Abstract
A new species of lepocreadiid, Opechonoides opisthoporus n. sp., is described infecting 12 pomacentrid fish species from the Great Barrier Reef, Australia, with Abudefduf whitleyi Allen & Robertson as the type-host. This taxon differs from the only other known member of the genus, Opechonoides gure Yamaguti, 1940, in the sucker width ratio, cirrus-sac length, position of the testes, position of the pore of Laurer’s canal, and relative post-testicular distance. The new species exhibits stenoxenic host-specificity, infecting pomacentrids from seven genera: Abudefduf Forsskål, Amphiprion Bloch & Schneider, Neoglyphidodon Allen, Neopomacentrus Allen, Plectroglyphidodon Fowler & Ball, Pomacentrus Lacépède and Stegastes Jenyns. Phylogenetic analyses of 28S rDNA sequence data demonstrate that O. opisthoporus n. sp. forms a strongly supported clade with Prodistomum orientale (Layman, 1930) Bray & Gibson, 1990. The life cycle of this new species is partly elucidated on the basis of ITS2 rDNA sequence data; intermediate hosts are shown to be three species of Ctenophora. New host records and molecular data are reported for Lepocreadium oyabitcha Machida, 1984 and Lepotrema amblyglyphidodonis Bray, Cutmore & Cribb, 2018, and new molecular data are provided for Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018 and Lepotrema adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996. Novel cox1 mtDNA sequence data showed intraspecific geographical structuring between Heron Island and Lizard Island for L. acanthochromidis but not for L. adlardi or O. opisthoporus n. sp.
Introduction
The Lepocreadiidae Odhner, 1905 is the largest family of the Lepocreadioidea Odhner, 1905, a group of digeneans primarily infecting fishes of shallow marine systems (Bray & Cribb, 2012). Eleven species in three lepocreadiid genera have been reported from the Pomacentridae (Table 1): Lepocreadium Stossich, 1904 (three species), Lepotrema Ozaki, 1932 (five species) and Preptetos Pritchard, 1960 (three species). Of these eleven species, it is likely that the infections of Lepocreadium album (Stossich, 1890) Stossich, 1904, Preptetos cannoni Barker, Bray & Cribb, 1993, P. trulla (Linton, 1907) Bray & Cribb, 1996, and P. xesuri (Yamaguti, 1940) Pritchard, 1960 are incidental infections, as each of these species has been reported primarily from fishes in other families: L. album in the Sparidae (Bartoli et al., 2005), P. cannoni in the Siganidae (Barker et al., 1993; Bray et al., 2022), P. trulla in the Lutjanidae (Claxton et al., 2017) and P. xesuri in the Acanthuridae (Bray et al., 1993; Bray & Cribb, 1996). On the Great Barrier Reef (GBR), there are reports of five lepocreadiid species occurring in pomacentrids (excluding infections of Preptetos spp.): Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018, L. adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996, L. amblyglyphidodonis Bray, Cutmore & Cribb, 2018, L. monile Bray & Cribb, 1998, and Lepocreadium oyabitcha Machida, 1984. Additionally, there is a report of an undescribed Lepotrema species from the banded scalyfin, Parma polylepis Günther (as Lepotrema sp. 4 in Bray et al., 2018b).
Table 1.
Species of lepocreadiids reported from pomacentrid fishes including information on host-specificity
| Species | Host-specificity | Host and locality | References |
|---|---|---|---|
| Lepocreadium album (Stossich, 1890) Stossich, 1904 | Euryxenic |
Chromis chromis (Linnaeus) Saronic Gulf, Greece |
Papoutsoglou (1976) |
| Blenniidae Rafinesque; Centracanthidae Gill; Sparidae Rafinesque | See Bartoli et al. (2005) | ||
| Lepocreadium oyabitcha Machida, 1984 | Stenoxenic |
Abudefduf vaigiensis (Quoy & Gaimard) Off Ryukyu Islands, Japan Abudefduf whitleyi Allen & Robertson Off Lizard Island, GBR, Australia |
Machida (1984); Bray & Cribb (1998) |
| Lepocreadium sogandaresi Nahhas & Powell, 1971 | Oioxenic |
Stegastes leucostictus (Müller & Troschel) Gulf of Mexico |
Nahhas & Powell (1971) |
| Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018 | Oioxenic |
Acanthochromis polyacanthus Bleeker Off Lizard and Heron Islands, GBR, Australia |
Barker et al. (1993); Barker et al. (1994); Bray et al. (2009); Bray et al. (2018b) |
| Lepotrema adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996 | Oioxenic |
Abudefduf bengalensis (Bloch) Off Lizard and Heron Islands, GBR, Australia Ningaloo Reef, WA, Australia |
Bray et al. (1993); Barker et al. (1994); Bray et al. (2018b) |
| Lepotrema amblyglyphidodonis Bray, Cutmore & Cribb, 2018 | Stenoxenic |
Amblyglyphidodon curacao (Bloch) Off Heron Island, GBR, Australia Amphiprion akindynos Allen Off Heron Island, GBR, Australia |
Bray et al. (1993); Barker et al. (1994); Bray et al. (2018b) |
| Lepotrema clavatum Ozaki, 1932 | Euryxenic |
Dascyllus albisella Gill Off Hawaii, United States |
Pritchard (1963) |
| Balistidae Rafinesque; Chaetodontidae Rafinesque; Monacanthidae Nardo; Paralichthyidae Regan; Pomacanthidae Jordan & Evermann | See Bray et al. (2018b) | ||
| Lepotrema monile Bray & Cribb, 1998 | Stenoxenic |
Pomacentrus amboinensis Bleeker Off Lizard Island, GBR, Australia Pomacentrus chrysurus Cuvier Off Lizard Island, GBR, Australia Stegastes apicalis (De Vis) Off Heron Island, GBR, Australia Pomacentrus wardi Whitley Off Heron Island, GBR, Australia |
Bray et al. (1993); Barker et al. (1994); Bray & Cribb (1998); Sun et al. (2012); Bray et al. (2018b) |
| Lepotrema sp. 4 of Bray et al. (2018b) | Unknown |
Parma polylepis Günther Off Heron Island, GBR, Australia |
Bray et al. (1993); Barker et al. (1994); Bray et al. (2018b) |
| Preptetos cannoni Barker, Bray & Cribb, 1993 | Stenoxenic |
Pomacentrus bankanensis Bleeker Off Heron Island, GBR, Australia |
Bray et al. (1993); Barker et al. (1994) |
| Siganidae Richardson | See Barker et al. (1993) and Bray et al. (2022) | ||
| Preptetos trulla (Linton, 1907) Bray & Cribb, 1996 | Stenoxenic |
Chromis multilineata (Guichenot) Off Puerto Rico |
Dyer et al. (1985) |
| Labridae Cuvier; Lutjanidae Gill; Sparidae Rafinesque | See Claxton et al. (2017) | ||
| Preptetos xesuri (Yamaguti, 1940) Pritchard, 1960 | Stenoxenic |
Parma polylepis Off Heron Island, GBR, Australia |
Bray et al. (1993); Barker et al. (1994) |
| Acanthuridae Bonaparte | See Bray & Cribb (1996) and Bray et al. (2022) |
Pomacentrid hosts are listed in full with locality information and other taxa are listed at family level
Abbreviations: GBR, Great Barrier Reef; WA, Western Australia
In this study, we describe a new species of lepocreadiid from GBR pomacentrid fishes using morphological and molecular data, and use molecular data to identify its second intermediate hosts. New hosts are reported for L. amblyglyphidodonis and Lepocreadium oyabitcha, and novel molecular data are reported for Lepotrema acanthochromidis, L. adlardi, and Lepocreadium oyabitcha.
Material and methods
Specimen collection
Pomacentrid fishes were collected at three Australian localities: off Heron Island, southern GBR (23°26'S, 151°54'E) and off Lizard Island, northern GBR (14°40'S, 145°28'E) and in Moreton Bay, southeast Queensland (27°24'S, 153°26'E). Fishes were collected by line fishing, spearfishing, anaesthetic (using a clove oil solution), and barrier netting, and were euthanised immediately prior to dissecting. The gastrointestinal tract of each fish was removed and examined for trematodes under a stereo-microscope; trematodes were collected, and the gut was re-examined after a gut-wash, following the protocols of Cribb & Bray (2010). Live trematodes were fixed in near boiling saline and immediately preserved in 80% ethanol.
Ctenophores were sampled along the east coast of Australia between Brisbane, Queensland and Hobart, Tasmania on the Research Vessel Investigator between the 8th and 27th of May 2021. Ctenophores were collected at night by towing a bongo net (mouth diameter = 0.7 m; mesh size = 500 μm) obliquely from the surface to approximately 30 m and back to the surface. The duration of each tow ranged from 10–15 minutes and tow speed ranged from 1–1.5 m-s. Ctenophores were immediately removed from the cod end, measured, and inspected for trematode metacercariae. Metacercariae were collected and preserved in 96% ethanol.
Morphological analysis
Specimens for morphological examination were rinsed with distilled water, overstained with Mayer’s haematoxylin, destained in a 1% hydrochloric acid solution and neutralised in a 1% ammonium hydroxide solution. Specimens were then dehydrated in a graded series of ethanol solutions (50%, 70%, 80%, 90%, 95% and twice in 100%) and cleared in methyl salicylate, before being mounted on glass slides in Canada balsam. Morphometric data were taken using a camera (Olympus SC50) mounted on a compound microscope (Olympus BX-53) and cellSens Standard imaging software. Measurements are in micrometres and are presented as a range, where length is followed by width, with the mean in parentheses. Drawings were made using a drawing tube attachment and digitised in Adobe Illustrator. Type- and voucher specimens are lodged in the Queensland Museum (QM), Brisbane, Australia, and the Natural History Museum (NHMUK), London, United Kingdom. To comply with the guidelines set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of the new species have been submitted to ZooBank and registered with a Life Science Identifier (LSID), which is provided in the taxonomic summary.
Molecular sequencing
Specimens for molecular analyses were either prepared as hologenophores, whereby a portion of the trematode is used for DNA sequencing and the remainder is used as a morphological voucher, or as paragenophores, whereby the trematode used for DNA sequencing is collected from the same individual host as the morphological voucher (Pleijel et al., 2008). Genomic DNA was extracted using a standard phenol/chloroform extraction method (Sambrook & Russell, 2001) and sequence data were generated for two ribosomal DNA (rDNA) markers, the large ribosomal subunit RNA coding region (28S) and the second internal transcribed spacer region (ITS2), and one mitochondrial DNA (mtDNA) marker, the cytochrome c oxidase subunit 1 (cox1). These regions were amplified using the following primers: LSU5 (5′-TAG GTC GAC CCG CTG AAY TTA AGC-3′, Littlewood, 1994) and 1500R (5′-GCT ATC CTG AGG GAA ACT TCG-3′, Snyder & Tkach, 2001) for 28S, 3S (5′-GGT ACC GGT GGA TCA CGT GGC TAG TG-3′, Morgan & Blair, 1995) and ITS2.2 (5′-CCT GGT TAG TTT CTT TTC CTC CGC-3′, Cribb et al., 1998) for ITS2, and Dig_cox1Fa (5′-ATG ATW TTY TTY TTY YTD ATG CC-3′, Wee et al., 2017) and Dig_cox1R (5′-TCN GGR TGH CCR AAR AAY CA AA-3′, Wee et al., 2017) for cox1.
A polymerase chain reaction (PCR) for each region was performed with a total of 20 μl comprising of 2 μl of DNA template for ITS2 or 4 μl for cox1 and 28S, 5 μl of MyTaq Reaction Buffer (Bioline), 0.75 μl of each primer for ITS2 and 28S or 2 μl for cox1, 0.25 μl of Taq DNA polymerase (Bioline MyTaqTM DNA Polymerase) and made up with InvitrogenTM ultraPURETM distilled water. A TaKaRa PCR Thermal Cycler was used to amplify each region using the following profiles: 28S: an initial 95°C denaturation for 4 minutes, 30 cycles of 95°C denaturation for 1 minute, 56°C annealing for 1 minute, 72°C extension for 2 minutes, 95°C denaturation for 1 minute, 55°C annealing for 45 seconds and a final 72°C extension for 4 minutes; ITS2: an initial 95°C denaturation for 3 minutes, 45°C annealing for 2 minutes, 72°C extension for 90 seconds, four cycles of 95°C denaturation for 45 seconds, 50°C annealing for 45 seconds, 72°C extension for 90 seconds, 30 cycles of 95°C denaturation for 20 seconds, 52°C annealing for 20 seconds, 72°C extension for 90 seconds and a final 72°C extension for 5 minutes; cox1: an initial 94°C denaturation for 3 minutes, 40 cycles of 94°C denaturation for 30 seconds, 50°C annealing for 30 seconds, 72°C extension for 30 seconds and a final 72°C extension for 10 minutes. Amplified DNA was sent to the Australian Genome Research Facility for purification and dual direction Sanger sequencing using the amplification primers for the ITS2 and cox1 regions, and the internal primers 300F (5′-CAA GTA CCG TGA GGG AAA GTT-3′, Littlewood et al., 2000) and ECD2 (5′-CTT GGT CCG TGT TTC AAG ACG GG-3′, Littlewood et al., 1997) for the 28S region. Sequences were assembled and edited in Geneious Prime version 2021.11.0.9 (https://www.geneious.com).
Phylogenetic analyses
Newly generated ITS2 rDNA and cox1 mtDNA sequences were aligned with sequences of other lepocreadiid taxa available on GenBank (Table 2) in MEGA X (Kumar et al., 2018) using MUSCLE, with UPGMA clustering for iterations 1 and 2. The cox1 alignment was translated (echinoderm/flatworm mitochondrial code) and examined in Mesquite version 3.61 (Maddison & Maddison, 2021) for internal stop codons and to determine the correct reading frame. The alignment was trimmed after the correct reading frame was determined. All codon positions were then tested for non-stationarity in PAUP* version 4.0a (Swofford, 2003), and substitution saturation using the “Test of substitution saturation by Xia et al.” function (Xia et al., 2003; Xia & Lemey, 2009) implemented in DAMBE version 7.2 (Xia, 2018); non-stationarity and substitution saturation were not detected, and as such, all codons were used in subsequent analyses. Neighbour-joining analyses were conducted for each alignment with the following parameters: “Test of Phylogeny = Bootstrap method”, “No. of Bootstrap Replications = 10,000”, “Model/Method = No. of differences”, “Substitutions to Include = d: Transitions + Transversions”, “Rates among Sites = Uniform rates” and “Gaps/Missing Data Treatment = Pairwise deletion”. Pairwise differences for each alignment were estimated using the following parameters: “Variance Estimation Method = None”, “Model/Method = No. of differences”, “Substitutions to Include = d: Transitions + Transversions”, “Rates among Sites = Uniform rates” and “Gaps/Missing Data Treatment = Pairwise deletion”.
Table 2.
Sequence data for species of Lepocreadium, Lepotrema and Opechonoides analysed in this study (GenBank accession numbers for cox1 mtDNA and ITS2 rDNA reference sequences and sequences generated in the present study, and host information)
| Species | Host | Locality | GenBank accession # | References | |
|---|---|---|---|---|---|
| cox1 mtDNA | ITS2 rDNA | ||||
| Lepocreadium oyabitcha Machida, 1984 | Abudefduf sexfasciatus (Lacépède) | LI | OM791389 | OM777007 | Present study |
| Abudefduf sordidus (Forsskål) | LI | OM791390 | OM777008 | Present study | |
| Abudefduf whitleyi Allen & Robertson | LI | OM791391 | OM777009 | Present study | |
| Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018 | Acanthochromis polyacanthus (Bleeker) |
HI LI |
Bray et al. (2018b) Present study Present study |
||
| Lepotrema adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996 | Abudefduf bengalensis (Bloch) |
HI LI |
MH730000 |
Bray et al. (2018b) Bray et al. (2018b) Present study |
|
| Lepotrema amansis Bray, Cutmore & Cribb, 2018 | Amanses scopas (Cuvier) | HI | Bray et al. (2018b) | ||
| Lepotrema amblyglyphidodonis Bray, Cutmore & Cribb, 2018 | Amphiprion akindynos Allen | HI | MH730033 | MH730002 | Bray et al. (2018b) |
| Amblyglyphidodon curacao (Bloch) | HI | MH730003 | Bray et al. (2018b) | ||
| Stegastes apicalis (De Vis) | HI | OM791399 | Present study | ||
| Plectroglyphidodon dickii (Liénard) | HI | OM791400 | OM777012 | Present study | |
| Lepotrema cirripectis Bray, Cutmore & Cribb, 2018 | Cirripectis chelomatus Williams & Maugé | HI | MH730036 | Bray et al. (2018b) | |
| Cirripectis filamentosus (Alleyne & MacLeay) |
HI LI |
Bray et al. (2018b) Bray et al. (2018b) |
|||
| Lepotrema hemitaurichthydis Bray, Cutmore & Cribb, 2018 | Hemitaurichthys polylepis (Bleeker) |
PA FP |
Bray et al. (2018b) Bray et al. (2018b) |
||
| Lepotrema melichthydis Bray, Cutmore & Cribb, 2018 | Melichthys vidua (Richardson) | PA | Bray et al. (2018b) | ||
| Lepotrema monile Bray & Cribb, 1998 | Pomacentrus wardi Whitley | HI | MH730009 | Bray et al. (2018b) | |
| Lepotrema moretonense Bray, Cutmore & Cribb, 2018 | Prionurus maculatus Ogilby | MB | MH730051 | Bray et al. (2018b) | |
| Prionurus microlepidotus Lacépède | MB | Bray et al. (2018b) | |||
| Selenotoca multifasciata (Richardson) | MB | MH730055 | Bray et al. (2018b) | ||
| Opechonoides opisthoporus n. sp. | Abudefduf septemfasciatus (Cuvier) | HI | OM791405 | OM777013 | Present study |
| Abudefduf sexfasciatus | HI | OM791401 | OM777014 | Present study | |
| Abudefduf whitleyi | LI | OM791403 | OM777015 | Present study | |
| Pomacentrus chrysurus Cuvier | HI | OM791402 | OM777016 | Present study | |
| Pomacentrus moluccensis Bleeker | HI | OM791404 | OM777017 | Present study | |
Abbreviations: FP, French Polynesia; HI, off Heron Island; LI, off Lizard Island; MB, Moreton Bay; PA, off Palau
Newly generated partial 28S rDNA sequences were aligned with sequences from GenBank of related lepocreadiids using MUSCLE version 3.7 (Edgar, 2004) through the CIPRES Portal (Miller et al., 2010) with UPGMB clustering for iterations 1 and 2. Using Mesquite, the alignment was then refined by trimming and removing indels (with three or more base pairs) affecting at least 5% of sequences. To estimate the best-fitting nucleotide substitution model for the dataset, the refined alignment was analysed in jModelTest 2.1.10 (Darriba et al., 2012). The model ‘GTR + I + Γ’ was predicted to be the best estimator by the corrected Akaike Information Criterion and Bayesian Information Criterion. Phylogenetic analysis of the 28S dataset were conducted using maximum likelihood and Bayesian inference analyses on the CIPRES Portal. The maximum likelihood analysis was run using RAxML version 8.2.12 (Stamatakis, 2014) with 1,000 bootstrap pseudoreplicates, and the Bayesian inference analysis was run using MrBayes version 3.2.7a (Ronquist et al., 2012) with the following parameters: “ngen = 10,000,000”, “nruns = 2”, “nchains = 4”, “samplefreq = 1,000”, “nst = 6”, “rates = invgamma”, “ngammacat = 4”, “ratepr = variable”, “sumt burnin value = 3,000”, “sump burnin value = 3,000” and “burninfrac = 0.3”. Species of the Aephnidiogenidae Yamaguti, 1934 and the Gorgocephalidae Manter, 1966 were designated as outgroup taxa (Table 3).
Table 3.
Sequence data for the Lepocreadioidea taxa analysed in this study (GenBank accession numbers for 28S rDNA reference sequences and sequences generated in the present study, and host information)
| Species | Host | GenBank accession # | References |
|---|---|---|---|
| Lepocreadiidae Odhner, 1905 | |||
| Bianium arabicum Sey, 1996 | Lagocephalus lunaris (Bloch & Schneider) | MH157076 | Bray et al. (2018a) |
| Bianium plicitum (Linton, 1928) Stunkard, 1931 | Torquigener pleurogramma (Regan) | MH157066 | Bray et al. (2018a) |
| Clavogalea trachinoti (Fischthal & Thomas, 1968) Bray & Gibson, 1990 | Trachinotus coppingeri Günther | FJ788471 | Bray et al. (2009) |
| Deraiotrema platacis Machida, 1982 | Platax pinnatus (Linnaeus) | MN073841 | Bray et al. (2019) |
| Diplocreadium tsontso Bray, Cribb & Barker, 1996 | Balistoides conspicillum (Bloch & Schneider) | FJ788472 | Bray et al. (2009) |
| Diploproctodaeum momoaafata Bray, Cribb & Barker, 1996 | Ostracion cubicum Linnaeus | FJ788474 | Bray et al. (2009) |
| Diploproctodaeum monstrosum Bray, Cribb & Justine, 2010 | Arothron stellatus (Anonymous) | FJ788473 | Bray et al. (2009) |
| Echeneidocoelium indicum Simha & Pershad, 1964 | Echeneis naucrates Linnaeus | FJ788475 | Bray et al. (2009) |
| Hypocreadium lamelliforme (Linton, 1907) Bravo Hollis & Manter, 1957 | Balistes capriscus Gmelin, 1789 | MZ345680 | Curran et al. (2021) |
| Hypocreadium cf. patellare Yamaguti, 1938 | Balistoides viridescens (Bloch & Schneider) | FJ788478 | Bray et al. (2009) |
| Hypocreadium picasso Bray, Cribb & Justine, 2009 | Rhinecanthus aculeatus (Linnaeus) | FJ788479 | Bray et al. (2009) |
| Hypocreadium toombo Bray & Justine, 2006 | Pseudobalistes fuscus (Bloch & Schneider) | FJ788480 | Bray et al. (2009) |
| Lepidapedoides angustus Bray, Cribb & Barker, 1996 | Epinephelus cyanopodus (Richardson) | FJ788482 | Bray et al. (2009) |
| Lepocreadium opsanusi Sogandares & Hutton, 1960 | Umbrina xanti Gill | MK648298 | Pérez-Ponce de León & Hernández-Mena (2019) |
| Lepocreadium oyabitcha Machida, 1984 | Abudefduf sordidus (Forsskål) | OM777006 | Present study |
| Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018 | Acanthochromis polyacanthus (Bleeker) | MH730014 | Bray et al. (2018b) |
| Lepotrema adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996 | Abudefduf bengalensis (Bloch) | MH730015 | Bray et al. (2018b) |
| Lepotrema amblyglyphidodonis Bray, Cutmore & Cribb, 2018 | Amphiprion akindynos Allen | MH730017 | Bray et al. (2018b) |
| Lepotrema monile Bray & Cribb, 1998 | Pomacentrus wardi Whitley | MH730024 | Bray et al. (2018b) |
| Lobatocreadium exiguum (Manter, 1963) Madhavi, 1972 | Pseudobalistes fuscus | FJ788484 | Bray et al. (2009) |
| Mobahincia teirae Bray, Cribb & Cutmore, 2018 | Platax teira (Forsskål) | MH157068 | Bray et al. (2018a) |
| Multitestis magnacetabulum Mamaev, 1970 | Platax teira | FJ788485 | Bray et al. (2009) |
| Neohypocreadium dorsoporum Machida & Uchida, 1987 | Chaetodon flavirostris Günther | FJ788487 | Bray et al. (2009) |
| Neomultitestis aspidogastriformis Bray & Cribb, 2003 | Platax teira | FJ788489 | Bray et al. (2009) |
| Neopreptetos arusettae Machida, 1982 | Pomacanthus sexstriatus (Cuvier) | FJ788490 | Bray et al. (2009) |
| Opechona austrobacillaris Bray & Cribb, 1998 | Pomatomus saltatrix (Linnaeus) | MH157073 | Bray et al. (2018a) |
| Opechona chloroscombri Nahhas & Cable, 1964 | Chloroscombrus chrysurus (Linnaeus) | MZ345679 | Curran et al. (2021) |
| Opechona kahawai Bray & Cribb, 2003 | Arripis trutta (Forster) | FJ788491 | Bray et al. (2009) |
| Opechona corkumi Curran, Martorelli & Overstreet, 2021 | Peprilus burti Fowler | MZ345683 | Curran et al. (2021) |
| Opechona olssoni (Yamaguti, 1934) Yamaguti, 1938 | Scomber japonicus Houttuyn | MT303947 | Sokolov et al. (2020) |
| Opechonoides opisthoporus n. sp. | Abudefduf whitleyi Allen & Robertson | OM777005 | Present study |
| Pelopscreadium spongiosum (Bray & Cribb, 1998) Dronen, Blend, Khalifa, Mohamadain & Karer, 2016 | Ostracion cubicum | FJ788469 | Bray et al. (2009) |
| Preptetos laguncula Bray & Cribb, 1996 | Naso unicornis (Forsskål) | MZ701988 | Bray et al. (2021) |
| Preptetos paracaballeroi Bray, Cutmore & Cribb, 2022 | Naso annulatus (Quoy & Gaimard) | MZ702003 | Bray et al. (2021) |
| Preptetos prudhoei Bray, Cutmore & Cribb, 2022 | Zebrasoma scopas (Cuvier) | MZ701995 | Bray et al. (2021) |
| Preptetos trulla (Linton, 1907) Bray & Cribb, 1996 | Ocyurus chrysurus (Bloch) | AY222237 | Olson et al. (2003) |
| Prodistomum alaskense (Ward & Fillingham, 1934) Bray & Merrett, 1998 | Aptocyclus ventricosus (Pallas) | MT303950 | Sokolov et al. (2020) |
| Prodistomum keyam Bray & Cribb, 1996 | Monodactylus argenteus (Linnaeus) | FJ788493 | Bray et al. (2009) |
| Prodistomum orientale (Layman, 1930) Bray & Gibson, 1990 | Scomber japonicus | MT299625 | Sokolov et al. (2020) |
| Prodistomum priedei Bray & Merrett, 1998 | Epigonus telescopus (Risso) | AJ405272 | Bray et al. (1999) |
| Aephnidiogenidae Yamaguti,1934 | |||
| Aephnidiogenes major Yamaguti, 1934 | Diagramma labiosum MacLeay | FJ788468 | Bray et al. (2009) |
| Austroholorchis sprenti (Gibson, 1987) Bray & Cribb, 1997 | Sillago ciliata Cuvier | MH157075 | Bray et al. (2018a) |
| Holorchis castex Bray & Justine, 2007 | Diagramma pictum (Thunberg) | FJ788476 | Bray et al. (2009) |
| Gorgocephalidae Manter, 1966 | |||
| Gorgocephalus graboides Huston, Cutmore, Miller, Sasal, Smit & Cribb, 2021 | Kyphosus cinerascens (Forsskål) | MW353905 | Huston et al. (2021) |
| Gorgocephalus yaaji Bray & Cribb, 2005 | Kyphosus cinerascens | KU951489 | Huston et al. (2016) |
Results
Overview
We have examined over 1,800 pomacentrids from Moreton Bay, off Heron Island and off Lizard Island since 1990 (Table 4). These specimens comprise 61 species from 16 genera: Abudefduf Forsskål, six species (570 individuals); Acanthochromis Gill, one species (169 individuals); Amblyglyphidodon Bleeker, two species (98 individuals); Amphiprion Bloch & Schneider, three species (28 individuals); Chromis Cuvier, six species (113 individuals); Chrysiptera Swainson, seven species (24 individuals); Dascyllus Cuvier, three species (117 individuals); Dischistodus Gill, three species (73 individuals); Hemiglyphidodon Bleeker, one species (five individuals); Neoglyphidodon Allen, two species (42 individuals); Neopomacentrus Allen, two species (32 individuals); Parma Günther, two species (9 individuals); Plectroglyphidodon Fowler & Bean, three species (31 individuals); Pomacentrus Lacépède, 15 species (429 individuals); Premnas Cuvier, one species (one individual); and Stegastes Jenyns, four species (128 individuals).
Table 4.
Prevalence of Opechonoides opisthoporus n. sp. in Great Barrier Reef and Moreton Bay pomacentrid fishes
| Genus | Species | Moreton Bay | Heron Island | Lizard Island |
|---|---|---|---|---|
| Abudefduf | bengalensis (Bloch) | 0/65 | 2/50 | 0/10 |
| septemfasciatus (Cuvier) | 1/2 | 1/5 | ||
| sexfasciatus (Lacépède) | 0/3 | 3/33 | 1/26 | |
| sordidus (Forsskål) | 0/3 | 0/3 | ||
| vaigiensis (Quoy & Gaimard) | 0/29 | 0/1 | 0/4 | |
| whitleyi Allen & Robertson | 0/29 | 1/290 | 2/17 | |
| Acanthochromis | polyacanthus (Bleeker) | 0/80 | 0/89 | |
| Amblyglyphidodon | curacao (Bloch) | 0/64 | 0/32 | |
| leucogaster (Bleeker) | 0/2 | |||
| Amphiprion | akindynos Allen | 0/14 | 1/2 | |
| melanopus Bleeker | 0/2 | |||
| perideraion Bleeker | 0/10 | |||
| Chromis | amboinensis (Bleeker) | 0/1 | ||
| atripectoralis Welander & Schultz | 0/17 | 0/31 | ||
| nitida (Whitley) | 0/10 | |||
| ternatensis (Bleeker) | 0/2 | |||
| viridis (Cuvier) | 0/11 | 0/31 | ||
| weberi Fowler & Bean | 0/10 | |||
| Chrysiptera | biocellata (Quoy & Gaimard) | 0/5 | ||
| cyanea (Quoy & Gaimard) | 0/6 | |||
| flavipinnis (Allen & Robertson) | 0/3 | |||
| rex (Snyder) | 0/1 | |||
| rollandi (Whitley) | 0/7 | |||
| taupou (Jordan & Seale) | 0/1 | |||
| unimaculata (Cuvier) | 0/1 | |||
| Dascyllus | aruanus (Linnaeus) | 0/54 | 0/19 | |
| reticulatus (Richardson) | 0/1 | 0/17 | 0/24 | |
| trimaculatus (Rüppell) | 0/2 | |||
| Dischistodus | melanotus (Bleeker) | 0/22 | 0/6 | |
| perspicillatus (Cuvier) | 0/24 | 0/11 | ||
| pseudochrysopoecilus (Allen & Robertson) | 0/5 | 0/5 | ||
| Hemiglyphidodon | plagiometapon (Bleeker) | 0/5 | ||
| Neoglyphidodon | melas (Cuvier) | 0/14 | 1/27 | |
| nigroris (Cuvier) | 0/1 | |||
| Neopomacentrus | azysron (Bleeker) | 3/30 | ||
| cyanomos (Bleeker) | 0/2 | |||
| Parma | oligolepis Whitley | 0/3 | ||
| polylepis Günther | 0/6 | |||
| Plectroglyphidodon | dickii (Liénard) | 0/6 | ||
| lacrymatus (Quoy & Gaimard) | 0/6 | 0/12 | ||
| leucozonus (Bleeker) | 0/5 | 1/2 | ||
| Pomacentrus | adelus Allen | 2/33 | ||
| amboinensis Bleeker | 0/1 | 0/4 | 0/43 | |
| bankanensis Bleeker | 0/12 | |||
| brachialis Cuvier | 0/1 | 0/2 | ||
| chrysurus Cuvier | 3/34 | 1/32 | ||
| coelestis Jordan & Starks | 0/1 | 0/16 | ||
| lepidogenys Fowler & Bean | 0/8 | |||
| moluccensis Bleeker | 2/58 | 0/27 | ||
| nagasakiensis Tanaka | 0/3 | 0/3 | ||
| nigromarginatus Allen | 0/1 | |||
| pavo (Bloch) | 0/10 | |||
| philippinus Evermann & Seale | 0/4 | |||
| tripunctatus Cuvier | 0/2 | |||
| vaiuli Jordan & Seale | 0/3 | |||
| wardi Whitley | 0/1 | 0/122 | 0/8 | |
| Premnas | biaculeatus (Bloch) | 0/1 | ||
| Stegastes | apicalis (De Vis) | 1/83 | 0/29 | |
| fasciolatus (Ogilby) | 0/5 | 0/1 | ||
| gascoynei (Whitley) | 0/1 | |||
| nigricans (Lacépède) | 0/9 |
Numbers for infected fish are shown in bold
We collected adult trematodes consistent with the Lepocreadiidae from pomacentrids off Heron Island and Lizard Island; no adult lepocreadiids were found in Moreton Bay pomacentrids. Of the pomacentrids collected since 1990, we found specimens of an unknown species of Opechonoides Yamaguti, 1940 in species of multiple pomacentrid genera off Heron Island and Lizard Island. In more recent collections (between July 2019 to April 2021, 662 individuals), new specimens of four known species of lepocreadiids were also collected. Lepotrema acanthochromidis and L. adlardi were collected from known hosts, Acanthochromis polyacanthus (Bleeker) and Abudefduf bengalensis (Bloch), respectively (off both Heron Island and Lizard Island). Lepotrema amblyglyphidodonis was collected from known hosts, Amblyglyphidodon curacao (Bloch) and Amphiprion akindynos Allen, and two new hosts, Plectroglyphidodon dickii (Liénard) and Stegastes apicalis (De Vis) off Heron Island. Lepocreadium oyabitcha was collected from a known host, Abudefduf whitleyi Allen & Robertson, and three new hosts, A. bengalensis (Bloch), A. sexfasciatus (Cuvier) and A. sordidus (Forsskål) at Lizard Island. Prevalence data in taxonomic summaries are based on new collections (July 2019 to April 2021) for all taxa except for the new species of Opechonoides.
Four ctenophore species were collected: Hormiphora (?) sp. (20 individuals), Ocyropsis (?) sp. (32 individuals) and Pukia falcata Gershwin, Zeidler & Davie (32 individuals) over the continental shelf of southeast Queensland and Bolinopsis (?) sp. (20 individuals) off the east coast of Tasmania. Metacercariae consistent with the Lepocreadiidae were collected from Bolinopsis sp., Ocyropsis sp. and P. falcata. Although we are confident that three species of ctenophore were infected, only P. falcata could be identified reliably to species. The other two species were unidentifiable due to the combination of damage to the specimens at the time of collection, inability to generate informative DNA sequences, and the unsettled nature of the taxonomy of ctenophores in Australian waters. As such, on the basis of their general morphology, we tentatively identified these specimens as relating to Bolinopsis and Ocyropsis. Unfortunately, the difficulties of preservation of ctenophores precludes the lodgement of morphologically informative voucher specimens in a museum.
Molecular results
ITS2 sequence data were generated for the new species of Opechonoides, Lepotrema acanthochromidis, L. amblyglyphidodonis, Lepocreadium oyabitcha and the lepocreadiid metacercariae. No intraspecific genetic variation was present for any of the species examined. The ITS2 sequences of the metacercariae collected from the ctenophores were identical to that of the new species of Opechonoides. cox1 data were generated for the new species of Opechonoides, Lepotrema acanthochromidis, L. adlardi, L. amblyglyphidodonis and Lepocreadium oyabitcha. Newly generated sequences of Lepotrema acanthochromidis from off Heron Island were identical to those available on GenBank, also from off Heron Island, and differed from sequences of Lizard Island specimens by up to 18 base pairs, showing clear geographical structuring (Fig. 1). In contrast, the cox1 sequences of L. adlardi from off Heron Island and Lizard Island differed by up to only two base pairs and showed no geographical structuring. New cox1 sequences of L. amblyglyphidodonis from the two new hosts from off Heron Island were identical to those collected from Amblyglyphidodon curacao and Amphiprion akindynos (available on GenBank and also from off Heron Island). The cox1 sequences for specimens of Lepocreadium oyabitcha and the new species of Opechonoides showed no intraspecific genetic variation.
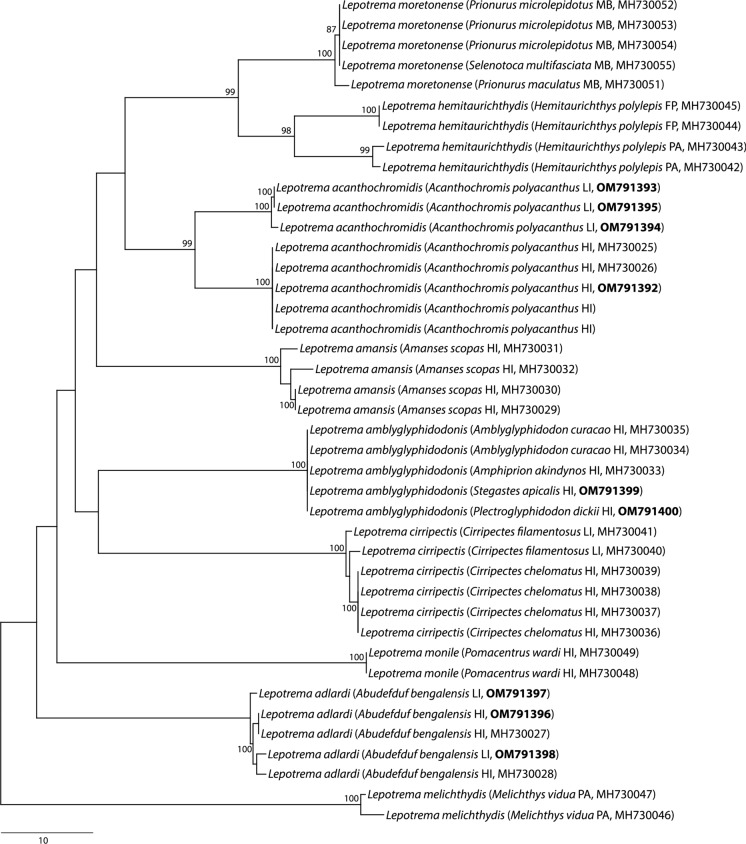
Fig. 1.
Phylogram from the unrooted neighbour-joining analysis of the cox1 mtDNA dataset for the genus Lepotrema (Lepocreadiidae). The newly generated sequences are shown in bold. Bootstrap support values are shown at the nodes; support values below 80 are not shown. Abbreviations: FP, French Polynesia; HI, off Heron Island; LI, off Lizard Island; MB, Moreton Bay; PA, Palau. The scale bar indicates the number of base differences
Family Lepocreadiidae Odhner, 1905
Genus Opechonoides Yamaguti, 1940
Type-species: Opechonoides gure Yamaguti, 1940, by original designation.
Opechonoides opisthoporus n. sp.
Type-host: Abudefduf whitleyi Allen & Robertson, Whitley’s sergeant (Pomacentridae).
Type-locality: Off Lizard Island, northern Great Barrier Reef, Australia.
Other hosts: Abudefduf bengalensis (Bloch), Bengal sergeant; A. septemfasciatus (Cuvier), Banded sergeant; A. sexfasciatus (Lacépède), Scissortail sergeant; Amphiprion akindynos Allen, Barrier Reef anemonefish; Neoglyphidodon melas (Cuvier), Bowtie damsel; Neopomacentrus azysron (Bleeker), Yellowtail demoiselle; Plectroglyphidodon leucozonus (Bleeker), Whiteband damsel; Pomacentrus adelus Allen, Obscure damsel; P. chrysurus Cuvier, Whitetail damsel; P. moluccensis Bleeker, Lemon damsel; Stegastes apicalis (De Vis), Australian gregory (Pomacentridae).
Other locality: Off Heron Island, southern Great Barrier Reef, Australia.
Site in host: Intestine.
Abundance and prevalence: Off Lizard Island: four specimens from one of five A. septemfasciatus; two specimens from one of 26 A. sexfasciatus; eight specimens from two of 17 A. whitleyi; two specimens from one of two A. akindynos; one specimen from one of 27 N. melas; seven specimens from three of 30 N. azysron; one specimen from one of two P. leucozonus; four specimens from two of 33 P. adelus; one specimen from one of 32 P. chrysurus. Off Heron Island: two specimens from two of 50 A. bengalensis; nine specimens from one of two A. septemfasciatus; six specimens from three of 33 A. sexfasciatus; six specimens from one of 290 A. whitleyi; four specimens from three of 34 P. chrysurus; two specimens from two of 58 P. moluccensis; one specimen from one of 83 S. apicalis.
Type-material: Holotype (QM G240016) and 28 paratypes (QM G240017–42, NHMUK 2022.2.25.1–2), including five hologenophores.
Representative DNA sequences: ITS2 rDNA, five identical sequences; 28S rDNA, one sequence; cox1 mtDNA, five identical sequences (See Tables 2 and 3 for GenBank accession numbers).
ZooBank LSID: urn:lsid:zoobank.org:act:1A4C1EB6-DE1D-4F2B-958C-0BD2AF3B7D9E.
Etymology: The epithet opisthoporus refers to the unusually posterior position of the dorsal pore of Laurer’s canal.
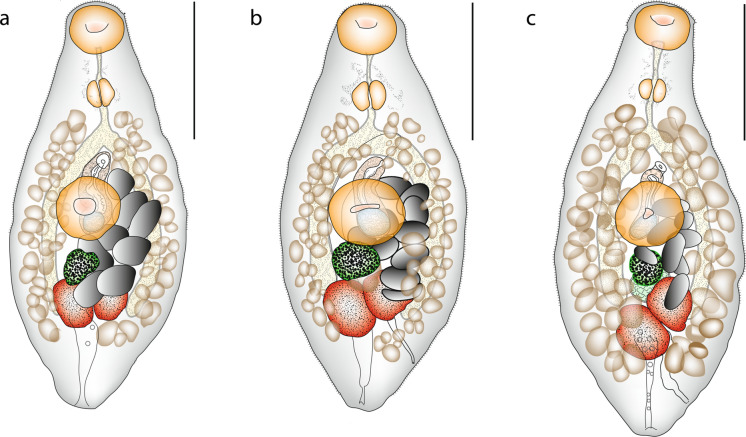
Description (Fig. 2)
Fig. 2.
Opechonoides opisthoporus n. sp. from off Lizard Island, Great Barrier Reef, Australia; a holotype ex Abudefduf whitleyi ventral view; b paratype ex Abudefduf septemfasciatus ventral view; c paratype ex Amphiprion akindynos ventral view. Scale bars: 200 µm
[Based on 24 whole mounts and five hologenophores.] Body small, fusiform, 502–706 × 233–324 (581 × 256). Tegument spined; extent of spines variable, to anterior hindbody, level of testes or close to posterior extremity. Eye-spot pigment scattered widely around prepharynx and pharynx. Oral sucker transversely oval, subterminal, 65–89 × 75–97 (72 × 85). Ventral sucker rounded, 84–121 × 94–115 (100 × 104), in mid-region of body. Prepharynx distinct, 20–40 (33), narrow. Pharynx oval, 37–50 × 42–53 (40 × 44). Oesophagus distinct, 14–34 (24). Intestinal bifurcation in posterior forebody. Caeca short, terminating at level of testes. Testes two, oval, entire, oblique, in mid-hindbody, 47–92 × 47–92 (66 × 65). External seminal vesicle not detected. Cirrus-sac claviform or dumbbell-shaped, 64–164 × 31–50 (118 × 36). Internal seminal vesicle oval. Pars prostatica vesicular, lined with anuclear cell-like bodies. Ejaculatory duct long. Genital atrium distinct. Genital pore sinistrally submedian, post-bifurcal, in posterior forebody. Ovary entire or slightly indented, 34–59 × 34–65 (40 × 44), often obscured by eggs. Laurer’s canal runs diagonally from dorsal to testes to sinistrally submarginal dorsal pore close to posterior extremity. Uterus from mid-testicular level to ventral sucker, mostly intracaecal. Eggs few, large, tanned, operculate, 30–49 × 57–79 (35 × 67). Metraterm muscular, short. Vitellarium follicular; follicles relatively large, with fields reaching from anterior oesophagus to anterior post-testicular region, not reaching close to posterior extremity. Excretory pore dorsally sub-terminal; vesicle narrow posteriorly, widens and reaches to intestinal bifurcation or just anterior, often contains corpuscles in highly characteristic single column.
Remarks
The features that suggest that this new species belongs in Opechonoides include the possession of a uterus that overlaps the testes, short caeca, a long excretory vesicle containing corpuscles, and oblique testes. It differs from the only other species of the genus, O. gure Yamaguti, 1940, in having a greater ventral to oral sucker width ratio (1:1.18–1.25 vs 1:0.80), a shorter cirrus-sac (118 μm vs 190 μm), contiguous testes (vs separated), and a longer post-testicular distance (14–19% vs 6% of body length) with a distinct posterior region lacking vitelline follicles. Additionally, the position and course of Laurer’s canal is unusual and not exactly as described for O. gure. In the latter species, Laurer’s canal does not reach the posterior extremity, but extends only to the level of the posterior testis, and opens ‘a little to the left of the median line’, not submarginally. Molecular data were generated for specimens collected from five of the 12 host species; the remaining specimens were identified based on morphology.
Intermediate hosts
Second intermediate hosts: Bolinopsis (?) sp. (Bolinopsidae); Ocyropsis (?) sp. (Ocyropsidae); Pukia falcata Gershwin, Zeidler & Davie (Pukiidae).
Localities: Off the continental shelf of southeast Queensland, Australia; off the east coast of Tasmania, Australia (see Table 5).
Table 5.
Second intermediate host information (ctenophore species, size, locality and ITS2 rDNA GenBank accession numbers) for Opechonoides opisthoporus n. sp.
| Host species | Diameter (mm) | Locality | Latitude | Longitude | GenBank accession # |
|---|---|---|---|---|---|
| Bolinopsis sp. | – | TAS | 42°36'S | 148°16'E | OM777004 |
| Ocyropsis sp. | – | SE QLD | 27°17'S | 153°35'E | OM776998 |
| Ocyropsis sp. | – | SE QLD | 26°50'S | 153°32'E | OM776999 |
| Ocyropsis sp. | – | SE QLD | 26°50'S | 153°31'E | OM777000 |
| Ocyropsis sp. | – | SE QLD | 26°42'S | 153°41'E | OM777001 |
| Pukia falcata Gershwin, Zeidler & Davie | 13 | SE QLD | 26°23'S | 153°47'E | OM777002 |
| P. falcata | 14 | SE QLD | 25°31'S | 153°48'E | OM777003 |
Abbreviations: SE QLD, off the coast of southeast Queensland; TAS, off the east coast of Tasmania
Site of infection: Mesogloea.
Representative DNA sequences: ITS2 rDNA, seven identical sequences (see Table 5 for GenBank accession numbers).
Remarks
Metacercariae recovered from three ctenophore species were genetically matched to O. opisthoporus n. sp. on the basis of identical ITS2 sequences. No morphological vouchers were available for examination as all specimens were used for molecular sequencing due to their small size.
Genus Lepocreadium Stossich, 1904
Type-species: Lepocreadium album (Stossich, 1890) Stossich, 1904, by original designation.
Lepocreadium oyabitcha Machida, 1984
Type-host: Abudefduf vaigiensis (Quoy & Gaimard), Indo-Pacific sergeant (Pomacentridae).
Type-locality: Off Okinawa, Japan.
Other host: Abudefduf whitleyi Allen & Robertson, Whitley’s sergeant (Pomacentridae).
Other locality: Off Lizard Island, northern Great Barrier Reef, Australia.
New material
New hosts: Abudefduf bengalensis (Bloch), Bengal sergeant; A. sexfasciatus (Cuvier), Scissortail sergeant; A. sordidus (Forsskål), Blackspot sergeant (Pomacentridae).
Known host: A. whitleyi.
Known locality: Off Lizard Island.
Site in host: Gall bladder.
Abundance and prevalence: Two specimens from two of 10 A. bengalensis; 21 specimens from eight of 26 A. sexfasciatus; five specimens from one of three A. sordidus; 18 specimens from six of 15 A. whitleyi.
Voucher material: Six specimens (QM G240043–8), including three hologenophores.
Representative DNA sequences: ITS2 rDNA, five identical sequences (three submitted to GenBank); 28S rDNA, one sequence; cox1 mtDNA, five identical sequences (three submitted to GenBank; see Tables 2 and 3 for GenBank accession numbers).
Remarks
Morphologically, the new specimens closely resemble the descriptions of Machida (1984) and Bray & Cribb (1998), especially in the distinctive tri-lobed ovary and the ramifying vitellarium. All specimens were collected from the gall bladder, which is consistent with the original report of Machida (1984); Bray & Cribb (1998) reported their specimen from the intestine, but we suspect that the worm had been relocated to the intestine from the gall bladder during dissection. Although Machida (1984) reported this worm to occur in pairs, Bray & Cribb (1998) reported a single worm from an individual host, and in the present study, up to five specimens were recovered from a single host.
Genus Lepotrema Ozaki, 1932
Type-species: Lepotrema clavatum Ozaki, 1932, by original designation.
Lepotrema acanthochromidis Bray, Cutmore & Cribb, 2018
Syn. L. clavatum of Bray et al. (1993), Barker et al. (1994) in part
Type-host: Acanthochromis polyacanthus (Bleeker, 1855), Spiny chromis (Pomacentridae).
Type-locality: Off Heron Island, southern Great Barrier Reef, Australia.
Other locality: Off Lizard Island, northern Great Barrier Reef, Australia.
Records: Bray et al. (1993); Barker et al. (1994); Bray et al. (2009); Bray et al. (2018b).
New material
Known host: A. polyacanthus.
Known localities: Off Heron Island; off Lizard Island.
Site in host: Intestine.
Abundance and prevalence: Off Heron Island: six specimens from two of 15 A. polyacanthus. Off Lizard Island: 12 specimens from eight of 15 A. polyacanthus.
Voucher material: Five specimens (QM G240049–53), including two hologenophores.
Representative DNA sequences: ITS2 rDNA, two identical sequences; cox1 mtDNA, six sequences (four submitted to GenBank; see Table 2 for GenBank accession numbers).
Remarks
In the present study, specimens of L. acanthochromidis were collected from the type-host in two known localities, off Heron and Lizard Islands. The new specimens from each locality are morphologically similar but show slight variation in some morphometrics, i.e., egg length [47–56 (51.2) vs 49–68 (55)] and body size [973–1,364 (1,165) vs 914–1,510 (1,194)]. However, comparable levels of morphological variation were also reported by Bray et al. (2018b). Newly generated cox1 sequence data for Heron Island specimens were identical to those published by Bray et al. (2018b). New sequences for Lizard Island specimens show that this species exhibits a strong intraspecific genetic variation that is structured geographically. Infection prevalences reported here differed from those reported by Bray et al. (2018b) in that fewer A. polyacanthus were infected at Heron Island (two of 15 vs 21 of 65) and more were infected at Lizard Island (eight of 15 vs 21 of 74).
Lepotrema adlardi (Bray, Cribb & Barker, 1993) Bray & Cribb, 1996
Syn. Lepocreadium adlardi Bray, Cribb & Barker, 1993
Type-host: Abudefduf bengalensis (Bloch), Bengal sergeant (Pomacentridae).
Type-locality: Off Heron Island, southern Great Barrier Reef, Australia.
Other localities: Off Lizard Island, northern Great Barrier Reef, Australia; Ningaloo Reef, Western Australia, Australia.
Records: Bray et al. (1993); Barker et al. (1994); Bray et al. (2018b).
New material
Known host: A. bengalensis.
Known localities: Off Heron Island; off Lizard Island.
Site in host: Intestine.
Abundance and prevalence: Off Heron Island: nine specimens from four of seven A. bengalensis. Off Lizard Island: six specimens from three of five A. bengalensis.
Voucher material: Four specimens (QM G240054–7), including two hologenophores.
Representative DNA sequences: cox1 mtDNA, three sequences (See Table 2 for GenBank accession numbers).
Remarks
New specimens of L. adlardi were collected from A. bengalensis at two known localities, Heron Island and Lizard Island. New cox1 sequences show no geographical structuring between these sites. In the present study, prevalences were similar to those reported by Bray et al. (2018b), who reported 20 of 43 A. bengalensis from off Heron Island and two of five from off Lizard Island to be infected.
Lepotrema amblyglyphidodonis Bray, Cutmore & Cribb, 2018
Syn. Lepocreadium sp. of Bray et al. (1993) and Barker et al. (1994)
Type-host: Amblyglyphidodon curacao (Bloch), Staghorn damselfish (Pomacentridae).
Type-locality: Off Heron Island, southern Great Barrier Reef, Australia.
Other host: Amphiprion akindynos Allen, Barrier Reef anemonefish (Pomacentridae).
Records: Bray et al. (1993); Barker et al. (1994); Bray et al. (2018b).
New material
New hosts: Plectroglyphidodon dickii (Liénard), Dick’s damsel; Stegastes apicalis (De Vis), Australian gregory (Pomacentridae).
Known hosts: A. curacao; Amphiprion akindynos.
Known locality: Off Heron Island.
Site in host: Intestine.
Abundance and prevalence: Five specimens from three of 23 A. curacao; one specimen from one of seven Amphiprion akindynos; one specimen from one of three P. dickii; one specimen from one of 55 S. apicalis.
Voucher material: Two specimens (QM G240058–9), both hologenophores.
Representative DNA sequences: ITS2 rDNA, one sequence; cox1 mtDNA, two identical sequences (See Table 2 for GenBank accession numbers).
Remarks
The specimens collected in the present study were genetically identical to those of Bray et al. (2018b). Although the single specimen collected from S. apicalis resembles L. monile, which was reported in the same host by Bray et al. (2018b), the new specimen differs slightly by having a muscular pad at the distal metraterm. The infection prevalences reported here are low and similar to those of Bray et al. (2018b), who reported low infection prevalences in A. curacao (five of 71) and Amphiprion akindynos (one of seven).
Phylogenetic results
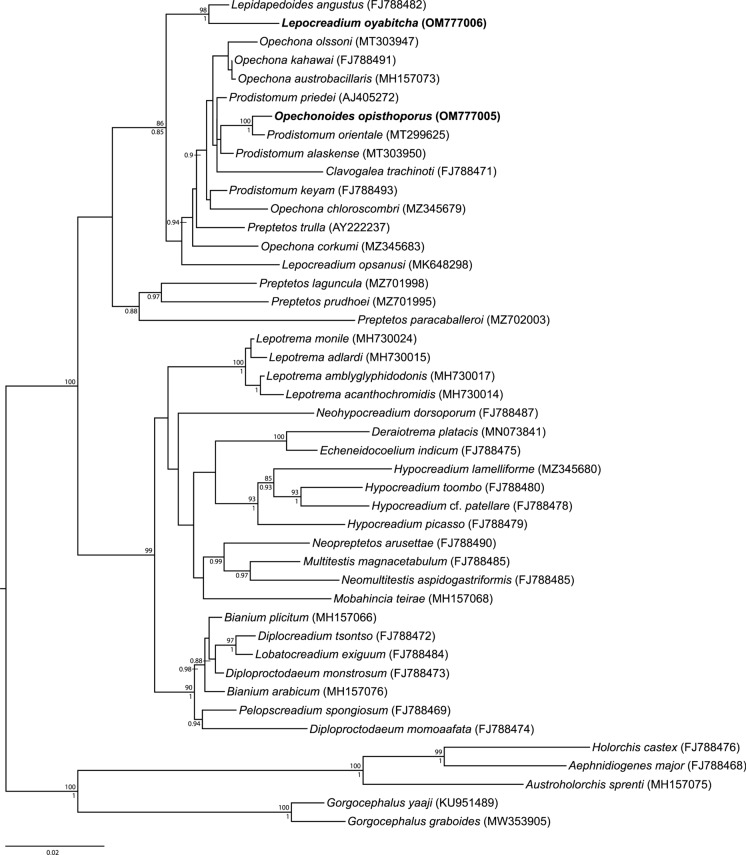
Maximum likelihood and Bayesian inference analyses of the partial 28S sequences produced phylogenetic trees with almost identical topologies but varying levels of nodal support (Fig. 3). While both analyses show strong support for two major clades within the Lepocreadiidae, the key difference between the two phylogenies is the placement of Mobahincia teirae Bray, Cribb & Cutmore, 2018 and Neohypocreadium dorsoporum Machida & Uchida, 1987 in the first major clade. In the maximum likelihood analysis, M. teirae is sister to a clade containing Neomultitestis aspidogastriformis Bray & Cribb, 2003, Multitestis magnacetabulum Mamaev, 1970 and Neopreptetos arusettae Machida, 1982. Neohypocreadium dorsoporum is sister to the previously mentioned clade, four species of Hypocreadium Ozaki, 1936, Echeneidocoelium indicum Simha & Pershad, 1964 and Deraiotrema platacis Machida, 1982. In the Bayesian inference analysis, M. teirae and N. dorsoporum formed a polytomy with all the taxa in this first clade. In both analyses, Lepocreadium oyabitcha and Opechonoides opisthoporus n. sp. fall within the second major clade. Opechonoides opisthoporus n. sp. is sister to Prodistomum orientale (Layman, 1930) Bray & Gibson, 1990 (with strong support) and both formed a polytomy with P. priedei Bray & Merrett, 1998, P. alaskense (Ward & Fillingham, 1934) Bray & Merrett, 1998 and Clavogalea trachinoti (Fischthal & Thomas, 1968) Bray & Gibson, 1990. Lepocreadium oyabitcha is sister to Lepidapedoides angustus Bray, Cribb & Barker, 1996 (with strong support), and is separated from Lepocreadium opsanusi Sogandares & Hutton, 1960.
Fig. 3.
Inferred relationships of Lepocreadium oyabitcha and Opechonoides opisthoporus n. sp. within the Lepocreadiidae based on a maximum likelihood analysis of the 28S rDNA dataset. The newly generated sequences are in bold. Bootstrap support values are shown above the nodes, and where relationships were replicated in the Bayesian inference analysis, posterior probabilities are shown below. Support values below 0.85 or 85 are not shown. Outgroup taxa are species of the Aephnidiogenidae and Gorgocephalidae. The scale bar indicates the number of substitutions per site
Discussion
Phylogeny
The phylogeny and systematics of the Lepocreadioidea have been extensively reviewed by Bray et al. (2009), Bray & Cribb (2012) and Bray et al. (2018a); these studies show that the Lepocreadiidae divides into two major clades, Clade VII and Clade VIII of Bray et al. (2009). Our analyses demonstrate that Lepocreadium oyabitcha and Opechonoides opisthoporus n. sp. both fit into Clade VII, which consists of multiple polyphyletic genera. The genus Lepocreadium appears to be polyphyletic, with L. oyabitcha separated from L. opsanusi, and sister to Lepidapedoides angustus. With genetic data for only two of 27 recognised Lepocreadium species and one of seven recognised Lepidapedoides species (WoRMS Editorial Board, 2021), it is difficult to interpret this relationship, especially given that L. oyabitcha and Lepidapedoides angustus are quite distinct morphologically and infect hosts from different families (L. angustus infects species of the Serranidae). It is noteworthy, however, that both species infect the gall bladders of their hosts (Bray et al., 1996; Justine et al., 2010). The only other species of Opechonoides, O. gure, was described from a single specimen collected from the large-scale blackfish, Girella punctata Gray (Kyphosidae) off Japan (Yamaguti, 1940), and, to our knowledge, it has not been reported elsewhere and no genetic data exist. Although the analyses show that O. opisthoporus n. sp. is embedded within a clade of Prodistomum species, it is clear that at best Prodistomum is paraphyletic and requires molecular study of its type-species P. gracile Linton, 1910.
Mitochondrial DNA
Interestingly, the two species of Lepotrema collected from two localities (Heron Island and Lizard Island), showed differing patterns of genetic variation. Based on cox1 sequences, specimens of L. acanthochromidis have a high level of intraspecific variation between samples collected from the two sites, differing by a minimum of 17 base pairs. A similar level of intraspecific variation was reported for specimens of L. hemitaurichthydis Bray, Cutmore & Cribb, 2018, which differed by 14 base pairs between samples from French Polynesian waters and off Palau (Bray et al., 2018b). Comparable intraspecific cox1 variation has been reported for several trematode species in Indo-Pacific fishes: six species of the Aporocotylidae Odhner, 1912 (see Cutmore et al., 2021), three species of the Gorgocephalidae Manter, 1966 (see Huston et al., 2021), three species of the Lepocreadiidae (see Bray et al., 2018b; 2022) and seven species of the Monorchiidae Odhner, 1911 (see McNamara et al., 2014). Conversely, sequences of L. adlardi collected from off Heron and Lizard Islands differed by up to two base pairs, but with no geographical structuring. A similarly low level of variation was reported for L. cirripectis Bray, Cutmore & Cribb, 2018 (see Bray et al., 2018b). Several trematode species also show little to no differences in cox1 sequences from sites at least as distant as Heron and Lizard Islands: one species of the Aporocotylidae (see Cutmore et al., 2021), two species of the Fellodistomidae Nicoll, 1909 (see Cribb et al., 2021), five species of the Lepocreadiidae (see Bray et al., 2018b; 2022) and three species of the Monorchiidae (see McNamara et al., 2014). These varying patterns of intraspecific genetic variation suggest that trematodes are likely differentially affected by geographical or host-related factors which either limit or benefit their dispersal; however, no synthesis of explanation has emerged so far.
In the case of L. acanthochromidis, it is possible that host-related factors play a role in the high level of intraspecific genetic variation that we see in the cox1 region [and the 28S region (Bray et al., 2018b)]. Populations of the host, Acanthochromis polyacanthus, exhibit strong levels of phenotypic and genetic variation across the GBR (Doherty et al., 1994; Planes et al., 2001; van Herwerden & Doherty, 2006). This species is unique among GBR pomacentrids as it lacks a dispersive pelagic larval stage (Robertson, 1973) and adults exhibit low rates of migration between reefs (Miller-Sims et al., 2008). Populations separated by large areas of deep water (e.g., the Swain Reefs and the Capricorn Bunker Group reefs, which are approximately 150 km apart) show strong levels of genetic structuring, whereas populations on patch reefs separated by shallow water (e.g., Heron Island reefs and Sykes reefs which are less than 2 km apart) show no population differentiation (Miller-Sims et al., 2008). Furthermore, groups of juvenile A. polyacanthus only travel between 50–100 metres between patch reefs (Kavanagh, 2000). These low rates of movement may be effective in limiting the gene flow of L. acanthochromidis, as parasite gene flow is expected to reflect or be determined by hosts with the highest dispersal ability (Prugnolle et al., 2005). Indeed, it is possible that L. acanthochromidis relies on intermediate hosts for dispersal, as there is evidence that gelatinous organisms (with high dispersal capabilities) are used by another species of Lepotrema (Kondo et al., 2016). However, given the distinct geographical genetic structuring, it is likely that L. acanthochromidis uses intermediate host taxa that also have limited dispersal capabilities. It is plausible that historic patterns of sea level resulting in the connectivity (via shallow water ‘crossings’) of distant present-day reefs allowed for the dispersal of A. polyacanthus (see Miller-Sims et al., 2008), L. acanthochromidis and the latter’s intermediate hosts. With changes in sea level, host and trematode populations have become isolated and likely show variation within the cox1 region due to the non-recombinant nature of mitochondrial genes (Morgan-Richards et al., 2017).
Host-specificity
The new collections show that Lepotrema amblyglyphidodonis, Lepocreadium oyabitcha and Opechonoides opisthoporus n. sp. all exhibit stenoxenic host-specificity (Miller et al., 2011); however, each pattern is quite distinct. Lepocreadium oyabitcha has been reported from species of a single genus (Abudefduf), Lepotrema amblyglyphidodonis has been reported from four species belonging to four genera (Amblyglyphidodonis, Amphiprion, Plectroglyphidodon and Stegastes), and O. opisthoporus n. sp. has been reported from 12 species belonging to seven genera (Abudefduf, Amphiprion, Neoglyphidodon, Neopomacentrus, Plectroglyphidodon, Pomacentrus and Stegastes). Such stenoxenicity is also exhibited by L. monile, which has been reported from four species belonging to two genera (Pomacentrus and Stegastes). Of the remaining pomacentrid-infecting lepocreadiids, three are oioxenic (L. acanthochromidis, L. adlardi and Lepocreadium sogandaresi) and one is euryxenic (Lepotrema clavatum). However, as Bray et al. (2018b) noted, it is unlikely that the reported euryxenicity of L. clavatum is valid, given the strict oioxenic-stenoxenic patterns of host-specificity seen for species of Lepotrema specifically, and other lepocreadiids generally.
Infections of O. opisthoporus n. sp. were never common in any pomacentrid species. They were most frequent in species of Abudefduf and Pomacentrus; only single species of the other five genera were infected. However, this could be an artefact of our sampling efforts; species of Abudefduf and Pomacentrus make up over 50% of the pomacentrids that we have examined. Given that we have collected O. opisthoporus n. sp. in other pomacentrid genera, albeit at lower prevalences and abundances, it is likely that with further sampling, more pomacentrid hosts will be discovered. In contrast, the host range of Lepocreadium oyabitcha is unlikely to increase to include other pomacentrid genera, as our current collection records indicate that this species is restricted to species of Abudefduf only. Although the second intermediate host of L. oyabitcha remains unknown, we have examined multiple pomacentrid species that share a similar habitat and diet (planktonic organisms and algae) without finding infections of L. oyabitcha. It is possible that the restriction of L. oyabitcha to species of Abudefduf is related to the size of these fishes and the trematode itself. Lepocreadium oyabitcha is substantially larger than other pomacentrid-infecting lepocreadiids and its size may restrict it from infecting the gall bladder of small pomacentrid species.
Geographical distribution
In the present study, specimens of Lepotrema acanthochromidis, L. adlardi and O. opisthoporus n. sp. were collected from off both Heron and Lizard Islands, which are almost 1,200 km apart. Bray et al. (2018b) also reported the same distribution for L. acanthochromidis, L. adlardi (with an additional locality: Ningaloo Reef in Western Australia) and L. monile. Lepotrema amblyglyphidodonis has only been found off Heron Island; however, the southern restriction of this species in the GBR may be an artefact of our sampling. To date, we have examined over 160 individuals of the host species, A. curacao, Amphiprion akindynos, P. dickii and S. apicalis at Heron Island whereas in comparison, fewer than 70 individuals have been examined at Lizard Island. Given the relatively broad geographical distribution of the other Lepotrema species, it is plausible that more sampling off Lizard Island and elsewhere will increase the geographic range of L. amblyglyphidodonis. In contrast, it is unlikely that the distribution of Lepocreadium oyabitcha includes Heron Island or Moreton Bay, as we have sampled extensively for species of Abudefduf in those regions without detecting it.
Second intermediate hosts
The finding of metacercariae of O. opisthoporus n. sp. in ctenophores represents the first report of intermediate hosts for a species of Opechonoides and indeed for pomacentrid-infecting lepocreadiids. Several studies have reported gelatinous organisms as second intermediate hosts of other lepocreadiids, including species of Lepocreadium, Lepotrema and Opechona Looss, 1907 (see Stunkard, 1972, 1980; Martorelli, 2001; Ohtsuka et al., 2010; Browne et al., 2020). Interestingly, adult O. opisthoporus n. sp. have been found only in GBR pomacentrid fishes, whereas the infections of metacercariae were found in ctenophores collected as far south as off the coast of Tasmania, more than 2,000 km south of the closest known locality for the adults. We can speculate that infected ctenophores are dispersed great distances by the southward flowing East Australian Current or that infections in southern fishes have not yet been detected due to limited sampling. The dispersal capabilities of ctenophores likely influences the distribution of O. opisthoporus n. sp. and explains the lack of genetic variation seen for specimens collected from off Heron and Lizard Islands. Given that Pukia falcata, and species of Bolinopsis L. Agassiz and Ocyropsis Mayer have been reported at various locations along the eastern coast of Australia (Gershwin et al., 2010), it is plausible that they act as intermediate hosts on the GBR. Whilst pomacentrids are not generally known to feed on ctenophores, most are at least partly planktivorous (Pratchett et al., 2016; Cowan et al., 2019; Emslie et al., 2019) and so may well consume ctenophores; the low prevalence and intensity of infection of O. opisthoporus n. sp. in pomacentrids suggests that the consumption may be opportunistic. As other lepocreadiid metacercariae collected from gelatinous host organisms have been linked to definitive hosts that would be expected to consume gelatinous organisms (Ohtsuka et al., 2010), this study has identified a potentially novel trophic link between pomacentrid fishes and ctenophores, highlighting the potential of parasites as novel trophic tracers.
Acknowledgements
We thank members of the Marine Parasitology Laboratory, the University of Queensland, Australia, for their support and assistance. We thank the staff and volunteers at Lizard Island, Heron Island, and Moreton Bay Research Stations, and the Marine National Facility and crew aboard the RV Investigator for their support in the field. We also thank chief scientist B. Slovan, C. Chapman, J. Arnold, N. Arafeh Dalmau, C. Cao, P. Lindholm, C. Olguin Jacobson, I. Suthers and M. Suthers for assistance with collecting and dissecting ctenophore samples.
Funding
This work was funded by the Holsworth Wildlife Research Endowment – Equity Trustees Charitable Foundation and the Ecological Society of Australia (awarded to BD), the PADI Foundation (awarded to BD), the Australian Biological Resources Study (ABRS National Taxonomy Research Grant RG19-37; awarded to SCC and THC) and the Sea World Research and Rescue Foundation (project number SWR12020; awarded to KAP, SCC and THC). Open Access funding enabled and organized by CAUL and its Member Institutions.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Barker SC, Bray RA, Cribb TH. Preptetos cannoni n. sp. (Digenea: Lepocreadiidae) from Siganus lineatus (Teleostei: Siganidae) from the southern Great Barrier Reef, Australia. Systematic Parasitology. 1993;26:151–155. doi: 10.1007/BF00009223. [DOI] [Google Scholar]
- Barker SC, Cribb TH, Bray RA, Adlard RD. Host-parasite associations on a coral reef: pomacentrid fishes and digenean trematodes. International Journal for Parasitology. 1994;24:643–647. doi: 10.1016/0020-7519(94)90116-3. [DOI] [PubMed] [Google Scholar]
- Bartoli P, Gibson DI, Bray RA. Digenean species diversity in teleost fish from a nature reserve off Corsica and a comparison with other Mediterranean regions. Journal of Natural History. 2005;39:47–70. doi: 10.1080/00222930310001613557. [DOI] [Google Scholar]
- Bray RA, Cribb TH. Preptetos and Neopreptetos (Digenea: Lepocreadiidae) from Australian marine fishes. Folia Parasitologica. 1996;43:209–226. doi: 10.1007/BF00009386. [DOI] [Google Scholar]
- Bray RA, Cribb TH. Lepocreadiidae (Digenea) of Australian coastal fishes: new species of Opechona Looss, 1907, Lepotrema Ozaki, 1932 and Bianium Stunkard, 1930 and comments on other species reported for the first time or poorly known in Australian waters. Systematic Parasitology. 1998;41:123–148. doi: 10.1023/A:1006055605808. [DOI] [Google Scholar]
- Bray RA, Cribb TH. Reorganistion of the superfamily Lepocreadioidea Odhner, 1905 based on an inferred molecular phylogeny. Systematic Parasitology. 2012;83:169–177. doi: 10.1007/s11230-012-9386-3. [DOI] [PubMed] [Google Scholar]
- Bray RA, Cribb TH, Barker SC. The Lepocreadiidae (Digenea) of pomacentrid fishes (Perciformes) from Heron Island, Queensland, Australia. Systematic Parasitology. 1993;26:189–200. doi: 10.1007/BF00009726. [DOI] [Google Scholar]
- Bray RA, Cribb TH, Barker SC. Four species of Lepidapedoides Yamaguti, 1970 (Digenea: Lepocreadiidae) from fishes of the southern Great Barrier Reef, with a tabulation of host-parasite data on the group. Systematic Parasitology. 1996;34:179–195. doi: 10.1007/BF00009386. [DOI] [Google Scholar]
- Bray RA, Cribb TH, Cutmore SC. Lepocreadiidae Odhner, 1905 and Aephnidiogenidae Yamaguti, 1934 (Digenea: Lepocreadioidea) of fishes from Moreton Bay, Queensland, Australia, with the erection of a new family and genus. Systematic Parasitology. 2018;95:479–498. doi: 10.1007/s11230-018-9803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray RA, Cutmore SC, Cribb TH. Lepotrema Ozaki, 1932 (Lepocreadidae: Digenea) from Indo-Pacific fishes, with the description of eight new species, characterised by morphometric and molecular features. Systematic Parasitology. 2018;95:693–741. doi: 10.1007/s11230-018-9821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray RA, Cutmore SC, Cribb TH. An anomalous phylogenetic position for Deraiotrema platacis Machida, 1982 (Lepocreadiidae) from Platax pinnatus on the Great Barrier Reef. Diversity. 2019;11:104. doi: 10.3390/d11070104. [DOI] [Google Scholar]
- Bray RA, Cutmore SC, Cribb TH. A paradigm for the recognition of cryptic trematode species in tropical Indo-west Pacific fishes: the problematic genus Preptetos (Trematoda: Lepocreadiidae) International Journal for Parasitology. 2022 doi: 10.1016/j.ijpara.2021.08.004. [DOI] [PubMed] [Google Scholar]
- Bray RA, Littlewood DTJ, Herniou EA, Williams B, Henderson RE. Digenean parasites of deep-sea teleosts: a review and case studies of intrageneric phylogenies. Parasitology. 1999;119:S125–S144. doi: 10.1017/S0031182000084687. [DOI] [PubMed] [Google Scholar]
- Bray RA, Waeschenbach A, Cribb TH, Weedall GD, Dyal P, Littlewood DTJ. The phylogeny of the Lepocreadioidea (Platyhelminthes, Digenea) inferred from nuclear and mitochondrial genes: Implications for their systematics and evolution. Acta Parasitologica. 2009;54:310–329. doi: 10.2478/s11686-009-0045-z. [DOI] [Google Scholar]
- Browne JG, Pitt KA, Cribb TH. DNA sequencing demonstrates the importance of jellyfish in life cycles of lepocreadiid trematodes. Journal of Helminthology. 2020;94:1–10. doi: 10.1017/S0022149X20000632. [DOI] [PubMed] [Google Scholar]
- Claxton AT, Fuehring AD, Andres MJ, Moncrief TD, Curran SS. Parasites of the Vermilion snapper, Rhomboplites aurorubens (Cuvier) from the Western Atlantic Ocean. Comparative Parasitology. 2017;84:1–14. doi: 10.1654/1525-2647-84.1.1. [DOI] [Google Scholar]
- Cowan ZL, Dworjanyn SA, Cabelles CF, Pratchett MS. Predation on crown-of-thorns starfish larvae by damselfishes. Coral Reefs. 2019;35:1253–1262. doi: 10.1007/s00338-016-1491-3. [DOI] [Google Scholar]
- Cribb TH, Anderson GR, Adlard RD, Bray RA. A DNA-based demonstration of a three-host life-cycle for the Bivesiculidae (Platyhelminthes: Digenea) International Journal for Parasitology. 1998;28:1791–1795. doi: 10.1016/S0020-7519(98)00127-1. [DOI] [PubMed] [Google Scholar]
- Cribb TH, Bray RA. Gut wash, body soak, blender and heat-fixation: approaches to the effective collection, fixation and preservation of trematodes of fishes. Systematic Parasitology. 2010;76:1–7. doi: 10.1007/s11230-010-9229-z. [DOI] [PubMed] [Google Scholar]
- Cribb TH, Martin SB, Diaz PE, Bray RA, Cutmore SC. Eight species of Lintonium Stunkard & Nigrelli, 1930 (Digenea: Fellodistomidae) in Australian tetraodontiform fishes. Systematic Parasitology. 2021;98:595–624. doi: 10.1007/s11230-021-10000-w. [DOI] [PubMed] [Google Scholar]
- Curran SS, Ksepka SP, Martorelli SR, Overstreet RM, Warren MB, Bullard SA. Opechona chloroscombri and Opechona corkumi n. sp. (Digenea: Lepocreadiidae) from the Nothern Gulf of Mexico with phylogenetic analysis based on 28S rDNA. Journal of Parasitology. 2021;107:606–620. doi: 10.1645/20-151. [DOI] [PubMed] [Google Scholar]
- Cutmore SC, Yong RQ-Y, Reimer JD, Shirakashi S, Nolan MJ, Cribb TH. Two new species of threadlike blood flukes (Aporocotylidae), with a molecular revision of the genera Ankistromeces Nolan & Cribb, 2004 and Phthinomita Bolan & Cribb, 2006. Systematic Parasitology. 2021 doi: 10.1007/s11230-021-10002-8. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PJ, Mather P, Planes S. Acanthochromis polyacanthus, a fish lacking larval dispersal, has genetically differentiated populations at local and regional scales on the Great Barrier Reef. Marine Biology. 1994;121:11–21. doi: 10.1007/BF00349469. [DOI] [Google Scholar]
- Dyer WG, Williams EH, Williams LB. Digenetic trematodes of marine fishes of the western and southwestern coasts of Puerto Rico. Proceedings of the Helminthological Society of Washington. 1985;52:85–94. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emslie MJ, Logan M, Cheal AJ. The distribution of planktivorous damselfishes (Pomacentridae) on the Great Barrier Reef and the relative influences of habitat and predation. Diversity. 2019;11:33. doi: 10.3390/d11030033. [DOI] [Google Scholar]
- Gershwin, L., Zeidler, W., & Davie, P. J. F. (2010). Ctenophora of Australia. In: Davie, P. J. F. & Phillips, J. A. (Eds) Proceedings of the Thirteenth International Marine Biological Workshops, the Marine Fauna and Flora of Moreton Bay, Queensland. Brisbane: Memoirs of the Queensland Museum, pp. 1–45.
- Huston DC, Cutmore SC, Cribb TH. The life-cycle of Gorgocephalus yaaji Bray & Cribb, 2005 (Digenea: Gorgocephalidae) with a review of the first intermediate hosts for the superfamily Lepocreadioidea Odhner, 1905. Systematic Parasitology. 2016;93:653–665. doi: 10.1007/s11230-016-9655-7. [DOI] [PubMed] [Google Scholar]
- Huston, D. C., Cutmore, S. C., Miller, T. L., Sasal, P., Smit, N. J., & Cribb, T. H. (2021). Gorgocephalidae (Digenea: Lepocreadioidea) in the Indo-West Pacific: new species, life-cycle data and perspectives on species delineation over geographic range. Zoological Journal of the Linnean Society, 193, 1416–1455. 10.1093/zoolinnean/zlab002
- ICZN. (2012). Amendment of articles 8 9 10 21 and 78 of the international code of zoological nomenclature to expand and refine methods of publication. ZooKeys, 219, 1–10. 10.3897/zookeys.219.3944 [DOI] [PMC free article] [PubMed]
- International Commission on Zoological Nomenclature. (2012). International Code of Zoological Nomenclature. International Trust for Zoological Nomenclature 1999, London. [DOI] [PMC free article] [PubMed]
- Justine, J.-L., Beveridge, I., Boxshall, G. A., Bray, R. A., Moravec, F., Trilles, J.-L., & Whittington, I. D. (2010). An annotated list of parasites (Isopoda, Copepoda, Monogenea, Digenea, Cestoda and Nematoda) collected in groupers (Serranidae, Epinephelinae) in New Caledonia emphasizes parasite biodiversity in coral reef fish. Folia Parasitologica, 57, 237–262. 10.14411/fp.2010.032 [DOI] [PubMed]
- Kavanagh KD. Larval brooding in the marine damselfish Acanthochromis polyacanthus (Pomacentridae) is correlated with highly divergent morphology, ontogeny and life-history traits. Bulletin of Marine Science. 2000;66:321–337. [Google Scholar]
- Kondo Y, Ohtsuka S, Hirabayashi T, Okada S, Ogawa NO, Ohkouchi N, Shimazu T, Nishikawa J. Seasonal changes in infection with trematode species utilizing jellyfish as hosts: evidence of transmission to definitive host fish via medusivory. Parasite. 2016;23:16. doi: 10.1051/parasite/2016016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood DTJ. Molecular phylogenetics of cupped oysters based on partial 28S ribosomal RNA gene sequences. Molecular Phylogenetics and Evolution. 1994;3:221–229. doi: 10.1006/mpev.1994.1024. [DOI] [PubMed] [Google Scholar]
- Littlewood DTJ, Curini-Galletti M, Herniou EA. The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Molecular Phylogenetics and Evolution. 2000;16:449–466. doi: 10.1006/mpev.2000.0802. [DOI] [PubMed] [Google Scholar]
- Littlewood DTJ, Rohde K, Clough KA. Parasite speciation within or between host species?—phylogenetic evidence from site-specific polystome monogeneans. International Journal for Parasitology. 1997;27:1289–1297. doi: 10.1016/S0020-7519(97)00086-6. [DOI] [PubMed] [Google Scholar]
- Machida M. Two new trematodes from gall bladder of tropical marine fishes, Myripristis and Abudefduf. Bulletin of the National Science Museum. Tokyo. Series a. Zoology. 1984;10:1–5. [Google Scholar]
- Maddison, W. P., & Maddison, D. R. 2021. Mesquite: a modular system for evolutionary analysis. Version 3.70. Available from: https://www.mesquiteproject.org
- Martorelli SR. Digenea parasites of jellyfish and ctenophores of the southern Atlantic. Hydrobiologia. 2001;451:305–310. doi: 10.1007/978-94-010-0722-1_25. [DOI] [Google Scholar]
- McNamara MKA, Miller TL, Cribb TH. Evidence for extensive cryptic speciation in trematodes of butterflyfishes (Chaetodontidae) of the tropical Indo-West Pacific. International Journal for Parasitology. 2014;44:37–48. doi: 10.1016/j.ijpara.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Miller-Sims VC, Gerlach G, Kingsford MJ, Atema J. Dispersal in the spiny damselfish, Acanthochromis polyacanthus, a coral reef fish species withou a larval pelagic stage. Molecular Ecology. 2008;17:5036–5048. doi: 10.1111/j.1365-294X.2008.03986.x. [DOI] [PubMed] [Google Scholar]
- Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, pp. 1–8.
- Miller TL, Bray RA, Cribb TH. Taxonomic approaches to and interpretation of host specificity of trematodes of fishes: lessons from the Great Barrier Reef. Parasitology. 2011;138:1710–1722. doi: 10.1017/S0031182011000576. [DOI] [PubMed] [Google Scholar]
- Morgan-Richards M, Bulgarella M, Sivyer L, Dowle EJ, Hale M, McKean NE, Trewick SA. Explaining large mitochondrial sequence differences within a population sample Royal Society Open. Science. 2017;4:170730. doi: 10.1098/rsos.170730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Blair D. Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar-spine group. Parasitology. 1995;111:609–615. doi: 10.1017/S003118200007709X. [DOI] [PubMed] [Google Scholar]
- Nahhas FM, Powell EC. Digenetic trematodes of marine fishes from the Floridian northern Gulf of Mexico. Tulane Studies in Zoology and Botany. 1971;17:1–9. [Google Scholar]
- Ohtsuka, S., Kondo, Y., Sakai, Y., Shimazu, T., Shimomura, M., Komai, T., Yanagi, K., Fujita, T., Nishikawa, J., Miyake, H., Venmathi Maran, B. A., Go, A., Nagagushi, K., Yamagushi, S., Dechsakulwatana, C., Srinui, K., Putchakarn, S., Mulyadi, M., Mujiono, N., Sutomo, & Yusoff, F. M. D. (2010). In-situ observations of symbionts on medusae occurring in Japan, Thailand, Indonesia and Malaysia. Bulletin of the Hiroshima University Museum, 2, 9–18. 10.15027/32060
- Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DTJ. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda) International Journal for Parasitology. 2003;33:733–755. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- Papoutsoglou SE. Metazoan parasites of fishes from Saronicos Gulf Athens - Greece. Thalassographica. 1976;1:69–102. [Google Scholar]
- Pérez-Ponce de León G, Hernández-Mena D. Testing the higher-level phylogenetic classification of Digenea (Platyhelminthes, Trematoda) based on nuclear rDNA sequences before entering the age of the 'next-generation' Tree of Life. Journal of Helminthology. 2019;93:260–276. doi: 10.1017/S0022149X19000191. [DOI] [PubMed] [Google Scholar]
- Planes S, Doherty PJ, Bernardi G. Strong genetic divergence among populations of a marine fish with limited dispersal, Acanthochromis polyacanthus, within the Great Barrier Reef and the Coral Sea. Evolution. 2001;55:2263–2273. doi: 10.1111/j.0014-3820.2001.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Pleijel F, Jondelius U, Norlinder E, Nygren A, Oxelman B, Schander C, Sundberg P, Thollesson M. Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Molecular Phylogenetics and Evolution. 2008;48:369–371. doi: 10.1016/j.ympev.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Pratchett, M. S., Hoey, A. S., Wilson, S. K., Hobbs, J. A., & Allen, G. R. (2016). Habitat-use and specialisation among coral reef damselfishes In: Parmentier, E. & Frédérich, B. (Eds) Biology of Damselfishes. Boca Raton: CRC Press LLC.
- Pritchard MH. Studies on digenetic trematodes of Hawaiian fishes, primarily families Lepocreadiidae and Zoogonidae. Journal of Parasitology. 1963;49:578–587. doi: 10.2307/3275763. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Théron A, Pointier JP, Jabbour-Zahab R, Jarne P, Durand P, de Meeûs T. Dispersal in a parasitic worm and its two hosts: consequence for local adaptation. Evolution. 2005;59:296–303. doi: 10.1111/j.0014-3820.2005.tb00990.x. [DOI] [PubMed] [Google Scholar]
- Robertson DR. Field observations on the reproductive behaviour of a pomacentrid fish, Acanthochromis polyacanthus. Zeitschrift Für Tierpsychologie. 1973;32:319–324. doi: 10.1111/j.1439-0310.1973.tb01108.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenk M, van der Mark P, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Snyder SD, Tkach VV. Phylogenetic and biogeographical relationships among some Holarctic frog lung flukes (Digenea: Haematoloechidae) Journal of Parasitology. 2001;87:1433–1440. doi: 10.1645/0022-3395(2001)087[1433:PABRAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Sokolov SG, Gordeev II, Atopkin DM. Phylogenetic affiliation of the lepocreadiid trematodes parasitizing some marine fishes in the North-western Pacific. Marine Biology Research. 2020;16:380–389. doi: 10.1080/17451000.2020.1758947. [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post–analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard HW. Observations on the morphology and life-history of the digenetic trematode, Lepocreadium setiferoides (Miller and Northup, 1926) Martin, 1938. Biology Bulletin. 1972;142:326–334. doi: 10.2307/1540235. [DOI] [PubMed] [Google Scholar]
- Stunkard HW. The morphology, life-history, and taxonomic relations of Lepocreadium areolatum (Linton, 1900) Stunkard, 1969 (Trematoda: Digenea) Biology Bulletin. 1980;158:154–163. doi: 10.2307/1540766. [DOI] [Google Scholar]
- Sun D, Blomberg SP, Cribb TH, McCormick MI, Grutter AS. The effects of parasites on the early life stages of a damselfish. Coral Reefs. 2012;31:1065–1075. doi: 10.1007/s00338-012-0929-5. [DOI] [Google Scholar]
- Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Available from: https://paup.phylosolutions.com/
- van Herwerden L, Doherty PJ. Contrasting genetic structures across two hybrid zones of a tropical reef fish, Acanthochromis polyacanthus (Bleeker 1855) Journal of Evolutionary Biology. 2006;19:239–252. doi: 10.1111/j.1420-9101.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- Wee NQ-X, Cribb TH, Bray RA, Cutmore SC. Two known and one new species of Proctoeces from Australian teleosts: Variable host-specificity for closely related species identified through multi-locus molecular data. Parasitology International. 2017;66:16–26. doi: 10.1016/j.parint.2016.11.008. [DOI] [PubMed] [Google Scholar]
- WoRMS. (2021). World Register of Marine Species. Available from: http://www.marinespecies.org at VLIZ. Accessed 27-09-2021. 10.14284/170
- Xia X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Molecular Biology and Evolution. 2018;35:1550–1552. doi: 10.1093/molbev/msy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Lemey P. Assessing substitution saturation with DAMBE. In: Lemey P, Salemi M, Vandamme AM, editors. Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny. University Press; 2009. pp. 615–630. [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen LU, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Yamaguti S. Studies on the helminth fauna of Japan, Part 31. Trematodes of fishes VII. Japanese Journal of Zoology. 1940;9:35–108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.