Summary
Multicellular organisms of various complexities self-organize in nature. Organoids are in vitro 3D structures that display important aspects of the anatomy and physiology of their in vivo counterparts and that develop from pluripotent or tissue-specific stem cells through a self-organization process. In this review, we describe the multidisciplinary concept of “synthetic developmental biology” where engineering approaches are employed to guide multicellular organization in an experimental setting. We introduce a novel classification of engineering approaches based on the extent of microenvironmental manipulation applied to organoids. In the final section, we discuss how engineering tools might help overcome current limitations in organoid construction.
Keywords: self-organization, engineering, organoid
In this article, Baharvand and colleagues describe synthetic developmental biology using a novel classification of engineering approaches based on the extent of microenvironmental manipulation applied to organoids
Introduction
Multicellular organisms of various complexities, ranging from algae to humans, are formed by common principles and phenomena that are collectively referred to as “self-organization.” Decades of scientific research have been dedicated to studying the physical, chemical, and genetic mechanisms that govern spontaneous organization and to manipulate these mechanisms for the reconstruction of cellular patterning in biological systems in vitro, particularly in organoids (see, for example, Laurent et al., 2017; Morales et al., 2021). Organoids are in vitro three-dimensional (3D) structures that display important aspects of the anatomy and physiology of their in vivo counterparts and that develop from pluripotent or tissue-specific stem cells through a self-organization process (Shiri et al., 2019). Recent developments in the field of organoids have led to the multidisciplinary concept of “synthetic developmental biology.” This term has recently been used to describe genetic engineering approaches for studying and constructing developmental systems (see, for example Ebrahimkhani and Ebisuya, 2019; Ho and Morsut, 2021). Here, we specifically review engineering approaches in synthetic biology that are used to explore principles of developmental biology in an effort to exert dynamic control over the organization of multicellular systems in vitro.
In this review, we briefly mention the cellular crosstalk that guides assembly, patterning, and morphogenetic events in multicellular organisms via physical contact and release of diffusible cues (Liu and Warmflash, 2021). We then elaborate on different engineering approaches that have been used to better reconstruct the artificial niche to guide organization of cells in an experimental setting, including controlled release systems, engineering of scaffolds, application of microfluidic chips, and bioreactors. These methods aim to simulate processes that occur in developing organs by modifying endogenous cues present in a cell’s microenvironment, including soluble factors, the extracellular matrix (ECM), intercellular interactions, and physical-mechanical properties (Gupta et al., 2021). We classify these engineering approaches based on the extent of microenvironmental manipulation that they impose on the formed structure. Each of these methods employs different means to trigger or steer natural processes of self-assembly, self-patterning, and self-morphogenesis (Figure 1). Furthermore, the complexity of formed structures ranges from simple two-dimensional (2D) patterns of various cell types to complex 3D constructs that resemble organs (i.e., organoids). In the final section of this review, we discuss how engineering tools may help to overcome organoid limitations.
Figure 1.
An overview of engineering methods in the field of synthetic developmental biology to guide the self-organization process
The co-culture method has been applied to guide self-organization and to generate complex multicellular structures. The presence of various cell types that respond to various signals at different developmental stages leads to the formation of more complex multicellular structures. The second and third methods include engineering ECM and 3D-patterning of the environment. These approaches are used to exert dynamic control over hydrogels and soluble factors that can change the physical and chemical properties of the microenvironment in a dynamic manner. The third and fourth methods include engineering cell surface and 3D-patterning of cells, which allow precise control over cell position within a multicellular structure. These approaches usually dictate the initial condition and thus lead to the generation of simpler structures in comparison with the previous approaches. Therefore, developing a dynamic system to control microenvironmental conditions in various developmental stages can provide more complex models to recapitulate development, function, and malfunction of human tissues.
Our evaluation of the self-organization phenomena as a dynamic system that changes in space and time suggests that the field of synthetic developmental biology will benefit from novel engineering approaches that can allow dynamic control of organoid formation over time. Such control might lead to construction of better models of human tissue, which in turn can provide unprecedented insights into the development, function, and malfunction of tissues.
Behind self-organization
In the context of developmental biology, self-organization is the process by which order emerges from an otherwise random ensemble of cells. Sasai (2013) classified self-organization into three overlapping categories of increasing levels of complexity: self-assembly, self-patterning, and self-driven morphogenesis. Together, these processes contribute to remarkable examples of self-organization that spontaneously occur in nature to create multicellular structures of various degrees of complexity.
The spontaneous appearance of pattern has been explained by two major models. One model, originally proposed by Turing (1952), uses reaction and diffusion of two or more chemicals to describe activation and inhibition of various signaling pathways that determine a cell’s fate. Signaling pathways and transcriptional networks regulate cell proliferation, migration, assembly, patterning, and morphogenesis in multicellular organisms. Indeed, organizing centers, morphogens, neurotransmitters, and extracellular vesicles contribute to the positional information that dictate a cell’s fate. A cell in a given position acquires a certain fate by detecting and responding to various spatial and temporal cues present in their environment (Wolpert, 1989). For an in-depth reading on the chemical influencers of development, see for example Grove and Monuki (2013) and Lyssiotis et al. (2011).
The second model uses mechanical and physical forces to describe pattern formation, suggesting that form arises from the equilibrium of various internal and external forces (Thompson, 1917). Intrinsic and extrinsic physical mechanisms that contribute to patterning of a multicellular aggregate include, but are not limited to, adhesive forces, intercalation forces, the ECM, and external mechanical cues. For further reading on the physical influencers of development, see for example Ladoux et al. (2016).
In biological systems, the combination of chemical and physical forces make up the microenvironment that shapes the overall organization of a multicellular aggregate (see for example Gilmour et al. [2017]). Indeed, it is the combination of various signals and forces received by a cell in a given point in space and time that mutually regulate one another to define the ultimate fate, structure, and interactions of a given cell (see for example Chan et al. [2017]).
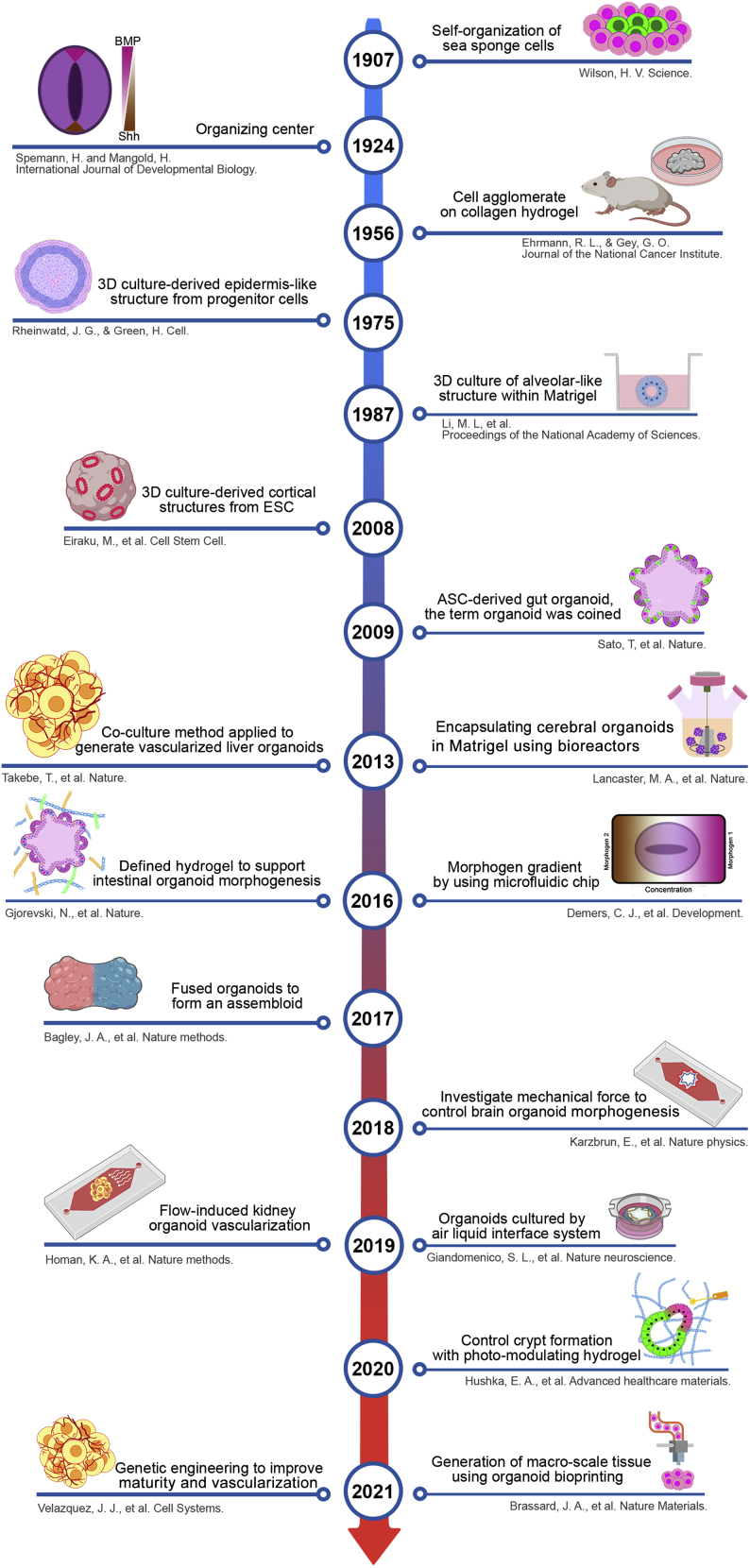
In the following section, a historical overview (Figure 2) of self-organization is given as a driving force for developing simple multicellular structures to complex engineered organoids.
Figure 2.
Advances in generation of complex multicellular structures from simple cell aggregate to engineered organoids throughout time
Historical overview on significant studies that impacted the organoid field. First evidence for self-organization was observed from the simple cell patterning that occurred in re-aggregation of adult cells in different species. Emergence of stem cells from diverse sources opened a new window into early stages of organ development. Furthermore, the engineering methods such as spatiotemporal delivery of soluble factors, manipulation of cell-cell or cell-ECM interactions, and mechanical forces led to increased control over multicellular complexity for guided self-organization, which has many applications in the field of biomedical engineering.
From simple multicellular structures to engineered organoids
An organoid is a complex 3D multicellular structure that can reflect the functional and architectural characteristics of counterpart organs in a dish (Rossi et al., 2018). This model is the result of years of research and efforts to simulate developmental events such as organogenesis in the laboratory. Initial efforts began with degeneration and regeneration assays, which greatly advanced our understanding of mechanisms of self-organization (Sasai, 2013). These studies date back to 1907 when Wilson (1907) first showed that dissociated sea sponge cells can re-organize via self-assembly. Other researchers conducted similar experiments on different species to assess the innate ability of cells to form self-organized structures (Holtfreter, 1943; Weiss and Taylor, 1960). Efforts were made to find the mechanisms behind this process and the positional cues that generate specific patterns within multicellular structures. Indeed, the transplantation of a group of cells (dorsal blastopore lip) of one newt gastrula embryo onto the ventral side of a host embryo at the same stage controlled cellular patterning and morphogenesis to form a second body axis (Spemann and Mangold, 1924). This experiment elegantly demonstrated that morphogenesis is a result of the release of signaling molecules from organizing centers.
Until the 1950s, researchers focused on the generation of organs based on re-aggregation of dissociated cells obtained from different organs via various methods. Early attempts to create a suitable substrate for guiding cell-ECM interactions among these cells dates back to 1956. Ehrmann and Gey (1956) developed a method to extract collagen from the rat’s tail and cultured 29 tissue fragments and cell lines to test cell growth, survival, and support of spontaneous agglomeration. Later, Mina Bissell and colleagues demonstrated the formation of a 3D lumen and duct structures with secretory function that grow on a basement membrane secreted from an Engelbreth-Holm-Swarm tumor (Matrigel) (Li et al., 1987). This study highlights the effects of ECM on gene expression, which led to the morphogenesis of tissue-like structures (Barcellos-Hoff et al., 1989; Petersen, 1992). Another study used matrix metalloproteinase as an active agent for ECM degradation to form branch-like structures in mammary glands (Simian et al., 2001).
Cell-cell interactions are also important in guiding the self-organization process. Precise control over cell-cell interactions enable control over cell sorting and patterning in different developmental stages (Gartner and Bertozzi, 2009). In 1975, Rheinwald and Green (1975) cultured epidermal keratinocytes on 3T3 fibroblasts, which increased formation of epithelial colonies similar to the human epidermis. These skin-like colonies had a squamous stratified structure with a dividing basal layer and a superficial layer containing differentiated keratinocytes. Improving cell-cell interactions can be achieved by fusing different multicellular structures or organoids for establishing cellular communication at the tissue or organ level. Another elegant study was conducted by Bagley et al. (2017) to show organoid-organoid (assembloids) interactions between dorsal and ventral brain organoids, which resulted in interneuron migration toward the dorsal portion. This approach enhanced brain organoid complexity and provided deeper insight into fetal brain development.
The self-organizing ability of organoids depends on the origin of the starting cells (Rossi et al., 2018). Pluripotent stem cell (PSC)-derived organoids mimic developmental pathways with a multi-step differentiation process and are able to form different cell lineages of different germ layers, while TSC-derived organoids mimic adult tissue signaling and often have a more limited developmental potential (Brassard and Lutolf, 2019). PSC-derived organoids are mostly used to study early developmental events in contrast with TSC-derived organoids, which are used to study the repair process and homeostasis of tissue (Kretzschmar and Clevers, 2016; McCracken et al., 2014). Therefore, the establishment of PSCs and isolation of TSCs are considered to be some of the most important events in this field (Evans and Kaufman, 1981; Takahashi and Yamanaka, 2006).
In 2008, Sasai and his colleagues (Eiraku et al., 2008) succeeded in forming 3D structures of stratified cortical tissue derived from self-patterning embryoid bodies (EB). In this study, the differentiation of EBs was based on the innate ability of ESCs to spontaneously differentiate toward a neural fate in the absence of external inhibitors. These cortical structures contain distinct zones along the apical-basal axis, including ventricles, deep and superficial layers of the cortical plate, and Cajal-Retzius cells. In this study, it was shown that secreting patterning factors (such as fibroblast growth factor, bone morphogenetic protein [BMP], and WNT) can dictate specific regional identity, including olfactory bulb, anterior-posterior cortices, hem organizing center, and choroid plexus (Eiraku et al., 2008). Later, Lancaster et al. (2013) produced brain organoids by embedding human PSC-derived EBs within a 3D Matrigel to increase tissue complexity and form brain organoids with different regions. During this process, rotational bioreactors increased the lifespan and size of organoids by increasing the amount of oxygen and nutrients diffused to the center of organoids, which resulted in a larger and more continuous neuroepithelial region.
In 2009, Sato et al. formed the first intestinal organoids from single LRG5+ TSCs obtained from the adult mouse intestine and coined the term “organoid.” Individual cells proliferated in vitro and differentiated to crypt-villus-like structures in Matrigel in the absence of a mesenchymal niche. These 3D organoids had a central lumen with villus-like epithelium and several surrounding crypt-like regions composed of different cell types (Sato et al., 2009).
Over the years, many organoids have been made via mechanisms of self-organization with little control over the cell-cell interactions, cell-ECM interactions, spatiotemporal pattern of soluble factors, and mechanical forces. Such uncontrolled conditions result in irreproducible generation of organoids that may lack certain cell types and cellular functions (Silva et al., 2019). Synthetic developmental biology has significant potential for developing the next generation organoids by advanced engineering methods to overcome previous limitations (see Table 1 for a list of engineered organoids).
Table 1.
Engineered organoids with different types of microenvironmental manipulation
| Engineering approach | Tissue or organ | Manipulation method | Result | References |
|---|---|---|---|---|
| Spatiotemporal delivery of soluble factors | Neural tube | Locally delivered agonists and antagonists of SHH signaling | Formation of neural tube-like structures | Yan et al. (2016) |
| Neural tube | Reconstructed primary signaling responsible for neural tube patterning using microfluidic chips | Spatiotemporal differentiation of motor neurons | Demers et al. (2016) | |
| Primitive streak | Exposed human ESC colonies to geometric gradients of BMP4 | Modulation of germ layer boundaries | Manfrin et al. (2019) | |
| Mammary gland | Delivered EGF to mammary organoids with meso-scale fluidic device | Multicellular structures respond to EGF gradient in contrast to single cells | Ellison et al. (2016) | |
| Colon | Exposed colon organoids to gradients of WNT-3a and R-spondin1 | Progenitor cells polarized according to the gradient of WNT | Attayek et al. (2016) | |
| Embryo | Controlled amnion and epiblast development by using microfluidic chips | Demonstrated the effect of signaling centers for triggering gastrulation | Zheng et al. (2019) | |
| Engineering cell-ECM interactions | Neural tube | Controlled physical and chemical properties of the ECM that impacted neural tube organization | Defined PEG-based hydrogels polarized the neural tube along the dorsal-ventral and apical-basal axes | Ranga et al. (2016) |
| Kidney | PAM hydrogels used as a CAM-like substrate for kidney organoid transplantation | Generation of nephron structures and renal vesicles | Garreta et al. (2019) | |
| Cerebral | PLGA microfiber scaffolds controlled organoid formation | PLGA microfibers promoted radial organization of cortical structures | Lancaster et al. (2017) | |
| Cerebral | Encapsulated cerebral organoid within HA-Na/chitosan hydrogels | Rosette and neural tube-like structure formation | Lindborg et al. (2016) | |
| Intestine | Functionalized fibrin hydrogels using factor XIII to covalently link different domains | Formation and long-term expansion of organoid | Broguiere et al. (2018) | |
| Kidney | Encapsulated kidney organoids within collagen hydrogels | Support kidney organoid growth | Cruz et al. (2017) | |
| Intestine | Designed fully synthetic and dynamic PEG-based hydrogels | Confined self-organization of organoid | Gjorevski et al. (2016) | |
| Liver | Encapsulated liver organoids within inverted colloidal crystal PEG | Mimic physiological condition for organoid formation | Ng et al. (2018) | |
| Engineering cell-cell interactions | Bone | Controlled cell assembly using airflow-assisted 3D bioprinting | Establish the spiral cell patterning inside microspheroidal organoids | Zhao et al. (2018) |
| – | Induced synthetic cell-cell signaling to guide assembly of multicellular structures | Organized cells in 3D with precise and robust patterning | Toda et al. (2018) | |
| Brain | Assembled organoid structures using 3D bioprinting | Created channel-like structures similar to the vascular system | Skylar-Scott et al. (2019) | |
| Cardiac | 3D bioprinting human iPSCs within photo-crosslinkable hydrogel | Developed bioink for differentiation of cardiomyocyte with electrochemical function | Kupfer et al. (2020) | |
| Brain | Incorporated human mesodermal progenitors within organoids | Formation of hierarchical structure of blood vessels | Wörsdörfer et al. (2019) | |
| Intestine | Fabricated a human intestinal structure using biopsy-derived organoids on a chip | Closely mimicked the human duodenum | Kasendra et al. (2018) | |
| Embryo | Co-culture of extra-embryonic and embryonic stem cell within 3D scaffold | Recapitulating spatiotemporal event of embryogenesis | Harrison et al. (2017) | |
| Pancreatic islet | Exposed pancreatic islet organoids to shear stress using perfusable microfluidic chips | Enhanced expression of genes related to pancreatic β-cell and β-cell hormone | Tao et al. (2019) | |
| Manipulation of mechanical forces | Brain | Designed a miniaturized bioreactor for optimization of culture conditions | Enhanced diffusion of soluble factors and reduced media volume | Qian et al. (2016) |
| Intestine | Established a liquid flow within intestinal organoids using millifluidic chips | Generation of a lumen within intestinal organoids to establish a flow | Sidar et al. (2019) | |
| Intestine | Exposed intestinal organoids to physical cues of the niche using compressed springs | Uniaxial strain enhanced maturation of human intestinal organoids | Poling et al. (2018) | |
| Brain | Controlled brain organoid wrinkling using internal mechanical forces on small molecules | Controlled cytoskeleton using small molecules to manipulate stiffness of different organoid regions | Karzbrun et al. (2018) | |
| Kidney | Developed millifluidic chips to apply shear stress onto organoids entrapped within collagen-based hydrogels | Observed flow-enhanced vascularization and maturation of kidney organoids | Homan et al. (2019) | |
| Intestine | Designed dynamic hydrogels with softening capabilities | Enhanced efficiency of crypt formation, its size and number per colony | Hushka et al. (2020) |
BMP4, bone morphogenetic protein four; CAM, chick chorioallantoic membrane; ECM, extracellular matrix; EGF, epidermal growth factor; HA-Na, hyaluronic acid sodium salt; hiPSC, human induced pluripotent stem cells; hESC, human embryonic stem cells; PAM, polyacrylamide; PEG, polyethylene glycol; PLGA, poly(lactic-co-glycolic-acid); SHH, Sonic hedgehog.
In the next section, we discuss different engineering approaches that allow manipulation of chemical and physical elements to guide organization of multicellular structures.
Bioengineering approaches facilitating guided organization
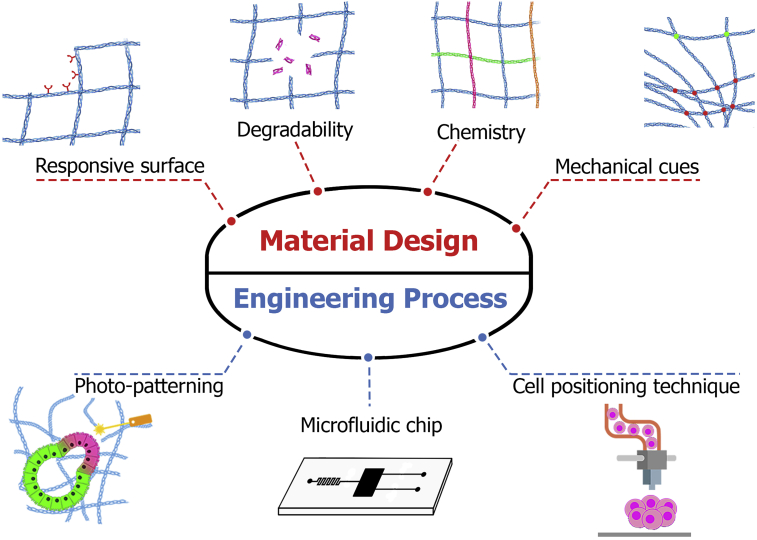
One approach to guiding organization in vitro, is the engineering of soluble factors using different techniques such as microfluidics and photopatterning (permit localized, sustained, smart or gradient-based deliveries of signaling factors). Since physical-mechanical cues in the cellular microenvironment direct cell assembly, scaffold engineering with different components (natural, synthetic, and hybrid) is another promising way to guide organization. In addition, various methods have been used to engineer cell-cell contacts, including manipulation of signaling molecules on the cell surface via genetic manipulation, cell patterning with bioprinting techniques, and encapsulation of different cell types on a chip. Finally, mechanical forces generated by flow of liquids are important inducers in organ development. These approaches are discussed in length below (Figure 3). The aim of designing engineering methods is to construct complex organoid structures with simple tools. The complexity of structures depends on the diversity of cell types, cellular organization, and functionality, which can range from a simple spheroid to an organoid with a function similar to the native organ (Ollé-Vila et al., 2016).
Figure 3.
Experimental design for engineering multicellular organization
Experimental design for guiding multicellular structure can be divided into two main categories: engineering process and material design. The material design consists of four subsections: (1) responsive surface; (2) degradability, including hydrolytic, enzymatic, and photo-degradation; (3) the chemistry of extracellular matrix (natural, synthetic, and composite); and (4) mechanical cues, which can be dictated by crosslinking density of the ECM. The engineering process consists of three subsections: (1) the 3D design of microenvironment with photopatterning technique, which includes (2) microfluidic chip to for cell encapsulation, control morphogen gradient and fluid shear through the time; and (3) cell positioning technique to define the cell’s position by using bioprinting, magnetic lavation, and acoustic assembly in a spatiotemporal manner.
Spatiotemporal delivery of soluble factors
Cellular organization during development is steered by a complex array of soluble factors. Morphogen gradients, in particular, play a crucial role in guiding the complex patterning of cells into tissues and organs. Insights from developmental studies have enabled engineering approaches to generate complex tissue structures via manipulation of morphogen gradients. Sophisticated engineering approaches allow a more precise control of the concentration, release site, and timing of morphogen delivery (Caballero Aguilar et al., 2019). Various approaches have been developed for the reconstruction of morphogen gradients, including 3D hydrogels and fluid-based methods. Fluid-based methods such as microfluidics and buoyancy forces (i.e., the tendency of an object to float when submerged in a fluid) essentially manipulate the presence of molecules using a dynamic fluid flow (Li et al., 2019). When using the buoyancy-driven method, a fluid with lower density moves from the bottom of a plate in opposite direction of denser material from upper layers, thereby producing a gradient of soluble factors (Li et al., 2019).
Microfluidics involves the manipulation of liquids at the micro- and nanoscales. Microfluidic chips are frequently made by “soft lithography,” which is a means of replicating patterns etched into silicon chips in biocompatible and flexible materials such as polydimethylsiloxane (PDMS) (Bhatia and Ingber, 2014; Convery and Gadegaard, 2019). Liquids at the micro- and nanoscales behave differently in comparison to liquids at a macroscale. In particular, laminar flow at the microfluidics dimension is similar to the flow of biological liquids in the human body. Laminar flow provides a low mixing rate of input streams, which allows establishment of sustained gradients of soluble factors. This feature allows one to mimic the spatiotemporal profile of morphogens during embryogenesis. Such approaches include source-sink diffusion devices, tree-like microfluidic gradient generators, mixing of input streams from syringe pumps, and microfluidic convection (i.e., the circular motion that occurs when warmer air or liquid rises while cooler air or liquid descends) (Sant et al., 2010). Droplet-based microfluidics can be used to encapsulate different cell populations in a predetermined pattern to engineer a multicellular structure of interest.
Traditional biological protocols for production of organoids do not provide a precise spatial control on morphogens. Thus, recapitulation of in vivo events is limited to daily refreshment of culture medium. Microfluidic chips have been used extensively for the reconstruction of morphogen gradients to overcome such limitations (Kim et al., 2012; O’Grady et al., 2019; Uzel et al., 2016). Reconstitution of morphogen gradients may help trigger critical events in morphogenetic processes such as gastrulation when extra-embryonic tissues or organizers are absent. In such an approach, a microfluidic technique was employed to expose human PSC colonies to a BMP4 gradient, which induced radial asymmetry and axially arranged domains. Counteracting sources of the BMP antagonist, Noggin, enhanced primitive streak-like patterning (Manfrin et al., 2019). Similarly, opposing gradients of sonic hedgehog (SHH) and its antagonist BMP were established in a microfluidic chip to generate neural tubes from mouse embryonic stem cells (Demers et al., 2016).
Induction of apical-basal polarization of human PSC aggregates, thereby mimicking early human post-implantation development, is also feasible, as demonstrated in a study based on microfluidic chips with a flow of BMP4 in parallel channels (Zhang et al., 2019). So far, microfluidic systems were mainly used to construct 2D patterns of soluble factors, but 3D patterning can also be achieved by designing complex multi-layered flow in such systems. The successful 3D reconstruction of morphogens in vitro may pave the way for complex development of embryos on a chip with determined body axes.
One of the major challenges for efficient production and maintenance of multicellular organoids is the use of appropriate media. Current methods for screening ECM and soluble factors in tissue culture plates is time-consuming and expensive. Microfluidic systems provide a platform for high-throughput screening of compounds to optimize culture media in a cost-effective manner due to the low volume of fluid used. In a recent study, the role of different inducing factors and signaling pathways in kidney organoid formation was investigated by testing more than 1,000 unique combinations on a microfluidic chip (Glass et al., 2020).
Microfluidic systems also present advantages for the long-term culture and maintenance of organ-like structures. The technology allows establishment of a precise flow route with a defined composition, flow rate, and frequency of media refreshment. Such precise interaction between fluid and cells (direct/indirect) allows optimization of the concentration of signaling molecules (including autocrine and paracrine), nutrients, and cell wastes, which is crucial for ex vivo tissue or organoid cultures (Lai et al., 2020). The development of techniques for organoid maintenance presents an avenue for body-on-chip platforms, in which different organoids can interact, offering opportunities for drug and toxicology screenings as well as disease modeling (Skardal et al., 2017).
Three-dimensional photopatterning of hydrogels is another method for manipulation of signaling molecules. Photopatterning of hydrogels can be achieved via three main techniques: mask-based photolithography, single-photon laser-scanning lithography, and multiphoton laser-scanning lithography. In mask-based photolithography, materials are exposed to light that is partially obstructed by a photomask, which contains 2D geometries of interest and is positioned between the sample and the light source. The method is affordable and enables 2D patterning with a resolution at the order of tens of microns. However, photolithography lacks the potential for 3D patterning. Laser-scanning lithography is a maskless method in which the laser light is focused on a specific area within a material to induce photoreactions near the focal point. The technique allows formation or cleavage of bonds in a 3D hydrogel via displacement of focal points in the x, y (with a resolution less than 1 μm), and z (with a resolution greater than 25 μm for a single photon and 2 μm for multiphoton laser-scanning) planes. Patterned arrangement of signaling molecules in biomaterials can exert spatial control on the fate of embedded cells and, therefore, manipulate cellular organization (Glass et al., 2020). Various photochemical tools have been developed for the formation or cleavage of chemical bonds based on light-responsive synthetic reactions. Studies have shown that through light exposure with single or multiphoton laser-scanning lithography, molecules are reversibly added or removed from a structure in a 3D space (Ruskowitz and DeForest, 2018). Biomolecule gradients have also been attached to hydrogels in a stepwise or continuous manner (Mosiewicz et al., 2013). In one study, the spatial distribution of SHH and ciliary neurotrophic factor was achieved by two-photon irradiation by establishing orthogonal physical binding pairs in 3D agarose hydrogels. The patterned hydrogel guides spatially controlled differentiation of retinal precursor cells into photoreceptors (Mosiewicz et al., 2013). In contrast to microfluidic systems, spatial control in 3D patterned hydrogels is usually determined by factors immobilized in the hydrogel structure. Although this may restrict the signaling of normally soluble morphogens to a matrix-bound form, it allows spatial control of anchoring ligands (such as adhesive proteins and peptides) via external stimulations (see engineering cell-ECM interactions section).
As discussed in the above section, soluble factors steer morphogenesis during development. In addition to deepening our understanding of the dynamic profile of these molecules in human embryogenesis, the establishment of high-resolution techniques such as microfluidics and photopatterning pave the way for successful reconstruction of in vivo events in culture systems. Alongside biomimicking, engineering high-throughput tools such as microfluidics provides the possibility to optimize the spatiotemporal profile of soluble factors in a cost-effective manner. However, despite the benefits of using engineering approaches for the spatiotemporal manipulation of soluble factors, we are far from achieving a user-friendly tool for application of such techniques in common biological protocols.
Engineering cell-ECM interactions
PSCs are potentially able to mimic early developmental stages of embryogenesis when the right composition of a 3D matrix is used. A comprehensive study demonstrated that 3D blastocyst-cultures in Matrigel enable human blastocyst development up to the formation of primitive streak-like structures, including the embryonic disc, amnion, basement membrane, primary and primate unique secondary yolk sac, as well as acquisition of the anterior-posterior polarity (Xiang et al., 2020). In contrast, traditional 2D cultures of blastocysts do not fully recapitulate in vivo events, including amnion formation (Deglincerti et al., 2016; Shahbazi et al., 2019). Furthermore, organoid structures have been shown to form both in 3D matrices and in suspension culture systems. For instance, self-organized optic cups have been generated in suspension cultures (Kuwahara et al., 2015) with a structure comparable to those forming in Matrigel (Eiraku et al., 2011). In certain cases, a combination of 2D and 3D systems may be advantageous for the development of complex structures in vitro. One study used a 2D culture of PSCs at early stages of hindgut differentiation and subsequently placed the aggregates in 3D Matrigel to improve differentiation and cellular patterning of intestinal organoids (Spence et al., 2011).
Current approaches for organoid culture rely on animal-derived ECMs such as Matrigel, collagen, and fibrin (Zahmatkesh et al., 2021). However, these media suffer from poorly defined composition, heterogeneity, and batch-to-batch variability. One approach for overcoming these limitations is to blend ECM components experimentally to attain a defined and reproducible matrix. The results of a recent study demonstrate that an artificial fibrin/laminin matrix supports long-term expansion of murine and human epithelial organoids, serving as a defined equivalent for Matrigel (Broguiere et al., 2018).
Alternatively, synthetic materials can be designed to mimic key features of natural ECMs. Motifs involved in cell adhesion as well as enzyme-responsive domains (i.e., matrix metalloproteinase and plasmin) that are crucial for guiding cell migration and organization can be freely combined. Another major advantage of synthetic matrices is the ability to manipulate its mechanical properties, including stiffness. Ranga et al. (2016) used polyethylene glycol (PEG) hydrogel with tailored mechanical properties for the development of neuroepithelial cysts. They showed that mechanotransduction pathways induce symmetry-breaking events and subsequently neural tube-like patterning along the dorsal-ventral axis (Ranga et al., 2016).
When developing a matrix, one must consider the mechanical, physical, and biochemical properties required for formation of target structures, since mechanical properties of substrates have striking effects during early embryogenesis. Mouse embryos grown on soft type 1 collagen hydrogels with stiffness similar to the uterine epithelium showed improved development to two-cell, blastocyst, and hatching blastocyst stages compared with tissue culture plates (Kolahi et al., 2012). Furthermore mechanical properties (i.e., stiffness) of fibrin are critical for 3D organization of mouse embryonic stem cells into the three germ layers (Poh et al., 2014). Soft matrix, which mimics the early embryonic microenvironment, accelerates differentiation of renal vesicles and nephron structures in human PSC-derived kidney organoids (Garreta et al., 2019).
As noted above, physical properties of a material, such as geometry, size, topography, and shape, influence cellular organization (Metzger et al., 2018). For instance, Lancaster et al. (2017) used polymeric microfibers as a substrate to generate elongated EBs. The microfilament structure induced cerebral organoid formation with enhanced cortical development and improved tissue organization.
Finally, biochemical properties of the ECM have been reconstructed using synthetic materials. For example, PEG was modified by the adhesive arginylglycylaspartic acid (RGD) peptide to mimic the adhesive domain of fibronectin and by a matrix metalloproteinase-sensitive peptide to emulate the Matrigel structure. Lutolf’s group used this synthetic hydrogel to mimic ECM properties, including active domains that promote cell adhesion and migration, allowing intestinal stem cell expansion and organoid formation (Gjorevski et al., 2016).
One of the current goals of ECM engineering is to design a fully synthetic and modular material that can both mimic the biological activity of the natural ECM (e.g., Matrigel and fibrin), and be customized for the production of different types of organoids. For this purpose, synthesis of novel materials with advanced properties such as dynamic presentation of adhesive ligands and mechanical properties may present an opportunity to completely substitute the natural ECM in organoid cultures.
Engineering cell-cell interactions
In the process of complex tissue formation, cell-cell interactions take place via secretion of soluble factors, cell adhesion events, and juxtacrine signaling. Application of co-culture systems is a simple method to recapitulate cell-cell interactions during development. For instance, embryo development led by extra-embryonic tissue implantation has inspired the application of extra-embryonic cells for PSC culture. During early mouse embryonic development, the crosstalk between embryonic and extra-embryonic stem cells is decisive for spatiotemporal patterning (Harrison et al., 2017) and development of extra-embryonic tissues (Rivron et al., 2018). In vitro studies on zebrafish embryos revealed that tensile forces among three germ layers induce assembly of structures when extra-embryonic tissues are absent (Krieg et al., 2008). Ectoderm cells without extra-embryonic tissues have the tendency to form inner layers within aggregates, while endodermal and mesodermal cells form the outer cell layer. In addition to biochemical signaling between cells, co-culture approaches may provide the mechanical forces required for cell migration and subsequent organization. Studies on the formation of vascularized tissue-specific buds from PSCs indicated that traction forces produced by mesenchymal stem cells play an important role for cell movements during formation of complex bud structure after transplantation (Takebe et al., 2015).
Surface cell-cell interactions also play a vital role in the organization of complex structures. Synthetic cellular networks have been used to manipulate the morphogenetic processes in multicellular systems (Toda et al., 2019). In a recent study, the modular juxtacrine signaling platform was used for artificial genetic programming of Notch receptors, where specific cell-cell contacts induce changes in cadherin cell adhesion and subsequently direct self-organization into multidomain structures (Toda et al., 2018).
Cells in an animal embryo receive positional cues from soluble and membrane-bound signaling molecules, which subsequently induce stepwise cells fate specification and organization (Lander, 2013). In addition, manipulating the position of different cell types in cell aggregates allows one to construct the micro-architecture with cell-cell interactions similar to native tissues (Brassard et al., 2021). In this regard, engineering approaches including bioprinting and application of remote forces determining the cell’s position have been used for organoid production.
One of the best-known methods for inducing cellular organization is bioprinting of different cell types. In the printing process, the initial position of cells is dictated by external mechanical forces in a point-by-point fashion. Therefore, this approach provides control over the cell placement to generate 2D or 3D patterns of one or more cell types in vitro (Heinrich et al., 2019). Three-dimensional bioprinting is used to fabricate complex cellular structures, such as tissues and solid organs, from different cell populations (Grigoryan et al., 2019). Guided 3D geometry and cell position within the organoids provide the possibility to produce constructs with enhanced complexity similar to native tissues. The technique involves the precise assembly of materials (i.e., hydrogels) and living cells in a layer-by-layer fashion with spatial control of active components in a predetermined layout. The printing resolution is known as a key parameter for 3D printing tissue-like structures. Recent attempts in bioprinting rely on development of novel methods that print cells individually. The main techniques used for bioprinting include inkjet (with a resolution of 50 μm), microextrusion (with a resolution of 5 μm to millimeters), and laser-assisted printing (microscale resolution). Because of technical limitations for high-resolution placement in common printing methods compared with 3D patterning, the printing method is usually used for cell placement, rather than spatially controlling the position of soluble or adhesive molecules. Current research is focused on the development of novel processing designs and synthesis of printable bioactive materials, which can be printed with high resolution while preserving cell viability (Murphy and Atala, 2014). Bioprinting has been used as a tool to engineer the structure of bone organoids (Zhao et al., 2018), salivary gland-like organoids (Adine et al., 2018), mammary epithelial organoids (Reid et al., 2019), mammary organoids (Mollica et al., 2019), and vascularized cortical organoids (Skylar-Scott et al., 2019).
In addition to contact force application, which is used in bioprinting, remote fields have recently emerged in the engineering of complex tissue structures where cellular structures can be formed without the role of ECM materials and are therefore scaffold-free (Armstrong and Stevens, 2020). Application of remote forces is a feasible approach for rapid production of 2D or 3D multicellular constructs. This method is used for forced formation of structures and is suitable for high-throughput generation of aggregates. In this approach, multicellular organization can be guided using remote forces such as acoustic streaming (i.e., the steady flow generated in a fluid by the absorption of high-amplitude sound oscillations) (Ma et al., 2019). Acoustic levitation provides a rapid platform for generation of multi-layered structures with defined number and thickness of layers, which could be a physiologically relevant structure such as the cortex or cerebellum (Bouyer et al., 2016). Using acoustic forces to induce packing density similar to native heart in human induced PSC (iPSC)-derived cardiomyocytes significantly improves the function of differentiated cells (Serpooshan et al., 2017).
Magnetic forces have also been used as versatile tools to levitate and subsequently guide cell populations into assembled structures (Souza et al., 2010; Tocchio et al., 2018; Zwi-Dantsis et al., 2019). Through patterning of magnetic fields, various shape of cell structures could be formed, including size-controlled spheres and rings (Jafari et al., 2019). Magnetic forces were used to construct a 3D model of breast cancer with organized fibroblasts and cancer cells (Yakavets et al., 2020). In another study, magnetic induction with nanoparticles promotes dental pulp stem cell differentiation to epithelial and neuronal lineages and subsequently the generation of salivary gland-like structure (Adine et al., 2018). Another method for remote control of cell populations is using optical tweezers. Optical tweezers could tailor the precise position of single mouse ESCs and their assembly in 3D structures (Kirkham et al., 2015).
As previously mentioned, cell-cell interactions play a vital role in formation of complex multicellular patterns, and a wide range of bioengineering approaches, including genetics, mechanical engineering, and materials science, have been used to manipulate cellular interactions in organoid culture. Approaches with different degrees of placement manipulation may play a significant role in triggering important biological aspects of organization.
Manipulation of mechanical forces
Mechanical forces fulfill a vital role in cellular migration, proliferation, differentiation, and organization during development (Jaalouk and Lammerding, 2009). The bulk properties of ECM (e.g., stiffness and degradability) and its interacting surface features (e.g., adhesive ligand density) are effective environmental factors that trigger mechanotransduction pathways in multicellular structures. For instance, in a study on intestinal organoids, dynamic mechanical properties were provided by exogenous degradation of the hydrogel through UV light exposure. The softening of the hydrogel resulted in a higher efficiency of crypt formation and an improvement in its size and number per colony (Hushka et al., 2020). Such studies introduce dynamic ECM hydrogels as a versatile tool for 3D application of external forces to cells.
Engineering studies have demonstrated that manipulation of external mechanical forces also influences the morphology and spatial patterning of cells. Various devices have been developed to apply tensile, compressive, or shear stresses onto multicellular aggregates. For instance, construction of a microfabricated chip with a confined geometry imitated compression forces in the early developing cortex, and induced wrinkling and folding in a human organoid (Karzbrun et al., 2018). Tensile forces induced by a lengthening spring promoted the in vivo formation of intestinal organoids. Poling et al. (2018) recreated mechanical forces affecting intestinal development to mimic the looped geometry of the gut, its vilification, and the localization of intestinal stem cells (ISCs) in the crypt. This study showed that induction of strain by compressed springs enhances the growth and maturation of these ESC-derived organoids.
To apply shear forces, flow-based devices such as micro/millifluidics and bioreactors have been used. Mechanical flow in a device is a biomimicking tool to apply force to cell populations. This tool was used to culture pancreatic islets, thereby promoting endocrine cell differentiation and maturation of organoids. Pancreatic islets derived from human iPSCs showed enhanced response to glucose under a dynamic flow condition (Tao et al., 2019). In addition to promoting improved organization of parenchymal cells, mechanical forces can enhance vascularization in organoid structures. In a recent study, shear stress in a millifluidic culture showed improved morphological maturation of the kidney and increased formation of vessels (Homan et al., 2019). The developing embryo experiences dynamic changes in mechanical forces. Hence, it will be critical to develop more sophisticated dynamic systems, in which mechanical forces can be triggered in a temporal and spatially controlled manner. Static systems exerting continuous mechanical force will most likely fail to recapitulate a changing environment that produces varying degrees of physical force over time.
Aiming for more complex tissues in vitro
Until recently, the study of human developmental processes had mostly been limited to animal models, which might not recapitulate all critical processes occurring in humans (Rossi et al., 2018). Organoids are now the focus of many research teams worldwide, as they hold the potential to reflect conditions in human tissues more accurately. The engineering approaches outlined in the previous section have been used extensively to guide organization of multicellular structures in vitro from a simple layer of cells to more complex tissues and organ-like structures (Figure 1). These engineering approaches can be classified according to the different types of manipulation applied. In fact, co-cultures of different cell types have been used to improve conditions for in vitro morphogenesis without exerting control over the cell microenvironment. The co-culture of mesenchymal stem cells, endothelial cells, and stem cells is one of the most common methods to produce vascularized organoids and organ buds (Takebe et al., 2015). Another class of microenvironmental manipulation can be achieved by controlling cell-ECM interactions and affecting multicellular structure development. For instance, synthetic hydrogels such as PEG provide physical-chemical signals that lead to neural tube patterning (Ranga et al., 2016). Furthermore, highly controlled provision of such signals on microfluidic chips allows better guidance of structure formation in vitro (Homan et al., 2019). More sophisticated engineering methods are applied when the environment or individual cells are patterned in a spatiotemporal manner. Three-dimensional patterning of ECM components, cell position, and surface ligands has been accomplished by 3D photopatterning, 3D bioprinting, and genetic modification techniques, respectively (Cakir et al., 2019; Heinrich et al., 2019).
Application of organoids for biomedical studies
The generation of near-physiological conditions in various organoids provides an opportunity to investigate different biological processes such as tissue development, repair, and homeostasis. Next generation organoids generated by advanced engineering methods will provide valuable new information for biomedical applications. Indeed, the study of organoid models and comparing them with animal models can identify developmental similarities and differences between humans and other species. For example, human optic cup organoids take longer to reach a similar stage of development compared with mouse organoids. These organoids are also larger and have a thicker neural retinal component in contrast to human PSC (hPSC)-derived optic cup organoids, which is confirmed by in vivo analysis of mouse and human embryonic tissues (Nakano et al., 2012).
Furthermore, organoids provide an accessible, easy to manipulate experimental system to investigate lineage specification and embryonic development in humans. For example, optic cup organoids have shed light on the mechanisms of optic cup morphogenesis, which relies on invagination of the neural retina (NR) within the retinal pigment epithelium (RPE). The use of drugs that inhibit cell proliferation revealed that proliferative cells within the NR generate a force for the invagination process that occurs within the RPE (Eiraku et al., 2011). The presence of different cell types is necessary in organogenesis and tissue development. The expression of Lgr5 gene by ISCs, which has an important effect on the colony’s self-renewal ability, depends on the presence of Paneth cells in its vicinity. Also, Paneth cells provide a high level of WNT signaling, which is required for ISC survival (Sato and Clevers, 2013). In an elegant study using intestinal organoids, it was shown that the WNT protein does not diffuse as a soluble factor but is rather transferred from Paneth cells near the ISC. Eventually by dividing the ISCs into daughter cells, a gradient of WNT forms along the crypt (Farin et al., 2016).
Organoids have also been used in evolutionary research by comparing various species-derived organoids. Indeed, the Neanderthal brain organoid, derived by engineering PSCs with a knockin NOVA1 gene, were compared with human brain organoids to allow the study of evolutionary mechanisms (Cohen, 2018).
Generation of fully functional and differentiated cells from PSCs that remain viable after engraftment is considered to be a main challenge in the field of regenerative medicine. Organoid technology might provide a superior microenvironment for proper stem cell differentiation, and organoids are a promising new source for transplantation (Fatehullah et al., 2016). For example, retinal sheets derived from retinal organoids have been transplanted into a retinal degeneration mouse model, which resulted in the establishment of synaptic connections leading to restoration of vision (Assawachananont et al., 2014). Another study focused on generation of mature T cells using artificial thymic organoids, since the current methods face different challenges in the differentiation of mature T cells from hPSCs. Sequential differentiation of T cells, induced by the 3D organoid structure, eventually led to the differentiation of embryonic mesoderm into CD4+CD3+ positive T cell with different T cell receptors for using immunotherapy applications (Montel-Hagen et al., 2019). In regenerative medicine, organoids can be used as a versatile tool to repair genetic abnormalities in patient-derived organoids and subsequently replace the malfunctioning tissue or organ. Stem cell-derived intestinal organoids reconstructed and repaired the intestinal epithelial function via grafting into injured colon site (Fordham et al., 2013). The process of transplantation and tissue regeneration has been also reported for different organoids, including the liver (Huch et al., 2013) and kidney (Taguchi et al., 2014).
In addition to providing the cells needed to restore normal tissue function, organoids can protect the graft from adverse pathological conditions (Assawachananont et al., 2014). Gene-editing technology can be used as a versatile tool to repair patient-derived organoids with a genetic disorder and replace it as an isogenic healthy tissue for regenerative applications. As a proof of concept, the Crisper-Cas9 system was used to correct cystic fibrosis transmembrane conductor receptor mutations and generate normal epithelia by restoring gene function in intestinal organoids (Dekkers et al., 2013).
Advances in organoid technology using patient-derived iPSCs have led to the development of disease models that reproduce physiological conditions of a disease, including the interaction of pathogens with host cells for drug discovery and screening applications. For example, organoids were used to select appropriate drugs to fight the Zika virus, which may lead to Guillain-Barré syndrome and congenital Zika syndrome (Russo et al., 2017). In this study, brain organoids were infected with the Zika virus, which caused apoptosis of neural progenitors and glial cells, and a subsequent decline in neural cell volume and restricted growth. Three types of drugs were used to counteract the flavivirus infection. Their effects were evaluated by quantification of the Zika virus mRNA in the brain organoids (Watanabe et al., 2017). Respiratory infections have been considered a major cause of death in recent years due to the coronavirus disease (COVID) pandemic. In this regard, different pathogens have been introduced to lung organoids to evaluate the changes in physiological condition of the infected organoid and their response to treatment with new drugs (Chen et al., 2017). In a recent study, hPSC-derived lung organoids were used in understanding COVID-19 pathogenesis and drug discovery. The authors found spike protein inhibitor, EK1 peptide, and TMPRSS2 inhibitors (camostat/nafamostat) block coronavirus entry into lung organoids (Tiwari et al., 2021). Moreover, human intestinal organoids have been used as in vitro model systems to study viral entry and replication of noroviruses (Ettayebi et al., 2016; Haga et al., 2020). Genetic engineering can be used to generate isogenic cell lines for disease modeling studies. Aicardi-Goutieres syndrome organoids were generated using a Crisper-Cas9 mutation in the TREX-1 gene. This mutation results in an accumulation of extrachromosomal DNA, which increases apoptosis and consequently reduces organoid size (Thomas et al., 2017).
Organoids have provided a platform for drug screening and personalized medicine, which is used to develop new treatment with available drugs. In this regard, patient-derived intestinal organoids were used to find a treatment for cystic fibrosis caused by a rare mutation by screening available drugs. The results showed that existing drugs used to treat other cystic fibrosis mutations were also effective for this rare mutation (Dekkers et al., 2016). The application of tumor organoids is an inevitable necessity to better understand the effect of different drugs on cancer cells. Organoids of different types and origins might be deposited in biobanks for off-the-shelf use to aid discovery of new drugs and facilitate investigation of disease pathologies (Dutta et al., 2017). Patient-derived tumor organoids can retain the tumor features, including genetic, epigenetic characteristic, and physiological phenotype. For instance, in vitro 3D culture of primary human pancreatic adenocarcinoma can retain the epigenetic feature of the original PDA tumor. In this regard, treating PDA-derived tumor organoids from five patients with histone methyltransferase EZH2 inhibitor (UNC 1999) reduced the proliferation of tumors in three patients with a H3K27me3 mark similar to the original tumor (Thress et al., 2015). This result suggests the potential treatment of PDA cancer patients with the same epigenetic state for clinical applications. The same strategy was used to identify a proper treatment for drug resistance cystic fibrosis that showed limited response to conventional drugs (Dekkers et al., 2016). Developing biobank derived from patient organoids is crucial to identify the most effective cancer treatments. In this regard, a study developed a biobank for high-throughput screening of 83 clinical drugs for colon cancer treatment (van de Wetering et al., 2015). A breast cancer biobank was also established for drug development and cancer research using well-characterized organoids (Sachs et al., 2018).
Concluding remarks
We provided examples from development to illustrate the basic principles of self-organization and described engineering tools that guide self-organization of multicellular structures for various applications. Most engineering methods employed thus far control the development of organoids in a static manner; however, the physiological environment for organ development changes over time and is dynamic. Engineering tools should therefore aim to control both initial culture conditions as well as later stages of the organization process. In co-culture systems, cells behave in a dynamic manner and can respond to different signals over time, thereby facilitating generation of more complex and representative structures in vitro. A promising strategy is to engineer the organoid microenvironment using dynamic hydrogels to alter the mechanical properties of organoids over time to mimic in vivo conditions (Gjorevski et al., 2016). Broader application and further refinement of such engineering tools will be useful to achieve more representative, reproducible, and mature models of different human organs. We hope that this view on the subject will be beneficial to promote biomedical applications.
Authors contributions
All authors contributed to the manuscript drafting and editing. All authors contributed to the design of figures. All authors read and approved the final manuscript.
Conflicts of interests
The authors declare no competing interests.
Acknowledgments
This study was funded by a grant from Royan Institute, the Iranian Cognitive Science and Technologies Council, the Iranian Council of Stem Cell Research and Technology, and the Ministry of Health and Medical Education (no. 56700/147) to H.B. We sincerely apologize to authors who have not been cited in this paper due to journal limitations.
References
- Adine C., Ng K.K., Rungarunlert S., Souza G.R., Ferreira J.N. Engineering innervated secretory epithelial organoids by magnetic three-dimensional bioprinting for stimulating epithelial growth in salivary glands. Biomaterials. 2018;180:52–66. doi: 10.1016/j.biomaterials.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Armstrong J.P.K., Stevens M.M. Using remote fields for complex tissue engineering. Trends Biotechnol. 2020;38:254–263. doi: 10.1016/j.tibtech.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assawachananont J., Mandai M., Okamoto S., Yamada C., Eiraku M., Yonemura S., Sasai Y., Takahashi M. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014;2:662–674. doi: 10.1016/j.stemcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attayek P.J., Ahmad A.A., Wang Y., Williamson I., Sims C.E., Magness S.T., Allbritton N.L. In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS ONE. 2016;11:e0153795. doi: 10.1371/journal.pone.0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley J.A., Reumann D., Bian S., Lévi-Strauss J., Knoblich J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff M.H., Aggeler J., Ram T.G., Bissell M.J. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Bouyer C., Chen P., Guven S., Demirtas T.T., Nieland T., Padilla F., Demirci U. A bio-acoustic levitational (BAL) assembly method for engineering of multilayered, 3D brain-like constructs, using human embryonic stem cell derived NeuroProgenitors. Adv. Mater. 2016;28:161–167. doi: 10.1002/adma.201503916. [DOI] [PubMed] [Google Scholar]
- Brassard J.A., Lutolf M.P. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Brassard J.A., Nikolaev M., Hübscher T., Hofer M., Lutolf M.P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021;20:22–29. doi: 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- Broguiere N., Isenmann L., Hirt C., Ringel T., Placzek S., Cavalli E., Ringnalda F., Villiger L., Züllig R., Lehmann R., et al. Growth of epithelial organoids in a defined hydrogel. Adv. Mater. 2018;30:1801621. doi: 10.1002/adma.201801621. [DOI] [PubMed] [Google Scholar]
- Caballero Aguilar L.M., Silva S.M., Moulton S.E. Growth factor delivery: defining the next generation platforms for tissue engineering. J. Control. Release. 2019;306:40–58. doi: 10.1016/j.jconrel.2019.05.028. [DOI] [PubMed] [Google Scholar]
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.-J., Chapeton K., Patterson B., Yuan Y., He C.-S., et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.J., Heisenberg C.-P., Hiiragi T. Coordination of morphogenesis and cell-fate specification in development. Curr. Biol. 2017;27:R1024–R1035. doi: 10.1016/j.cub.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Chen Y.-W., Huang S.X., de Carvalho A.L.R.T., Ho S.-H., Islam M.N., Volpi S., Notarangelo L.D., Ciancanelli M., Casanova J.-L., Bhattacharya J., et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Neanderthal brain organoids come to life. Science. 2018;360:1284. doi: 10.1126/science.360.6395.1284. [DOI] [PubMed] [Google Scholar]
- Convery N., Gadegaard N. 30 years of microfluidics. Micro Nano Eng. 2019;2:76–91. [Google Scholar]
- Cruz N.M., Song X., Czerniecki S.M., Gulieva R.E., Churchill A.J., Kim Y.K., Winston K., Tran L.M., Diaz M.A., Fu H., et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017;16:1112–1119. doi: 10.1038/nmat4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., Brivanlou A.H. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W.M., Bijvelds M.J.C., Scholte B.J., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E.S., Houwen R.H.J., et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- Demers C.J., Soundararajan P., Chennampally P., Cox G.A., Briscoe J., Collins S.D., Smith R.L. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development. 2016;143:1884–1892. doi: 10.1242/dev.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Grove E.A., Monuki E.S. Patterning and Cell Type Specification in the Developing CNS and PNS: Comprehensive Developmental Neuroscience. Elsevier Inc.); 2013. Chapter 2 - morphogens, patterning centers, and their mechanisms of action; p. 20. [Google Scholar]
- Ebrahimkhani M.R., Ebisuya M. Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol. 2019;52:9–15. doi: 10.1016/j.cbpa.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Ehrmann R.L., Gey G.O. The growth of cells on a transparent gel of reconstituted rat-tail collagen. J. Natl. Cancer Inst. 1956;16:1375–1403. [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Ellison D., Mugler A., Brennan M.D., Lee S.H., Huebner R.J., Shamir E.R., Woo L.A., Kim J., Amar P., Nemenman I., et al. Cell–cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. U S A. 2016;113:E679–E688. doi: 10.1073/pnas.1516503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.-L., Qu L., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Farin H.F., Jordens I., Mosa M.H., Basak O., Korving J., Tauriello D.V.F., de Punder K., Angers S., Peters P.J., Maurice M.M., et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R.F., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T., et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta E., Prado P., Tarantino C., Oria R., Fanlo L., Martí E., Zalvidea D., Trepat X., Roca-Cusachs P., Gavaldà-Navarro A., et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019;18:397–405. doi: 10.1038/s41563-019-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner Z.J., Bertozzi C.R. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl. Acad. Sci. U S A. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D., Rembold M., Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordóñez-Morán P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- Glass N.R., Takasako M., Er P.X., Titmarsh D.M., Hidalgo A., Wolvetang E.J., Little M.H., Cooper-White J.J. Multivariate patterning of human pluripotent cells under perfusion reveals critical roles of induced paracrine factors in kidney organoid development. Sci. Adv. 2020;6:eaaw2746. doi: 10.1126/sciadv.aaw2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan B., Paulsen S.J., Corbett D.C., Sazer D.W., Fortin C.L., Zaita A.J., Greenfield P.T., Calafat N.J., Gounley J.P., Ta A.H., et al. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science. 2019;364:458–464. doi: 10.1126/science.aav9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Lutolf M.P., Hughes A.J., Sonnen K.F. Bioengineering in vitro models of embryonic development. Stem Cell Rep. 2021;16:1104–1116. doi: 10.1016/j.stemcr.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Ettayebi K., Tenge V.R., Karandikar U.C., Lewis M.A., Lin S.-C., Neill F.H., Ayyar B.V., Zeng X.-L., Larson G., et al. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional Fucosyltransferase 2 gene for secretor-dependent human Norovirus infection. MBio. 2020;11:e00251-20. doi: 10.1128/mBio.00251-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Heinrich M.A., Liu W., Jimenez A., Yang J., Akpek A., Liu X., Pi Q., Mu X., Hu N., Schiffelers R.M., et al. 3D bioprinting: from benches to translational applications. Small. 2019;15:1805510. doi: 10.1002/smll.201805510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C., Morsut L. Novel synthetic biology approaches for developmental systems. Stem Cell Rep. 2021;16:1051–1064. doi: 10.1016/j.stemcr.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtfreter J. Experimental studies on the development of the pronephros. Rev. Can. Biol. 1943:220–250. [Google Scholar]
- Homan K.A., Gupta N., Kroll K.T., Kolesky D.B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M.T., Ferrante T., Bonventre J.V., et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S.W., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J., et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hushka E.A., Yavitt F.M., Brown T.E., Dempsey P.J., Anseth K.S. Relaxation of extracellular matrix forces directs crypt formation and architecture in intestinal organoids. Adv. Healthc. Mater. 2020;9:1901214. doi: 10.1002/adhm.201901214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaalouk D.E., Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari J., Han X., Palmer J., Tran P.A., O’Connor A.J. Remote control in formation of 3D multicellular assemblies using magnetic forces. ACS Biomater. Sci. Eng. 2019;5:2532–2542. doi: 10.1021/acsbiomaterials.9b00297. [DOI] [PubMed] [Google Scholar]
- Karzbrun E., Kshirsagar A., Cohen S.R., Hanna J.H., Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasendra M., Tovaglieri A., Sontheimer-Phelps A., Jalili-Firoozinezhad S., Bein A., Chalkiadaki A., Scholl W., Zhang C., Rickner H., Richmond C.A., et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Kreppenhofer K., Kashef J., Gradl D., Herrmann D., Schneider M., Ahrens R., Guber A., Wedlich D. Diffusion- and convection-based activation of Wnt/β-catenin signaling in a gradient generating microfluidic chip. Lab. Chip. 2012;12:5186. doi: 10.1039/c2lc40172j. [DOI] [PubMed] [Google Scholar]
- Kirkham G.R., Britchford E., Upton T., Ware J., Gibson G.M., Devaud Y., Ehrbar M., Padgett M., Allen S., Buttery L.D., et al. Precision assembly of complex cellular microenvironments using holographic optical tweezers. Sci. Rep. 2015;5:8577. doi: 10.1038/srep08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolahi K.S., Donjacour A., Liu X., Lin W., Simbulan R.K., Bloise E., Maltepe E., Rinaudo P. Effect of substrate stiffness on early mouse embryo development. PLoS ONE. 2012;7:e41717. doi: 10.1371/journal.pone.0041717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K., Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev. Cell. 2016;38:590–600. doi: 10.1016/j.devcel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Krieg M., Arboleda-Estudillo Y., Puech P.-H., Käfer J., Graner F., Müller D.J., Heisenberg C.-P. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- Kupfer M., Lin W., Ravikumar V., Qiu K., Wang L., Gao L., Bhuiyan D., Lenz M., Ai J., Mahutga R., et al. In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ. Res. 2020;127:207–224. doi: 10.1161/CIRCRESAHA.119.316155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015;6:6286. doi: 10.1038/ncomms7286. [DOI] [PubMed] [Google Scholar]
- Ladoux B., Mège R.-M., Trepat X. Front–rear polarization by mechanical cues: from single cells to tissues. Trends Cell Biol. 2016;26:420–433. doi: 10.1016/j.tcb.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B.F.L., Lu R.X.Z., Hu Y., Davenport Huyer L., Dou W., Wang E.Y., Radulovich N., Tsao M.S., Sun Y., Radisic M. Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv. Funct. Mater. 2020;30:2000545. doi: 10.1002/adfm.202000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander A.D. How cells know where they are. Science. 2013;339:923–927. doi: 10.1126/science.1224186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent J., Blin G., Chatelain F., Vanneaux V., Fuchs A., Larghero J., Théry M. Convergence of microengineering and cellular self-organization towards functional tissue manufacturing. Nat. Biomed. Eng. 2017;1:939–956. doi: 10.1038/s41551-017-0166-x. [DOI] [PubMed] [Google Scholar]
- Li C., Ouyang L., Pence I.J., Moore A.C., Lin Y., Winter C.W., Armstrong J.P.K., Stevens M.M. Buoyancy-driven gradients for biomaterial fabrication and tissue engineering. Adv. Mater. 2019;31:1900291. doi: 10.1002/adma.201900291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Aggeler J., Farson D.A., Hatier C., Hassell J., Bissell M.J. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. U S A. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindborg B.A., Brekke J.H., Vegoe A.L., Ulrich C.B., Haider K.T., Subramaniam S., Venhuizen S.L., Eide C.R., Orchard P.J., Chen W., et al. Rapid induction of cerebral organoids from human induced pluripotent stem cells using a chemically defined hydrogel and defined cell culture medium: cerebral organoid induction using defined reagents. Stem Cells Transl. Med. 2016;5:970–979. doi: 10.5966/sctm.2015-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Warmflash A. Self-organized signaling in stem cell models of embryos. Stem Cell Rep. 2021;16:1065–1077. doi: 10.1016/j.stemcr.2021.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis C.A., Lairson L.L., Boitano A.E., Wurdak H., Zhu S., Schultz P.G. Chemical control of stem cell fate and developmental potential. Angew. Chem. Int. Ed. 2011;50:200–242. doi: 10.1002/anie.201004284. [DOI] [PubMed] [Google Scholar]
- Ma Z., Holle A.W., Melde K., Qiu T., Poeppel K., Kadiri V.M., Fischer P. Acoustic holographic cell patterning in a biocompatible hydrogel. Adv. Mater. 2019;32:1904181. doi: 10.1002/adma.201904181. [DOI] [PubMed] [Google Scholar]
- Manfrin A., Tabata Y., Paquet E.R., Vuaridel A.R., Rivest F.R., Naef F., Lutolf M.P. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods. 2019;16:640–648. doi: 10.1038/s41592-019-0455-2. [DOI] [PubMed] [Google Scholar]
- McCracken K.W., Catá E.M., Crawford C.M., Sinagoga K.L., Schumacher M., Rockich B.E., Tsai Y.-H., Mayhew C.N., Spence J.R., Zavros Y., et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger J.J., Simunovic M., Brivanlou A.H. Synthetic embryology: controlling geometry to model early mammalian development. Curr. Opin. Genet. Dev. 2018;52:86–91. doi: 10.1016/j.gde.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica P.A., Booth-Creech E.N., Reid J.A., Zamponi M., Sullivan S.M., Palmer X.-L., Sachs P.C., Bruno R.D. 3D bioprinted mammary organoids and tumoroids in human mammary derived ECM hydrogels. Acta Biomater. 2019;95:201–213. doi: 10.1016/j.actbio.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel-Hagen A., Seet C.S., Li S., Chick B., Zhu Y., Chang P., Tsai S., Sun V., Lopez S., Chen H.-C., et al. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell. 2019;24:376–389.e8. doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J.S., Raspopovic J., Marcon L. From embryos to embryoids: how external signals and self-organization drive embryonic development. Stem Cell Rep. 2021;16:1039–1050. doi: 10.1016/j.stemcr.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosiewicz K.A., Kolb L., van der Vlies A.J., Martino M.M., Lienemann P.S., Hubbell J.A., Ehrbar M., Lutolf M.P. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 2013;12:1072–1078. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- Murphy S.V., Atala A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Ng S.S., Saeb-Parsy K., Blackford S.J.I., Segal J.M., Serra M.P., Horcas-Lopez M., No D.Y., Mastoridis S., Jassem W., Frank C.W., et al. Human iPS derived progenitors bioengineered into liver organoids using an inverted colloidal crystal poly (ethylene glycol) scaffold. Biomaterials. 2018;182:299–311. doi: 10.1016/j.biomaterials.2018.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady B., Balikov D.A., Wang J.X., Neal E.K., Ou Y.-C., Bardhan R., Lippmann E.S., Bellan L.M. Spatiotemporal control and modeling of morphogen delivery to induce gradient patterning of stem cell differentiation using fluidic channels. Biomater. Sci. 2019;7:1358–1371. doi: 10.1039/c8bm01199k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollé-Vila A., Duran-Nebreda S., Conde-Pueyo N., Montañez R., Solé R. A morphospace for synthetic organs and organoids: the possible and the actual. Integr. Biol. 2016;8:485–503. doi: 10.1039/c5ib00324e. [DOI] [PubMed] [Google Scholar]
- Petersen O.W. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Cell Biol. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh Y.-C., Chen J., Hong Y., Yi H., Zhang S., Chen J., Wu D.C., Wang L., Jia Q., Singh R., et al. Generation of organized germ layers from a single mouse embryonic stem cell. Nat. Commun. 2014;5:4000. doi: 10.1038/ncomms5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poling H.M., Wu D., Brown N., Baker M., Hausfeld T.A., Huynh N., Chaffron S., Dunn J.C.Y., Hogan S.P., Wells J.M., et al. Mechanically induced development and maturation of human intestinal organoids in vivo. Nat. Biomed. Eng. 2018;2:429–442. doi: 10.1038/s41551-018-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A., Girgin M., Meinhardt A., Eberle D., Caiazzo M., Tanaka E.M., Lutolf M.P. Neural tube morphogenesis in synthetic 3D microenvironments. Proc. Natl. Acad. Sci. U S A. 2016;113:E6831–E6839. doi: 10.1073/pnas.1603529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J.A., Palmer X.-L., Mollica P.A., Northam N., Sachs P.C., Bruno R.D. A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Sci. Rep. 2019;9:7466. doi: 10.1038/s41598-019-43922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald J., Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.-C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Rossi G., Manfrin A., Lutolf M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- Ruskowitz E.R., DeForest C.A. Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater. 2018;3:17087. [Google Scholar]
- Russo F.B., Jungmann P., Beltrão-Braga P.C.B. Zika infection and the development of neurological defects. Cell. Microbiol. 2017;19:e12744. doi: 10.1111/cmi.12744. [DOI] [PubMed] [Google Scholar]
- Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Sant S., Hancock M.J., Donnelly J.P., Iyer D., Khademhosseini A. Biomimetic gradient hydrogels for tissue engineering. Can. J. Chem. Eng. 2010;88:899–911. doi: 10.1002/cjce.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Serpooshan V., Chen P., Wu H., Lee S., Sharma A., Hu D.A., Venkatraman S., Ganesan A.V., Usta O.B., Yarmush M., et al. Bioacoustic-enabled patterning of human iPSC-derived cardiomyocytes into 3D cardiac tissue. Biomaterials. 2017;131:47–57. doi: 10.1016/j.biomaterials.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., Siggia E.D., Zernicka-Goetz M. Self-organization of stem cells into embryos: a window on early mammalian development. Science. 2019;364:948–951. doi: 10.1126/science.aax0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiri Z., Simorgh S., Naderi S., Baharvand H. Optogenetics in the era of cerebral organoids. Trends Biotechnol. 2019;37:1282–1294. doi: 10.1016/j.tibtech.2019.05.009. [DOI] [PubMed] [Google Scholar]
- Sidar B., Jenkins B.R., Huang S., Spence J.R., Walk S.T., Wilking J.N. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip) Lab. Chip. 2019;19:3552–3562. doi: 10.1039/c9lc00653b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva T.P., Cotovio J.P., Bekman E., Carmo-Fonseca M., Cabral J.M.S., Fernandes T.G. Design principles for pluripotent stem cell-derived organoid engineering. Stem Cells Int. 2019;2019:1–17. doi: 10.1155/2019/4508470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simian M., Hirai Y., Navre M., Werb Z., Lochter A., Bissell M.J. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.-W., Seol Y.-J., Shrike Zhang Y., Shin S.-R., Zhao L., Aleman J., et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar-Scott M.A., Uzel S.G.M., Nam L.L., Ahrens J.H., Truby R.L., Damaraju S., Lewis J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019;5:eaaw2459. doi: 10.1126/sciadv.aaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza G.R., Molina J.R., Raphael R.M., Ozawa M.G., Stark D.J., Levin C.S., Bronk L.F., Ananta J.S., Mandelin J., Georgescu M.-M., et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat. Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spemann H., Mangold H. über Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Roux Arch. Entwmech. Org. 1924;100:599–638. [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebe T., Enomura M., Yoshizawa E., Kimura M., Koike H., Ueno Y., Matsuzaki T., Yamazaki T., Toyohara T., Osafune K., et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Tao T., Wang Y., Chen W., Li Z., Su W., Guo Y., Deng P., Qin J. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab. Chip. 2019;19:948–958. doi: 10.1039/c8lc01298a. [DOI] [PubMed] [Google Scholar]
- Thomas C.A., Tejwani L., Trujillo C.A., Negraes P.D., Herai R.H., Mesci P., Macia A., Crow Y.J., Muotri A.R. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell. 2017;21:319–331.e8. doi: 10.1016/j.stem.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. Cambridge University Press; 1917. On Growth and Form. [Google Scholar]
- Thress K.S., Paweletz C.P., Felip E., Cho B.C., Stetson D., Dougherty B., Lai Z., Markovets A., Vivancos A., Kuang Y., et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat. Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.K., Wang S., Smith D., Carlin A.F., Rana T.M. Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 2021;16:437–445. doi: 10.1016/j.stemcr.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchio A., Durmus N.G., Sridhar K., Mani V., Coskun B., El Assal R., Demirci U. Magnetically guided self-assembly and coding of 3D living architectures. Adv. Mater. 2018;30:1705034. doi: 10.1002/adma.201705034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Blauch L.R., Tang S.K.Y., Morsut L., Lim W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Frankel N.W., Lim W.A. Engineering cell–cell communication networks: programming multicellular behaviors. Curr. Opin. Chem. Biol. 2019;52:31–38. doi: 10.1016/j.cbpa.2019.04.020. [DOI] [PubMed] [Google Scholar]
- Turing A. The chemical basis of morphogenesis. Philos. Trans. R. Soc. B Biol. Sci. 1952;237:37–72. [Google Scholar]
- Uzel S.G.M., Amadi O.C., Pearl T.M., Lee R.T., So P.T.C., Kamm R.D. Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small. 2016;12:612–622. doi: 10.1002/smll.201501905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D., et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., Taylor A.C. Reconstitution of complete organs from single-cell suspensions of Chick embryos in advanced stages of differentiation. Proc. Natl. Acad. Sci. U S A. 1960;46:1177–1185. doi: 10.1073/pnas.46.9.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H.V. A new method by which sponges may Be artificially reared. Sci. New Ser. 1907;25:912–915. doi: 10.1126/science.25.649.912. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Cell to Cell Signalling. Elsevier; 1989. Positional information and prepattern in the development of pattern; pp. 133–143. [Google Scholar]
- Wörsdörfer P., Dalda N., Kern A., Krüger S., Wagner N., Kwok C.K., Henke E., Ergün S. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep. 2019;9:15663. doi: 10.1038/s41598-019-52204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Yin Y., Zheng Y., Ma Y., Li Y., Zhao Z., Guo J., Ai Z., Niu Y., Duan K., et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. 2020;577:537–542. doi: 10.1038/s41586-019-1875-y. [DOI] [PubMed] [Google Scholar]
- Yakavets I., Francois A., Benoit A., Merlin J.-L., Bezdetnaya L., Vogin G. Advanced co-culture 3D breast cancer model for investigation of fibrosis induced by external stimuli: optimization study. Sci. Rep. 2020;10:21273. doi: 10.1038/s41598-020-78087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Bejoy J., Xia J., Guan J., Zhou Y., Li Y. Neural patterning of human induced pluripotent stem cells in 3-D cultures for studying biomolecule-directed differential cellular responses. Acta Biomater. 2016;42:114–126. doi: 10.1016/j.actbio.2016.06.027. [DOI] [PubMed] [Google Scholar]
- Zahmatkesh E., Ghanian M.H., Zarkesh I., Farzaneh Z., Halvaei M., Heydari Z., Moeinvaziri F., Othman A., Ruoß M., Piryaei A., et al. Tissue-specific microparticles improve organoid microenvironment for efficient maturation of pluripotent stem-cell-derived hepatocytes. Cells. 2021;10:1274. doi: 10.3390/cells10061274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zwick S., Loew E., Grimley J.S., Ramanathan S. Mouse embryo geometry drives formation of robust signaling gradients through receptor localization. Nat. Commun. 2019;10:4516. doi: 10.1038/s41467-019-12533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Chen Y., Shao L., Xie M., Nie J., Qiu J., Zhao P., Ramezani H., Fu J., Ouyang H., et al. Airflow-assisted 3D bioprinting of human heterogeneous microspheroidal organoids with microfluidic nozzle. Small. 2018;14:1802630. doi: 10.1002/smll.201802630. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L., et al. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwi-Dantsis L., Wang B., Marijon C., Zonetti S., Ferrini A., Massi L., Stuckey D.J., Terracciano C.M., Stevens M.M. Remote magnetic nanoparticle manipulation enables the dynamic patterning of cardiac tissues. Adv. Mater. 2019;32:e1904598. doi: 10.1002/adma.201904598. [DOI] [PMC free article] [PubMed] [Google Scholar]