Abstract
Species of Sporocadaceae have often been reported as plant pathogens, endophytes or saprophytes and are commonly isolated from a wide range of plant hosts. The isolated fungi were studied through a complete examination, based on multilocus phylogenies from combined datasets of ITS/tub2/tef1, in conjunction with morphological characteristics. Nine strains were isolated from Ficusmicrocarpa, Ilexchinensis and Schimasuperba in China which represented four species, viz., Monochaetiaschimaesp. nov., Neopestalotiopsishaikouensissp. nov., Neopestalotiopsispiceana and Pestalotiopsislicualicola. Neopestalotiopsispiceana was a new country record for China and first host record from Ficusmacrocarpa. Pestalotiopsislicualicola was first report from Ilexchinensis in China.
Keywords: Monochaetia , multigene phylogeny, Neopestalotiopsis , Pestalotiopsis
Introduction
The family Sporocadaceae was established by Corda in 1842 (type genus: Sporocadus). Species of Sporocadaceae are endophytic, plant pathogenic or saprobic, and associated with a wide range of host plants (Maharachch. et al. 2013; Jayawardena et al. 2015; Liu et al. 2019). Currently, the family comprises 35 genera including Monochaetia (Sacc.) Allesch., Neopestalotiopsis Maharachch. et al., Pestalotiopsis Steyaert, Pseudopestalotiopsis Maharachch.et al., and etc. Most genera have multi-septate and more or less fusiform conidia with appendages at one or both ends, frequently with some melanised cells. Also known as pestalotioid fungi, resembling those taxa having affinities with Pestalotia (Liu et al. 2019).
Steyaert (1949) segregated two novel genera from Pestalotia, namely Pestalotiopsis (with 5-celled conidia) and Truncatella (with 4-celled conidia) based on the conidial forms. This resulted in apparent controversy from Guba (1956, 1961). He emphasised that there was no point in assembling species with similar numbers of conidial septa into distinct genera. Subsequently, Steyaert (1953, 1961, 1963) provided further evidence in support of splitting Pestalotia. Sutton (1980) accepted most of the genera discussed here (Pestalotia, Pestalotiopsis, Truncatella) which fitted into fairly well-defined groups and cited the electron microscope investigation of Griffiths and Swart (1974), which examined the conidial wall of Pestalotiapezizoides and two species of Pestalotiopsis (P.funerea and P.triseta) to support Steyaert’s division of Pestalotiopsis. Maharachch. et al. 2014 segregated two novel genera from Pestalotiopsis, namely Neopestalotiopsis and Pseudopestalotiopsis, based on conidia pigment colour, conidiophores and molecular phylogeny. Neopestalotiopsis can be easily distinguished from Pseudopestalotiopsis and Pestalotiopsis by its versicolourous median cells (Maharachch. et al. 2014). Saccardo (1884) introduced Monochaetia as a subgenus of Pestalotia (as Pestolozzia). The genus Monochaetia was introduced by Allescher (1902), which included 23 species. Allescher (1902) designated the type Monochaetiamonochaeta which has a single apical appendage (Guba 1961; Maharachch. et al. 2014; Senanayake et al. 2015). Steyaert (1949) transferred numerous Monochaetia species to Pestalotiopsis or Truncatella. More than 40 species of Monochaetia were recognised by the monograph of Guba (1961). There are 127 Monochaetia epithets in the Index Fungorum (accession date: 31 March 2022) and most have been transferred to other genera such as Sarcostroma, Seimatosporium and Seiridium (Nag Raj 1993; Maharachch. et al. 2011, 2014, 2016). Seridium and Monochaetia have obvious morphological differences and show separate clades (de Silva et al. 2017).
To date, most phylogenetic studies addressing genera of Sporocadaceae have been based solely on ITS and LSU sequences (Barber et al. 2011; Tanaka et al. 2011; Jaklitsch et al. 2016), or on concatenated datasets of more genes but with incomplete datasets (Senanayake et al. 2015; Wijayawardene et al. 2016). In this study, we made a collection of the established genera Monochaetia, Neopestalotiopsis and Pestalotiopsis species from leaves of Ficusmicrocarpa, Ilexchinensis and Schimasuperba in Hainan Province, China. The inventories allowed establishing two new species that are described here.
Materials and methods
Isolation and morphological studies
The samples were collected from Hainan Province, China. The strains were isolated from diseased leaves of Ficusmicrocarpa, Ilexchinensis and Schimasuperba using surface disinfected tissue fragments (0.5 × 0.5 cm) taken from the margin of leaf lesions (Gao et al. 2014; Jiang et al. 2021a). Surface disinfection consisted of steps including immersion in 75% ethanol for 30 s, 5% sodium hypochlorite (Aladdin, Shanghai, China) for 1 min, and sterile distilled water for 30 s. The pieces were dried with sterilized paper towels and placed on potato dextrose agar (PDA). All plates were incubated at 25 °C for 3–4 days. Then, hyphae were picked out of the periphery of the colonies and inoculated onto new PDA plates. Photographs of the colonies were taken at 7 and 15 days using a Powershot G7X mark II digital camera. Micromorphological characters were observed using an Olympus SZX10 stereomicroscope and Olympus BX53 microscope, all fitted with Olympus DP80 high definition colour digital cameras to photo-document fungal structures. The size of conidia was measured by software Digimizer (https://www.digimizer.com/), and thirty individual measurements were obtained for each character. All fungal strains were stored in 10% sterilised glycerin at 4 °C for further studies. The holotype specimens were deposited in the Herbarium of Plant Pathology, Shandong Agricultural University (HSAUP). Ex-type cultures were deposited in theShandong Agricultural University Culture Collection (SAUCC). Taxonomic information on the new taxa was submitted to MycoBank (http://www.mycobank.org).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelium grown on PDA using cetyltrimethylammonium bromide (CTAB) protocol as described in Guo et al. (2000). The internal transcribed spacer regions with intervening 5.8S nrRNA gene (ITS) and partial beta-tubulin (tub2) and translation elongation factor 1-alpha (tef1) genes were amplified and sequenced by using primers pairs ITS5/ITS4 (White et al. 1990), T1/Bt2b (Glass and Donaldson 1995; O’Donnell and Cigelnik 1997), and EF1-728F/EF-2 (O’Donnell et al. 1998; Carbone and Kohn 1999).
PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification reactions were performed in a 50 μL reaction volume, which contained 25 μL Green Taq Mix (Vazyme, Nanjing, China), 2 μL of each forward and reverse primer (10 μM) (Tsingke, Beijing, China), and 2 μL template genomic DNA, to which distilled deionized water was added. PCR parameters were as follows: 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at a suitable temperature for 30 s, extension at 72 °C for 1 min and a final elongation step at 72 °C for 7 min. Annealing temperature was 55 °C for ITS, 54 °C for tub2, 52 °C for tef1. The PCR products were visualised on 1% agarose electrophoresis gel. Sequencing was done bi-directionally, conducted by the Tsingke Biotechnology Company Limited (Qingdao, China). Consensus sequences were obtained using MEGA 7.0 or MEGA-X (Kumar et al. 2016). All sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
Species and GenBank accession numbers of DNA sequences used in this study. New sequences are in bold.
| Species | Strain | Host/substrate | Country | GenBank accession number | Reference | ||
|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | |||||
| Bartaliniarobillardoides | CBS 122705 T | Leptoglossusoccidentalis | Italy | LT853104 | LT853202 | LT853252 | Bonthond et al. 2018 |
| Ciliochorellaphanericola | MFLUCC 14-0984 T | Phanerapurpurea | Thailand | KX789680 | – | KX789682 | Jiang et al. 2021b |
| MFLUCC 12-0310 | Phanerapurpurea | Thailand | KF827444 | KF827477 | KF827478 | Jiang et al. 2021b | |
| Monochaetiacastaneae | CFCC 54354 = SM9-1 T | Castaneamollissima | China | MW166222 | MW199741 | MW218515 | Jiang et al. 2021b |
| SM9-2 | Castaneamollissima | China | MW166223 | MW199742 | MW218516 | Jiang et al. 2021b | |
| M.dimorphospora | NBRC 9980 | Castaneapubinervis | Japan | LC146750 | – | – | Liu et al. 2019 |
| M.ilicis | KUMCC 15-0520 T | Ilex sp. | China | KX984153 | – | – | de Silva et al. 2017 |
| CBS 101009 | Air | Japan | MH553953 | MH554371 | MH554612 | Liu et al. 2019 | |
| M.junipericola | CBS 143391 T | Juniperuscommunis | Germany | MH107900 | MH108021 | MH108045 | Crous et al. 2018 |
| M.kansensis | PSHI2004Endo1030 | Cyclobalaopsismyrsinaefolia | China | DQ534044 | – | DQ534047 | Liu et al. 2006 |
| PSHI2004Endo1031 | Cyclobalaopsismyrsinaefolia | China | DQ534045 | – | DQ534048 | Liu et al. 2006 | |
| M.monochaeta | CBS 546.80 | Culture contaminant | Netherlands | MH554056 | MH554491 | MH554732 | Liu et al. 2019 |
| CBS 199.82 T | Quercuspubescens | Italy | MH554018 | – | MH554694 | Liu et al. 2019 | |
| CBS 115004 | Quercusrobur | Netherlands | AY853243 | MH554398 | MH554639 | Liu et al. 2019 | |
| M.quercus | CBS 144034 T | Quercuseduardi | Mexico | MH554171 | MH554606 | MH554844 | Liu et al. 2019 |
| M.schimae | SAUCC212201 T | Schimasuperba | China | MZ577565 | OK104874 | OK104867 | This study |
| SAUCC212202 | Schimasuperba | China | MZ577566 | OK104875 | OK104868 | This study | |
| SAUCC212203 | Schimasuperba | China | MZ577567 | OK104876 | OK104869 | This study | |
| M.sinensis | HKAS 10065 T | Quercus sp. | China | MH115995 | – | MH115999 | de Silva et al. 2018 |
| Neopestalotiopsisacrostichi | MFLUCC 17-1754 T | Acrostichumaureum | Thailand | MK764272 | MK764316 | MK764338 | Norphanphoun et al. 2019 |
| N.alpapicalis | MFLUCC 17-2544 T | Rhizophoramucronata | Thailand | MK357772 | MK463547 | MK463545 | Kumar et al. 2019 |
| N.aotearoa | CBS 367.54 T | Canvas | New Zealand | KM199369 | KM199526 | KM199454 | Maharachch. et al. 2014 |
| N.asiatica | MFLUCC 12-0286 T | Unidentified tree | China | JX398983 | JX399049 | JX399018 | Maharachch. et al. 2012 |
| CFCC 54339 = SM32 | Castaneamollissima | China | MW166224 | MW199743 | MW218517 | Jiang et al. 2021b | |
| N.brachiata | MFLUCC 17-1555 T | Rhizophoraapiculata | Thailand | MK764274 | MK764318 | MK764340 | Norphanphoun et al. 2019 |
| N.brasiliensis | COAD 2166 T | Psidiumguajava | Brazil | MG686469 | MG692402 | MG692400 | Bezerra et al. 2018 |
| CFCC 54341 = ZY4 | Castaneamollissima | China | MW166229 | MW199748 | MW218522 | Jiang et al. 2021b | |
| ZY4-2D | Castaneamollissima | China | MW166230 | MW199749 | MW218523 | Jiang et al. 2021b | |
| N.chiangmaiensis | MFLUCC 18-0113 T | Dead leaves | Thailand | – | MH388404 | MH412725 | Tibpromma et al. 2018 |
| N.chrysea | MFLUCC 12-0261 T | Pandanus sp. | China | JX398985 | JX399051 | JX399020 | Maharachch. et al. 2012 |
| N.clavispora | MFLUCC 12-0281 T | Magnolia sp. | China | JX398979 | JX399045 | JX399014 | Maharachch. et al. 2012 |
| N.cocoes | MFLUCC 15-0152 T | Cocosnucifera | Thailand | KX789687 | KX789689 | – | Norphanphoun et al. 2019 |
| N.coffea-arabica | HGUP 4019 T | Coffeaarabica | China | KF412649 | KF412646 | KF412643 | Song et al. 2013 |
| N.cubana | CBS 600.96 T | Leaf litter | Cuba | KM199347 | KM199521 | KM199438 | Maharachch. et al. 2014 |
| N.dendrobii | MFLUCC 14-0106 T | Dendrobiumcariniferum | Chiang Rai, Thailand | MK993571 | MK975829 | MK975835 | Ma et al. 2019 |
| N.egyptiaca | CBS 140162 T | Mangiferaindica | Egypt | KP943747 | KP943748 | KP943746 | Crous et al. 2015 |
| N.ellipsospora | MFLUCC 12-0283 T | Dead plant materials | China | JX398980 | JX399047 | JX399016 | Maharachch. et al. 2012 |
| N.eucalypticola | CBS 264.37 T | Eucalyptusglobulus | – | KM199376 | KM199551 | KM199431 | Maharachch. et al. 2014 |
| N.foedans | CGMCC 3.9123 T | Mangrove plant | China | JX398987 | JX399053 | JX399022 | Maharachch. et al. 2012 |
| N.formicidarum | CBS 362.72 T | Dead ant | Ghana | KM199358 | KM199517 | KM199455 | Maharachch. et al. 2014 |
| CBS 115.83 | Plant debris | Cuba | KM199344 | KM199519 | KM199444 | Maharachch. et al. 2014 | |
| N.hadrolaeliae | COAD 2637 T | Hadrolaeliajongheana | Minas Gerais, Brazil | MK454709 | MK465122 | MK465120 | Freitas et al. 2019 |
| N.haikouensis | SAUCC212271 T | Ilexchinensis | China | OK087294 | OK104877 | OK104870 | This study |
| SAUCC212272 | Ilexchinensis | China | OK087295 | OK104878 | OK104871 | This study | |
| N.honoluluana | CBS 114495 T | Telopea sp. | USA | KM199364 | KM199548 | KM199457 | Maharachch. et al. 2014 |
| N.iraniensis | CBS 137768 T | Fragariaananassa | Iran | KM074048 | KM074051 | KM074057 | Ayoubi et al. 2016 |
| N.javaensis | CBS 257.31 T | Cocosnucifera | Indonesia | KM199357 | KM199543 | KM199437 | Maharachch. et al. 2014 |
| N.macadamiae | BRIP 63737c T | Macadamiaintegrifolia | Australia | KX186604 | KX186627 | KX186654 | Akinsanmi et al. 2017 |
| N.magna | MFLUCC 12-0652 T | Pteridium sp. | France | KF582795 | KF582791 | KF582793 | Maharachch. et al. 2012 |
| N.mesopotamica | CBS 336.86 T | Pinusbrutia | Iraq | KM199362 | KM199555 | KM199441 | Maharachch. et al. 2014 |
| N.musae | MFLUCC 15-0776 T | Musa sp. | Thailand | KX789683 | KX789685 | KX789686 | Norphanphoun et al. 2019 |
| N.natalensis | CBS 138.41 T | Acaciamollissima | South Africa | KM199377 | KM199552 | KM199466 | Maharachch. et al. 2014 |
| N.pandanicola | KUMCC 17-0175 T | Pandanaceae | China | – | MH388389 | MH412720 | Tibpromma et al. 2018 |
| N.pernambucana | URM 7148-01 T | Vismiaguianensis | Brazil | KJ792466 | KU306739 | – | Silvério et al. 2016 |
| N.petila | MFLUCC 17-1738 T | Rhizophoramucronata | Thailand | MK764276 | MK764320 | MK764342 | Norphanphoun et al. 2019 |
| N.phangngaensis | MFLUCC 18-0119 T | Pandanaceae | Thailand | MH388354 | MH388390 | MH412721 | Tibpromma et al. 2018 |
| N.piceana | CBS 394.48 T | Picea sp. | UK | KM199368 | KM199527 | KM199453 | Maharachch. et al. 2014 |
| CBS 254.32 | Cocosnucifera | Indonesia | KM199372 | KM199529 | KM199452 | Maharachch. et al. 2014 | |
| SAUCC210112 | Ficusmicrocarpa | China | OK149224 | OK206436 | OK206434 | This study | |
| SAUCC210113 | Ficusmicrocarpa | China | OK149225 | OK206437 | OK206435 | This study | |
| N.protearum | CBS 114178 T | Leucospermumcuneiforme cv. “Sunbird” | Zimbabwe | JN712498 | KM199542 | KM199463 | Maharachch. et al. 2014 |
| N.rhizophorae | MFLUCC 17-1550 T | Rhizophoramucronata | Thailand | MK764278 | MK764322 | MK764344 | Norphanphoun et al. 2019 |
| N.rosae | CBS 124745 | Paeoniasuffruticosa | USA | KM199360 | KM199524 | KM199430 | Maharachch. et al. 2014 |
| CBS 101057 T | Rosa sp. | New Zealand | KM199359 | KM199523 | KM199429 | Maharachch. et al. 2014 | |
| N.rosicola | CFCC 51992 T | Rosachinensis | China | KY885239 | KY885243 | KY885245 | Norphanphoun et al. 2019 |
| CFCC 51993 | Rosachinensis | China | KY885240 | KY885244 | KY885246 | NNorphanphoun et al. 2019 | |
| N.samarangensis | MFLUCC 12-0233 T | Syzygiumsamarangense | Thailand | JQ968609 | JQ968611 | JQ968610 | Maharachch. et al. 2012 |
| N.saprophytica | MFLUCC 12-0282 T | Magnolia sp. | China | KM199345 | KM199538 | KM199433 | Maharachch. et al. 2014 |
| N.sichuanensis | CFCC 54338 = SM15-1 T | Castaneamollissima | China | MW166231 | MW199750 | MW218524 | Jiang et al. 2021b |
| N.sonneratae | MFLUCC 17-1745 T | Sonneronataalba | Thailand | MK764280 | MK764324 | MK764346 | Norphanphoun et al. 2019 |
| N.steyaertii | IMI 192475 T | Eucalytpusviminalis | Australia | KF582796 | KF582792 | KF582794 | Maharachch. et al. 2012 |
| N.surinamensis | CBS 450.74 T | soil under Elaeisguineensis | Suriname | KM199351 | KM199518 | KM199465 | Maharachch. et al. 2014 |
| N.thailandica | MFLUCC 17-1730 T | Rhizophoramucronata | Thailand | MK764281 | MK764325 | MK764347 | Norphanphoun et al. 2019 |
| N.umbrinospora | MFLUCC 12-0285 T | unidentified plant | China | JX398984 | JX399050 | JX399019 | Maharachch. et al. 2012 |
| N.vitis | MFLUCC 15-1265 T | Vitisvinifera cv. “Summer black” | China | KU140694 | KU140676 | KU140685 | Jayawardena et al. 2016 |
| N.zimbabwana | CBS 111495 T | Leucospermumcunciforme cv. “Sunbird” | Zimbabwe | JX556231 | KM199545 | KM199456 | Maharachch. et al. 2014 |
| Nonappendiculataquercina | CBS 116061 T | Quercussuber | Italy | MH553982 | MH554400 | MH554641 | Liu et al. 2019 |
| CBS 270.82 | Quercuspubescens | Italy | MH554025 | MH554459 | MH554701 | Liu et al. 2019 | |
| Pestalotiopsisaustralasiae | CBS 114126 T | Knightia sp. | New Zealand | KM199297 | KM199499 | KM199409 | Maharachch. et al. 2014 |
| P.australis | CBS 114193 T | Grevillea sp. | Australia | KM199332 | KM199475 | KM199383 | Maharachch. et al. 2014 |
| P.grevilleae | CBS 114127 T | Grevillea sp. | Australia | KM199300 | KM199504 | KM199407 | Maharachch. et al. 2014 |
| P.hollandica | CBS 265.33 T | Sciadopitysverticillata | The Netherlands | KM199328 | KM199481 | KM199388 | Maharachch. et al. 2014 |
| P.kenyana | CBS 442.67 T | Coffea sp. | Kenya | KM199302 | KM199502 | KM199395 | Maharachch. et al. 2014 |
| P.knightiae | CBS 114138 T | Knightia sp. | New Zealand | KM199310 | KM199497 | KM199408 | Maharachch. et al. 2014 |
| P.licualicola | HGUP4057 T | Licualagrandis | China | KC492509 | KC481684 | KC481683 | Geng et al. 2013 |
| SAUCC210087 | Ilexchinensis | China | OK087323 | OK104879 | OK104872 | This study | |
| SAUCC210088 | Ilexchinensis | China | OK087324 | OK104880 | OK104873 | This study | |
| P.oryzae | CBS 353.69 T | Oryzasativa | Denmark | KM199299 | KM199496 | KM199398 | Maharachch. et al. 2014 |
| P.parva | CBS 278.35 | Leucothoefontanesiana | – | KM199313 | KM199509 | KM199405 | Maharachch. et al. 2014 |
| P.portugalica | CBS 393.48 T | – | Portugal | KM199335 | KM199510 | KM199422 | Maharachch. et al. 2014 |
| P.spathuliappendiculata | CBS 144035 T | Phoenixcanariensis | Australia | MH554172 | MH554607 | MH554845 | Liu et al. 2019 |
| Pseudopestalotiopsiscocos | CBS 272.29 T | Cocosnucifera | Indonesia | KM199378 | KM199553 | KM199467 | Maharachch. et al. 2014 |
| Pse.elaeidis | CBS 413.62 T | Elaeisguineensis | Nigeria | MH554044 | MH554479 | MH554720 | Liu et al. 2019 |
| Pse.indica | CBS 459.78 T | Rosasinensis | India | KM199381 | KM199560 | KM199470 | Maharachch. et al. 2014 |
| Seiridiumpapillatum | CBS 340.97 T | Eucalyptusdelegatensis | Australia | LT853102 | MH554468 | LT853250 | Bonthond et al. 2018 |
| Seir.phylicae | CBS 133587 T | Phylicaarborea | Tristan da Cunha | LT853091 | LT853188 | LT853238 | Bonthond et al. 2018 |
Isolates marked with “T” are ex-type or ex-epitype strains.
Phylogeny
Newly generated sequences in this study were aligned with additional related sequences downloaded from GenBank (Table 1) using MAFFT 7 online service with the Auto strategy (Katoh et al. 2019, http://mafft.cbrc.jp/alignment/server/). To establish the identity of the isolates at the species level, phylogenetic analyses were conducted first individually for each locus and then as combined analyses of three loci (ITS, tub2 and tef1). Phylogenetic analyses were based on maximum likelihood (ML) and Bayesian inference (BI) for the multi-locus analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses. ML and BI were run on the CIPRES Science Gateway portal (https://www.phylo.org/) (Miller et al. 2012) using RaxML-HPC2 on XSEDE v. 8.2.12 (Stamatakis 2014) and MrBayes on XSEDE v. 3.2.7a (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012), respectively. Four Markov chains were run for two runs from random starting trees for 10,000,000 generations (ITS + tub2 + tef1) until the split deviation frequency value < 0.01, and trees were sampled every 1000 generation. The first quarter generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated. The resulting trees were plotted using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) and edited with Adobe Illustrator CC 2019. New sequences generated in this study were deposited at GenBank (https://www.ncbi.nlm.nih.gov; Table 1). The final concatenated sequence alignments were deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S29480).
Result
Phylogenetic analyses
Nine strains of Sporocadaceae isolated from plant hosts from Hainan, China, were grown in culture and used for analyses of molecular sequence data. The combined dataset of ITS-tub2-tef1 has an aligned length of 2285 total characters (ITS: 1–638, tub2: 639–1558, tef1: 1559–2285) including gaps, of which 869 characters are constant, 292 variable and parsimony-uninformative, and 1124 parsimony-informative. For the BI and ML analyses, the substitution model GTR+G for ITS, HKY+I+G for tub2 and GTR+I+G for tef1 were selected and incorporated into the analyses. The MCMC analysis of the three concatenated genes run for 7,795,000 generations, resulting in 7796 trees. The ML tree topology confirmed the tree topologies obtained from the BI analyses, and therefore, only the ML tree is presented (Fig. 1).
Figure 1.
Phylogram of Sporocadaceae based on combined ITS, tub2 and tef1 sequences. The BI and ML bootstrap support values above 0.90 and 70% are shown at the first and second position, respectively. The tree is rooted to Bartaliniarobillardoides (CBS 122705), ex-type or ex-epitype cultures are indicated in bold face. Strains from the current study are in red. Some branches were shortened according to the indicated mulipliers.
Bayesian posterior probability (≥ 0.90) and ML bootstrap support values (≥ 70%) are shown as first and second position above nodes. The 96 strains were assigned to 75 species clades based on the three gene loci phylogeny (Fig. 1). Based on the multi-locus phylogeny and morphology, nine isolates were assigned to four species, including Monochaetiaschimae sp. nov., Neopestalotiopsishaikouensis sp. nov., Neopestalotiopsispiceana and Pestalotiopsislicualicola.
Taxonomy
. Monochaetia schimae
Z. X. Zhang, J. W. Xia & X. G. Zhang sp. nov.
5ACD3395-C104-58D5-8EC2-53136E31A7FE
MycoBank No: 841381
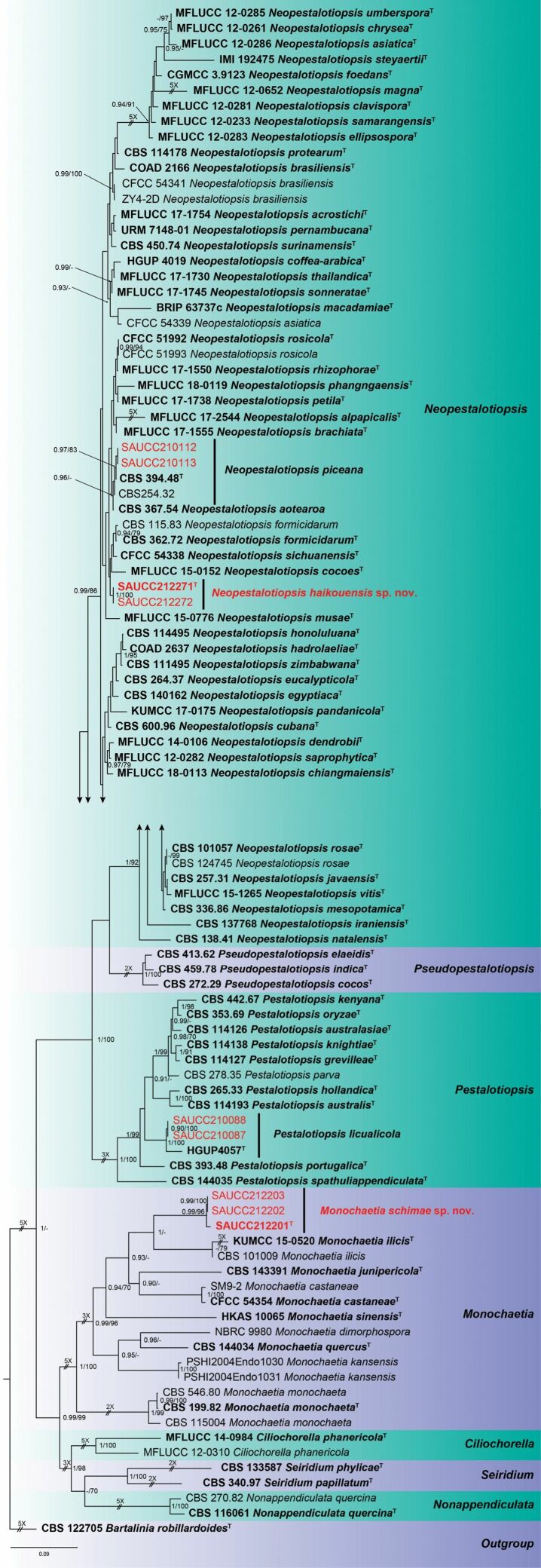
Figure 2.
Monochaetiaschimae (SAUCC212201, ex-type) a diseased leaf of Schimasuperbab surface of colony after 15 days on PDAc reverse of colony after 15 days on PDAd conidiomata e, f conidiogenous cells with conidia g–j conidia. Scale bars: 10 μm (e–j).
Type.
China, Hainan Province: East Harbour National Nature Reserve, on diseased leaves of Schimasuperba, 23 May 2021, Z.X. Zhang (holotype HSAUP212201; ex-type living culture SAUCC212201).
Etymology.
Name refers to the genus of the host plant Schimasuperba.
Description.
Leaf spots irregular, pale brown in centre, brown to tan at margin. Sexual morph not observed. Asexual morph on PDA: Conidiomata solitary, scattered, black, raising above surface of culture medium, subglobose, exuding black conidial droplets from central ostioles after 10 days in light at 25 °C. Conidiophores cylindrical, hyaline, smooth-walled. Conidiogenous cells 9.0–16.5 × 1.2–2.2 μm, phialidic, ampulliform, discrete, hyaline, smooth, thin-walled. Conidia 18–24 × 4.5–6.0 μm, mean ± SD = 20.5 ± 1.1 × 5.5 ± 0.4 μm, fusiform, tapering at both ends, 4-septate; apical cell 2.0–4.0 μm long, conical, hyaline and smooth-walled; three median cells doliiform, 12.5–15.5 μm long, mean ± SD = 14.2 ± 0.7 μm, olivaceous, rough-walled, upper second cell 3.8–5.3 μm long, upper third cell 3.4–5.0 μm long, upper fourth cell 4.4–5.4 μm long; basal cell 2.2–4.5 μm long, conical, hyaline and smooth-walled; apical appendage 7.0–12.5 μm long (mean = 9.2 μm), single, unbranched, central, tubular, filiform; basal appendage 2.5–5.0 μm long, single, unbranched tubular, filiform.
Culture characteristics.
Colonies on PDA 39.0–45.0 mm in diameter after 15 days at 25 °C in darkness, growth rate 2.5–3.0 mm/day, irregularly circular, raised, dense surface with lobate edge, zonate in different sectors, light brown at the margin, brown at the centre; reverse brown at the margin, dark brown at the centre.
Additional specimen examined.
China, Hainan Province: East Harbour National Nature Reserve, 23 May 2021, Z.X. Zhang. On diseased leaves of Schimasuperba, paratype HSAUP212202, living culture SAUCC212202; on diseased leaves of Schimasuperba, paratype HSAUP212203, living culture SAUCC212203.
Notes.
Monochaetiaschimae is introduced based on the multi-locus phylogenetic analysis, with three isolates clustering separately in a well-supported clade (BI/ML = 0.99/96). Monochaetiaschimae is phylogenetically close to M.castaneae from leaves of Castaneamollissima, M.ilicis from leaves of Ilex sp., and M.junipericola from twigs of Juniperuscommunis. However, Monochaetiaschimae differs from M.castaneae by 148 nucleotides (11/463 in ITS, 89/743 in tub2 and 48/403 in tef1), from M.ilicis by 94 nucleotides (18/526 in ITS, 32/698 in tub2 and 44/456 in tef1), and from M.junipericola by 91 nucleotides (10/524 in ITS, 40/411 in tub2 and 41/304 in tef1). Furthermore, they are distinguished by hosts and conidial sizes (18.0–24.0 × 4.5–6.0 μm in M.schimae vs. 18.8–27.3 × 4.7–6.6 μm in M.castaneae vs. 20.0–27.0 × 5.0–8.0 μm in M.ilicis vs. 22.0–28.0 × 5.0–7.0 μm in M.junipericola). In morphology, Monochaetiacastaneae differs from M.schimae by the colour of colonies (cinnamon vs. brown), Monochaetiailicis differs from M.schimae by the colour of median cells (brown vs. olivaceous), and M.junipericola differs from M.schimae by longer conidiogenous cells (10.0–30.0 μm vs. 9.0–16.5 μm) (de Silva et al. 2017; Crous et al. 2018; Jiang et al. 2021b).
. Neopestalotiopsis haikouensis
Z. X. Zhang, J. W. Xia & X. G. Zhang sp. nov.
258515C1-6141-59F9-8114-3C1274776E97
MycoBank No: 841382
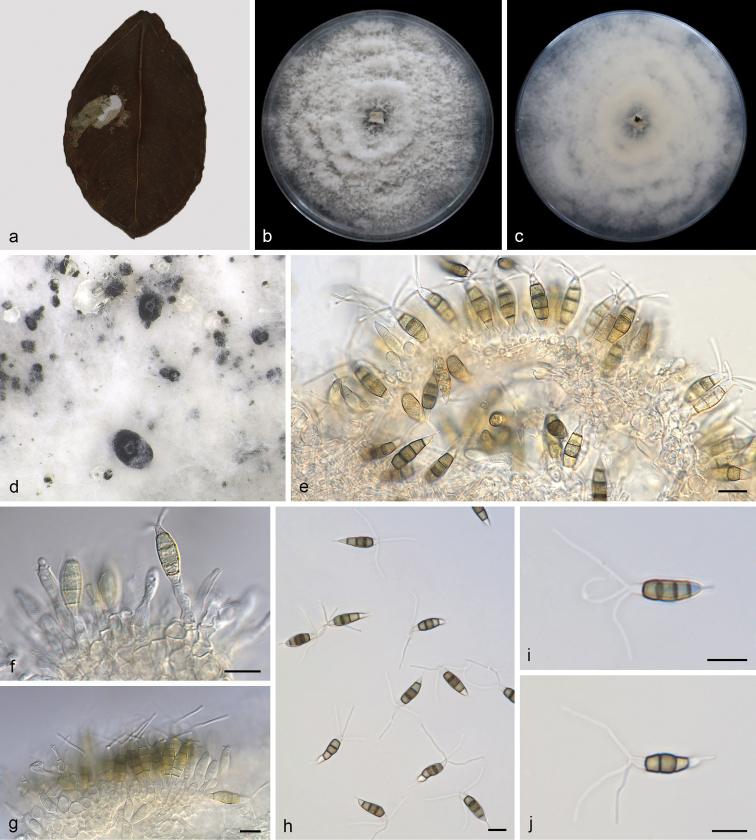
Figure 3.
Neopestalotiopsishaikouensis (SAUCC212271, ex-type) a diseased leaf of Ilexchinensisb surface of colony after 7 days on PDAc reverse of colony after 7 days on PDAd conidiomata e–g conidiogenous cells with conidia h–j conidia. Scale bars: 10 μm (e–j).
Type.
China, Hainan Province, Haikou City: East Harbour National Nature Reserve, on diseased leaves of Ilexchinensis. 23 May 2021, Z.X. Zhang (holotype HSAUP212271; ex-type living culture SAUCC212271).
Etymology.
Named after the host location, Haikou City.
Description.
Leaf spots irregular, grey white in centre, brown to tan at margin. Sexual morph not observed. Asexual morph on PDA: Conidiomata globose to clavate, solitary or confluent, embedded or semi-immersed to erumpent, dark brown, exuding globose, dark brown to black conidial masses. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells discrete, subcylindrical to ampulliform, hyaline, 5.0–10.0 × 2.0–6.0 μm, apex 1.0–2.0 μm diam. Conidia fusoid, ellipsoid, straight to slightly curved, 4-septate, 16.0–22.0 × 4.5–7.0 μm, mean ± SD = 20.0 ± 1.8 × 5.5 ± 0.4 μm; basal cell conical with a truncate base, hyaline, rugose and thin-walled, 3.0–4.5 μm long; three median cells doliiform, 11.5–15.0 μm long, mean ± SD = 13.2 ± 1.0 μm, wall rugose, septa darker than the rest of the cell, second cell from the base pale brown, 3.5–5.5 μm long; third cell honey-brown, 4.0–6.0 μm long; fourth cell brown, 3.8–5.7 μm long; apical cell 2.5–5.5 μm long, hyaline, cylindrical to subcylindrical, thin- and smooth-walled; with 2–3 tubular apical appendages (mostly 3), arising from the apical crest, unbranched, filiform, 13.5–24.0 μm long, mean ± SD = 19.1 ± 3.5 μm; basal appendage 2.0–7.0 μm long, single, tubular, unbranched, centric.
Culture characteristics.
Colonies on PDA occupying an entire 90 mm petri dish in 7 days at 25 °C in darkness, growth rate of 7.0–14.0 mm/day, edge undulate, white to grey white, with moderate aerial mycelium on the surface, with black, gregarious conidiomata; reverse similar in colour.
Additional specimen examined.
China, Hainan Province: East Harbour National Nature Reserve, 23 May 2021, Z.X. Zhang. On diseased leaves of Ilexchinensis, paratype HSAUP212272, living culture SAUCC212272.
Notes.
Phylogenetic analysis of a combined three-gene ITS-tub2-tef1 showed that Neopestalotiopsishaikouensis formed an independent clade with full-supported (BI/ML = 1/100, Fig. 1) and is phylogenetically distinct from N.cocoes (MFLUCC 15-0152), N.formicidarum (CBS 362.72) and N.sichuanensis (CFCC 54338). Neopestalotiopsishaikouensis can be distinguished from the phylogenetically most closely related species N.cocoes by narrower conidia (4.5–7.0 vs. 7.5–9.5 μm), N.formicidarum by smaller conidia (16.0–22.0 × 4.5–7.0 vs. 20.0–29.0 × 7.5–9.5 μm), and N.sichuanensis by shorter conidia (16.0–22.0 vs. 23.2–32.8 μm). Furthermore, some species were reported from the same host genus Ilex, including Pestalotianeglecta, Pestalotiopsisannulata, P.humicola and P.ilicis. After comparison, P.humicola was closest to N.haikouensis in morphology, but with 78/588 differences in the ITS region (Maharachch. et al. 2014; Liu et al. 2019; Jiang et al. 2021b).
. Neopestalotiopsis piceana
S.S.N. Maharachch., K.D. Hyde & P.W. Crous, Studies in Mycology 79:146. (2014)
6B8AB555-F733-5E02-8311-780850EB95F6
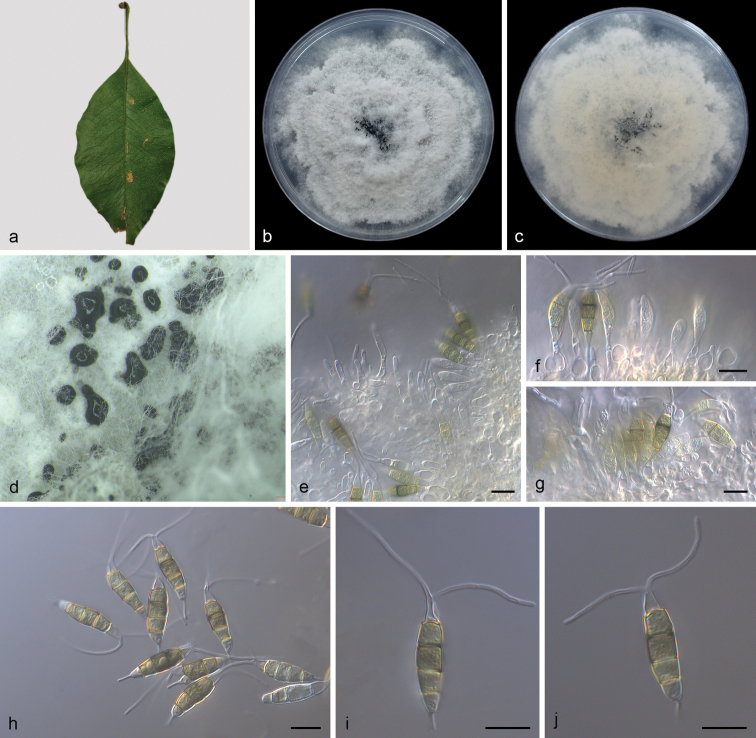
Figure 4.
Neopestalotiopsispiceana (SAUCC210112) a diseased leaf of Ficusmicrocarpab surface of colony after 7 days on PDAc reverse of colony after 7 days on PDAd conidiomata e–g conidiogenous cells with conidia h–j conidia. Scale bars: 10 μm (e–j).
Description.
Leaf spots irregular, pale brown in centre, brown to tan at margin. Asexual morph on PDA: Conidiomata solitary, globose to clavate, semi-immersed, brown to black; exuding globose, dark brown to black conidial masses. Conidiophores reduced to conidiogenous cells. Conidiogenous cells discrete, ampulliform to lageniform, hyaline, smooth and thin walled, simple, 4.0–12.0 × 2.0–10.0 μm, apex 2.0–5.0 μm diam. Conidia ellipsoid to clavate, straight to slightly curved, 4-septate, 19.5–26.5 × 5.5–7.0 μm, mean ± SD = 22.7 ± 0.8 × 6.1 ± 0.4 μm; somewhat constricted at septa; basal cell obconic with truncate base, rugose and thin-walled, 2.7–5.0 μm long; three median cells 12.0–16.0 μm long, mean ± SD = 14.7 ± 0.9 μm, doliiform, verruculose, versicoloured, septa darker than the rest of the cell, second cell from base pale brown, 4.0–5.7 μm long; third cell dark brown, 3.5–5.2 μm long; fourth cell brown, 3.8–5.8 μm long; apical cell obconic, hyaline, thin and smooth-walled, 2.5–5.2 μm long; with 1–3 tubular apical appendages, arising from the apical crest, flexuous, unbranched, 21.0–32.0 μm long, mean ± SD = 24.8 ± 3.5 μm; basal appendage single, tubular, unbranched, centric, 2.7–6.5 μm long.
Culture characteristics.
Colonies on PDA incubated at 25 °C in the dark with an average radial growth rate of 9.0–14.0 mm/day and occupying an entire 90 mm petri dish in 7 d, with edge undulate, whitish, aerial mycelium on surface, fruiting bodies black, concentric; reverse of culture yellow to pale brown.
Specimen examined.
China, Hainan Province: Five Fingers Group Scenic Area, 20 May 2021, Z.X. Zhang. On diseased leaves of Ficusmicrocarpa, HSAUP210112, living culture SAUCC210112; on diseased leaves of Ficusmicrocarpa, HSAUP210113, living culture SAUCC210113.
Notes.
In the present study, two strains (SAUCC210112 and SAUCC210113) from symptomatic leaves of Ficusmicrocarpa were clustered with Neopestalotiopsispiceana clade (Maharachch. et al. 2014) based on phylogeny (Fig. 1). Morphologically, our strains were the same as N.piceana, which was originally described with an asexual morph on wood of Picea sp., Cocosnucifera and fruit of Mangiferaindica. The sexual morph of N.piceana was undetermined yet. Neopestalotiopsispiceana was a new record for China and first reported from Ficusmacrocarpa (Moraceae).
. Pestalotiopsis licualicola
K. Geng, Y. Song, K.D. Hyde & Yong Wang bis, Phytotaxa 88 (3):51. (2013)
CE5D32D9-599D-5EF1-BF8F-16BB35C4CE4F
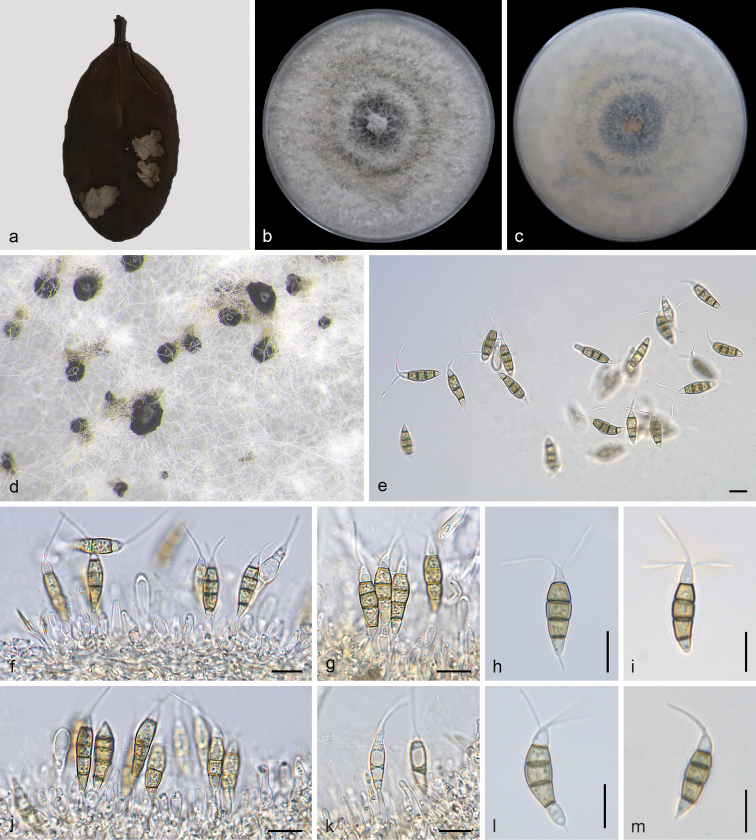
Figure 5.
Pestalotiopsislicualicola (SAUCC210087) a diseased leaf of Ilexchinensisb surface of colony after 7 days on PDAc reverse of colony after 7 days on PDAd conidiomata f, g, j, k conidiogenous cells with conidia e, h, i, l, m conidia. Scale bars: 10 μm (e–m).
Description.
Leaf spots irregular, pale brown in centre, brown to tan at margin. Asexual morph on PDA: Conidiomata solitary, scattered, black, raising above surface of culture medium, subglobose. Conidiophores cylindrical, hyaline, smooth-walled. Conidiophores often indistinct. Conidiogenous cells discrete, hyaline, simple, filiform, 5.5–10.0 μm long. Conidia 18.0–24.5 × 4.0–5.5 μm, mean ± SD = 20.5 ± 1.9 × 5.3 ± 0.3 μm, fusiform, straight to slightly curved, 4-septate, smooth, greyish brown; basal cell conical, hyaline, thin-walled, 2.8–6.0 μm long; with three median cells, dark brown, concolorous, septa and periclinal walls darker than the rest of the cell, together 11.5–16.0 μm long, mean ± SD = 13.2 ± 1.2 μm; second cell from base 3.4–5.5 μm; third cell 3.3–4.7 μm; fourth cell 3.5–5.1 μm; apical cell hyaline, conic to subcylindrical, 3.1–5.3 μm; with 1–3 tubular apical appendages (mostly 1) without knobs, arising from the apex of the apical cell, 10.0–20.5 μm long, mean ± SD = 16.0 ± 4.0 μm; basal appendage filiform, short.
Culture characteristics.
Colonies on PDA reaching 70.0–80.0 mm diam after 7 d at 25 °C, growth rate 9.0–12.0 mm/day, edge entire, whitish to pale honey coloured, with sparse aerial mycelium on the surface, with black, gregarious conidiomata; reverse similar in colour.
Specimen examined.
China, Hainan Province: East Harbour National Nature Reserve, 23 May 2021, Z.X. Zhang. On diseased leaves of Ilexchinensis, HSAUP210087, living culture SAUCC210087; on diseased leaves of Ilexchinensis, HSAUP210088, living culture SAUCC210088.
Notes.
In the present study, two strains (SAUCC210087 and SAUCC210088) from symptomatic leaves of Ilexchinensis were clustered to Pestalotiopsislicualicola clade (Geng et al. 2013) based on phylogeny (Fig. 1). Morphologically, our strains were the same as P.licualicola, which was originally described with an asexual morph on leaves of Licualagrandis in China. The sexual morph of P.licualicola was undetermined yet. This is the first time this species has been reported in Ilexchinensis (Aquifoliaceae) in China.
Discussion
Based on phylogeny and morphology, nine strains from three host species (Ficusmicrocarpa, Ilexchinensis and Schimasuperba) were described as well as two new species (Monochaetiaschimae sp. nov. and Neopestalotiopsishaikouensis sp. nov.) and two known species (Neopestalotiopsispiceana and Pestalotiopsislicualicola). In the genus Monochaetia, most species were found on Fagaceae hosts, including Castaneapubinervis (Monochaetiadimorphospora), Castaneamollissima (Monochaetiacastaneae), Quercuspubescens (Monochaetiamonochaeta) and etc. In our study, the species of Monochaetia (M.schimae) was first reported from Schimasuperba (Theaceae). Ilex was widely grown as an evergreen tree all over the world and isolated many pathogens, endophytes or saprophytes (Alfieri et al. 1984; Maharachch. et al. 2014; de Silva et al. 2017; Solarte et al. 2018). More than 100 strains (Xylariales) have been isolated from the genus Ilex. Among these, there was 13 pestalotia-like fungi, and we compare morphology with my new collection. In morphology, the conidia size of Pestalotiopsishumicola is similar to Neopestalotiopsishaikouensis. Phylogenetic analyses of Maharachch. et al. (2014) and the current study show Neopestalotiopsis and Pestalotiopsis are different genus. The known species Neopestalotiopsispiceana was described from Picea sp. (Pinaceae) in United Kingdom (Maharachch. et al. 2014) and Pestalotiopsislicualicola was described from Licualagrandis (Palmaceae) in China (Geng et al. 2013). In this study, Neopestalotiopsispiceana was a new record for China and first reported from Ficusmacrocarpa (Moraceae), Pestalotiopsislicualicola was first reported from Ilexchinensis (Aquifoliaceae) in China, so we described and illustrated N.piceana and P.licualicola again. Species in genera have multi-septate and more or less fusiform conidia with a single apical and basal appendage (Monochaetia, Seiridium); other genera do not form appendages (Nonappendiculata) or have 2–4 appendages (Pestalotiopsis, Ciliochorella, Neopestalotiopsis, Pseudopestalotiopsis) (Maharachch. et al. 2014; Bonthond et al. 2018; Liu et al. 2019). Our study supported this phenomenon.
As many pestalotioid species have overlapping morphological traits, sequence data is essential to resolve these three genera and introduce new species (Jeewon et al. 2002; de Silva et al. 2017; Norphanphoun et al. 2019). Combined gene sequences of ITS, tub2 and tef1 can provide a better resolution for Monochaetia. However, more genes are needed to provide better resolution and support in Neopestalotiopsis. In the previous studies, members of Sporocadaceae are of particular interest with regard to the production of secondary metabolites, e.g. Bartalinia, Morinia and Pestalotiopsis (Collado et al. 2006; Gangadevi and Muthumary 2008; Liu et al. 2009). Pestalotiopsisfici was shown to possess a very high number of gene clusters involved in bioactive compound synthesis (Wang et al. 2016). Owing to Pestalotiopsis and other genus in this family sharing the same evolutionary history, it is important to report novel species and screen for novel metabolites in future studies.
Supplementary Material
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (nos. 31900014, U2002203, 31750001) and National Science and Technology Fundamental Resources Investigation Program of China (2019FY100704).
Citation
Zhang Z, Liu R, Liu S, Mu T, Zhang X, Xia J (2022) Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 88: 171–192. https://doi.org/10.3897/mycokeys.88.82229
Funding Statement
This work was supported by the National Natural Science Foundation of China (nos. 31900014, U2002203, 31750001).
Supplementary materials
The combined ITS, tub2 and tef1 sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhaoxue Zhang, Rongyu Liu, Shubin Liu, Taichang Mu, Xiuguo Zhang, Jiwen Xia
Data type
phylogenetic
Explanation note
The combined ITS, tub2 and tef1 sequences.
References
- Akinsanmi OA, Nisa S, Jeff-Ego OS, Shivas RG, Drenth A. (2017) Dry Flower Disease of Macadamia in Australia Caused by Neopestalotiopsismacadamiae sp. nov. and Pestalotiopsismacadamiae sp. nov. Plant Disease 101(1): 45–53. 10.1094/PDIS-05-16-0630-RE [DOI] [PubMed] [Google Scholar]
- Alfieri Jr SA, Langdon KR, Wehlburg C, Kimbrough JW. (1984) Index of Plant Diseases in Florida (Revised). Florida Dept. Agric. And Consumer Serv., Div. Plant Ind. Bull. 11: 1–389. [Google Scholar]
- Allescher A. (1902) Fungi Imperfecti: Gefärbt-sporige Sphaerioideen. Rabenhorst’s Kryptogamen-Flora von Deutschland. Österreich und der Schweiz. 2nd edn. Kummer, Leipzig, 65–128.
- Ayoubi N, Soleimani MJ. (2016) Strawberry Fruit Rot Caused by Neopestalotiopsisiranensis sp. nov., and N.mesopotamica. Current Microbiology 2016(72): 329–336. 10.1007/s00284-015-0955-y [DOI] [PubMed]
- Barber PA, Crous PW, Groenewald JZ, Pascoe IG, Keane P. (2011) Reassessing Vermisporium (Amphisphaeriaceae), a genus of foliar pathogens of eucalypts. Persoonia 27(1): 90–118. 10.3767/003158511X617381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra JDP, Machado AR, Firmino AL, Rosado AWC, Souza CAF, Souza-Motta CM, Freire KTLS, Paiva LM, Magalhaes OMC, Pereira OL, Crous PW, Oliveira TGL, Abreu VP, Fan XL. (2018) Mycological Diversity Description I. Acta Botanica Brasílica 32(4): 656–666. 10.1590/0102-33062018abb0154 [DOI] [Google Scholar]
- Bonthond G, Sandoval-Denis M, Groenewald JZ, Crous PW. (2018) Seiridium (Sporocadaceae): An important genus of plant pathogenic fungi. Persoonia 40(1): 96–118. 10.3767/persoonia.2018.40.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 91(3): 553–556. 10.2307/3761358 [DOI] [Google Scholar]
- Collado J, Platas G, Bills GF, Basilio Á, Vicente F, Rubén Tormo J, Hernández P, Teresa Díez M, Peláez F. (2006) Studies on Morinia: Recognition of Morinialongiappendiculata sp. nov. as a new endophytic fungus, and a new circumscription of Moriniapestalozzioides. Mycologia 98(4): 616–627. 10.1080/15572536.2006.11832665 [DOI] [PubMed]
- Crous PW, Wingfield MJ, Le RJJ, Richardson DM, Strasberg D, Shivas RG, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane MS, Tan YP, Altés A, Barasubiye T, Barnes CW, Blanchette RA, Boertmann D, Bogo A, Carlavilla JR, Cheewangkoon R, Daniel R, de Beer ZW, de Jesús Yáñez-Morales M, Duong TA, Fernández-Vicente J, Geering ADW, Guest DI, Held BW, Heykoop M, Hubka V, Ismail AM, Kajale SC, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque CA, Lombard L, Magista D, Manjón JL, Marincowitz S, Mohedano JM, Nováková A, Oberlies NH, Otto EC, Paguigan ND, Pascoe IG, Pérez-Butrón JL, Perrone G, Rahi P, Raja HA, Rintoul T, Sanhueza RMV, Scarlett K, Shouche YS, Shuttleworth LA, Taylor PWJ, Thorn RG, Vawdrey LL, Solano-Vidal R, Voitk A, Wong PTW, Wood AR, Zamora JC, Groenewald JZ. (2015) Fungal planet description sheets: 371–399. Persoonia 35(1): 264–327. 10.3767/003158515X690269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, Akulov A, Denman S, Roux J, Braun U, Burgess T, Carnegie AJ, Vaczy KZ, Guatimosim E, Schwartsburd PB, Barreto RW, Hernandez-Restrepo M, Lombard L, Groenewald JZ. (2018) New and Interesting Fungi. 1. Fungal Systematics and Evolution 1(1): 169–215. 10.3114/fuse.2018.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva N, Phookamsak R, Maharachchikumbura SSN, Thambugala KM, Jayarama Bhat D, Al-Sadi AM, Lumyong S, Hyde KD. (2017) Monochaetiailexae sp. nov. (Pestalotiopsidaceae) from Yunnan Province in China. Phytotaxa 291(2): 123–132. 10.11646/phytotaxa.291.2.3 [DOI] [Google Scholar]
- de Silva N, Maharachchikumbura SSN, Thambugala KM, Jayarama Bhat D, Phookamsak R, Al-Sadi AM, Lumyong S, Hyde KD. (2018) Monochaetiasinensis sp. nov. from Yunnan Province in China. Phytotaxa 375(1): 59–69. 10.11646/phytotaxa.375.1.2 [DOI] [Google Scholar]
- Freitas EFS, de Silva N, Barros MVP, Kasuya MCM. (2019) Neopestalotiopsishadrolaeliae sp. nov., a new endophytic species from the roots of the endangered orchid Hadrolaeliajongheana in Brazil. Phytotaxa 416(3): 211–220. 10.11646/phytotaxa.416.3.2 [DOI] [Google Scholar]
- Gangadevi V, Muthumary J. (2008) Taxol, an anticancer drug produced by an endophytic fungus Bartaliniarobillardoides Tassi, isolated from a medicinal plant, Aeglemarmelos Correa ex Roxb. World Journal of Microbiology & Biotechnology 24(5): 717–724. 10.1007/s11274-007-9530-4 [DOI] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai L. (2014) Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13(1): 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Geng K, Zhang B, Song Y, Hyde KD, Kang JC, Wang Y. (2013) A new species of Pestalotiopsis from leaf spots of Licualagrandis from Hainan, China. Phytotaxa 88(3): 49–54. 10.11646/phytotaxa.88.3.2 [DOI] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths DA, Swart HJ. (1974) Conidial structure in two species of Pestalotiopsis. Transactions of the British Mycological Society 62(2): 295–304. 10.1016/S0007-1536(74)80038-0 [DOI]
- Guba EF. (1956) Monochaetia and Pestalotia vs. Truncatella, Pestalotiopsis and Pestalotia. Annals of Microbiology 7: 74–76.
- Guba EF. (1961) Monograph of Pestalotia and Monochaetia. Harvard University Press, Cambridge.
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistonachinensis based on morphology and rDNA sequences. The New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics (Oxford, England) 17(17): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Gardiennet A, Voglmayr H. (2016) Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37(1): 82–105. 10.3767/003158516X690475 [DOI] [PMC free article] [PubMed]
- Jayawardena RS, Zhang W, Liu M, Maharachchikumbura SSN, Zhou Y, Huang JB, Nilthong S, Wang ZY, Li XH, Yan JY, Hyde KD. (2015) Identification and characterization of Pestalotiopsis-like fungi related to grapevine diseases in China. Fungal Biology 119(5): 348–361. 10.1016/j.funbio.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Jayawardena RS, Liu M, Maharachchikumbura SSN, Zhang W, Xing QK, Hyde KD, Nilthong S, Li XH, Yan JY. (2016) Neopestalotiopsisvitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 258(1): 63–74. 10.11646/phytotaxa.258.1.4 [DOI] [Google Scholar]
- Jeewon R, Liew ECY, Hyde KD. (2002) Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Molecular Phylogenetics and Evolution 25(3): 378–392. 10.1016/S1055-7903(02)00422-0 [DOI] [PubMed] [Google Scholar]
- Jiang N, Voglmayr H, Bian DR, Piao CG, Wang SK, Li Y. (2021a) Morphology and Phylogeny of Gnomoniopsis (Gnomoniaceae, Diaporthales) from Fagaceae Leaves in China. Journal of Fungi (Basel, Switzerland) 7(10): e792. 10.3390/jof7100792 [DOI] [PMC free article] [PubMed]
- Jiang N, Fan XL, Tian CM. (2021b) Identification and Characterization of Leaf-Inhabiting Fungi from Castanea Plantations in China. Journal of Fungi (Basel, Switzerland) 7(1): e64. 10.3390/jof7010064 [DOI] [PMC free article] [PubMed]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Cheewangkoon R, Gentekaki E, Maharachchikumbura SSN, Brahmanage RS, Hyde KD. (2019) Neopestalotiopsisalpapicalis sp. nov. a new endophyte from tropical mangrove trees in Krabi Province (Thailand). Phytotaxa 393(3): 251–262. 10.11646/phytotaxa.393.3.2 [DOI] [Google Scholar]
- Liu L, Li Y, Liu SC, Zheng ZH, Chen XL, Zhang H, Guo LD, Che YS. (2009) Chloropestolide A, an antitumor metabolite with an unprecedented spiroketal skeleton from Pestalotiopsisfici. Organic Letters 11(13): 2836–2839. 10.1021/ol901039m [DOI] [PubMed]
- Liu F, Bonthond G, Groenewald JZ, Cai L, Crous PW. (2019) Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Studies in Mycology 92(1): 287–415. 10.1016/j.simyco.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XY, Maharachchikumbura SSN, Chen BW, Hyde KD, Mckenzie EHC, Chomnunti P, Kang JC. (2019) Endophytic pestalotiod taxa in Dendrobium orchids. Phytotaxa 419(3): 268–286. 10.11646/phytotaxa.419.3.2 [DOI] [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Chukeatirote E, Bahkali AH, Hyde KD. (2011) Pestalotiopsis – morphology, phylogeny, biochemistry and diversity. Fungal Diversity 50(1): 167–187. 10.1007/s13225-011-0125-x [DOI] [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Cai L, Chukeatirote E, Wu WP, Sun X, Crous PW, Jayarama Bhat D, McKenzie EHC, Bahkali AH, Hyde KD. (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 2012(56): 95–129. 10.1007/s13225-012-0198-1 [DOI] [Google Scholar]
- Maharachchikumbura SSN, Guo LD, Chukeatirote E, McKenzie EHC, Hyde KD. (2013) A destructive new disease of Syzygiumsamarangense in Thailand caused by the new species Pestalotiopsissamarangensis. Tropical Plant Pathology 38(3): 227–235. 10.1590/S1982-56762013005000002 [DOI]
- Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu J, Crous PW. (2014) Pestalotiopsis revisited. Studies in Mycology 79(1): 121–186. 10.1016/j.simyco.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Jayarama Bhat D, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, Shang QJ, Xiao Y, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang WY, Jeewon R, Phillips AJL, Wahab MAA, Sadi AMA, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li QR, Liu JK, Liu XZ, Liu ZY, Luangsa-ard JJ, Pang KL, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen TC, Wijayawardene NN. (2016) Families of Sordariomycetes. Fungal Diversity 79(1): 1–317. 10.1007/s13225-016-0369-6 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, 1–8. 10.1145/2335755.2335836 [DOI]
- Nag Raj TR. (1993) Coelomycetous Anamorphs with Appendage-Bearing Conidia. Mycologue Publications, Waterloo, Ontario.
- Norphanphoun C, Jayawardena RS, Chen Y, Wen TC, Meepol W, Hyde KD. (2019) Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere: Journal of Fungal Biology 10(1): 531–578. 10.5943/mycosphere/10/1/9 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModelTest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo PA. (1884) Sylloge fungorum omnium hucusque cognitorum 3: 797.
- Senanayake IC, Maharachchikumbura SSN, Hyde KD, Jayarama Bhat D, Gareth Jones EB, McKenzie EHC, Dai DQ, Daranagama DA, Dayarathne MC, Goonasekara ID, Konta S, Li WJ, Shang QJ, Stadler M, Wijayawardene NN, Xiao YP, Norphanphoun C, Li Q, Liu XZ, Bahkali AH, Kang JC, Wang Y, Wen TC, Wendt L, Xu JC, Camporesi E. (2015) Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Diversity 73(1): 73–144. 10.1007/s13225-015-0340-y [DOI] [Google Scholar]
- Silvério ML, de Cavalcanti MA. (2016) A new epifoliar species of Neopestalotiopsis from Brazil. Agrotópica 28(2): 151–158. 10.21757/0103-3816.2016v28n2p151-158 [DOI] [Google Scholar]
- Solarte F, Munoz CG, Maharachchikumbura SSN, Alvarez E. (2018) Diversity of Neopestalotiopsis and Pestalotiopsis spp., causal agents of guava scab in Colombia. Plant Disease 102(1): 49–59. 10.1094/PDIS-01-17-0068-RE [DOI] [PubMed] [Google Scholar]
- Song Y, Geng K, Zhang B, Hyde KD, Zhao WS, Wei JG, Kang JC, Wang Y. (2013) Two new species of Pestalotiopsis from Southern China. Phytotaxa 126(1): 22–30. 10.11646/phytotaxa.126.1.2 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert RL. (1949) Contribution a l’etude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bulletin Jardin Botanique Etat Bruxelles 19(3): 285–354. 10.2307/3666710 [DOI] [Google Scholar]
- Steyaert RL. (1953) New and old species of Pestalotiopsis. Transactions of the British Mycological Society 36(2): 81–89. 10.1016/S0007-1536(53)80052-5 [DOI]
- Steyaert RL. (1961) Type specimens of Spegazzini’s collections in the Pestalotiopsis and related genera (Fungi Imperfecti: Melanconiales). Darwinia (Buenos Aires) 12: 157–190. [Google Scholar]
- Steyaert RL. (1963) Complementary informations concerning Pestalotiopsisguepini (Desmazieres) Steyaert and designation of its lectotype. Bulletin Jardin Botanique l’Etat Bruxelles 33(3): 369–373. 10.2307/3667200 [DOI] [Google Scholar]
- Sutton BC. (1980) The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew, Surrey.
- Tanaka K, Endo M, Hirayama K, Okane I, Hosoya T, Sato T. (2011) Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 26(1): 85–98. 10.3767/003158511X576666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibpromma S, Hyde KD, Mckenzie E, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu J, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC. (2018) Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Diversity 93(1): 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Wang B, Zhang ZW, Guo LD, Liu L. (2016) New cytotoxic meroterpenoids from the plant endophytic fungus Pestalotiopsisfici. Helvetica Chimica Acta 99(2): 151–156. 10.1002/hlca.201500197 [DOI]
- White TJ, Bruns T, Lee S. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press Inc, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Wanasinghe DN, Papizadeh M, Goonasekara ID, Camporesi E, Jayarama Bhat D, McKenzie EHC, Phillips AJL, Diederich P, Tanaka K, Li WJ, Tangthirasunun N, Phookamsak R, Dai DQ, Dissanayake AJ, Weerakoon G, Maharachchikumbura SSN, Hashimoto A, Matsumura M, Bahkali AH, Wang Y. (2016) Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity 77(1): 1–316. 10.1007/s13225-016-0360-2 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The combined ITS, tub2 and tef1 sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Zhaoxue Zhang, Rongyu Liu, Shubin Liu, Taichang Mu, Xiuguo Zhang, Jiwen Xia
Data type
phylogenetic
Explanation note
The combined ITS, tub2 and tef1 sequences.