Abstract
Understanding the neural basis of human movement in naturalistic scenarios is critical for expanding neuroscience research beyond constrained laboratory paradigms. Here, we describe our Annotated Joints in Long-term Electrocorticography for 12 human participants (AJILE12) dataset, the largest human neurobehavioral dataset that is publicly available; the dataset was recorded opportunistically during passive clinical epilepsy monitoring. AJILE12 includes synchronized intracranial neural recordings and upper body pose trajectories across 55 semi-continuous days of naturalistic movements, along with relevant metadata, including thousands of wrist movement events and annotated behavioral states. Neural recordings are available at 500 Hz from at least 64 electrodes per participant, for a total of 1280 hours. Pose trajectories at 9 upper-body keypoints were estimated from 118 million video frames. To facilitate data exploration and reuse, we have shared AJILE12 on The DANDI Archive in the Neurodata Without Borders (NWB) data standard and developed a browser-based dashboard.
Subject terms: Brain-machine interface, Neural decoding, Motor cortex
| Measurement(s) | Brain activity measurement • Body Position • Behavior labels • Brain electrode locations |
| Technology Type(s) | Electrocorticography • Video Recording |
| Sample Characteristic - Organism | Homo sapiens |
| Sample Characteristic - Environment | hospital |
| Sample Characteristic - Location | Harborview Medical Center |
Background & Summary
Natural human movements are complex and adaptable, involving highly coordinated sensorimotor processing in multiple cortical and subcortical areas1–4. However, many experiments focusing on the neural basis of human upper-limb movements often study constrained, repetitive motions such as center-out reaching within a controlled laboratory setup5–9. Such studies have greatly increased our knowledge about the neural correlates of movement, but it remains unclear how well these findings generalize to the natural movements that we often make in everyday situations10,11. Human upper-limb movement studies have incorporated self-cued and less restrictive movements12–16, but focusing on unstructured, naturalistic movements can enhance our knowledge of the neural basis of motor behaviors17, help us understand the role of neurobehavioral variability18,19, and aid in the development of robust brain-computer interfaces for real-world use20–26.
Here, we present synchronized intracranial neural recordings and upper body pose trajectories opportunistically obtained from 12 human participants while they performed unconstrained, naturalistic movements over 3–5 recording days each (55 days total). Intracranial neural activity, recorded via electrocorticography (ECoG), involves placing electrodes directly on the cortical surface, beneath the skull and dura, to provide high spatial and temporal resolution27–29. Pose trajectories were obtained from concurrent video recordings using computer vision to automate the often-tedious annotation procedure that has previously precluded the creation of similar datasets30,31. Along with these two core datastreams, we have added extensive metadata, including thousands of wrist movement initiation events previously used for neural decoding32,33, 10 quantitative event-related features describing the type of movement performed and any relevant context18, coarse labels describing the participant’s behavioral state based on visual inspection of videos34, and 14 different electrode-level features18. This dataset, which we call AJILE12 (Annotated Joints in Long-term Electrocorticography for 12 human participants), builds on our previous AJILE dataset35 and is depicted in Fig. 1.
Fig. 1.
Schematic overview of our Annotated Joints in Long-term Electrocorticography for 12 human participants (AJILE12) dataset. AJILE12 includes ECoG recordings and upper body pose trajectories for 12 participants across 55 total recordings days, along with a variety of behavioral, movement event-related, and electrode-level metadata. All data is stored on The DANDI Archive in the NWB data standard, and we have created a custom browser-based dashboard in Jupyter Python to facilitate data exploration without locally downloading the data files.
AJILE12 has high reuse value for future analyses because it is large, comprehensive, well-validated, and shared in the NWB data standard. We have included 55 days of semi-continuous intracranial neural recordings along with thousands of verified wrist movement events, which both greatly exceed the size of typical ECoG datasets from controlled experiments36 as well as other long-term naturalistic ECoG datasets34,35,37,38. Such a wealth of data improves statistical power and enables large-scale exploration of more complex behaviors than previously possible, especially with modern machine learning techniques such as deep learning32,39–42. In addition, AJILE12 contains comprehensive metadata, including coarse behavior labels, quantitative event features, and localized electrode positions in group-level coordinates that enable cross-participant comparisons of neural activity. We have also pre-processed the neural data and visually validated all 6931 wrist movement events to ensure high-quality data, which have been already used in multiple studies18,32,33. In addition, we have released AJILE12 in the NWB data standard (Table 1)43 to adhere to the FAIR data principles of findability, accessibility, interoperability, and reusability44. Unified, open-source data formats such as NWB enable researchers to easily access the data and apply preexisting, reusable workflows instead of starting from scratch. Furthermore, we have developed an accessible and interactive browser-based dashboard that visualizes neural and pose activity, along with relevant metadata. This dashboard can access AJILE12 remotely to visualize the data without requiring local data file downloads, improving AJILE12’s accessibility.
Table 1.
The main variables contained in each data file.
| Data file variable | Description |
|---|---|
| \acquisition\ElectricalSeries | ECoG recordings |
| \processing\behavior\data_interfaces\Position | Upper body poses |
| \processing\behavior\data_interfaces\ReachEvents | Wrist move events |
| \intervals\reaches | Quantitative event features |
| \intervals\epochs | Coarse behavior labels |
| \electrodes | Electrode features |
Files are named sub-##_ses-#_behavior+ecephys.nwb, with ## indicating the participant and # denoting the day of recording.
Methods
Participants
We collected data from 12 human participants (8 males, 4 females; 29.4 ± 7.6 years old [mean ± SD]) during their clinical epilepsy monitoring at Harborview Medical Center (Seattle, USA). See Table 2 for individual participant details. Each participant had been implanted with electrocorticography (ECoG) electrodes placed based on clinical need. We selected these participants because they were generally active during their monitoring and had ECoG electrodes located near motor cortex. All participants provided written informed consent. Our protocol was approved by the University of Washington Institutional Review Board.
Table 2.
Individual participant characteristics.
| Participant | Gender | Age (years) | Recording days used | Hemisphere implanted | Surface electrodes: # good/total | Depth electrodes: # good/total |
|---|---|---|---|---|---|---|
| P01 | M | 44 | 4 | L | 79/86 | 6/8 |
| P02 | M | 20 | 4 | R | 69/70 | 16/16 |
| P03 | M | 33 | 4 | L | 79/80 | 0/16 |
| P04 | F | 19 | 5 | R | 67/84 | 0/0 |
| P05 | F | 31 | 3 | R | 104/106 | 0/0 |
| P06 | M | 37 | 5 | L | 70/80 | 0/0 |
| P07 | M | 26 | 5 | R | 63/64 | 0/0 |
| P08 | F | 33 | 5 | R | 83/92 | 0/0 |
| P09 | M | 20 | 5 | L | 96/98 | 28/28 |
| P10 | M | 34 | 5 | L | 82/86 | 39/40 |
| P11 | F | 34 | 5 | L | 103/106 | 0/0 |
| P12 | M | 22 | 5 | L | 88/92 | 24/32 |
Gender is denoted as male (M) or female (F), and implantation hemisphere is either left (L) or right (R). Surface electrodes denote grid and strip electrodes placed on the cortical surface, while depth electrodes reach deep cortical and subcortical areas. We identified bad/noisy electrodes based on high standard deviation or kurtosis values relative to that participant’s other electrodes. Good electrode counts are shown for each participant’s first available day of recording.
Data collection
Semi-continuous ECoG and video were passively recorded from participants during 24-hour clinical monitoring for epileptic seizures. Recordings lasted 7.4 ± 2.2 days (mean ± SD) for each participant with sporadic breaks in monitoring (on average, 8.3 ± 2.2 breaks per participant each lasting 1.9 ± 2.4 hours). For all participants, we only included recordings during days 3–7 following the electrode implantation surgery to avoid potentially anomalous neural and behavioral activity immediately after the surgery. We excluded recording days with corrupted or missing data files, as noted in Table 2, and stripped all recording dates to de-identify participant data. These long-term, clinical recordings include various everyday activities, such as eating, sleeping, watching television, and talking while confined to a hospital bed. ECoG and video sampling rates were 1 kHz and 30 FPS (frames per second), respectively.
ECoG data processing
We used custom MNE-Python scripts to process the raw ECoG data45. First, we removed DC drift by subtracting out the median voltage at each electrode. We then identified high-amplitude data discontinuities, based on abnormally high electrode-averaged absolute voltage (>50 interquartile ranges [IQRs]), and set all data within 2 seconds of each discontinuity to 0.
With data discontinuities removed, we then band-pass filtered the data (1–200 Hz), notch filtered to minimize line noise at 60 Hz and its harmonics, downsampled to 500 Hz, and re-referenced to the common median for each grid, strip, or depth electrode group. For each recording day, noisy electrodes were identified based on abnormal standard deviation (>5 IQRs) or kurtosis (>10 IQRs) compared to the median value across electrodes. Using this procedure, we marked on average 7.3 ± 5.6 ECoG electrodes as bad during each participant’s first available day of recording (Table 2).
Electrode positions were localized using the Fieldtrip toolbox in MATLAB. This process involved co-registering preoperative MRI and postoperative CT scans, manually selecting electrodes in 3D space, and warping electrode positions into MNI space (see Stolk et al.46 for further details).
Markerless pose estimation
We performed markerless pose estimation on the raw video footage using separate DeepLabCut models for each participant31. First, one researcher manually annotated the 2D positions of 9 upper-body keypoints (nose, ears, wrists, elbows, and shoulders) during 1000 random video frames for each participant (https://tinyurl.com/human-annotation-tool). Frames were randomly selected across all recording days, with preference towards frames during active, daytime periods. These 1000 frames correspond to ∼0.006% of the total frames from each participant’s video recordings. These manually annotated frames were used to train a separate DeepLabCut neural network model for each participant (950 frames for training, 50 frames for validation). The model architecture was a convolutional neural network that was 50 layers deep (ResNet-50). We then applied the trained model to every video frame for that participant to generate estimated pose trajectories.
We synchronized ECoG data and pose trajectories using video timestamps and combined multiple recording sessions so that each file contained data from one entire 24-hour recording day that started and ended at midnight47.
Wrist movement event identification
We used the estimated pose trajectories in order to identify unstructured movement initiation events of the wrist contralateral to the implanted hemisphere. To identify movement events, a first-order autoregressive hidden semi-Markov model was applied to the pose trajectory of the contralateral wrist. This model segmented the contralateral wrist trajectory into discrete move or rest states. Movement initiation events were identified as state transitions where 0.5 seconds of rest was followed by 0.5 seconds of wrist movement (see Singh et al.33 for further details).
Next, we selected the movement initiation events that most likely corresponded to actual reaching movements. We excluded arm movements during sleep, unrelated experiments, and private times based on coarse behavioral labels, which are described in the next section. In addition, we only retained movement events that (1) lasted between 0.5–4 seconds, (2) had DeepLabCut confidence scores >0.4, indicating minimal marker occlusion, and (3) had parabolic wrist trajectories, as determined by a quadratic fit to the wrist’s radial movement (). We used this quadratic fit criterion to eliminate outliers with complex movement trajectories. For each recording day, we selected up to 200 movement events with the highest wrist speeds during movement onset. Finally, we visually inspected all selected movement events and removed those with occlusions or false positive movements (17.8% ± 9.9% of events [meanSD]).
For each movement event, we also extracted multiple, quantitative behavioral and environmental features. To quantify movement trajectories, we defined a reach as the maximum radial displacement of the wrist during the identified movement event, as compared to wrist position at movement onset. Movement features include reach magnitude, reach duration, 2D vertical reach angle (90 for upward reaches, −90 for downward reaches), and radial speed during movement onset. We also include the recording day and time of day when each movement event occurred, as well as an estimate of speech presence during each movement using audio recordings.
In addition, we quantified the amount of bimanual movement for event based on ipsilateral wrist movement. These features include a binary classification of bimanual/unimanual based on temporal lag between wrist movement onsets, the ratio of ipsilateral to contralateral reach magnitude, and the amount of each contralateral move state that temporally overlapped with an ipsilateral move state. The binary feature was bimanual if at least 4 frames (0.13 seconds) of continuous ipsilateral wrist movement began either 1 second before contralateral wrist movement initiation or anytime during the contralateral wrist move state. Please see Peterson et al.18 for further methodological details.
Coarse behavioral labels
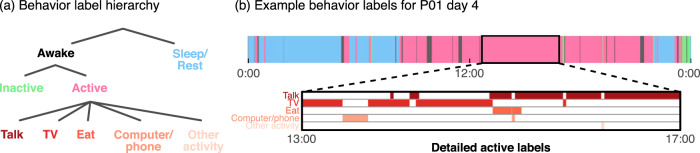
To improve wrist movement event identification, we performed coarse annotation of the video recordings every ∼3 minutes. These behavioral labels were either part of a blocklist to avoid during event detection or general activities/states that the participant was engaged in at the time. Identified activities include sleep/rest, inactive, and active behaviors, which were further subdivided into activities such as talking, watching TV, and using a computer or phone (Fig. 2). Blocklist labels include times where event detection would likely be inaccurate, such as camera movement and occlusion, as well as private times and unrelated research experiments. Some participants also have clinical procedure labels, indicating times when the clinical staff responded to abnormal participant behavior. We upsampled all labels to match the 30 Hz sampling rate of the pose data. Tables 3 and 4 show the duration of each label across participants for activity and blocklist labels, respectively.
Fig. 2.
Coarse behavior labelling. (a) We annotated participant behavior in the video recordings using hierarchical labels to detail common awake and active behaviors. These annotations also include blocklist labels, which indicate times to potentially avoid during data exploration. (b) We show an example of the behavior labels for participant P01 during the entirety of recording day 4. Sleep/rest occurs in the morning and night times, as expected, with predominantly active periods during the day (8:00–20:00). Bottom row shows detailed active labels during a 4-hour active period that is dominated mostly by talk and TV behaviors. Note that these detailed active labels can overlap in time.
Table 3.
Coarse activity label durations (in hours) for each participant.
| Participant | Sleep/rest | Inactive | Talk | TV | Computer/phone | Eat | Other activity | Total |
|---|---|---|---|---|---|---|---|---|
| P01 | 29.2 | 1.9 | 15.6 | 22.0 | 7.8 | 2.6 | 0.8 | 75.7 |
| P02 | 53.2 | 2.1 | 9.2 | 7.7 | 2.9 | 2.1 | 4.8 | 79.8 |
| P03 | 11.9 | 0.3 | 21.8 | 1.9 | 19.8 | 3.5 | 0.9 | 51.1 |
| P04 | 34.0 | 3.1 | 28.3 | 11.0 | 8.2 | 2.0 | 1.2 | 78.9 |
| P05 | 35.4 | 2.8 | 8.9 | 12.1 | 5.8 | 1.0 | 1.2 | 62.5 |
| P06 | 37.8 | 0.1 | 2.8 | 5.7 | 2.6 | 0.3 | 0.2 | 45.3 |
| P07 | 46.8 | 0.3 | 5.0 | 0.3 | 1.9 | 0.4 | 0.2 | 53.6 |
| P08 | 47.8 | 0.8 | 6.8 | 5.3 | 1.3 | 0.6 | 2.2 | 61.8 |
| P09 | 87.3 | 8.1 | 3.8 | 0.0 | 0.0 | 0.6 | 1.9 | 100.6 |
| P10 | 67.5 | 2.1 | 5.8 | 0.1 | 5.6 | 0.0 | 1.8 | 81.4 |
| P11 | 36.0 | 1.4 | 6.6 | 0.0 | 0.1 | 0.0 | 0.8 | 44.9 |
| P12 | 32.4 | 0.3 | 1.5 | 0.1 | 0.5 | 0.0 | 0.6 | 35.4 |
These labels describe various participant behaviors such as talking, eating, and watching television. Labels were generated by manual reviewing videos every 3 minutes. While sleep is by far the most common, several activity labels appear over multiple hours for each participant. Note that multiple activity labels can co-occur (ex. eating while watching television). Therefore, the total duration of any activity label (last column) may be less than the sum of individual label durations for each participant.
Table 4.
Coarse blocklist label durations (in hours) for each participant.
| Participant | Data break | Camera move/zoom | Camera occluded | Experiment | Private time | Tether/ bandage | Hands under blanket | Clinical procedure | Total |
|---|---|---|---|---|---|---|---|---|---|
| P01 | 8.8 | 0.5 | 0.0 | 6.7 | 2.8 | 1.3 | 0.0 | 0.1 | 20.3 |
| P02 | 0.8 | 1.2 | 0.0 | 3.6 | 7.0 | 3.7 | 0.0 | 0.0 | 16.2 |
| P03 | 24.4 | 0.7 | 0.0 | 18.1 | 1.1 | 0.6 | 0.0 | 0.0 | 44.9 |
| P04 | 27.9 | 1.4 | 0.0 | 7.2 | 2.8 | 1.7 | 0.0 | 0.0 | 41.1 |
| P05 | 0.6 | 0.8 | 0.3 | 3.4 | 2.4 | 2.0 | 0.0 | 0.0 | 9.5 |
| P06 | 0.7 | 0.3 | 0.0 | 7.0 | 1.1 | 0.8 | 0.0 | 0.0 | 9.9 |
| P07 | 7.8 | 1.4 | 0.2 | 4.4 | 2.6 | 0.6 | 0.0 | 0.1 | 17.0 |
| P08 | 0.9 | 0.4 | 0.2 | 6.2 | 2.6 | 0.6 | 0.0 | 0.0 | 10.8 |
| P09 | 0.8 | 1.3 | 0.0 | 5.5 | 1.9 | 0.9 | 0.3 | 0.6 | 11.2 |
| P10 | 1.2 | 1.0 | 0.0 | 2.1 | 3.5 | 2.9 | 0.0 | 0.0 | 10.7 |
| P11 | 0.7 | 1.7 | 5.1 | 2.5 | 4.8 | 1.4 | 0.0 | 0.0 | 16.2 |
| P12 | 0.6 | 1.0 | 0.1 | 3.6 | 3.3 | 0.7 | 0.0 | 0.0 | 9.3 |
These labels indicate times to avoid when extracting wrist movement events due to camera movements, unrelated experiments, and private times. Labels were generated by manual review of videos with 3 minute resolution.
Data Records
The data files are available on The DANDI Archive (10.48324/dandi.000055/0.220127.0436)47, in the Neurodata Without Borders: Neurophysiology 2.0 (NWB:N) format43. All datastreams and metadata have been combined into a single file for each participant and day of recording, as indicated by the file name. For example, sub-01_ses-3_behavior+ecephys.nwb contains data from participant P01 on recording day 3. We used PyNWB 1.4.0 to load and interact with these data files. Table 1 shows the location of all main variables within each data file.
Each file contains continuous ECoG and pose data over a 24-hour period, with units of and pixels, respectively. ECoG data is located under\acquisition\ElectricalSeries as a pynwb.ecephys.ElectricalSeries variable. Pose data can be found under\processing\behavior\data_interfaces\Position as an pynwb.behavior.Position variable. Pose data is provided for the left/right ear (L_Ear, R_Ear), shoulder (L_Shoulder, R_Shoulder), elbow (L_Elbow, R_Elbow), and wrist (L_Wrist, R_Wrist), as well as the nose (Nose).
In addition to these core datastreams, each file contains relevant metadata. Contralateral wrist movement events are located in\processing\behavior\data_interfaces\ReachEvents as an ndx_events.events.Events variable. Quantitative neural and behavioral features for each event can be found in\intervals\reaches as a pynwb.epoch.TimeIntervals table with columns for each feature. Coarse behavioral labels are included in\intervals\epochs as a pynwb.epoch.TimeIntervals table. Each row contains the label along with the start and stop time in seconds.
We also include electrode-specific metadata in\electrodes as a hdmf.common.table.DynamicTable. Columns contain different metadata features, such as Montreal Neurological Institute (MNI) x, y, z coordinates and electrode group names. Electrode groups were named by clinicians based on their location in the brain. This table also contains the standard deviation, kurtosis, and median absolute deviation for each electrode computed over the entire recording file (excluding non-numeric values). Electrodes that we identified as noisy based on abnormal standard deviation and kurtosis are marked as False under the ‘good’ column. Table 2 shows the number of good electrodes that remain for each participant during the first available day of recording. We have also included the scores obtained from regressing ECoG spectral power on the 10 quantitative event features for each participant’s wrist movement events18. Low-frequency power (used for low_freq_R2) indicates power between 8–32 Hz, while high-frequency power (used for high_freq_R2) denotes power between 76–100 Hz.
Technical Validation
In this section, we assess the technical quality of AJILE12 by validating our two core datastreams: intracranial neural recordings and pose trajectories. In addition to this assessment, we have previously validated the quality and reliability of AJILE12 in multiple published studies18,32,33. We validated ECoG data quality by assessing spectral power projected into common brain regions48. This projection procedure enables multi-participant comparisons despite heterogeneous electrode coverage and reduces the dimensionality of the ECoG data from 64 or more electrodes (Fig. 3(a)) to a few brain regions of interest18,32. For this analysis, we focused on 4 sensorimotor and temporal regions in the left hemisphere defined using the AAL2 brain atlas48,49: precentral gyrus, postcentral gyrus, middle temporal gyrus, and inferior temporal gyrus. For participants with electrodes implanted primarily in the right hemisphere, we mirrored electrode positions into the left hemisphere. We divided the neural data into 30-minute windows and applied Welch’s method to compute the median spectral power over non-overlapping 30-second sub-windows50. We excluded 30-minute windows with non-numeric data values, likely due to data breaks. On average, we used 160.4 ± 30.6 windows per participant (80.2 ± 15.3 hours) across all recording days. Spectral power was interpolated to integer frequencies and projected into the 4 predefined brain regions (see Peterson et al.18 for further methodological details).
Fig. 3.
Validation of intracranial neural signal quality. (a) Electrocorticography (ECoG) electrode positions are shown in MNI coordinates for each participant. ECoG power spectra is shown for (b) all 12 participants (shading denotes standard deviation) and (c) participant P01 over all available half-hour time windows. We projected spectral power into sensorimotor and temporal brain regions, excluding time windows with non-numeric values that likely indicated a data break. Lines for participant P01 denote power in each window ( total, or 65 hours). The power spectra shape (exponential decrease for increasing frequencies) and consistency over time demonstrate the cleanliness and stability of our neural recordings across multiple recording days.
Figure 3(b) shows the average spectral power across time windows, separated by participant. In general, power spectra remain quite consistent across participants with tight standard deviations across time windows, indicating that much of the ECoG data is good to use51,52. We also plotted the power spectra of each individual window for participant P01, as shown in Fig. 3(c). Again, the variation among time windows appears small, and we see clear differences in spectral power between sensorimotor (pre/postcentral gyri) and temporal areas, as expected. Additionally, we retained 92.3% ± 6.3% ECoG electrodes per participant (Table 2), further demonstrating the quality of our neural data53,54.
We validated pose trajectories by comparing each pose estimation model’s output to our manual annotations of each participant’s pose (Table 5). While manual annotations are susceptible to human error55, they are often used to evaluate markerless pose estimation performance when marker-based motion capture is not possible30,56. We used root-mean-square (RMS) error averaged across all keypoints to evaluate model performance for the 950 frames used to train the model as well as 50 annotated frames that were withheld from training. RMS errors for the holdout set (5.71 ± 1.90 pixels) are notably larger than the train set errors (1.52 ± 0.12 pixels), as expected, but are still within an acceptable tolerance given that 3 pixels are approximately equal to just 1 cm33.
Table 5.
Pose estimation model errors.
| Participant | Train set error (pixels) | Holdout set error (pixels) |
|---|---|---|
| P01 | 1.45 | 4.27 |
| P02 | 1.44 | 3.58 |
| P03 | 1.58 | 6.95 |
| P04 | 1.63 | 6.02 |
| P05 | 1.43 | 3.42 |
| P06 | 1.43 | 6.63 |
| P07 | 1.51 | 5.45 |
| P08 | 1.84 | 10.35 |
| P09 | 1.40 | 4.05 |
| P10 | 1.48 | 7.59 |
| P11 | 1.51 | 5.45 |
| P12 | 1.52 | 4.73 |
Root-mean-square reconstruction errors of our automated markerless pose models are shown for each participant’s train (950 frames) and holdout (50 frames) sets. We used manual annotations as ground truth when computing the error, which was averaged across all 9 upper-body keypoints. For reference3,pixels are approximately equal to 1 cm.
Usage Notes
We have developed a Jupyter Python dashboard that can be run online to facilitate data exploration without locally downloading the data files (https://github.com/BruntonUWBio/ajile12-nwb-data). Our dashboard includes visualizations of electrode locations, along with ECoG and wrist pose traces for a user-selected time window (Fig. 4). Users can also visualize the average contralateral wrist trajectory during identified movement events for each file. The dashboard streams from The DANDI Archive only the data needed for visualization, enabling efficient renderings of time segments from the large, 24-hour data files. Our code repository also includes all scripts necessary to create Figs. 2, 3 and Tables 2–4. In addition, we have previously used AJILE12 to decode and analyze the neurobehavioral variability of naturalistic wrist movements and have publicly released multiple workflows that can be modified for use on this dataset18,32,33.
Fig. 4.
Browser-based Jupyter Python dashboard for dataset exploration. We designed a browser-based dashboard, available at https://github.com/BruntonUWBio/ajile12-nwb-data, to facilitate exploration of AJILE12 without needing to download any data files locally. (a) Participant keypoint positions are displayed for the first sample of a user-defined time window, with the option to animate keypoint positions across the entire window. We included a virtual neck marker for this visualization at the midpoint between the left and right shoulders. (b) Time-series traces of horizontal (x) and vertical (y) wrist positions are displayed over the same selected time window. (c) Electrode coverage is shown in MNI coordinates on a standardized brain model. This visualization is interactive, allowing three-dimensional rotations, alterations of hemisphere opacity to inspect depth electrodes, and the ability to visualize various electrode-level metadata such as electrode groups and identified bad electrodes. (d) Raw ECoG signals are visualized over the same user-selected time window, color-coded by electrode group.
Supplementary information
Acknowledgements
We thank Nancy Wang for contributing to the data collection, John So for generating the coarse behavior annotations, and the clinical staff at the Harborview Hospital Neurosurgery department for their assistance collecting and analyzing the data, especially Leigh Weber, Jeffrey G. Ojemann, and Andrew Ko. This research was supported by funding from the National Science Foundation (1630178 and EEC-1028725), the Defense Advanced Research Projects Agency (FA8750-18-2-0259), the Sloan Foundation, the Washington Research Foundation, and the Weill Neurohub.
Author contributions
R.P.N.R. and B.W.B. conceived the study; S.M.P. and S.H.S. performed the data analysis; S.M.P., S.H.S., R.P.N.R., and B.W.B. interpreted the results; S.M.P., B.D., and M.S. created the public dataset and corresponding analysis dashboard; S.M.P. and B.W.B. wrote the paper; all authors reviewed and approved the final draft of the paper.
Code availability
Code to run our Jupyter Python dashboard and recreate all results in this paper can be found at https://github.com/BruntonUWBio/ajile12-nwb-data. We used Python 3.8.5 and PyNWB 1.4.0. A requirements file listing the Python packages and versions necessary to run the code is provided in our code repository. Our code is publicly available without restriction other than attribution.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01280-y.
References
- 1.Sober SJ, Sponberg S, Nemenman I, Ting LH. Millisecond spike timing codes for motor control. Trends in Neurosciences. 2018;41:644–648. doi: 10.1016/j.tins.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalaska, J. F. From Intention to Action: Motor Cortex and the Control of Reaching Movements, 139–178 (Springer US, Boston, MA, 2009). [DOI] [PubMed]
- 3.Truccolo W, Friehs GM, Donoghue JP, Hochberg LR. Primary motor cortex tuning to intended movement kinematics in humans with tetraplegia. The Journal of Neuroscience. 2008;28:1163. doi: 10.1523/JNEUROSCI.4415-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller KJ, et al. Spectral changes in cortical surface potentials during motor movement. Journal of Neuroscience. 2007;27:2424–2432. doi: 10.1523/JNEUROSCI.3886-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakanishi Y, et al. Prediction of three-dimensional arm trajectories based on ecog signals recorded from human sensorimotor cortex. PloS one. 2013;8:e72085. doi: 10.1371/journal.pone.0072085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Decoding onset and direction of movements using electrocorticographic (ecog) signals in humans. Frontiers in neuroengineering. 2012;5:15. doi: 10.3389/fneng.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schalk G, et al. Two-dimensional movement control using electrocorticographic signals in humans. Journal of neural engineering. 2008;5:75. doi: 10.1088/1741-2560/5/1/008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgopoulos AP, Merchant H, Naselaris T, Amirikian B. Mapping of the preferred direction in the motor cortex. Proceedings of the National Academy of Sciences. 2007;104:11068. doi: 10.1073/pnas.0611597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leuthardt EC, Schalk G, Wolpaw JR, Ojemann JG, Moran DW. A brain–computer interface using electrocorticographic signals in humans. Journal of neural engineering. 2004;1:63. doi: 10.1088/1741-2560/1/2/001. [DOI] [PubMed] [Google Scholar]
- 10.Umeda T, Koizumi M, Katakai Y, Saito R, Seki K. Decoding of muscle activity from the sensorimotor cortex in freely behaving monkeys. NeuroImage. 2019;197:512–526. doi: 10.1016/j.neuroimage.2019.04.045. [DOI] [PubMed] [Google Scholar]
- 11.Fried I, Haggard P, He BJ, Schurger A. Volition and action in the human brain: Processes, pathologies, and reasons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2017;37:10842–10847. doi: 10.1523/JNEUROSCI.2584-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornhuber HH, Deecke L. Brain potential changes in voluntary and passive movements in humans: readiness potential and reafferent potentials. Pflügers Archiv - European Journal of Physiology. 2016;468:1115–1124. doi: 10.1007/s00424-016-1852-3. [DOI] [PubMed] [Google Scholar]
- 13.Jackson A, Mavoori J, Fetz EE. Correlations between the same motor cortex cells and arm muscles during a trained task, free behavior, and natural sleep in the macaque monkey. Journal of Neurophysiology. 2007;97:360–374. doi: 10.1152/jn.00710.2006. [DOI] [PubMed] [Google Scholar]
- 14.Lee IH, Assad JA. Putaminal activity for simple reactions or self-timed movements. Journal of Neurophysiology. 2003;89:2528–2537. doi: 10.1152/jn.01055.2002. [DOI] [PubMed] [Google Scholar]
- 15.Romo R, Schultz W. Neuronal activity preceding self-initiated or externally timed arm movements in area 6 of monkey cortex. Experimental Brain Research. 1987;67:656–662. doi: 10.1007/BF00247297. [DOI] [PubMed] [Google Scholar]
- 16.Pistohl T, Schulze-Bonhage A, Aertsen A, Mehring C, Ball T. Decoding natural grasp types from human ecog. Neuroimage. 2012;59:248–260. doi: 10.1016/j.neuroimage.2011.06.084. [DOI] [PubMed] [Google Scholar]
- 17.Dastjerdi M, Ozker M, Foster BL, Rangarajan V, Parvizi J. Numerical processing in the human parietal cortex during experimental and natural conditions. Nature Communications. 2013;4:2528. doi: 10.1038/ncomms3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson, S. M., Singh, S. H., Wang, N. X., Rao, R. P. & Brunton, B. W. Behavioral and neural variability of naturalistic arm movements. Eneuro (2021). [DOI] [PMC free article] [PubMed]
- 19.Basu I, et al. Consistent linear and non-linear responses to invasive electrical brain stimulation across individuals and primate species with implanted electrodes. Brain stimulation. 2019;12:877–892. doi: 10.1016/j.brs.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbaspourazad H, Choudhury M, Wong YT, Pesaran B, Shanechi MM. Multiscale low-dimensional motor cortical state dynamics predict naturalistic reach-and-grasp behavior. Nature communications. 2021;12:1–19. doi: 10.1038/s41467-020-20197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson NR, et al. Cortical topography of error-related high-frequency potentials during erroneous control in a continuous control brain–computer interface. Frontiers in Neuroscience. 2019;13:502. doi: 10.3389/fnins.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omedes J, Schwarz A, Müller-Putz GR, Montesano L. Factors that affect error potentials during a grasping task: toward a hybrid natural movement decoding bci. Journal of Neural Engineering. 2018;15:046023. doi: 10.1088/1741-2552/aac1a1. [DOI] [PubMed] [Google Scholar]
- 23.Gilja V, et al. Challenges and opportunities for next-generation intracortically based neural prostheses. IEEE Transactions on Biomedical Engineering. 2011;58:1891–1899. doi: 10.1109/TBME.2011.2107553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor, D. M., Tillery, S. I. H. & Schwartz, A. B. Direct cortical control of 3d neuroprosthetic devices. Science296, 1829–1832, 10.1126/science.1070291, https://science.sciencemag.org/content/296/5574/1829.full.pdf (2002). [DOI] [PubMed]
- 25.Meisler SL, Kahana MJ, Ezzyat Y. Does data cleaning improve brain state classification? Journal of neuroscience methods. 2019;328:108421. doi: 10.1016/j.jneumeth.2019.108421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anumanchipalli GK, Chartier J, Chang EF. Speech synthesis from neural decoding of spoken sentences. Nature. 2019;568:493–498. doi: 10.1038/s41586-019-1119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parvizi J, Kastner S. Promises and limitations of human intracranial electroencephalography. Nature neuroscience. 2018;21:474–483. doi: 10.1038/s41593-018-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs J, Kahana MJ. Direct brain recordings fuel advances in cognitive electrophysiology. Trends in cognitive sciences. 2010;14:162–171. doi: 10.1016/j.tics.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ball T, Kern M, Mutschler I, Aertsen A, Schulze-Bonhage A. Signal quality of simultaneously recorded invasive and non-invasive eeg. NeuroImage. 2009;46:708–716. doi: 10.1016/j.neuroimage.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 30.Mathis A, Schneider S, Lauer J, Mathis MW. A primer on motion capture with deep learning: principles, pitfalls, and perspectives. Neuron. 2020;108:44–65. doi: 10.1016/j.neuron.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Mathis, A. et al. DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Tech. Rep., Nature Publishing Group (2018). [DOI] [PubMed]
- 32.Peterson SM, Steine-Hanson Z, Davis N, Rao RP, Brunton BW. Generalized neural decoders for transfer learning across participants and recording modalities. Journal of Neural Engineering. 2021;18:026014. doi: 10.1088/1741-2552/abda0b. [DOI] [PubMed] [Google Scholar]
- 33.Singh, S. H., Peterson, S. M., Rao, R. P. & Brunton, B. W. Mining naturalistic human behaviors in long-term video and neural recordings. Journal of Neuroscience Methods 109199 (2021). [DOI] [PubMed]
- 34.Alasfour A, et al. Coarse behavioral context decoding. Journal of neural engineering. 2019;16:016021. doi: 10.1088/1741-2552/aaee9c. [DOI] [PubMed] [Google Scholar]
- 35.Wang, N., Farhadi, A., Rao, R. & Brunton, B. Ajile movement prediction: Multimodal deep learning for natural human neural recordings and video. In Proceedings of the AAAI Conference on Artificial Intelligence, vol. 32 (2018).
- 36.Miller KJ. A library of human electrocorticographic data and analyses. Nature human behaviour. 2019;3:1225–1235. doi: 10.1038/s41562-019-0678-3. [DOI] [PubMed] [Google Scholar]
- 37.Gabriel PG, et al. Neural correlates of unstructured motor behaviors. Journal of neural engineering. 2019;16:066026. doi: 10.1088/1741-2552/ab355c. [DOI] [PubMed] [Google Scholar]
- 38.Wang NX, Olson JD, Ojemann JG, Rao RP, Brunton BW. Unsupervised decoding of long-term, naturalistic human neural recordings with automated video and audio annotations. Frontiers in human neuroscience. 2016;10:165. doi: 10.3389/fnhum.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy Y, et al. Deep learning-based electroencephalography analysis: a systematic review. Journal of neural engineering. 2019;16:051001. doi: 10.1088/1741-2552/ab260c. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, et al. A survey on deep learning-based non-invasive brain signals: recent advances and new frontiers. Journal of neural engineering. 2021;18:031002. doi: 10.1088/1741-2552/abc902. [DOI] [PubMed] [Google Scholar]
- 41.Tan, C. et al. A survey on deep transfer learning. In International conference on artificial neural networks, 270–279 (Springer, 2018).
- 42.Craik A, He Y, Contreras-Vidal JL. Deep learning for electroencephalogram (eeg) classification tasks: a review. Journal of neural engineering. 2019;16:031001. doi: 10.1088/1741-2552/ab0ab5. [DOI] [PubMed] [Google Scholar]
- 43.Teeters JL, et al. Neurodata without borders: creating a common data format for neurophysiology. Neuron. 2015;88:629–634. doi: 10.1016/j.neuron.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson MD, et al. The fair guiding principles for scientific data management and stewardship. Scientific data. 2016;3:1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gramfort A, et al. Meg and eeg data analysis with mne-python. Frontiers in Neuroscience. 2013;7:267. doi: 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stolk A, et al. Integrated analysis of anatomical and electrophysiological human intracranial data. Nature Protocols. 2018;13:1699–1723. doi: 10.1038/s41596-018-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson SM, 2022. Ajile12: Long-term naturalistic human intracranial neural recordings and pose. The DANDI Archive. [DOI] [PMC free article] [PubMed]
- 48.Bigdely-Shamlo N, Mullen T, Kreutz-Delgado K, Makeig S. Measure projection analysis: a probabilistic approach to eeg source comparison and multi-subject inference. NeuroImage. 2013;72:287–303. doi: 10.1016/j.neuroimage.2013.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tzourio-Mazoyer N, et al. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 50.Cole, S., Donoghue, T., Gao, R. & Voytek, B. Neurodsp: A package for neural digital signal processing. Journal of Open Source Software4, 10.21105/joss.01272 (2019).
- 51.Cohen, M. X. Analyzing Neural Time Series Data: Theory and Practice, 10.7551/mitpress/9609.001.0001 (2014).
- 52.Keil A, et al. Committee report: publication guidelines and recommendations for studies using electroencephalography and magnetoencephalography. Psychophysiology. 2014;51:1–21. doi: 10.1111/psyp.12147. [DOI] [PubMed] [Google Scholar]
- 53.Pedroni A, Bahreini A, Langer N. Automagic: Standardized preprocessing of big eeg data. NeuroImage. 2019;200:460–473. doi: 10.1016/j.neuroimage.2019.06.046. [DOI] [PubMed] [Google Scholar]
- 54.Bigdely-Shamlo N, Mullen T, Kothe C, Su K-M, Robbins KA. The prep pipeline: standardized preprocessing for large-scale eeg analysis. Frontiers in neuroinformatics. 2015;9:16. doi: 10.3389/fninf.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karashchuk P, et al. Anipose: a toolkit for robust markerless 3d pose estimation. Cell reports. 2021;36:109730. doi: 10.1016/j.celrep.2021.109730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nath T, et al. Using deeplabcut for 3d markerless pose estimation across species and behaviors. Nature protocols. 2019;14:2152–2176. doi: 10.1038/s41596-019-0176-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Peterson SM, 2022. Ajile12: Long-term naturalistic human intracranial neural recordings and pose. The DANDI Archive. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Code to run our Jupyter Python dashboard and recreate all results in this paper can be found at https://github.com/BruntonUWBio/ajile12-nwb-data. We used Python 3.8.5 and PyNWB 1.4.0. A requirements file listing the Python packages and versions necessary to run the code is provided in our code repository. Our code is publicly available without restriction other than attribution.