(G) between d22 and d21 were compared between AE-related matched and unmatched groups. Matched group, n = 18; unmatched group, n = 16 (AE 3-parameter). GMFI, geometric mean fluorescence intensity.
(H and I) Representative histogram plot for expression levels of CXCR3 (H) and CCR2 (I) on DC3s and CD11c- AS-DCs at day 21. Plasmacytoid DCs (pDCs) were plotted as CXCR3 positive control (H) and classical monocytes (cMos) were plotted as CCR2 positive control (I). Bars represent the median, and each dot represents data for an individual participant in (C)–(G).
Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, Mann-Whitney test in (C)–(G) (matched versus unmatched groups), Spearman’s rank-order coefficient test in (B) and (E). Spearman’s r values are indicated above the plots in (B).
See also Figure S6.
The two parameters (DC3s and CD11c− AS-DCs) linked to the AEs, but not to NT titers, were combined (Figure S6A). The combined Z score correlated with systemic-symptom scores but not with NT titers (Figures S6B). Also, the subgroups of the vaccinees that were matched or unmatched with two selected parameters mounted equivalent NT titers; however, the matched group had higher systemic-symptom scores by 1.8-fold compared with those of the unmatched group (Figure S6C). Again, no significant difference was observed in gender distribution between the classified groups (matched group: male, n = 10; female, n = 20, and unmatched group: male, n = 12; female, n = 18, p = 0.7892, Fisher’s exact test).
IFN-γ-inducible chemokine responses and peripheral decreases of DC subsets
To gain mechanistic insights for the decreasing numbers of indicated cell subsets in peripheral blood, the fold increases of vaccine-elicited cytokines/chemokines were compared between the matched and unmatched groups (Figure 5D). The secondary vaccination elicited IFN-γ and IFN-γ-inducible chemokines (IP-10, MCP-1, and MIG) at significantly higher levels in the matched group than in the unmatched group (Figure 5D), revealing the elevation of IFN-γ and the inducible chemokines as the early immune signatures in the matched group.
The expression levels of activation markers (CD69 and CD86), chemokine receptors (CXCR3 and CCR2), and pre-apoptotic Annexin V markers were quantitated in three cell subsets (NKT-like cells, DC3, and CD11c− AS-DC) of AE correlates. CD69 and CD86 were chosen as the activation markers for NKT-like cells and DC subsets, respectively. CCR2 and CXCR3 expression were examined as the receptors for MCP and IP-10/MIG, respectively.25, 26, 27, 28 It should be mentioned that CXCR3 is also downregulated in activated T cells.29 All available samples from matched and unmatched groups were subjected to the phenotypic analysis using these markers.
CD69 expression was more profoundly upregulated in NKT-like cells from the matched group by the vaccination, but CD86 expression was equivalent in both matched and unmatched groups (Figure 5E), suggesting more prominent activation of NKT-like cells, but not DC subsets, in the matched group. Downregulated CXCR3 expression in NKT-like cells from the matched group further support the activation status and imply the possible contribution of this cell subset as the IFN-γ-producer cells (Figure 5F). CXCR3 was constitutively expressed in the CD11c− AS-DC, but not the DC3, subset, independently of the vaccination status (Figures 5F and 5H). Likewise, CCR2 was constitutively expressed in DC subsets from both groups (Figures 5G and 5I), suggesting that these DC subsets respond chemotactically to IFN-γ-inducible MCP, IP-10, and MIG that are secreted upon the secondary vaccination. Comparable Annexin V binding in three cell subsets between the matched and unmatched groups reduces the likelihood that decreasing numbers of the subsets in the matched group are mediated by apoptotic cell death (Figure S6D).
Peripheral decrease of NK and monocyte subsets links to NT antibody titers
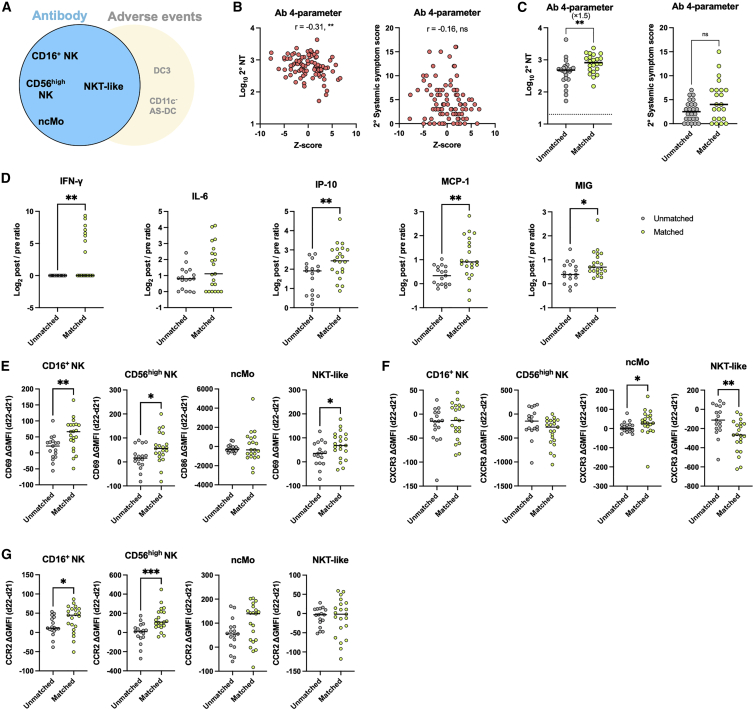
Similar to cell correlates for systemic AEs, we next sought to evaluate the cell correlates for NT titers following secondary vaccination. The four (CD16+ NK cells, CD56high NK cells, non-classical monocytes, and NKT-like cells) or three (CD16+ NK cells, CD56high NK cells, and non-classical monocytes) parameters linked to high NT titers, but not to systemic symptom scores, were combined (Figures 6A and S7A). The combined Z scores showed a stronger correlation with NT titers than individual parameters (Figures 6B and S7B; r = −0.31 to −0.29 versus r = −0.27 to −0.22 in Figures 2C–2F) but not with systemic-symptom scores (Figures 6B and S7B). The matched groups with 4 or 3 selected parameters had significantly higher NT titers by 1.5-fold compared with those of the unmatched groups, while the systemic-symptom scores remained equivalent (Figures 6C and S7C). There was no significant difference in gender distribution between the matched and unmatched groups by 4 parameters (matched group: male, n = 6; female, n = 16, and unmatched group: male, n = 8; female, n = 16, p = 0.7539, Fisher’s exact test) and by 3 parameters (matched group: male, n = 6; female, n = 18, and unmatched group: male, n = 8; female, n = 16, p = 0.7516, Fisher’s exact test).
Figure 6.
The combination of NK, NKT, and monocyte subset dynamics classify a subgroup of vaccinees with high neutralizing-antibody responses
(A) Scheme of the 4-parameter model related to antibody responses.
(B) Z score calculated based on four parameters (i.e., the post/pre ratios of NKT-like, CD16+ NK, CD56high NK, and non-cMOs [ncMos]). Correlation of Z score and NT titers at days 47–51 and systemic-symptom-severity scores. n = 92.
(C) Participants were stratified into matched and unmatched groups for the 4-parameter model related to antibody responses. NT titers and systemic-symptom-severity scores compared between matched and unmatched groups. Matched group, n = 22; unmatched group, n = 24.
(D) Cytokine dynamics (d22–d21 post/pre ratios) are compared between Ab-related matched and unmatched groups. Matched group, n = 21 (d21), n = 22 (d22); unmatched group, n = 17 (d21), n = 18 (d22) (Ab 4-parameter).
(E–G) Differences in expression levels of CD69 or CD86 (E), CXCR3 (F), and CCR2 (G) between d22 and d21 were compared between Ab-related matched and unmatched groups. Matched group, n = 21 (d21), n = 22 (d22); unmatched group, n = 17 (d21), n = 18 (d22) (Ab 4-parameter). GMFI, geometric mean fluorescence intensity. Bars represent the median, and each dot represents data for an individual participant in (C)–(G).
Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Mann-Whitney test in (C)–(G) (matched versus unmatched groups), Spearman’s rank-order coefficient test in (B). Spearman’s r values are indicated above the plots in (B).
See also Figure S7.
The mechanistic basis behind the cell correlates for NT titers was evaluated by a similar approach with Figure 5. Higher amounts of IFN-γ and IFN-γ-inducible chemokines were detected in the matched group following the secondary vaccination (Figure 6D). CD69 expression in NK subsets as well as NKT-like cells was highly elevated in the matched group (Figure 6E), again suggesting the contribution of these cell subsets on the IFN-γ provision. NKT-like cells downregulated CXCR3 expression as indicative of the activation status, and non-classical monocytes upregulated the expression in the matched group (Figure 6F). In stark contrast to DC subsets with constitutive CCR2 expression, NK subsets and non-classical monocytes upregulated CCR2 expression in the matched group after the secondary vaccination (Figure 6G), although the increased level failed to generate a statistical difference in non-classical monocytes. Thus, these subsets correlating with NT titers likely acquire the chemotactic ability to IFN-γ-inducible chemokines upon vaccination, a feature distinct from the AE-related subsets that constitutively expressed CCR2 in pre-vaccination status.
Discussion
By profiling 18 immune cell dynamics at day 1 post-vaccination, we identified six cellular dynamics that correlated with the severity of AEs as well as the amounts of NT antibodies at later time points. These include the innate immune cell dynamics that have been identified as the correlates of vaccine-induced Ab responses via the systems vaccinology approach.2,3,6 Our study identified the NKT-like cell dynamics in peripheral blood as an early correlate of high antibody titers and severity of systemic symptoms. Furthermore, more profound reductions in the numbers of DC subsets were identified as specific correlates for AEs and those in NK/monocyte subsets as correlates for NT titers. Importantly, based on the cellular parameters, we were able to classify the vaccinees who suffered from more severe AEs or those who mounted more NT titers after the secondary vaccination.
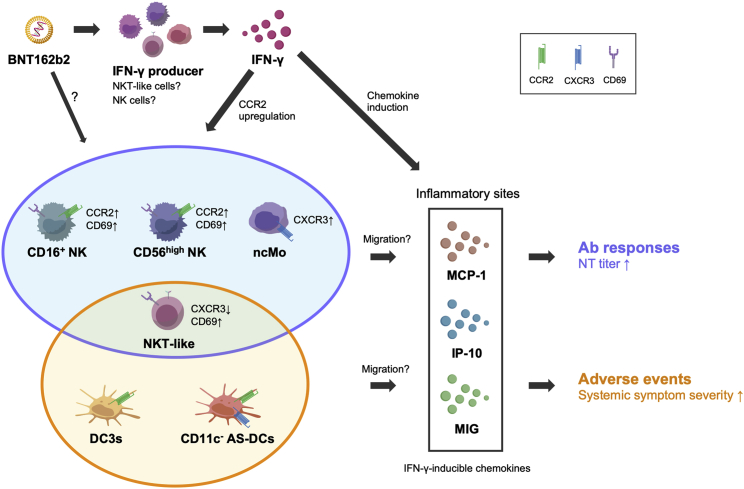
The exact mechanism behind the decreasing numbers of indicated cell subsets in the peripheral blood remains to be determined, but the available evidence suggest the following model (Figure 7). The prompt IFN-γ production and subsequent activation of IFN-γ-inducible chemotaxis are common triggers for the observed cell correlates for NT titers or systemic AEs. Vaccine-induced activation of NKT-like cells and NK subsets may contribute to IFN-γ production, as is supported by the activated phenotypes of these cell subsets. The vaccine-induced CCR2 or CXCR3 upregulation on NK/monocyte subsets enhances their chemotactic ability and helps to migrate from the peripheral blood to inflammatory sites upon MCP-1/IP-10/MIG guidance. In contrast, the DC subsets related to severe AEs constitutively express the CCR2 and CXCR3 receptors and migrate to the inflammatory sites in accordance with the MCP-1/IP-10/MIG chemotaxis. Thus, the cellular dynamics behind high NT titers or severe AEs following the second dose of mRNA vaccine share IFN-γ and the inducible chemokines as the initial trigger but are distinct not only in the involved cell types but also in the regulation of chemokine receptor expression.
Figure 7.
Model for immune cell dynamics relevant to neutralizing Ab responses or AEs after secondary mRNA vaccination
Second dose of BNT162b2 vaccine promotes IFN-γ secretion from producer cells, including activated NKT-like cells and NK cell subsets, that upregulate CD69 and downregulate CXCR3 (NKT-like cells only). IFN-γ then enhances the secretion of IFN-γ-inducible chemokines (MCP-1, IP-10, and MIG) at the inflammatory sites. Following vaccination, NK cell subsets (CD16+ NK and CD56high NK) and ncMos upregulate CCR2 and CXCR3, respectively, increase the chemotactic activity to MCP-1, IP-10, and MIG, and migrate into the inflammatory sites, contributing to the enhancement of neutralizing Ab responses. DC3 and CD11c- AS-DC subsets also migrate into the inflammatory sites in response to the same chemokines, owing to their constitutive expression of CCR2 and CXCR3 (CD11c- AS-DC only), the dynamics correlating with the systemic AEs.
Supporting our model, intramuscular vaccination induces local inflammation at the injection site and recruits multiple cell subsets, including NK cells, monocytes, and DCs, into the tissue or draining lymph nodes via the chemokine, its receptor axis,30,31 resulting in a corresponding decrease in their abundance within the circulation in response to the secondary vaccination. Furthermore, NK cells are shown to be recruited into lymph nodes following vaccination and are suggested to contribute to efficient T/B cell responses via the upregulated IFN-γ or IL-6.32, 33, 34
Considering that DC3s and CD11c− AS-DCs are both recently classified subsets via high-resolution single-cell analysis,35, 36, 37, 38, 39, 40, 41 their biological functions remain largely unknown. DC3s are considered a distinct subset that is specialized to produce pro-inflammatory cytokines (TNF, IL-1β, IL-6, IL-12, IL-23, etc.).35 Some of these cytokines possess pyrogenic effects and differentiate antigen-stimulated T cells into Th1 or Th17 subsets to accelerate inflammation,42, 43, 44, 45 possibly contributing to the exacerbation of AEs. Meanwhile, functional analysis remains limited for CD11c- AS-DCs, and it is not yet clear how this DC subset links with AEs.
IFN-γ production was highly elevated after the secondary vaccination and was identified as the key response that governs both NT antibody responses and systemic AEs in this study. Lower IFN-γ production after the primary vaccination indicates the contribution of immunological memory, including innate and adaptive immune memory, that probably enhances NT antibodies as well as systemic AEs via IFN-γ and other regulatory molecules. In this context, more severe AEs in vaccinees with previous infection histories are noteworthy.13,46 The data suggest that such memory responses can be primed by SARS-CoV-2 antigens without the formulation of mRNA vaccines. Moreover, the memory responses are unlikely to wane in a short period of time because symptom enhancement is observed in individuals who have experienced a previous infection greater than 6 months prior.13 Identification of the memory response behind IFN-γ production, NT antibody titers, and systemic AEs may be important for a comprehensive understanding of vaccine-induced immunity and for developing vaccine strategies that can balance the immunogenicity and reactogenicity of SARS-CoV-2 mRNA vaccines.
Limitations of the study
Certain limitations are noted in this study. First, no analysis beyond day 1 post-vaccination was performed to track extended immune cell dynamics. Moreover, our analysis was limited to peripheral blood cells and did not include cells within the local injection sites where the immune responses for eliciting NT antibodies and AEs take place. Further investigation is warranted to dissect the biological mechanisms of the correlations, especially related to AEs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022 | Meulen et al., 2006 | DQ 168569.1, DQ168570.1 |
| HRP-conjugated goat anti-human IgG | Southern Biotech | Cat#2040-05; RRID: AB_2795644 |
| Anti-human CD45-APC-Alexa Fluor 700 | BioLegend | Cat#304024; RRID: AB_493761 |
| Anti-human CD3-BV605 | BD Biosciences | Cat# 563217; RRID: AB_2714001 |
| Anti-human CD19-Brilliant Violet 605 | BioLegend | Cat# 302244; RRID: AB_2562015 |
| Anti-human CD56-Brilliant Violet 605 | BioLegend | Cat# 362538; RRID: AB_2565856 |
| Anti-human HLA-DR-PE/Cy7 | BD Biosciences | Cat# 560651; RRID: AB_1727528 |
| Anti-human CD11b-FITC | BioLegend | Cat# 301330; RRID: AB_2561703 |
| Anti-human CD33-PE | BioLegend | Cat# 366608; RRID: AB_2566107 |
| Anti-human CD14 BV421 | BD Biosciences | Cat# 565283; RRID: AB_2739154 |
| Anti-human CD15-APC | BioLegend | Cat# 323007; RRID: AB_756013 |
| Anti-human CD45-BV570 | BD Biosciences | Cat# 304034; RRID: AB_2563426 |
| Anti-human CD3-BUV661 | BD Biosciences | Cat# 741596; RRID: AB_2871005 |
| Anti-human CD4-PE/Cy5 | BD Biosciences | Cat# 555348; RRID: AB_395753 |
| Anti-human CD8-BB660 | BD Biosciences, custom | Cat#624295; no RRID |
| Anti-human CD-BV786 | BD Biosciences | Cat# 563823; RRID: AB_2687487 |
| Anti-human CD19-BUV563 | BD Biosciences | Cat# 612916; RRID: AB_2870201 |
| Anti-human CD56 BB515 | BD Biosciences | Cat# 564488; RRID: AB_2744428 |
| Anti-human HLA-DR-APC/H7 | BD Biosciences | Cat# 561358; RRID: AB_10611876 |
| Anti-human CD16-BUV395 | BD Biosciences | Cat# 563784; RRID: AB_2744293 |
| Anti-human CD11c-APC-R700 | BD Biosciences | Cat# 566610; RRID: AB_2869792 |
| Anti-human CD88-PE/Cyanine7 | BioLegend | Cat# 344308; RRID: AB_11126750 |
| Anti-human CD1c-BUV737 | BD Biosciences | Cat# 748723; RRID: AB_2873127 |
| Anti-human CD123-BUV496 | BD Biosciences | Cat# 564196; RRID: AB_2738662 |
| Anti-human CD141-BV605 | BD Biosciences | Cat# 740421; RRID: AB_2740151 |
| Anti-human CD5-BV480 | BD Biosciences | Cat# 566122; RRID: AB_2739524 |
| Anti-human CD163-PE/CF594 | BD Biosciences | Cat# 562670; RRID: AB_2737711 |
| Anti-human CD163-BV786 | BD Biosciences | Cat# 741003; RRID: AB_2740626 |
| Anti-human Siglec-6-BV650 | BD Biosciences | Cat# 747911; RRID: AB_2872373 |
| Anti-human Axl-BUV615 | BD Biosciences | Cat# 751050; RRID: AB_2875090 |
| Anti-human CD45-BUV805 | BD Biosciences | Cat# 612891; RRID: AB_2870179 |
| Anti-human CCR2-Brilliant Violet 785 | BioLegend | Cat# 150621; RRID: AB_2721565 |
| Anti-human CXCR3-PE/Cyanine5 | BioLegend | Cat# 353755; RRID: AB_2904375 |
| Anti-human CD86-PE | BD Biosciences | Cat# 555665; RRID: AB_396019 |
| Anti-human CD69-BV711 | BD Biosciences | Cat# 563836; RRID: AB_2738443 |
| Annexin V Alexa Fluor™647 conjugated | Thermo Fisher Scientific | Cat# A23204; RRID: AB_2341149 |
| Briliant Violet 785 Mouse IgG2a, κ Isotype control | BioLegend | Cat#400647; no RRID |
| PE/Cyanine5 Mouse IgG1, κ Isotype control | BioLegend | Cat#400118; no RRID |
| Human TruStain FcX (Fc receptor blocking solution) | BioLegend | Cat#422302 |
| Bacterial and virus strains | ||
| VSV pseudovirus bearng SARS-CoV-2 spike protein | Tani et al., 2021 | N/A |
| Biological samples | ||
| Human venous blood | Tokyo-Shinagawa Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RBD (amino acids: 331-529) | In-house | MN994467 |
| HiTrap Protein G HP Columns | Cytiva | 17-0404-01 |
| Tween-20 | Fujifilm | 167-11515 |
| Can Get Signal #2 | TOYOBO | NKB-301 |
| OPD substrate | Sigma-Aldrich | P8287 |
| D-MEM (Low Glucose) with L-Glutamine and Phenol Red | Fujifilm Wako Pure Chemicals | 041-29775 |
| D-MEM (High Glucose) with L-Glutamine and Phenol Red | Fujifilm Wako Pure Chemicals | 044-29765 |
| Bovine serum albumin | Sigma-Aldrich | A2153 |
| Fetal bovine serum | Biowest | S1780-500 |
| Fetal bovine serum | Nichirei Biosciences | 171012 |
| Geneticin | Thermo Fisher Scientific | 10131-027 |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15140-122 |
| Phosphate buffered saline (-) | Fujifilm Wako Pure Chemicals | 166-23555 |
| RPMI-1640 with L-glutamine and phenol red | Fujifilm Wako Pure Chemicals | 189-02025 |
| Penicillin-Streptomycin solution | Fujifilm Wako Pure Chemicals | 168-23191 |
| 0.5M EDTA | Thermo Fisher Scientific | 15575 |
| Bovine serum albumin fraction V | Roche | 10735086001 |
| CELLBANKER 1 plus | ZENOAQ RESOURCE | CD021 |
| Brilliant Stain Buffer Plus | BD Biosciences | 566385 |
| 7-amino-actinomycin D | Sigma-Aldrich | A9400 |
| Critical commercial assays | ||
| Bright-Glo luciferase assay system | Promega | E2620 |
| Expi293 expression system | Thermo Fisher Scientific | A29133 |
| Elecsys Anti-SARS-CoV-2 | Roche Diagnostics | 518316181 |
| Cytometric Bead Array Human IL-1β | BD Biosciences | 558279 |
| Cytometric Bead Array Human IL-2 | BD Biosciences | 558270 |
| Cytometric Bead Array Human IL-4 | BD Biosciences | 558272 |
| Cytometric Bead Array Human IL-5 | BD Biosciences | 558278 |
| Cytometric Bead Array Human IL-6 | BD Biosciences | 558276 |
| Cytometric Bead Array Human IL-8 | BD Biosciences | 558277 |
| Cytometric Bead Array Human IL-10 | BD Biosciences | 558274 |
| Cytometric Bead Array Human IL-12p70 | BD Biosciences | 558283 |
| Cytometric Bead Array Human IL-17F | BD Biosciences | 562151 |
| Cytometric Bead Array Human G-CSF | BD Biosciences | 558326 |
| Cytometric Bead Array Human GM-CSF | BD Biosciences | 558335 |
| Cytometric Bead Array Human IFN-α | BD Biosciences | 560379 |
| Cytometric Bead Array Human IFN-γ | BD Biosciences | 558269 |
| Cytometric Bead Array Human IP-10 | BD Biosciences | 558280 |
| Cytometric Bead Array Human MCP-1 | BD Biosciences | 558287 |
| Cytometric Bead Array Human MIG | BD Biosciences | 558286 |
| Cytometric Bead Array Human MIP-1α | BD Biosciences | 558325 |
| Cytometric Bead Array Human TNF-α | BD Biosciences | 560112 |
| Enhanced Sensitivity Cytometric Bead Array Human IFN-γ | BD Biosciences | 561515 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-1β | BD Biosciences | 561509 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-2 | BD Biosciences | 561517 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-4 | BD Biosciences | 561510 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-5 | BD Biosciences | 561511 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-6 | BD Biosciences | 561512 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-8 | BD Biosciences | 561513 |
| Enhanced Sensitivity Cytometric Bead Array Human IL-10 | BD Biosciences | 561514 |
| Enhanced Sensitivity Cytometric Bead Array Human IL- IL-12p70 | BD Biosciences | 561518 |
| Enhanced Sensitivity Cytometric Bead Array Human IL- TNF-α | BD Biosciences | 561516 |
| Human Soluble Protein Master Buffer Kit | BD Biosciences | 558265 |
| Human Enhanced Sensitivity Master Buffer Kit | BD Biosciences | 562248 |
| Experimental models: Cell lines | ||
| VeroE6/TMPRSS2 cells | JCRB Cell Bank | JCRB1819 |
| Software and algorithms | ||
| FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| FCAP Array Software Version 3.0 | BD Biosciences | https://www.bdbiosciences.com/us/applications/research/bead-based-immunoassays/analysis-software/fcap-array-software-v30/p/652099 |
| FlowJo v10.8.0 | BD Biosciences | http://www.flowjo.com/ |
| GraphPad Prism version 9 | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Python -Version 3.7.3 | Python Software Foundation. | https://www.python.org/ |
| UMAP (Python) - Version 0.5.2 | Mclnnes et al., 2018 | https://umap-learn.readthedocs.io/en/latest/ |
| Other | ||
| BD Vacutainer® CPT™ Tube | BD Biosciences | 362761 |
| iMark microplate reader | Bio-Rad | 168-1130J1 |
| Eposh2 | Biotek | EPOCH2NS |
| GroMax Navigator Microplate Luminometer | Promega | GM2000 |
| Cobas e 411 plus | Roche Diagnostics | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yoshimasa Takahashi (ytakahas@niid.go.jp).
Materials availability
This study did not generate new or unique reagents.
Experimental model and subject details
Human subjects
Healthcare workers without a prior history of SARS-CoV-2 infection at Tokyo-Shinagawa Hospital were enrolled. All participants were examined anti-nucleocapsid antibody titers at pre-vaccination (day 0) and post-vaccination (days 47-51) and excluded the samples from further analysis if they showed >0.1 antibody titer, which is lower than 1.0 cut-off recommended by manufacturer (Roche kit Elecsys® Anti-SARS-CoV-2 RUO) and even lower than the cut-off (0.128) described in previous report.47 All volunteers provided written informed consent prior to enrollment. The participants received two doses of the Pfizer/BioNTech BNT162b2 vaccine. Secondary vaccination was conducted on day 21 after primary vaccination. Blood samples were collected at five time points for longitudinal analyses: day 0 (the day of first vaccination), day 1, day 21 (the day of secondary vaccination), day 22, and day 47–51. The following information was collected for each participant: gender, age, BMI, pre-existing medical conditions (hypertension, diabetes mellitus, and dyslipidemia), and history of PCR-confirmed SARS-CoV-2 infection. All studies were approved by the Institutional Review Board of the National Institute of Infectious Diseases and Tokyo Shinagawa Hospital (Permit numbers: 1292 and 20-A-33). This study was performed in accordance with the principles of the Declaration of Helsinki.
Method details
Sample processing and cell isolation
Blood samples were collected in Vacutainer CPT tubes (BD Biosciences), and peripheral blood mononuclear cells (PBMCs) and plasma samples were isolated via centrifugation at 1800 × g for 20 min. PBMCs and plasma samples were separated by centrifugation at 300 × g for 15 min. Plasma samples were isolated via additional centrifugation at 800 × g for 15 min and stored at −80°C until further analysis. For ELISA and pseudotyped virus-neutralizing assays, plasma samples were used after heat-inactivation at 56°C for 30 min. For cytokine/chemokine quantification, plasma samples were used immediately after thawing at room temperature. PBMCs were centrifuged at 300 × g for 10 min and washed twice with phosphate-buffered saline (PBS; FUJIFILM Wako Chemicals). PBMCs were used immediately after isolation for analysis of MDSCs (e-MDSCs, M-MDSCs, and PMN-MDSCs), or cryopreserved at −135°C using CELLBANKER 1 plus (ZENOGEN PHARMA) for additional analysis.
Adverse events questionnaire
All participants completed two questionnaires on vaccine-related adverse events (AEs) that occurred during days 1–7 (primary vaccination-related AEs) and days 22–29 (secondary vaccination-related AEs). Questionnaires enquired about the presence of three local symptoms (pain, redness, and swelling) and eight systemic symptoms (fever, fatigue, headache, chills, vomiting, diarrhea, muscle pain, and joint pain), and the symptom severities were evaluated on five grades (grade 0–4, see Table S1). The criteria used for the clinical trial of the BNT162b2 mRNA vaccine were applied.1 Local 3 symptom severity scores and systemic 8 symptom severity scores were summed for the total local symptom severity grades and total systemic symptom severity grades, respectively.
ELISA
The recombinant RBD protein of SARS-CoV-2 isolate (GenBank: MN994467) was produced previously described.7 Briefly, the human codon-optimized nucleotide sequence encoding the RBD (amino acids: 331-529) with the signal peptide (amino: acids: 1-20; MIHSVFLLMFLLTPTESYVD) with a C-terminal histidine tag was cloned into the mammalian expression vector pCAGGS. The cloned expression vector was transfected into Expi293F cells according to the manufacturer’s instructions (Thermo Fisher Scientific) and the RBD protein was purified using Ni-NTA agarose (QIAGEN) from the culture supernatant of the transfected cells harvested for 5 days.
A recombinant reference monoclonal antibody (CR3022) was produced as previously described.48,49 Briefly, the VH and VL genes of CR3022 were cloned into expression vectors with human IgG1 heavy chain and kappa light chain, respectively. The prepared expression vectors were transfected into Expi293F and the recombinant monoclonal antibody was purified from the culture supernatant using a protein G column (Thermo Fisher Scientific).
RBD protein was coated at 2 μg/mL in F96 Maxisorp Nunc-Immuno plates (Thermo Fisher Scientific) overnight at 4°C. The plates were washed with PBS, and then blocked with 1% bovine serum albumin (BSA) in PBS for 1.5 h at room temperature. Heat-inactivated plasma samples and monoclonal antibodies were diluted in PBS containing 1% BSA and 0.1% Tween 20 (eight 4-fold serial dilutions, starting at 1:20 dilution for plasma; starting at 1 μg/mL for monoclonal antibody), then incubated overnight at 4°C. The following day, after washing the plates with PBS containing 0.05% Tween 20, HRP-conjugated goat anti-human IgG (Southern Biotech) in Can Get Signal Immunoreaction Enhancer Solution 2 (TOYOBO) was incubated for 1.5 h at room temperature. After washing the plates with PBS containing 0.05% Tween 20 and PBS, OPD substrate (Sigma) was added and OD490 was measured using an Epoch2 microplate reader (Biotek) and iMark microplate reader (Bio-Rad). IgG titers were quantified using CR3022 as a reference antibody in each plate. Threshold of RBD IgG titers for seropositive was determined in a previous publication (Moriyama et al., 2021) and shown as dotted lines in Figure 1B.
RBD IgG titers of COVID-19 convalescent patients were partially referred from data used in Figure 1C of a previous publication.7 The criteria for data selection were (1) time points 23–64 days after symptom onset, and (2) severe symptom severity defined by the WHO.
Pseudotyped virus neutralization assay
SARS-CoV-2 pseudotyped virus was generated as described previously.50 Briefly, the pCAGGS expression vector was generated with commercially synthesized cDNA of the SARS-CoV-2 spike protein (Integrated DNA Technologies Inc.) and the plasmid (pCAG-SARS-CoV-2) containing a 19 aa truncation at the C-terminus of the spike protein was constructed. The expression vector was transfected into 293T cells on collagen-coated tissue culture plates. After incubating for 24 h, the cells were infected with G-complemented VSVΔG/Luc51 at a multiplicity of infection of 0.5. After 24 h of incubation, VSV pseudotyped virus were collected from the centrifuged culture supernatants and stored at −80°C.
Pseudotyped virus neutralization assay was performed as previously described.7 Briefly, VeroE6/TMPRSS2 were maintained in DMEM (Fujifilm Wako Pure Chemical) containing 10% heat-inactivated fetal bovine serum (Biowest), 1 mg/mL geneticin (Thermo Fisher Scientific), and 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific). SARS-CoV-2 pseudotyped virus was incubated with heat-inactivated plasma sample (five 5-fold serial dilutions starting at 1:10 dilution) for 1 h at 37°C. After the incubation, the mixture was inoculated into VeroE6/TMPRSS2 cells seeded in 96-well solid white flat-bottom plates (Corning), and then incubated for 24 h at 37°C with 5% CO2. Luciferase activity in cultured cells was measured using the Bright-Glo Luciferase Assay System (Promega) with a GroMax Navigator Microplate Luminometer (Promega). Half-maximal inhibitory concentration (IC50) was calculated as neutralization titers. NT titers under the detection limit (IC50 = 20) were set to 20 and detection limit was indicated as dotted line in Figure 1C.
NT titers of COVID-19 convalescent patients were partially referred from data used in Figure 1D of a previous publication.7 The criteria of data selection were the same as described in ELISA section.
Flow cytometry
For MDSC analysis, after blocking non-specific antibody binding using Human TruStain FcX (1:200, BioLegend) for 5 min at room temperature, cells were stained for 2 h at 4°C with the following antibodies: CD45-APC-Alexa Fluor 700 (HI30, 1:300; BioLegend), CD3-BV605 (SK7, 1:300, BD Biosciences), CD19-BV605 (HIB19, 1:300, BioLegend), CD56-BV605 (5.1H11, 1:300 BioLegend), HLA-DR-PE-Cy7 (G46-6, 1:300, BD Biosciences), CD11b-FITC (ICRF44, 1:300 BioLegend), CD33-PE (P67.6, 1:300 BioLegend), CD14-BV421 (M5E2, 1:300, BD Biosciences), and CD15-APC (W6D3, 1:300 BioLegend). To analyze the 15 immune cell types of interest, cryopreserved PBMCs were thawed at 37°C and washed twice with RPMI 1640 (FUJIFILM Wako Pure Chemical Corporation) containing 10% heat-inactivated fetal bovine serum (Nichirei Biosciences), 2 mM glutamine (FUJIFILM Wako Pure Chemical Corporation), 100 U/mL penicillin (FUJIFILM Wako Pure Chemical Corporation), and 100 μg/mL streptomycin (FUJIFILM Wako Pure Chemical Corporation) before use. After blocking non-specific antibody binding as described above, cells were stained for 2 h at 4°C with the following antibodies: CD45-Brilliant Violet 570 (HI30, 1:150, BioLegend), CD3-BUV661 (HIT3a, 1:300, BD Biosciences), CD4-PE-Cy5 (RPA-T4, 1:100, BD Biosciences), CD8-BB660 (RPA-T8, 1:300, BD Biosciences), CD8-BV786 (RPA-T8, 1:300, BD Biosciences), CD19-BUV563 (SJ25C1, 1:300, BD Biosciences), CD56-BB515 (B159, 1:300, BD Biosciences), HLA-DR-APC-H7 (G46-6, 1:300, BD Biosciences), CD14-BV421 (M5E2, 1:300, BD Biosciences), CD16-BUV395 (3G8, 1:300, BD Biosciences), CD11c-APC-R700 (BU15, 1:300, BD Biosciences), CD88-PE-Cyanine7 (S5/1, 1:300 BioLegend), CD1c-BUV737 (F10/21A3, 1:300, BD Biosciences), CD123-BUV496 (6H6, 1:300, BD Biosciences), CD141-BV605 (1A4, 1:300, BD Biosciences), CD5-BV480 (UCHT2, 1:300, BD Biosciences), CD163-PE-CF594 (GHI/61, 1:300, BD Biosciences), CD163-BV786 (GHI/61, 1:300, BD Biosciences), Siglec-6-BV650 (767329, 1:300, BD Biosciences), Axl-BUV615 (108724, 1:300, BD Biosciences). To examine the expression of CCR2, CXCR3, CD86, and CD69, and the binding of Annexin V, cells were stained with the following antibodies: CD45-BUV805 (HI30, 1:150, BD Biosciences), CD3-BUV661 (HIT3a, 1:300, BD Biosciences), CD19-BUV563 (SJ25C1, 1:300, BD Biosciences), CD56-BB515 (B159, 1:300, BD Biosciences), HLA-DR-APC-H7 (G46-6, 1:300, BD Biosciences), CD14-BV421 (M5E2, 1:300, BD Biosciences), CD16-BUV395 (3G8, 1:300, BD Biosciences), CD11c-APC-R700 (BU15, 1:300, BD Biosciences), CD88-PE-Cyanine7 (S5/1, 1:300 BioLegend), CD1c-BUV737 (F10/21A3, 1:300, BD Biosciences), CD123-BUV496 (6H6, 1:300, BD Biosciences), CD141-BV605 (1A4, 1:300, BD Biosciences), CD5-BV480 (UCHT2, 1:300, BD Biosciences), CD163-PE-CF594 (GHI/61, 1:300, BD Biosciences), Siglec-6-BV650 (767329, 1:300, BD Biosciences), Axl-BUV615 (108724, 1:300, BD Biosciences), CCR2-Brilliant Violet 785 (K036C2, 1:300, BioLegend), CXCR3-PE/Cyanine5 (G025H7, 1:300, BioLegend), CD86-PE (IT.2, 1:50, BD Biosciences), CD69-BV711 (FN50, 1:300, BD Biosciences), Annexin V-Alexa647 (1:50, Thermo Fisher Scientific) Brilliant Violet 785 Mouse IgG2a, κ Iso-type control (MOPC-173, 1:300, BioLegend), PE/Cyanine5 Mouse IgG1, κ Iso-type control (MOPC-21, 1:300, BioLegend). Subsequently, the cells were washed twice and resuspended in PBS containing 0.5% BSA, 5 mM EDTA (Thermo Fisher Scientific), and 0.25 μg/mL 7-AAD (Sigma) to detect dead cells.
MDSCs were analyzed using a FACSCanto II (BD Biosciences) or FACSAria III cytometer (BD Biosciences) and FACSDiva v.9.0 software (BD Biosciences), and 15 immune cells other than MDSCs were analyzed using a FACSymphony S6 cytometer (BD Biosciences) and FACSDiva v.9.1.2 software (BD Biosciences). Expression of CCR2, CXCR3, CD86, and CD69, and binding of Annexin V were analyzed using a FACSymphony A3 cytometer (BD Biosciences) and FACSDiva v.9.1 software (BD Biosciences).
FCS files were analyzed using FlowJo software (v.10.8.0, BD Biosciences). Gating strategies for identifying immune cells are shown in Figure S1. The frequency of each immune cell population was calculated as a proportion of CD45+ 7-AAD- cells (live CD45+ cells). Geometric mean fluorescence intensity (GMFI) was calculated using FlowJo software. The frequency of apoptotic cells (Annexin V+) in each cell type was calculated as a proportion to each cell. Immune cell dynamics were expressed as the fold increases or decreases from pre- (day 0 or day 21) to post-vaccination (day 1 or day 22). Z-scores were generated to validate AE-related cellular parameters by log-2 transforming each immune cell post/pre ratio, which were normalized to have a mean of 0 and a variance of 1. Composite scores were then calculated by directly summing these normalized values. All other parameters were analyzed without additional data transformation.
High-dimensional analysis was performed using the scikit-learn python library. Frequency data of 18 immune cell types at four time points (day 0, 1, 21, and 22) were standardized by removing the mean and scaling to unit variance, and dimensionality reduction to a two-dimensional space was performed using UMAP (Figures 1J and S3).
In cases that the cell post/pre ratios inversely correlated with AEs or NT antibody responses, the cell post/pre ratios below the median of the cohort were identified as "positive”, and vice versa for "negative”. In Figures 5 and 6, and Figure S6, participants whose cellular parameters were all "positive” (all parameters were below the median of the cohort) were defined as the matched group, and vice versa.
Cytokine/chemokine quantification
The plasma levels of 18 cytokines/chemokines were quantified using a cytometric bead array kit (BD Biosciences) according to the manufacturer’s instructions. Plasma samples were diluted 3–4 fold for analysis. Data were acquired using a FACSCanto II cytometer (BD Biosciences) and analyzed using FCAP Array Software Version 3.0 (BD Biosciences). For plasma cytokine/chemokine levels below the detection limit (40 pg/mL for standard CBA kit, and 0.822 pg/mL for enhanced sensitivity CBA kit), the value was set to 40 pg/mL or 0.822 pg/mL.
Quantification and statistical analysis
Statistical analyses and visual representation were performed using Prism 9 (GraphPad). Age, BMI, RBD IgG, pseudotyped virus NT titer, local or systemic symptom severity scores, cytokine/chemokine post/pre ratios, cytokine/chemokine concentration and expression of cell surface molecules were compared using the Mann-Whitney test (Figures 1A, 5C–5H, 6C–6H, S6C and S7C) or Kruskal-Wallis test followed by Dunn’s post hoc test (Figures 1B–1D, 3B–3E and 3G–3J).
Longitudinal analysis of local and systemic symptom scores (Figure 1E), immune cell frequencies (Figure S2) and plasma cytokine/chemokine concentrations (Figure S4A) were performed using the Wilcoxon test. Fisher exact test was performed to examine the statistical difference in the incidence of symptoms between primary and secondary vaccinations (Figures 1F and 1G) and gender distributions in Ab response 3 or 4-prameter matched and unmatched group, and AE 2 or 3-parameter matched group (described in the Results section).
Spearman correlation analysis was performed between primary and secondary symptom severity scores (Figures 1H and 1I), cell post/pre ratios, antibody responses, symptom severity scores, plasma cytokine/chemokine post/pre ratios, and Z-scores combining cell post/pre ratio (Figures 2, 3A, 3F, 3K, 4, 5B, 6B, S4B, S5, S6B and S7B). Correlations were considered significant at p < 0.05. Benjamini-Hochberg correction was performed in correlation analysis to adjust false discovery rate (FDR).
In all statistical analyses, significance was considered as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Ryutaro Iwabuchi, Akira Dosaka, Eriko Izumiyama, Emi Kohda, Rieko Iwaki, Tami Kobayashi, Chiyuki Asano, Naoka Yoshida, and Kayoko Mizuno for technical support and Ryoko Itami for secretarial assistance. This work was supported by Japan Agency for Medical Research and Development grants (grant numbers JP20fk0108534 and JP22mk0101224 to T.M. and Y.T.).
Author contributions
Conceptualization, T.M. and Y.T.; methodology, T.T., K. Tomii, K.Y., and T.M.; investigation, T.T., M.M., Y.A., K.K., N.S., S.M., L.S., M.I., A.N., T.O., K. Terahara, K. Tonouchi, M.N., K. Tomii, K.Y., and T.M.; funding acquisition, T.M. and Y.T.; project administration, K.K., T.M., M.S., and Y.T.; supervision: M.S. and Y.T.; writing – original draft, T.T., T.M., and Y.T.; writing – review & editing, T.T., Y.A., N.S., S.M., M.I., K. Terahara, K. Tomii, K.Y., T.M., M.S., and Y.T.
Declaration of interests
N.S. is an employee of KOTAI Biotechnologies, Inc. K.Y. is a founder, shareholder, and board director of KOTAI Biotechnologies, Inc. All other authors declare that they have no competing interests.
Published: April 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100631.
Contributor Information
Takayuki Matsumura, Email: matt@niid.go.jp.
Masaharu Shinkai, Email: shinkai050169@gmail.com.
Yoshimasa Takahashi, Email: ytakahas@niid.go.jp.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakaya H.I., Hagan T., Duraisingham S.S., Lee E.K., Kwissa M., Rouphael N., Frasca D., Gersten M., Mehta A.K., Gaujoux R., et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wimmers F., Donato M., Kuo A., Ashuach T., Gupta S., Li C., Dvorak M., Foecke M.H., Chang S.E., Hagan T., et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell. 2021;184:3915–3935.e21. doi: 10.1016/j.cell.2021.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natrajan M.S., Rouphael N., Lai L., Kazmin D., Jensen T.L., Weiss D.S., Ibegbu C., Sztein M.B., Hooper W.F., Hill H., et al. Systems vaccinology for a live attenuated tularemia vaccine reveals unique transcriptional signatures that predict humoral and cellular immune responses. Vaccines (Basel) 2019;8:4. doi: 10.3390/vaccines8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriyama S., Adachi Y., Sato T., Tonouchi K., Sun L., Fukushi S., Yamada S., Kinoshita H., Nojima K., Kanno T., et al. Temporal maturation of neutralizing antibodies in COVID-19 convalescent individuals improves potency and breadth to circulating SARS-CoV-2 variants. Immunity. 2021;54:1841–1852.e4. doi: 10.1016/j.immuni.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modenese A., Paduano S., Bargellini A., Bellucci R., Marchetti S., Bruno F., Grazioli P., Vivoli R., Gobba F. Neutralizing anti-SARS-CoV-2 antibody titer and reported adverse effects, in a sample of Italian nursing home personnel after two doses of the BNT162b2 vaccine administered four weeks apart. Vaccines (Basel). 2021;9:652. doi: 10.3390/vaccines9060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michos A., Tatsi E.B., Filippatos F., Dellis C., Koukou D., Efthymiou V., Kastrinelli E., Mantzou A., Syriopoulou V. Association of total and neutralizing SARS-CoV-2 spike -receptor binding domain antibodies with epidemiological and clinical characteristics after immunization with the 1(st) and 2(nd) doses of the BNT162b2 vaccine. Vaccine. 2021;39:5963–5967. doi: 10.1016/j.vaccine.2021.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coggins S.A.A., Laing E.D., Olsen C.H., Goguet E., Moser M., Jackson-Thompson B.M., Samuels E.C., Pollett S.D., Tribble D.R., Davies J., et al. Adverse effects and antibody titers in response to the BNT162b2 mRNA COVID-19 vaccine in a prospective study of healthcare workers. Open Forum Infect. Dis. 2021;9:ofab575. doi: 10.1093/ofid/ofab575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda K., Amano M., Uemura Y., Tsuchiya K., Matsushima T., Noda K., Shimizu Y., Fujiwara A., Takamatsu Y., Ichikawa Y., et al. Correlates of neutralizing/SARS-CoV-2-S1-binding antibody response with adverse effects and immune kinetics in BNT162b2-vaccinated individuals. medRxiv. 2021 doi: 10.1101/2021.07.27.21261237. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naaber P., Tserel L., Kangro K., Sepp E., Jurjenson V., Adamson A., Haljasmagi L., Rumm A.P., Maruste R., Karner J., et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg. Health Eur. 2021;10:100208. doi: 10.1016/j.lanepe.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menni C., Klaser K., May A., Polidori L., Capdevila J., Louca P., Sudre C.H., Nguyen L.H., Drew D.A., Merino J., et al. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: a prospective observational study. Lancet Infect. Dis. 2021;21:939–949. doi: 10.1016/s1473-3099(21)00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 18.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slauenwhite D., Johnston B. Regulation of NKT cell localization in homeostasis and infection. Front. Immunol. 2015;6:255. doi: 10.3389/fimmu.2015.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaya M., Barral P., Burbage M., Aggarwal S., Montaner B., Warren Navia A., Aid M., Tsui C., Maldonado P., Nair U., et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell. 2018;172:517–533.e20. doi: 10.1016/j.cell.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingaropoli M.A., Perri V., Pasculli P., Cogliati Dezza F., Nijhawan P., Savelloni G., La Torre G., D'Agostino C., Mengoni F., Lichtner M., et al. Major reduction of NKT cells in patients with severe COVID-19 pneumonia. Clin. Immunol. 2021;222:108630. doi: 10.1016/j.clim.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox A., Cevik H., Feldman H.A., Canaday L.M., Lakes N., Waggoner S.N. Targeting natural killer cells to enhance vaccine responses. Trends Pharmacol. Sci. 2021;42:789–801. doi: 10.1016/j.tips.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergamaschi C., Terpos E., Rosati M., Angel M., Bear J., Stellas D., Karaliota S., Apostolakou F., Bagratuni T., Patseas D., et al. Systemic IL-15, IFN-γ, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep. 2021;36:109504. doi: 10.1016/j.celrep.2021.109504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto S., Fukunaga A., Tanaka A., Takeuchi J.S., Inoue Y., Kimura M., Maeda K., Ueda G., Mizoue T., Ujiie M., et al. Association between reactogenicity and SARS-CoV-2 antibodies after the second dose of the BNT162b2 COVID-19 vaccine. Vaccine. 2022;40:1924–1927. doi: 10.1016/j.vaccine.2022.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azuma M., Ito D., Yagita H., Okumura K., Phillips J.H., Lanier L.L., Somoza C. B70 antigen is a second ligand for CTLA-4 and CD28. Nature. 1993;366:76–79. doi: 10.1038/366076a0. [DOI] [PubMed] [Google Scholar]
- 26.McLellan A.D., Starling G.C., Williams L.A., Hock B.D., Hart D.N.J. Activation of human peripheral blood dendritic cells induces the CD86 co-stimulatory molecule. Eur. J. Immunol. 1995;25:2064–2068. doi: 10.1002/eji.1830250739. [DOI] [PubMed] [Google Scholar]
- 27.Clausen J., Vergeiner B., Enk M., Petzer A.L., Gastl G., Gunsilius E. Functional significance of the activation-associated receptors CD25 and CD69 on human NK-cells and NK-like T-cells. Immunobiology. 2003;207:85–93. doi: 10.1078/0171-2985-00219. [DOI] [PubMed] [Google Scholar]
- 28.Gorski K.S., Waller E.L., Bjornton-Severson J., Hanten J.A., Riter C.L., Kieper W.C., Gorden K.B., Miller J.S., Vasilakos J.P., Tomai M.A., Alkan S.S. Distinct indirect pathways govern human NK-cell activation by TLR-7 and TLR-8 agonists. Int. Immunol. 2006;18:1115–1126. doi: 10.1093/intimm/dxl046. [DOI] [PubMed] [Google Scholar]
- 29.Chen J., Vistica B.P., Takase H., Ham D.I., Fariss R.N., Wawrousek E.F., Chan C.C., DeMartino J.A., Farber J.M., Gery I. A unique pattern of up- and down-regulation of chemokine receptor CXCR3 on inflammation-inducing Th1 cells. Eur. J. Immunol. 2004;34:2885–2894. doi: 10.1002/eji.200425318. [DOI] [PubMed] [Google Scholar]
- 30.Cagigi A., Lore K. Immune responses induced by mRNA vaccination in mice, monkeys and humans. Vaccines (Basel). 2021;9:61. doi: 10.3390/vaccines9010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Rohss J., John S., Hassett K., Yuzhakov O., Bahl K., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farsakoglu Y., Palomino-Segura M., Latino I., Zanaga S., Chatziandreou N., Pizzagalli D.U., Rinaldi A., Bolis M., Sallusto F., Stein J.V., Gonzalez S.F. Influenza vaccination induces NK-Cell-Mediated type-II IFN response that regulates humoral immunity in an IL-6-dependent manner. Cell Rep. 2019;26:2307–2315.e5. doi: 10.1016/j.celrep.2019.01.104. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Fontecha A., Thomsen L.L., Brett S., Gerard C., Lipp M., Lanzavecchia A., Sallusto F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat. Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 34.Rydyznski C.E., Waggoner S.N. Boosting vaccine efficacy the natural (killer) way. Trends Immunol. 2015;36:536–546. doi: 10.1016/j.it.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourdely P., Anselmi G., Vaivode K., Ramos R.N., Missolo-Koussou Y., Hidalgo S., Tosselo J., Nunez N., Richer W., Vincent-Salomon A., et al. Transcriptional and functional analysis of CD1c(+) human dendritic cells identifies a CD163(+) subset priming CD8(+)CD103(+) T cells. Immunity. 2020;53:335–352.e8. doi: 10.1016/j.immuni.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cytlak U., Resteu A., Pagan S., Green K., Milne P., Maisuria S., McDonald D., Hulme G., Filby A., Carpenter B., et al. Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. 2020;53:353–370.e8. doi: 10.1016/j.immuni.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutertre C.A., Becht E., Irac S.E., Khalilnezhad A., Narang V., Khalilnezhad S., Ng P.Y., van den Hoogen L.L., Leong J.Y., Lee B., et al. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity. 2019;51:573–589.e8. doi: 10.1016/j.immuni.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Alcantara-Hernandez M., Leylek R., Wagar L.E., Engleman E.G., Keler T., Marinkovich M.P., Davis M.M., Nolan G.P., Idoyaga J. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. 2017;47:1037–1050.e6. doi: 10.1016/j.immuni.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leylek R., Alcantara-Hernandez M., Lanzar Z., Ludtke A., Perez O.A., Reizis B., Idoyaga J. Integrated cross-species analysis identifies a conserved transitional dendritic cell population. Cell Rep. 2019;29:3736–3750.e8. doi: 10.1016/j.celrep.2019.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.See P., Dutertre C.A., Chen J., Gunther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Handler K., Duan K., et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:eaag3009. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manetti R., Parronchi P., Giudizi M.G., Piccinni M.P., Maggi E., Trinchieri G., Romagnani S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J. Exp. Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh C.S., Macatonia S.E., Tripp C.S., Wolf S.F., O'Garra A., Murphy K.M. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 44.Harrington L.E., Hatton R.D., Mangan P.R., Turner H., Murphy T.L., Murphy K.M., Weaver C.T. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 45.Revu S., Wu J., Henkel M., Rittenhouse N., Menk A., Delgoffe G.M., Poholek A.C., McGeachy M.J. IL-23 and IL-1β drive human Th17 cell differentiation and metabolic reprogramming in absence of CD28 costimulation. Cell Rep. 2018;22:2642–2653. doi: 10.1016/j.celrep.2018.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales-Nunez J.J., Munoz-Valle J.F., Meza-Lopez C., Wang L.F., Machado Sulbaran A.C., Torres-Hernandez P.C., Bedolla-Barajas M., De la O.G.B., Balcazar-Felix P., Hernandez-Bello J. Neutralizing antibodies titers and side effects in response to BNT162b2 vaccine in healthcare workers with and without prior SARS-CoV-2 Infection. Vaccines (Basel) 2021;9:742. doi: 10.3390/vaccines9070742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Public Health England Evaluation of sensitivity and specificity of four commercially available SARS-CoV-2 antibody immunoassays. 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/898437/Evaluation__of_sensitivity_and_specificity_of_4_commercially_available_SARS-CoV-2_antibody_immunoassays.pdf
- 48.Adachi Y., Tonouchi K., Nithichanon A., Kuraoka M., Watanabe A., Shinnakasu R., Asanuma H., Ainai A., Ohmi Y., Yamamoto T., et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat. Commun. 2019;10:3883. doi: 10.1038/s41467-019-11821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tani H., Kimura M., Tan L., Yoshida Y., Ozawa T., Kishi H., Fukushi S., Saijo M., Sano K., Suzuki T., et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein. Virol. J. 2021;18:16. doi: 10.1186/s12985-021-01490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tani H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., Matsuura Y. Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 2010;84:2798–2807. doi: 10.1128/jvi.02499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.