Abstract
Background
It has been found that patients recovered from COVID 19 may still test Reverse Transcriptase- Polymerase Chain Reaction (RT- PCR) positive without being infectious; the reasons are unclear. The occurrence of false-negative results of RT- PCR interferes with a proper diagnosis. The objectives of that work were to determine factors associated with persistently detectable SARS-CoV-2 RNA among recovered hospitalized patients and to determine the incidence of false-negative RT-PCR results and associated factors.
Methods
Relevant data were collected from 482 COVID 19 patients hospitalized in six referral centers from four countries.
Results
The median duration of RT- PCR conversion to negative was 20 days. Out of 482 studied patients, 8.7% tested positive after more than four weeks and were considered prolonged convertors. Binary logistic regression analysis revealed headache as an independent risk factor for short conversion time while fever, hypertension, chronic obstructive pulmonary disease, lymphopenia, elevated erythrocyte sedimentation rate, and the number of lobes affected, and bilateralism were found to be independent risk factors for prolonged positivity. Eighteen patients had initial negative results then turned positive after 24–48 h. Associated factors and outcomes were identified.
Conclusion
Identifying patients with a high likelihood of COVID-19 despite a negative RT-PCR is critical for effective clinical care. However, patient isolation resumption depending on positive RT-PCR despite clinical and radiological recovery is an overrating that greatly burdens the health sector.
Keywords: RT-PCR, Conversion time, COVID 19, False-negative
1. Introduction
Since the beginning of the Coronavirus disease 2019 (COVID- 19) pandemic, the Center for Disease Control and Prevention (CDC) and medical researchers worldwide have been conducting studies to understand better the rapidly evolving issues related to SARS- CoV-2. They continue to share their conclusions whenever new findings are available. Existing information proposes that patients with mild-to-moderate COVID-19 continue to test RT- PCR positive for about ten days behind symptom commencement. Most patients with more severe-to-critical illness or those who are severely immunocompromised presumably stay positive for approximately 20 days after symptom commencement. Such prolonged positivity does not imply that the people are a transmission risk [1,2]. However, several reports have documented severely immunocompromised people shedding replication-competent viruses for more than 20 days [[3], [4], [5], [6], [7]]. Based on these data, CDC has updated isolation and quarantine recommendations for patients recovered from COVID- 19 in December 2021 [8].
Among several diagnostic tests for SARS-CoV-2 infection, RT-PCR of upper respiratory tract samples, including a nasopharyngeal (NSP), nasal, or oropharyngeal (OP) specimen, has obtained Emergency Use Authorizations from the Food and Drug Administration, but not diagnostic approval [9]. However, due to the simplicity of procedures, RT-PCR-based diagnostic tests are regarded as the gold standard for detecting SARS-CoV-2 infection. The occurrence of false-negative results of RT- PCR is an obstacle to proper diagnosis and consequently optimal patient management Initial false-negative NPS result is a negative test result despite a consistent clinical illness, compatible thoracic imaging, with or without pursuing positive COVID-19 antibodies testing [10].
The present work aimed to determine factors associated with persistently detectable SARS-CoV-2 RNA among hospitalized patients recovering from severe to critically ill or those who are severely immunocompromised and to identify the incidence of false-negative RT-PCR results together with chaperoning factors.
2. Materials and methods
2.1. Ethical approval
The study has been approved by the Institutional Review Board (IRB) of the Faculty of Medicine, Port Said University, Port Said, Egypt.
2.2. Data collection
The present work retrospectively explored the demographic criteria, clinical features, laboratory and radiological parameters, treatments, and outcomes of 482 hospitalized COVID-19 patients confirmed by RT-PCR NSP and OP swabs testing. Information about RT-PCR included results at admission and after one-, two-, three-, and four-weeks intervals.
Data were collected via an online Google Form Questionnaire from six referral centers in four participating countries: (1) Port Said University Hospital, (2) Zagazig University Hospital, and (3) Altaiseer Private Hospital, Egypt, (4) Kazemia Teaching Hospital, Iraq, (5) Sancaktepe Sehit Prof Dr. Ilhan Varank Training and Research Hospital, Turkey, and (6) Clinical Hospital for Infectious Diseases, Romania from July 1st to August 8th, 2021.
2.3. Hospital admission criteria
The admission decision of COVID-19 patients in each center followed the COVID-19 guidelines of the Ministry of Health in each country. In Egypt, mild symptomatic cases (fever, sore throat, dry cough, malaise, and body aches or nausea, vomiting, abdominal pain, and loose stools) without radiologic findings of pneumonia and moderate cases with positive radiologic findings of pneumonia were treated in a hospital only if associated with high risk for severe disease or mortality. Risk factors include age ≥65 years, temperature >38, uncontrolled comorbidities (diabetes mellitus, hypertension, coronary artery disease, chronic kidney, liver, and lung diseases, or immunocompromised states), pregnancy, obesity (BMI>40), O2 saturation ≤92% at room air, heart rate ≥110/min, respiratory rate >24/min, neutrophil/lymphocyte ratio on CBC ≥3.1. Severe cases (with O2 saturation <92% at room air, respiratory rate >30/min, PaO2/FiO2 ratio <300, chest radiologic findings showing >50% lesion or progressive lesion within 24–48 h) and critically ill cases (with O2 saturation <92%, respiratory rate >30/min, PaO2/FiO2 ratio <200 despite oxygen therapy) were admitted to intermediate care and intensive care, respectively [11]. In Turkey, patients admitted to hospital were (a) those who had mild-to-moderate pneumonia (fever, muscle/joint pains, cough, sore throat, and nasal congestion) with respiratory rate ≥24 and O2 saturation ≤93%, or poor blood tests on admission (blood lymphocyte count <800/μl or serum CRP> 10 mg/L, serum ferritin >500 ng/mL or D-Dimer >1000 ng/mL), (b) those who had severe pneumonia (respiratory ≥30/min, O2 ≤ 90% at room air, bilateral diffuse lung involvement >50% on imaging); however, (c) patients considered to need intensive care were those who had dyspnea, respiratory distress, change of consciousness, respiratory ≥30/min, PaO2/FiO2 ratio <300, increased need of oxygen on follow up, hypotension (<90/60 mmHg, mean arterial blood pressure <65 mmHg), heart rate >100/min, acute organ dysfunction (renal damage, liver dysfunction, confusion, hemorrhage, and immunosuppression), arrythmia, increased troponin level, or lactate > 2 mmol, sepsis, and septic shock are admitted to intensive care [12]. In Iraq and Romania, symptomatic cases were admitted to the hospital and intensive care unit according to WHO COVID-19 related guidelines [13].
2.4. Sample collection, real time RT-PCR testing and virus strain
The sample collection procedures in all the participating centers were established on the recommendation from the CDC to limit the use of test resources and maximize the sensitivity. Two simultaneous NSP and OP swabs (except for Iraq, only NSP swabs) were taken from each study participant by a trained healthcare provider and admixed in a viral transport medium [14]. The real-time PCR kits, the real-time platforms, and the negative cycle threshold (Ct) values in the participating centers are summarized in Table 1 . A positive control template, negative extraction control, and a negative amplification control with RNase-free water were included in each run.
Table 1.
RT- PCR platforms, RT-PCR kits, and cycle threshold for negative sample in different referral canters.
| Country | RT- PCR Platforms | Real time PCR kit | Negative Ct value |
|---|---|---|---|
| Egypt | -Applied Biosystems ABI 7500 Fasta | - LiliF™ COVID-19 Multi Real-Time RT-PCR Kit (Intron Biotechnology) | -No Ct values reported or Ct value > 35 |
| -QuantStudio 5 Dxb | -TaqPath™ COVID-19 Combo Kit (Thermo Fischer Scientific) | -No Ct values reported or Ct value ≥ 40 | |
| -BioRad CFX96 Touchc | -Allplex™ SARS-CoV-2 Assay (Seegene) | -No Ct values reported or Ct value > 40 | |
| Iraq | -BioRad iCycler IQ | SARS-CoV-2 Nucleic Acid Detection Kit (Zybio Inc) | No Ct values reported or Ct value ≥ 40 |
| Turkey | -BioRad iCycler IQ | -Bio-Speedy SARS CoV-2 Double Gene RT-qPCR (Bioeksen R&D Technologies Ltd.) | -No Ct values reported or Ct value ≥ 38 |
| Romania | -Applied Biosystems 7500 Fast Dx | -PowerCheck 2019-nCoV Real-Time PCR Kit (Kogene Biotech) | -No Ct values reported or Ct value ≥ 40 |
RT-PCR, reverse transcription polymerase chain reaction; Ct, cycle threshold.
Port Said University Hospital.
Zagazig University Hospital.
Altaiseer Private Hospital.
The data regarding the virus strains predominated when patients admitted to hospitals were dependent on WHO Weekly Epidemiological Update during the study period [15]. According to WHO Weekly Epidemiological Update, the Delta variant has emerged and predominated in almost all countries.
2.5. Statistical analysis
Statistical Package for the Social Sciences (SPSS) software version 24 was employed to execute data analysis. As indicated, continuous variables were expressed as mean and standard deviation (±SD) or median and interquartile range (IQR). They were compared using an independent t-test or Mann–Whitney U test when appropriate. Categorical variables were expressed as number and percentage (n%) and were compared using Chi-square and Fisher exact tests when appropriate. Multivariable binary logistic regression analysis was applied to identify independent risk factors associated with prolonged RT-PCR conversion time. Probability (P) value ≤ 0.05 was considered significant.
3. Results
3.1. Patient characteristics and their effect on duration of persistence of SARS-CoV-2 RNA by univariate analysis
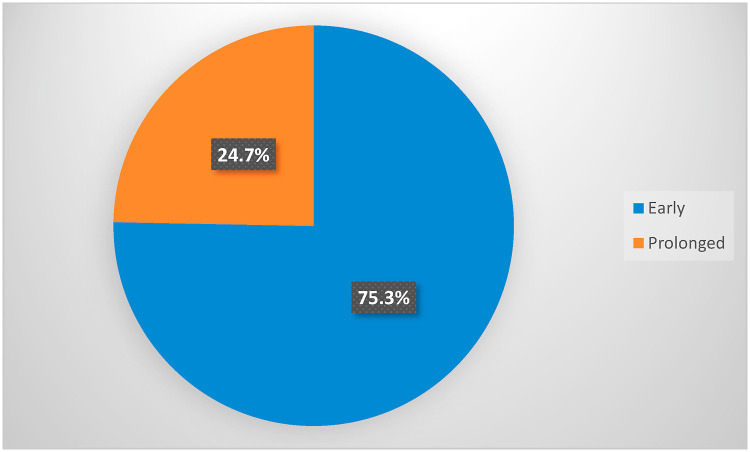
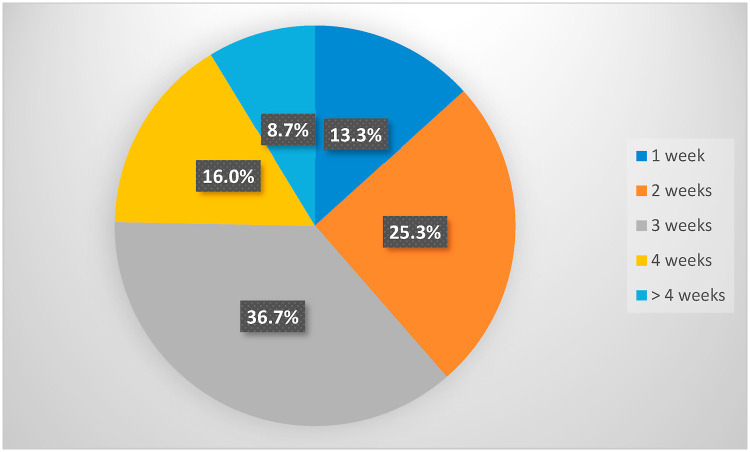
The present study included a total of 482 hospitalized COVID-19 patients. The diagnosis was confirmed by clinical, radiological, and laboratory investigations. The median of PCR conversion to negative was 20 days. Patients were dichotomized into two groups based on the median RT-PCR conversion time (3 weeks) to negative. Patients with persistent positive RT- PCR at ≥3 weeks intervals were considered prolonged RT-PCR negative converters. After initial positive, nearly three-quarters of patients were early converters, and nearly one-quarter was prolonged converter, Fig. 1 . The detailed distribution of conversion time to negativity among the study population is shown in Fig. 2 . The univariate analysis of sociodemographic characteristics and clinical features with their impact on the length of persistence of COVID-19 RNA is demonstrated in Table 2 .
Fig. 1.
Distribution of early and prolonged RT-PCR conversion time among the study participants.
Fig. 2.
Detailed distribution of RT-PCR conversion time among the study participants.
Table 2.
Sociodemographic characteristics and clinical features and their effect on length of persistence of COVID- 19 RNA.
| Variable | ≤20 days (n = 363) n (%) | >20 days (n = 119) n (%) | P value |
|---|---|---|---|
| Age | 0.25 | ||
| - ≤29 years | 21(5.8) | 3(2.5) | |
| - 30–39 years | 52(14.3) | 14(11.8) | |
| - 40–49 years | 62(17.1) | 16(13.4) | |
| - 50–59 years | 84(23.1) | 28(23.5) | |
| - 60–69 years | 80(22.0) | 32(26.9) | |
| - 70–79 years | 46(12.7) | 14(11.8) | |
| - ≥80 years |
18(5.0) |
12(10.1) |
|
| Contact of COVID-19 | 0.001a | ||
| - No | 139(38.3) | 26(21.8) | |
| - Yes |
224(61.7) |
93 (78.2) |
|
| Smoking | 0.57 | ||
| - No | 245 (67.5) | 77 (64.7) | |
| - Yes |
118(32.5) |
42 (35.3) |
|
| Symptoms | |||
| - Cough | 293(80.7) | 100(84.0) | 0.41 |
| - Body aches | 273 (75.2) | 84 (70.6) | 0.31 |
| - Fever | 246(67.8) | 99 (83.2) | 0.001a |
| - Dyspnea | 198 (54.5) | 73 (61.3) | 0.19 |
| - Headache | 198(54.5) | 44(37.0) | 0.001a |
| - Expectoration | 89(24.5) | 26(21.8) | 0.55 |
| - Sore throat | 127(35.0) | 28 (23.5) | 0.02a |
| - Digestive symptoms: | |||
|
14(3.9) | 8(6.7) | 0.19 |
|
273(75.2) | 88(73.9) | 0.78 |
| - Loss of smell | 145(39.9) | 20(16.8) | <0.001a |
| - Loss of taste |
120(33.1) |
16 (13.4) |
<0.001a |
| Days of symptoms before admission | <0.001a | ||
| - < 7 days | 253(69.7) | 60(50.4) | |
| - ≥ 7 days |
110(30.3) |
59(49.6) |
|
| Comorbidity | |||
| - Hypertension | 158(43.5) | 67(56.3) | 0.01a |
| - Diabetes | 95(26.2) | 45 (37.8) | 0.015a |
| - Ischemic heart disease | 56(15.4) | 18(15.1) | 0.93 |
| - COPD | 28(7.7) | 24(20.2) | <0.001a |
| - Othersb | 71 (22.7) | 45(26.6) | 0.33 |
Coronavirus disease 2019, COVID-2019; RNA, ribonucleic acid; COPD, chronic obstructive pulmonary disease.
Significant difference.
Others included heart failure (5.6%), renal disease (8.7%), hepatic disease (8.9%), bronchial asthma (9.3%), immunosuppressive illness (2.7%), HIV (0.4%) and other illnesses (2.1%).
3.2. Laboratory parameters, CT imaging, medications, and patient outcome
Table 3 shows laboratory parameters, chest CT imaging, medications, and patient outcome and their association with length of persistence of COVID- 19 RNA by univariate analysis.
Table 3.
Laboratory parameters, CT imaging, medications, and patient outcome and their association with length of persistence of COVID- 19 RNA.
| Variable | ≤20 days (n = 363) n (%) | >20 days (n = 119) n (%) | P value |
|---|---|---|---|
| Total Leucocytic count | |||
| - Leukopenia | 72 (19.8) | 17(14.3) | 0.03a |
| - Leukocytosis | 105 (28.9) | 49(41.2) | |
| Neutrophils | |||
| - Neutropenia | 73 (19.1) | 23(19.3) | 0.03a |
| - Neutrophilia | 113 (31.2) | 51(42.8) | |
| Lymphocytes | |||
| - Lymphopenia | 253 (69.7) | 92(77.3) | 0.002a |
| - Lymphocytosis | 23 (6.3) | 5(4.2) | |
| Platelets | |||
| - Thrombocytopenia |
34 (9.4) |
18 (15.1) |
0.07 |
| ESR | <0.001a | ||
| - Normal | 124 (34.2) | 16(13.8) | |
| - Elevated |
239 (65.8) |
103 (86.6) |
|
| CRP | 0.04a | ||
| - Normal | 23 (6.3) | 2(1.7) | |
| - Elevated |
340(93.7) |
117(98.3) |
|
| LDH a (U/L)b |
259(188–403.75) |
366(217–610) |
0.007a |
| D dimer a (ng/mL)b |
585(0.94–935.0) |
807.5(0.81–.1987) |
0.5 |
| Ferritin a (ng/mL)b |
332(150–552) |
540(345–540) |
0.04a |
| ALT serum level | 0.15 | ||
| - Normal | 234(64.5) | 68(57.1) | |
| - Elevated |
129(35.5) |
51(42.9) |
|
| AST serum level | 0.25 | ||
| - Normal | 261(71.9) | 79(66.4) | |
| - Elevated |
102(28.1) |
40(33.6) |
|
| Serum creatinine level | 0.001a | ||
| - Normal | 296(81.5) | 79(66.4) | |
| - Elevated |
67(18.5) |
40 (33.6) |
|
| Blood glucose level | 0.006a | ||
| - Hypoglycemia | 7(1.9) | 0(0.0) | |
| - Hyperglycemia | 125(34.4) | 59 (49.6) | |
| - Euglycemia |
231(63.6) |
60(50.4) |
|
| Radiological findings | <0.001a | ||
| CT pattern | |||
| - Presence of GGO and consolidation | 146(40.2) | 76(93.6) | |
| - Presence of GGO without consolidation | 119(32.8) | 36(30.3) | |
| - Presence of consolidation without GGO |
52(14.3) |
6(5.0) |
|
| Number of affected lobes | <0.001a | ||
| - One lobe | 15(4.1) | 4(3.4) | |
| - Two lobes | 80(22.0) | 15(12.6) | |
| - Three lobes | 115(31.7) | 42(35.3) | |
| - Four lobes | 83(22.9) | 43(36.1) | |
| - Five lobes |
24(6.6) |
14(11.8) |
|
| Bilateralism | 0.003a | ||
| - Unilateral | 29(8.0) | 9(7.6) | |
| - Bilateral |
289(79.6) |
108(90.8) |
|
| Medications | |||
| -Steroids | 229(70.2) | 97(29.8) | <0.001a |
| -Anticoagulants | 292(71.2) | 118(28.8) | <0.001a |
| -Antibiotics |
292(71.2) |
113(27.9) |
<0.001a |
| -Antiviral | |||
|
147 (75.4) | 48 (24.6) | 0.97 |
|
26(61.9) | 16(38.1) | 0.03a |
|
114 (76.5) | 35(23.5) | 0.68 |
| -Othersc |
108(92.3) |
9(7.7) |
<0.001a |
| Outcome | 0.01a | ||
| - Discharged | 327(90.1) | 97(81.5) | |
| - Died | 36(9.9) | 22(17.5) |
CT; computed topography, COVID-19; Coronavirus disease 2019, RNA; ribonucleic acid, COPD; chronic obstructive pulmonary disease.
Significant difference.
Median (Interquartile range).
Others include analgesics and antipyretics, antioxidants, lactoferrin, vitamin D, vitamin C, and Zinc.
Leukocytosis, neutrophilia, and lymphopenia were the blood cell indices significantly linked to the prolonged existence of COVID- 19 RNA. Erythrocyte sedimentation rate and acute phase reactants including CRP, serum LDH and ferritin were significantly elevated in patients with prolonged RT- PCR conversion time to negativity. Short RT-PCR conversion time was significantly more prevalent among patients with normal serum creatinine and good control of blood glucose levels. Other laboratory parameters demonstrated did not significantly affect RT-PCR conversion time. The presence of ground glace opacities (GGO) and consolidation, multiple lung lobes affection, and bilateral lung disease visualized by CT scans were significantly associated with prolonged conversion time. Sixty to more than 90% of the patients with a short clearance time of viral RNA have received steroids, anticoagulants, antibiotics, remdesivir, and other medications such as analgesics and antipyretics, antioxidants, lactoferrin, vitamin D, vitamin C, and Zinc. The outcome was that most of the patients with short conversion time were discharged from the hospital.
3.3. Predictors of prolonged RT-PCR conversion time by binary logistic regression analysis
When introducing all variables found to be significant in univariate analysis, Table 3, into binary logistic regression analysis, Table 4 , headache was found to be an independent factor for short conversion time [OR = 0.45, CI (0.25–0.78), P= 0.005], while fever [OR = 3.42, CI (1.66–7.05), P= 0.001], hypertension [OR = 2.88, CI (1.37–4.37), P = 0.001], COPD [OR = 2.54, CI (1.15–5.63), P= 0.02], lymphopenia [OR = 0.83, CI (0.70–0.99), P= 0.05], ESR [OR = 2.3, CI (1.19–4.43), P = 0.01], number of lobes affected [OR = 1.28, CI (1.07–2.70), P= 0.04] and bilateralism [OR = 1.5, CI (1.03–3.2), P= 0.035] remained as independent risk factors for prolonged existence of COVID 19 RNA in RT- PCR samples.
Table 4.
Multivariable binary logistic regression analysis to detect predictors of conversion time.
| Variable | B | S.E. | P-value | Odds ratio | 95% CI |
|
|---|---|---|---|---|---|---|
| lower | upper | |||||
| Demographic characteristics | 0.93 | 3.11 | ||||
| -Recent contact to covid-19 case |
0.53 |
0.30 |
0.07 |
1.71 |
||
| Symptoms | ||||||
| -Fever |
1.23 |
0.36 |
0.001a |
3.43 |
1.66 |
7.05 |
| -Headache |
−0.83 |
0.29 |
0.005a |
0.45 |
0.25 |
0.78 |
| History of illness | ||||||
| -Hypertension |
1.06 |
0.31 |
0.001a |
2.88 |
1.37 |
4.37 |
| -Diabetes |
0.60 |
0.34 |
0.07 |
1.83 |
0.93 |
3.58 |
| -Ischemic heart disease |
−0.60 |
0.37 |
0.11 |
0.54 |
0.26 |
1.13 |
| -COPD | 0.93 | 0.40 | 0.02a | 2.54 | 1.15 | 5.63 |
| Laboratory findings | ||||||
| -Leukocytosis |
0.105 |
0.190 |
0.57 |
1.11 |
0.76 |
1.61 |
| -Neutrophilia |
0.09 |
0.10 |
0.36 |
1.10 |
0.89 |
1.36 |
| -Lymphopenia |
−0.15 |
0.09 |
0.05a |
0.83 |
0.70 |
0.99 |
| -Thrombocytopenia |
−0.16 |
0.19 |
0.47 |
0.84 |
0.57 |
1.25 |
| -ESR |
0.83 |
0.33 |
0.01a |
2.30 |
1.19 |
4.43 |
| -Elevated ALT |
0.083 |
0.38 |
0.82 |
1.08 |
0.51 |
2.03 |
| -Elevated AST |
−0.28 |
0.43 |
0.51 |
0.75 |
0.31 |
1.75 |
| -Elevated serum creatinine |
0.22 |
0.34 |
0.52 |
1.24 |
0.63 |
2.45 |
| -Hyperglycaemia |
0.35 |
0.33 |
0.28 |
1.43 |
0.74 |
2.73 |
| Radiologic findings | ||||||
| -CT chest pattern |
0.02 |
0.2 |
0.89 |
1.02 |
0.68 |
1.54 |
| -Number of lobes affected |
0.73 |
0.34 |
0.04a |
1.28 |
1.07 |
2.70 |
| -Lung disease (Bilateralism) | 0.65 | 0.38 | 0.035a | 1.5 | 1.03 | 3.2 |
S.E.; standard error, CI; Confidence interval, COVID-19; Coronavirus disease 2019, COPD; chronic obstructive pulmonary disease, ESR; erythrocyte sedimentation rate, ALT; alanine transaminase, AST; aspartate transaminase, CT; computed topography.
Significant difference.
3.4. Initial false-negative RT-PCR testing
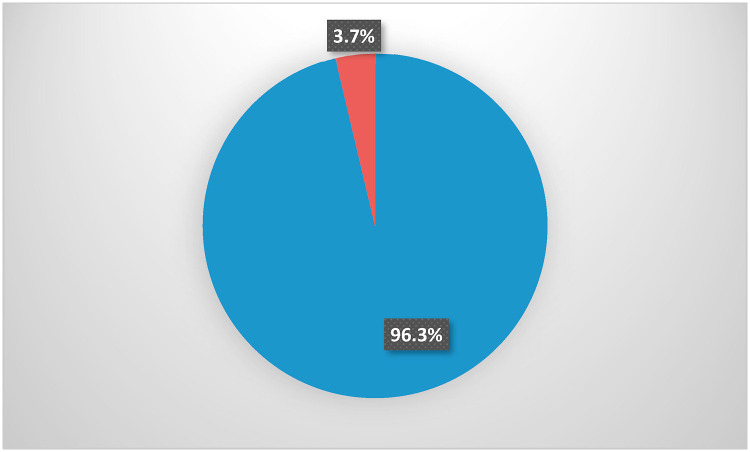
According to Fig. 3 , 464 (96.3%) of studied patients showed positive RT-PCR from the 1st sample taken upon admission to the hospital; on the other hand, 18 patients (3.7%) had initial false-negative results. As demonstrated in Table 5 , all those patients turned positive when RT-PCR was performed 24–48 h later and had bilateral lung affection on chest CT imaging. Almost all of them were admitted to the hospital within one week after onset of symptoms, only one third had recent contact with COVID-19 cases, less than half had prolonged conversion time, and no one died.
Fig. 3.
The distribution of the initial RT-PCR test results among the study participants.
Table 5.
Initial false-negative RT-PCR.
| Variable | n = 18 | % |
|---|---|---|
| - ≤29 years | 1 | 5.6 |
| - 30–39 years | 5 | 27.8 |
| - 40–49 years | 2 | 11.1 |
| - 50–59 years | 7 | 39.8 |
| - 60–69 years |
3 |
16.7 |
| Recent Contact to COVID 19 case | ||
| - No | 12 | 66.7 |
| - Yes |
6 |
33.3 |
| RT-PCR conversion time | ||
| - <3 weeks | 10 | 55.6 |
| - ≥ 3 weeks |
8 |
44.4 |
| Days of symptoms before admission | ||
| - < 7 days | 17 | 94.4 |
| - ≥ 7 days |
1 |
5.6 |
| Radiological findings | ||
| CT pattern | ||
| - Presence of GGO and consolidation | 6 | 33.3 |
| - Presence of GGO without consolidation | 10 | 55.6 |
| - Presence of consolidation without GGO |
2 |
11.1 |
| Number of affected lobes | ||
| - One lobe | 0 | 0 |
| - Two lobes | 2 | 11.1 |
| - Three lobes | 11 | 61.1 |
| - Four lobes | 5 | 27.8 |
| - Five lobes |
0 |
0 |
| Bilateralism | ||
| -Unilateral | 0 | 0 |
| -Bilateral |
18 |
100.0 |
| 2nd RT- PCR | ||
| - Positive | 18 | 100.0 |
| - Negative |
0 |
0.0 |
| Outcome | ||
| - Discharged | 17 | 94.4 |
| - Died | 1 | 5.6 |
RT-PCR, reverse transcription polymerase chain reaction, CT; computed topography, GGO; ground glass opacity.
4. Discussion
Two consecutive negative SARS-CoV-2 RT-PCR test results 24 h apart have been among the requirements for the discontinuation of isolation. However, those who have recovered from the severe form of the disease may have prolonged RNA detection for up to 3 months (12 weeks) [[16], [17], [18]]. Nevertheless, there is no possibility of isolation of competent viruses capable of infection from these people.
In the current work, the cut-off value of three weeks proposed for long versus short RT-PCR conversion time was chosen based on the average duration agreed upon by most workers; 21 days [16,17,[19], [20], [21]]. he median duration of RT- PCR conversion to negative (20 days) identified in the present work was comparable to that reported in many studies and is more than the median identified by Li ng et al. (2020) [22]. Significantly prolonged RT- PCR conversion to negative is not a rare phenomenon, with the lengthiest duration being 83 days [22,23]. The variation of sample size, characteristics of the study population, severity of COVID-19 infection, and the vaccination status might justify data variation.
The predictors of continuation of viral RNA in RT- PCR URT samples from recovered COVID-19 patients remain blurred. Even though RT- PCR test results conversion to negative are no more a criterion for termination of isolation, factors leading to such prolonged conversion need to be identified. One purpose, as explained by Zhang et al. (2020) [24] allows for a personalized treatment strategy for COVID 19 patients who tested positive after convalescing, and the cause is either reinfection or reactivation. Since it is not easy to differentiate between reinfection and prolonged viral clearance and because causal mechanisms and patient features of these conditions may be quite different, identifying predictors of prolonged viral clearance will help properly manage cases [25,26]. Another reason is linked to the immunocompromised status of patients, particularly if healthcare workers. There are concerns that an immunocompromised individual may remain infectious for more than 20 days from the start of SARS-CoV-2 infection, indicating the implementation of additional medical and infection control procedures. It may also necessitate delayed return of HCWs to work, so authorities of healthcare facilities can optimize the carrying capacity in a specified period and the required medical resources.
It is worth considering that COVID-19 is a brand-new illness that is only a few years old, and many aspects relevant to it are still incomprehensible. The long-term impact cannot be identified for the time being. Apart from very few microorganisms, cure of illnesses requires, as we all know and as it seems logical, ridding of the causing microbe [27]. Generally, the host-microorganism relationship in cases of very long carriage of microorganisms may lead to later untoward consequences. So, studying the reasons for that persistence will help develop optimal strategies to combat the disease.
Previous studies revealed considerable risk elements associated with prolonged viral persistence, including sex, chest tightness at admission, the period from the start of symptoms to hospitalization, albumin level, antiviral treatment, and length of hospital stay [[17], [18], [19], [20]].
The present work identified headache as an independent predictor of early RT-PCR conversion to negative. Savtale et al. (2021) [28] have reported that ear, nose, and throat symptoms, including headache, could be used as biomarkers for early identification of COVID-19 infection despite their lower incidence than fever and cough. Therefore, at their onset, these symptoms may urge patients, especially during the pandemic, to promptly seek medical advice and help early identification and diagnosis of COVID 19 infection with early intervention and control of the infection, hence shortening the duration of PCR positivity [19,28]. Fever, as reported before, has been associated in both children and adult COVID-19 patients with prolonged PCR conversion to negativity [[29], [30], [31]]. These patients were assumed to be more harshly influenced by the SARS-CoV-2 in their lungs [32]. At the beginning of an illness, fever may be due to a cytokine storm, or a higher viral load associated with more intense COVID-19 and a longer virus-shedding period [33].
Comorbidities of COVID 19 patients (manifested in this work by hypertension and COPD) were independent risk factors for the persistence of the virus. This effect could be due to the impact on increasing disease severity. Patients suffering from chronic illnesses comparatively experience low immunity, which in turn might prolong the duration of virus existence [34]. Trump et al. (2021) [35] have noticed a delay in SARS-CoV-2 virus clearance and a worsening of lung inflammation in hypertensive patients. Hypertension has been associated with endothelial dysfunction [31], which is the common denominator in most COVID-19 comorbidities. Moreover, hypertensive patients have dysregulated CD4+ T helper cells and CD8+ cytotoxic T cells. In addition, CD8+ T cells fail to be activated in hypertensive patients during viral infection [36]. These observations might explain the severity of COVID-19 associated with hypertension. On the other side, COPD can blunt the immune response to viral infections via suppressing the mucocilliary clearance mechanism and the activity of immune cells, including alveolar macrophages, dendritic cells, CD8+ cytotoxic T cells, B lymphocytes, and antibody production [37]. While smoking remains the main cause of COPD, active smokers' antiviral immune response, including interferons, is seriously compromised [38].
Lymphopenia indicated prolonged clearance of COVID-19 RNA. Since the first descriptive study in China regarding the COVID-19 infection [39], lymphocyte count has been linked to severe COVID-19 [40,41], and non-survivors of this disease were noted to have a remarkably lower lymphocyte count than survivors [42]. It has been noted that the level of lymphocytes during COVID-19 gradually increases with the recovery and by the negative conversion of the PCR test leading to conclude that delayed eradication of the virus might be due to the profound disruption of the immune system. The precise immunological mechanism warrants additional investigation [43]. One limitation of this retrospective study was that neutrophils and lymphocytes values were collected from symptomatic patients at admission time, and we did not analyze their values at different time points. These findings might support the worth of monitoring neutrophil to lymphocyte ratio [44] and the more powerful neutrophil to CD4+ lymphocyte ratio [45] as predictors of conversion time to negative.
Consistent with previous reports [46,47], most of our COVID-19 patients presented with high values of ESR. To a large extent, it could be assigned to multiple organ impairment or failure to be compatible with the acute respiratory distress syndrome course of severe and critical COVID-19 infection [48,49].
Previous studies have confirmed the direct correlation between chest CT severity scores and clinical harshness in COVÍD 19 patients and proposed chest CT scan score as a useful tool in predicting the progression of the disease and patient outcome [50,51]. In the current study, the presence of consolidation together with GGO, multiple lung lobe affection, and bilateralism implied a prolonged RT-PCR conversion time.
One of the interesting observations in the present study is the first negative RT-PCR testing among 18 of the studied patients. Nevertheless, all patients turned RT-PCR positive on the second test. Clinically, there may be a window period of 5 days after exposure and before detecting viral nucleic acids in nasopharyngeal swabs. Otherwise, false-negative results may happen when a mutation occurs in the virus genome in areas targeted by PCR primers or improper collection, handling, and processing of specimens [52,53]. The intermittent detection of viral RNA is another cause for false-negative initial results [53]. The findings that; 1) two-thirds of these patients had no contact to COVID-19 case, 2) almost all of them have been admitted to hospital within one week of symptoms onset, 3) more than half have short RT-PCR conversion time, and 4) all, except one of them, had a favorable outcome, encourage early hospital admission and adequate management of COVD-19 patients regardless of their age and pour into the favorable outcome of the disease.
A simple tripartite approach to the interpretation of prolonged PCR conversion time will result in first, isolation of a large number of people who are no longer infectious associated with exaggerated and unnecessary infection control measures; second, exhaustion of the healthcare sector, materially and humanly; and finally, the strict mitigation strategies adopted by different countries. Hence, an essential question is proposed, is a person with positive RT-PCR always infectious or infectious if the Ct is below a certain value? The answer to this question settles in detecting a cultivable virus in upper respiratory tract samples, the reliable proxy for infectivity [54]. Nevertheless, according to Singanayagam et al. (2020) [54], 8% of the samples with Ct value above 35 were positive for the viable virus. Serial Ct values may offer a good utility for interpretation; however, Ct values between assays are not directly comparable and may not even be reported by some PCR platforms. In addition, the interpretation of a single positive Ct value should be used in parallel with clinical history to help in diagnosis and prognosis or to use as an indicator for infectivity or recovery [55].
5. Conclusions
Effective clinical care must identify patients with a high probability of COVID-19 despite a negative RT-PCR. The decision of hospital admission, isolation, management protocols, and discharge should be guided by integrating clinical presentation, laboratory testing, and radiologic evidence considering the estimated RT-PCR conversion time among COVID-19 patients in the present study. Relying solely on RT-PCR conversion for discharge decision would be Sisyphus analogy. Moreover, the study's findings are ringing an alarm bell for healthcare authorities in different countries relying on positive PCR results to reconsider the application of quarantine procedures to incoming travelers before granting them access to their territories.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Noha M. Hammad: Methodology, Software, Data curation, Writing – original draft, preparation, and, Visualization, Writing – review & editing. Maysaa A. Saeed: Investigation. Shaker Wagih Shaltout: Investigation. Hanaa A. Nofal: Data curation, Validation, and. Ramadan M. Nafae: Investigation. Kadem Arslan: Investigation. Alpaslan Tanoglu: Investigation. Mihai Nechifor: Investigation. Catalina Luca: Investigation. Zaid Hashim Ali Al-kadhim: Investigation. Ahmed Mosallem: Investigation. Fatma A. Amer: Conceptualization, Supervision, and, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that there are no conflicts of interests.
Acknowledgement
The authors would like to thank the staff members at the “Zagazig Scientific and Medical Research Center, Zagazig University” for their assistance during the laboratory work. This work did not receive any funding.
References
- 1.Liu K., Chen Y., Lin R., Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. 2020;80(6):e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao S., Lian J., Lu Y., Jia H., Hu J., Yu G., et al. Decreased B cells on admission associated with prolonged viral RNA shedding from the respiratory tract in coronavirus disease 2019: a case-control study. J Infect Dis. 2020;222(3):367–371. doi: 10.1093/infdis/jiaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aydillo T., Gonzalez-Reiche A.S., Aslam S., van de Guchte A., Khan Z., Obla A., et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med. 2020;383(26):2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183(7):1901–1912. doi: 10.1016/j.cell.2020.10.049. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baang J.H., Smith C., Mirabelli C., Valesano A.L., Manthei D.M., Bachman M.A., et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis. 2021;223(1):23–27. doi: 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarhini H., Recoing A., Bridier-Nahmias A., Rahi M., Lambert C., Martres P., et al. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis. 2021;223(9):1522–1527. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Center for Disease Control and Prevention COVID 19, ending isolation and precautions for people with COVID-19: interim guidance. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html [cited 2022 January 12]; Available from:
- 9.National Institutes of Health . 2021. Testing for SARS-CoV-2 infection.https://www.covid19treatmentguidelines.nih.gov/overview/sars-cov-2-testing/ [cited 2021 December 27 ]; Available from: [Google Scholar]
- 10.Gupta-Wright A., Macleod C.K., Barrett J., Filson S.A., Corrah T., Parris V., et al. False-negative RT-PCR for COVID-19 and a diagnostic risk score: a retrospective cohort study among patients admitted to hospital. BMJ Open. 2021;11(2) doi: 10.1136/bmjopen-2020-047110. e047110-e047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masoud H., Elassal G., Hassany M., Shawky A., Abdel Hakim M., Zaky S. 2020. Egyptian Ministry of Health and Population. Management protocol for COVID-19 patients MoHP protocol for COVID19.https://www.researchgate.net/publication/345813633_Management_Protocol_for_COVID-19_Patients_MoHP_Protocol_for_COVID19_November_2020 [cited 2022 March 17]; Available from: [Google Scholar]
- 12.Republic of Turkey Ministry of Health . 2020. COVID-19 (SARS-CoV-2 infection) guide.https://hsgm.saglik.gov.tr/depo/birimler/goc_sagligi/covid19/rehber/COVID-19_Rehberi20200414_eng_v4_002_14.05.2020.pdf [cited 2022 March 17]; Available from: [Google Scholar]
- 13.World Health Organization. COVID-19 Clinical management. Living guidance. 2021 [cited 2022 March 16]; Available from: file:///C:/Users/Noha/Desktop/WHO-2019-nCoV-clinical-2021.1-eng.pdf.
- 14.Center for Disease Control and Prevention . 2020. Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19.https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html Available from: [Google Scholar]
- 15.World Health Organization Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [cited 2022 March 10]; Available from:
- 16.Kanji J.N., Zelyas N., MacDonald C., Pabbaraju K., Khan M.N., Prasad A., et al. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 2021;18(1):1–6. doi: 10.1186/s12985-021-01489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C., Zhang T., Ren H., Sun S., Yu X., Sheng J., et al. Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging (Albany NY) 2020;12(22):22399. doi: 10.18632/aging.104114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu K., Chen Y., Yuan J., Yi P., Ding C., Wu W., et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19) Clin Infect Dis. 2020;71(15):799–806. doi: 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X., Xing Y., Jia J., Ni W., Liang J., Zhao D., et al. Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi L., Yang Y., Jiang D., Tu C., Wan L., Chen X., et al. Factors associated with the duration of viral shedding in adults with COVID-19 outside of Wuhan, China: a retrospective cohort study. Int J Infect Dis. 2020;96:531–537. doi: 10.1016/j.ijid.2020.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan D., Liu X.-Y., Zhu Y.-n., Huang L., Dan B.-t., Zhang G.-j., et al. Factors associated with prolonged viral shedding and impact of lopinavir/ritonavir treatment in hospitalised non-critically ill patients with SARS-CoV-2 infection. Eur Respir J. 2020;56(1) doi: 10.1183/13993003.00799-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling Y., Xu S.-B., Lin Y.-X., Tian D., Zhu Z.-Q., Dai F.-H., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133(9):1039. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N., Wang X., Lv T. Prolonged SARS‐CoV‐2 RNA shedding: not a rare phenomenon. J Med Virol. 2020 doi: 10.1002/jmv.25952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Wang X., Jia X., Li J., Hu K., Chen G., et al. Risk factors for disease severity, unimprovement, and mortality in COVID-19 patients in Wuhan, China. Clin Microbiol Infect : Off Publ Eur Soc Clin Microbiol Infect Dis. 2020;26(6):767–772. doi: 10.1016/j.cmi.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M., Zhang J., Shi H., Liu B., Zeng F. Viral shedding prolongation in a kidney transplant patient with COVID‐19 pneumonia. Am J Transplant. 2020 doi: 10.1111/ajt.15996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao A.T., Tong Y.X., Zhang S. False‐negative of RT‐PCR and prolonged nucleic acid conversion in COVID‐19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savtale S., Hippargekar P., Bhise S., Kothule S. Prevalence of otorhinolaryngological symptoms in Covid 19 patients. Indian J Otolaryngol Head Neck Surg. 2021:1–7. doi: 10.1007/s12070-021-02410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni L., Cheng M.-L., Feng Y., Zhao H., Liu J., Ye F., et al. Impaired cellular immunity to SARS-CoV-2 in severe COVID-19 patients. Front Immunol. 2021:59. doi: 10.3389/fimmu.2021.603563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y., Li Y., Deng W., Liu M., He Y., Huang L., et al. Symptomatic infection is associated with prolonged duration of viral shedding in mild coronavirus disease 2019: a retrospective study of 110 children in Wuhan. Pediatr Infect Dis J. 2020;39(7):e95. doi: 10.1097/INF.0000000000002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T.Z., Cao Z.H., Chen Y., Cai M.T., Zhang L.Y., Xu H., et al. Duration of SARS‐CoV‐2 RNA shedding and factors associated with prolonged viral shedding in patients with COVID‐19. J Med Virol. 2021;93(1):506–512. doi: 10.1002/jmv.26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennasrallah C., Zemni I., Dhouib W., Sriha H., Mezhoud N., Bouslama S., et al. Factors associated with a prolonged negative conversion of viral RNA in patients with COVID-19. Int J Infect Dis. 2021;105:463–469. doi: 10.1016/j.ijid.2021.02.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi D., Wu W., Wang Q., Xu K., Xie J., Wu J., et al. Clinical characteristics and factors associated with long-term viral excretion in patients with severe acute respiratory syndrome coronavirus 2 infection: a single-center 28-day study. J Infect Dis. 2020;222(6):910–918. doi: 10.1093/infdis/jiaa388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattey‐Mora P.P., Begle C.A., Owusu C.K., Chen C., Parker M.A. Hospitalised versus outpatient COVID‐19 patients' background characteristics and comorbidities: a systematic review and meta‐analysis. Rev Med Virol. 2021:e2306. doi: 10.1002/rmv.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trump S., Lukassen S., Anker M.S., Chua R.L., Liebig J., Thürmann L., et al. Hypertension delays viral clearance and exacerbates airway hyperinflammation in patients with COVID-19. Nat Biotechnol. 2021;39(6):705–716. doi: 10.1038/s41587-020-00796-1. [DOI] [PubMed] [Google Scholar]
- 36.Batiha G.E.-S., Gari A., Elshony N., Shaheen H.M., Abubakar M.B., Adeyemi S.B., et al. Hypertension and its management in COVID-19 patients: the assorted view. Int J Cardiol Cardiovasc Risk Prev. 2021;11 doi: 10.1016/j.ijcrp.2021.200121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polverino F., Kheradmand F. COVID-19, COPD, and AECOPD: immunological, epidemiological, and clinical aspects. Front Med. 2021;7:1121. doi: 10.3389/fmed.2020.627278g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Valero J., Olloquequi J., Montes J.F., Rodríguez E., Martín-Satué M., Texidó L., et al. Deficient pulmonary IFN‐β expression in COPD patients. PLoS One. 2019;14(6):e0217803. doi: 10.1371/journal.pone.0217803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavakolpour S., Rakhshandehroo T., Wei E.X., Rashidian M. Lymphopenia during the COVID-19 infection: what it shows and what can be learned. Immunol Lett. 2020;225:31–32. doi: 10.1016/j.imlet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucirka L.M., Lauer S.A., Laeyendecker O., Boon D., Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173(4):262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., et al. Clinical characteristics of refractory coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2021;73(11):e4208–e4213. doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao D., Yao F., Wang L., Zheng L., Gao Y., Ye J., et al. A comparative study on the clinical features of coronavirus 2019 (COVID-19) pneumonia with other pneumonias. Clin Infect Dis. 2020;71(15):756–761. doi: 10.1093/cid/ciaa247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 50.Saeed G.A., Gaba W., Shah A., Al Helali A.A., Raidullah E., Al Ali A.B., et al. Correlation between chest CT severity scores and the clinical parameters of adult patients with COVID-19 pneumonia. Radiol Res Pract. 2020:2021. doi: 10.1155/2021/6697677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zayed N.E., Bessar M.A., Lutfy S. CO-RADS versus CT-SS scores in predicting severe COVID-19 patients: retrospective comparative study. Egypt J Bronchol. 2021;15(1):1–10. doi: 10.1186/s43168-021-00060-3. [DOI] [Google Scholar]
- 52.Administration F.a.D. 2021. Genetic variants of SARS-CoV-2 may lead to false negative results with molecular tests for detection of SARS-CoV-2 - letter to clinical laboratory staff and health care providers.https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2 [cited 2021 December 27]; Available from: [Google Scholar]
- 53.Lascarrou J.-B., Colin G., Le Thuaut A., Serck N., Ohana M., Sauneuf B., et al. Predictors of negative first SARS-CoV-2 RT-PCR despite final diagnosis of COVID-19 and association with outcome. Sci Rep. 2021;11(1):1–7. doi: 10.1038/s41598-021-82192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singanayagam A., Patel M., Charlett A., Bernal J.L., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Public Health England Understanding cycle threshold (Ct) in SARS-CoV-2 RT-PCR. Guide Health Protect Team. 2020 https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/926410/Understanding_Cycle_Threshold__Ct__in_SARS-CoV-2_RT-PCR_.pdf [cited March 24; Available from: [Google Scholar]