Summary
Doxorubicin is a commonly used chemotherapeutic drug, but its use is limited by doxorubicin-induced cardiotoxicity (DIC), which can lead to irreversible heart failure and death. A missense variant rs2229774 (p.S427L) in the retinoic acid receptor gamma (RARG) gene is associated with increased susceptibility to DIC, but the precise mechanism underlying this association is incompletely understood. We performed molecular dynamic simulations to determine the effect of this variant on RARG structure and then validated these predictions using CRISPR-Cas9-genome-edited, induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). We found that this variant leads to reduced activation of its target genes in response to doxorubicin, including gene pathways involved in DNA repair and consequently an inability to mediate DNA repair after exposure to doxorubicin. Our findings establish a role of RARG p.S427L in attenuating DNA repair in DIC and provide insight into the pathogenesis of this cardiotoxic effect.
Keywords: RARG, doxorubicin, pharmacogenomics, cardiomyocyte, iPSC, cardiotoxicity, DNA repair
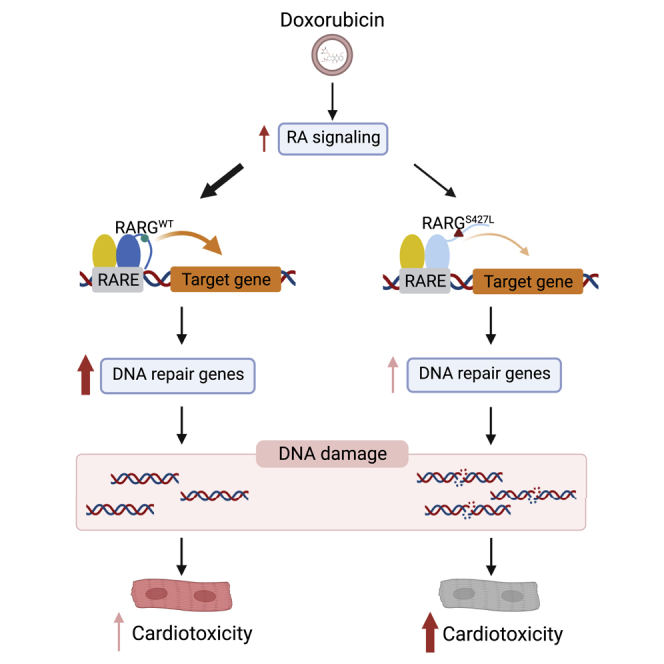
Graphical abstract
Highlights
-
•
RARG p.S427L is predicted to alter the stability of the C terminus of the protein
-
•
The RARG p.S427L variant has impaired ability to activate its target genes
-
•
This variant attenuates the DNA repair response to doxorubicin
In this report, Huang and colleagues show that the RARG p.S427L variant, associated with doxorubicin-induced cardiotoxicity, acts by reducing the activation of its target genes in iPSC-derived cardiomyocytes. This leads to attenuation of the DNA repair response to doxorubicin and increased cardiomyocyte cell death. These findings identify a mechanism by which this variant increases the risk for cardiotoxicity.
Introduction
Doxorubicin is a well-established and highly effective anthracycline chemotherapy drug. However, the use of doxorubicin is limited by dose-dependent and cumulative cardiotoxicity, leading to left ventricular dysfunction, arrhythmias, and heart failure (Singal et al., 1997; Volkova and Russell, 2011). The mechanisms of doxorubicin-induced cardiotoxicity (DIC) are incompletely understood, and we lack the ability to predict the risk of the cardiotoxic effect in individual patients.
Over 20 genetic association studies have identified genetic variants that are associated with the risk of DIC (Linschoten et al., 2018). Among these, a missense variant rs2229774 (p.S427L) in the retinoic acid receptor gamma (RARG) gene was identified in a genome-wide association study as being associated with increased risk of DIC (odds ratio [OR] = 4.7; p = 5.9 × 10−8; Aminkeng et al., 2015; Schneider et al., 2017). Induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) from individuals with DIC who harbor RARG-S427L have increased susceptibility to doxorubicin-induced double-strand DNA beaks, reactive oxygen species production, and cell death compared with isogenic control cells (Christidi et al., 2020). These findings suggest a direct and causal role of RARG-S427L in the pathogenesis of DIC. However, the specific molecular mechanisms by which RARG-S427L leads to DIC remain to be elucidated.
Here, we examined the functional consequences of RARG-S427L on retinoic-acid (RA)-responsive element (RARE) target gene expression in genome-edited iPSC-CMs. Our findings reveal a role for RARG-S427L in transcriptional response to doxorubicin in cardiomyocytes, leading to impaired activation of signaling pathways that are essential for protection against DIC.
Results
Substitution of serine to leucine in residue 427 decreases F domain stability of RARG
RARG is a ligand-dependent transcription factor that plays critical roles in various biological processes, including heart development, skeletal growth, and matrix homeostasis (di Masi et al., 2015; Wiesinger et al., 2021). RA receptors (RARs) form heterodimers with retinoid X receptors (RXRs) and bind to specific RARE on the promoter of their target genes (Rochette-Egly and Germain, 2009). RARs can either repress or activate target gene transcription depending on the ligation with RA (Farboud et al., 2003; Farboud and Privalsky, 2004). Unliganded RARs repress gene transcription, while liganded RARs can activate target genes under conformational changes of helix 12 that release co-repressor and promote co-activator binding (Cordeiro et al., 2019). Since RARG-S427L is located in the F-domain, we hypothesized that it may impact the ability of the adjacent helix 12 to mediate transcriptional activation or repression of doxorubicin-responsive pathways. To test this, we used molecular dynamic (MD) simulations to predict the consequences of RARG-S427L on RARG protein structure and function. The crystal structure of wild-type (WT) RARG was obtained from PDB: 2LBD (Renaud et al., 1995) and modified at residue 427 from serine to leucine. MD simulations were performed on both RARG-WT and RARG-S427L models that docked with all-trans retinoic acid (ATRA) as the ligand.
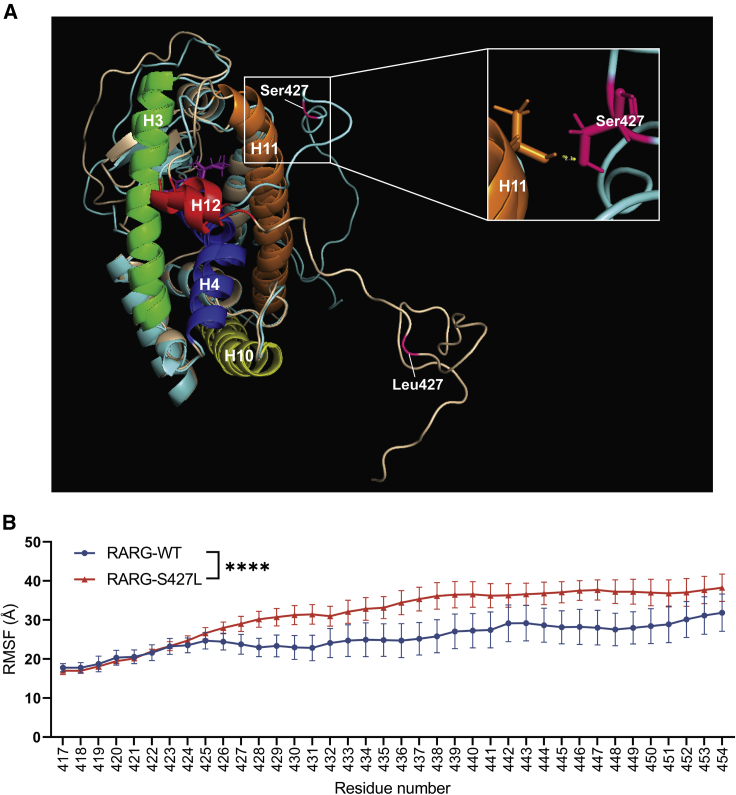
The predicted RARG protein structures were clustered from each model system (Figures S1A–S1D). Among these predicted structures, the representative structures with the highest cluster coverage in each RARG-WT and RARG-S427L system were selected and aligned for comparison. As shown in Figure 1A, the WT serine 427 in the F domain of RARG forms a hydrogen bond with threonine 399 in helix 11, maintaining the F domain in close proximity to the protein surface. However, in the presence of leucine 427, the F domain was more disordered and did not form hydrogen bonds with other protein regions, including helix 12. Comparing the F domain root-mean-square fluctuation (RMSF) between two model systems (Figures S1E and S1F), RARG-WT showed a significantly (p < 0.0001) lower average RMSF value than RARG-S427L starting at residue 422, which suggests that p.S427L impairs the stability of the F domain of RARG (Figure 1B). These observations indicate that RARG-S427L is predicted to decrease the stability of the F domain, which may impair the activation function of RARG.
Figure 1.
MD simulation of RARG with ligand
(A) Molecular-dynamics-predicted RARG structure with ATRA (purple), highlighting interaction between residue Thr399 (on helix 11) and Ser427 (on F domain) of RARG-WT. Protein structure of RARG-WT is shown in blue and RARG-S427L in yellow. Key components are helix 3 (green), helix 4 (deep blue), helix 10 (light yellow), helix 11 (orange), helix 12 (red), and the variant site 427 (pink).
(B) Cα RMSF of F domain (residues 417–454) of RARG-ATRA complex. MD experiments were run as independent triplicates. Analyses are shown as mean ± SEM; n = 3; ∗∗∗∗p < 0.0001; t test.
Doxorubicin treatment activates RA signaling in iPSC-CMs
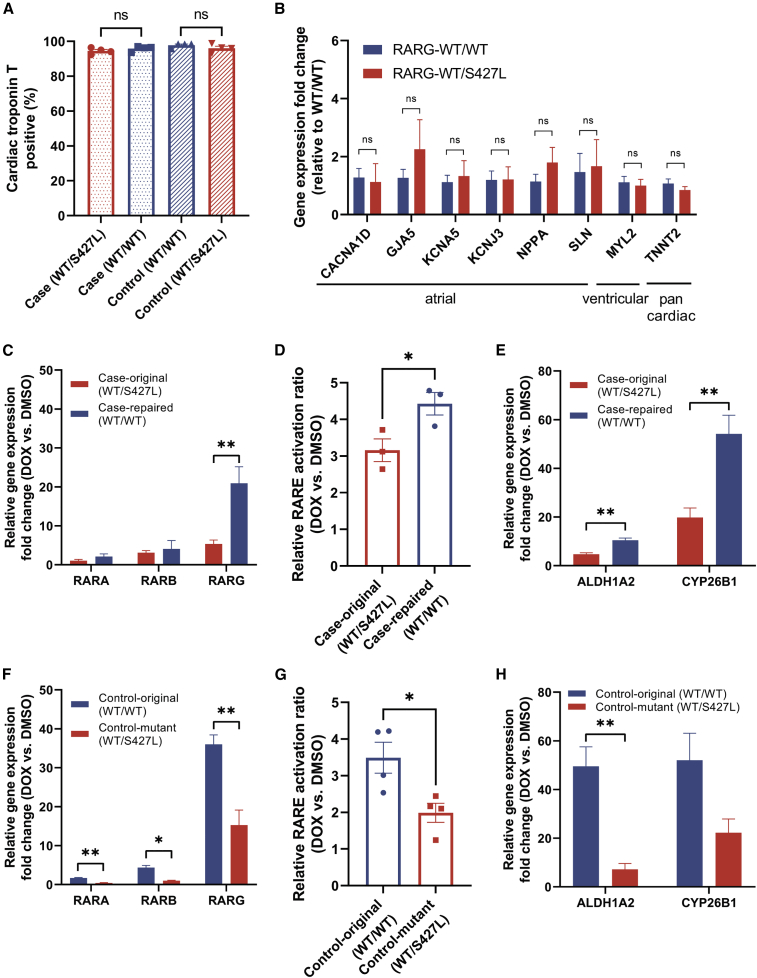
To further investigate the effect of RARG-S427L on the ability of RARG to activate target genes in response to doxorubicin, we generated iPSC-CMs from patients treated with doxorubicin who did (case) or did not (control) develop DIC and performed CRISPR-Cas9-mediated genome editing to correct or introduce the RARG-S427L variant (Christidi et al., 2020). All genotypes were validated by Sanger sequencing (Figure S2A). The iPSC-CMs displayed high levels of cardiac troponin T (cTnT) positivity (>95% of cell cTnT positive), with no significant differences between genotypes, indicating that the RARG variant did not impact differentiation efficiency (Figures 2A and S2B). We further characterized the iPSC-CMs by immunostaining and observed high levels of cTnT and sarcomeric alpha-actinin expression (Figure S2C). Because RARG has been implicated in cardiac development (Wiesinger et al., 2021), we examined the expression of atrial and ventricular markers in the iPSC-CMs by qPCR. We observed no difference in the expression of these transcripts between genotypes, suggesting that the RARG variant did not influence cardiac cell sub-type specification (Figure 2B).
Figure 2.
Assessment of retinoic acid signaling in cellular stress response to doxorubicin
(A) Flow cytometry for cardiac troponin T (cTnT), indicating the percentage of cardiomyocytes present in different differentiation batches (n = 4). ns, not significant.
(B) Gene expression of atrial, ventricular, and pan cardiac markers in two genotypes (n = 8).
(C–H) Gene expression of RA receptors RARA, RARB, and RARG measured by qPCR in (C) DIC-case and (F) DIC-control cell lines (n = 3). Luciferase assay results of (D) DIC-case (n = 3) and (G) DIC-control (n = 4) cell lines in response to doxorubicin are shown. Gene expression of RA signaling genes ALDH1A2 and CYP26B1 measured by qPCR in (E) DIC-case and (H) DIC-control cell lines (n = 3). All experiments are biological, independent replicates and are shown as mean ± SEM, ∗p < 0.05; ∗∗p < 0.01; t test.
To test which RAR sub-types play the dominant role in doxorubicin response, we measured gene expression of RARA, RARB, and RARG by qPCR in both DIC-case and control lines (Figures 2C and 2F). In response to 1 μM doxorubicin treatment in the DIC-case cell line, gene expression of RARA and RARB displayed minor differences between genotypes, whereas expression of RARG increased 21.0-fold in RARG-WT/WT versus 5.4-fold in RARG-WT/S427L iPSC-CMs (p = 0.009; Figure 2C). The same trend was seen in the DIC-control cell line, with 32.0-fold increase of RARG gene expression in RARG-WT/WT versus 15.3-fold increase in RARG-WT/S427L iPSC-CMs (p = 0.004; Figure 2F).
To further study the activity of RA signaling, we measured RARE activation by luciferase assay. RARE binding activity was increased in response to 1 μM doxorubicin compared with no treatment control in iPSC-CMs (Figure 2D). Genetic correction of RARG from S427L to WT in a DIC case led to higher activation of RARE (3.2-fold versus 4.4-fold activation in RARG-WT/S427L versus RARG-WT/WT; p = 0.04). Similarly, introducing the S427L variant in a DIC control cell line resulted in less RARE activation (3.5-fold versus 2.0-fold activation in RARG-WT/WT versus RARG-WT/S427L; p = 0.02; Figure 2G).
We next measured the gene expression of two key molecular targets in RA signaling, aldehyde dehydrogenase one family member 2 (ALDH1A2) and cytochrome P450 26B1 (CYP26B1), in response to 1 μM doxorubicin treatment (Figures 2E and 2H). The expression of both genes was increased upon doxorubicin treatment. ALDH1A2 expression was increased to a significantly greater extent in RARG-WT/WT than RARG-WT/S427L in DIC-case (10.5-fold versus 4.7-fold; p = 0.005) and DIC-control (49.6-fold versus 7.2-fold; p = 0.007) iPSC-CMs. CYP26B1 expression was induced to a significantly greater extent in iPSC-CMs from DIC cases with the RARG-WT/WT as compared with RARG-WT/S427L (54.2-fold versus 19.8-fold; p = 0.007). The difference was not statistically significant in DIC-control cells (52.0-fold versus 22.3-fold in RARG-WT/WT versus RARG-WT/S427L; p = 0.08). This finding indicates that increased RAR transcriptional activation activity in response to doxorubicin treatment is mainly modulated by increased expression of RARG and that the presence of RARG-S427L impairs the up-regulation of RARG and other key RA signaling genes in response to doxorubicin.
RARG-S427L leads to reduced target gene activation in response to doxorubicin
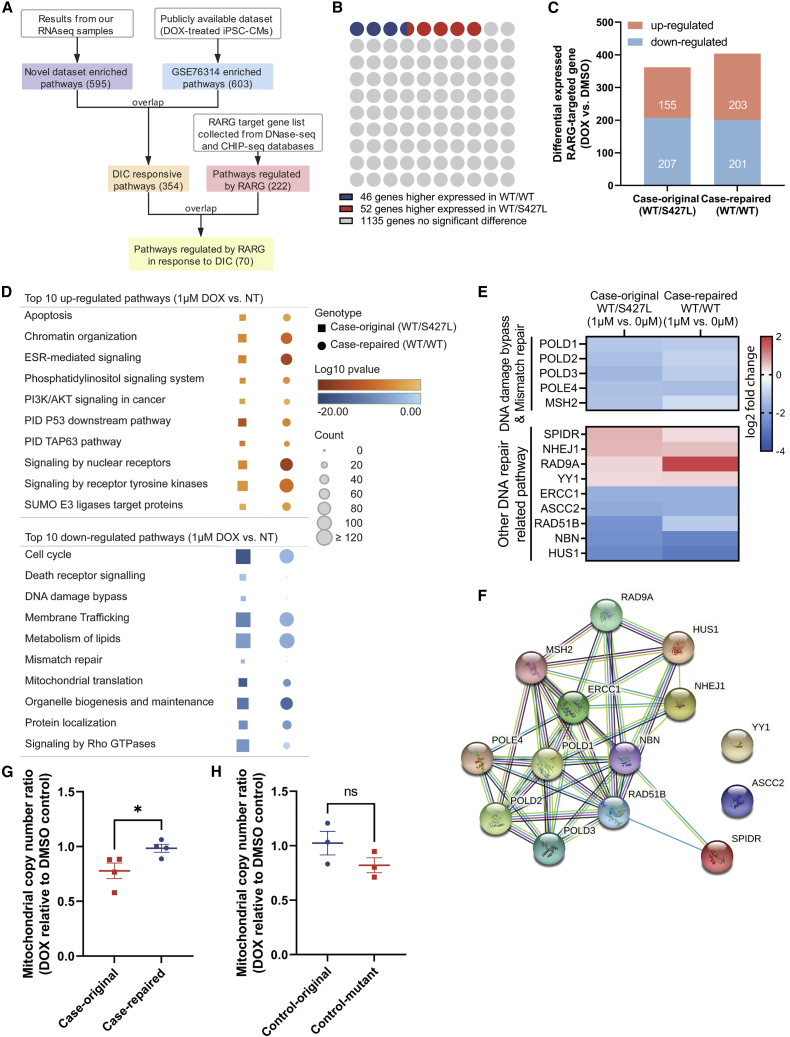
To more comprehensively assess the genes and pathways that are activated by RARG in response to doxorubicin, we performed RNA sequencing (RNA-seq) in DIC-case isogenic iPSC-CMs with and without RARG-S427L (Figure S3A). We co-analyzed a publicly available RNA-seq dataset (GEO: GSE76314) that was also generated from doxorubicin-treated, patient-specific iPSC-CMs for comparison (Burridge et al., 2016; Figure 3A). At baseline (vehicle control), less than 10% of RARG target genes were differentially expressed between RARG-WT/S427L and RARG-WT/WT (Figure 3B). When treated with 1 μM doxorubicin, 29.4% (362/1,233) of RARG target genes in RARG-WT/S427L and 32.8% (404/1,233) in RARG-WT/WT were significantly differentially regulated, with 42 more RARG target genes up-regulated in the WT cell line (Figures 3C, S3B, and S3C). We observed similar patterns of up-regulation by doxorubicin of selected genes in the control isogenic lines, as measured by qPCR (Figures S4A–S4E).
Figure 3.
Differentially expressed genes and pathways measured by RNA-seq in doxorubicin-treated iPSC-CMs
(A) Summary of enriched pathways and selection process. ChIP, chromatin immunoprecipitation; DOX, doxorubicin.
(B) Differentially expressed gene ratio comparing non-treated case-original and case-repaired samples.
(C) Summary of up- and down-regulated RARG target genes expression under doxorubicin treatment.
(D) Top 10 up and down-regulated pathways that targeted by RARG and responded to doxorubicin. ESR, estrogen receptor; NT, no treatment; PI3K, phosphatidylinositol 3-kinase.
(E) Heatmap of RARG-targeted gene expression in DNA-repair-related pathways.
(F) Protein association network analysis of 11 highlighted RARG-targeted, DNA-repair-related genes.
(G and H) Mitochondrial copy number ratio in iPSC-CM quantified by qPCR after doxorubicin treatment (normalized to DMSO control). All experiments are biological, independent replicates (n = 3-4) and are shown as mean ± SEM, ∗p < 0.05; ns, not significant; t test.
We performed over-represented pathway analysis of doxorubicin-induced differentially expressed genes and RARG-targeted genes (collected from GTRD and hTFtarget databases; Kolmykov et al., 2021; Zhang et al., 2020). Pathways represented in both our dataset and GEO: GSE76314 were considered as common pathways that respond to doxorubicin in cardiomyocytes. Overlapping pathways between DIC responsive and RARG targeted were then investigated in greater detail (Figure 3D). Within these pathways, signaling of nuclear receptor, estrogens-mediated signaling, and signaling by receptor tyrosine kinases were significantly more activated in RARG-WT/WT group, while genes in the pathways of DNA repair, mitochondria functions, and Rho guanosine triphosphatases (GTPases) signaling were more down-regulated in RARG-WT/S427L group. We further investigated the genes presented in DNA damage bypass and mismatch repair pathways that have predicted RARE-binding sites in their promoter regions as well as other RARG-targeted genes with over five RARE-binding sites in DNA-repair-related pathways (Figure 3E). The association network of these RARG-targeted, DNA-repair-related proteins is shown (Figure 3F). Since mitochondria-related pathway was more down-regulated in RARG-WT/S427L group, we also studied mitochondrial function between genotypes. Mitochondrial copy number was significantly increased in case RARG-WT/WT group (Figures 3G and 3H), but mitochondrial membrane potential between genotypes displayed no significant difference (Figures S4F and S4G). These findings indicate that S427L disrupts RARG target signaling pathway activation, potentially decreasing DNA repair activity following doxorubicin exposure.
The impact of RARG-S427L on doxorubicin-induced DNA damage and repair
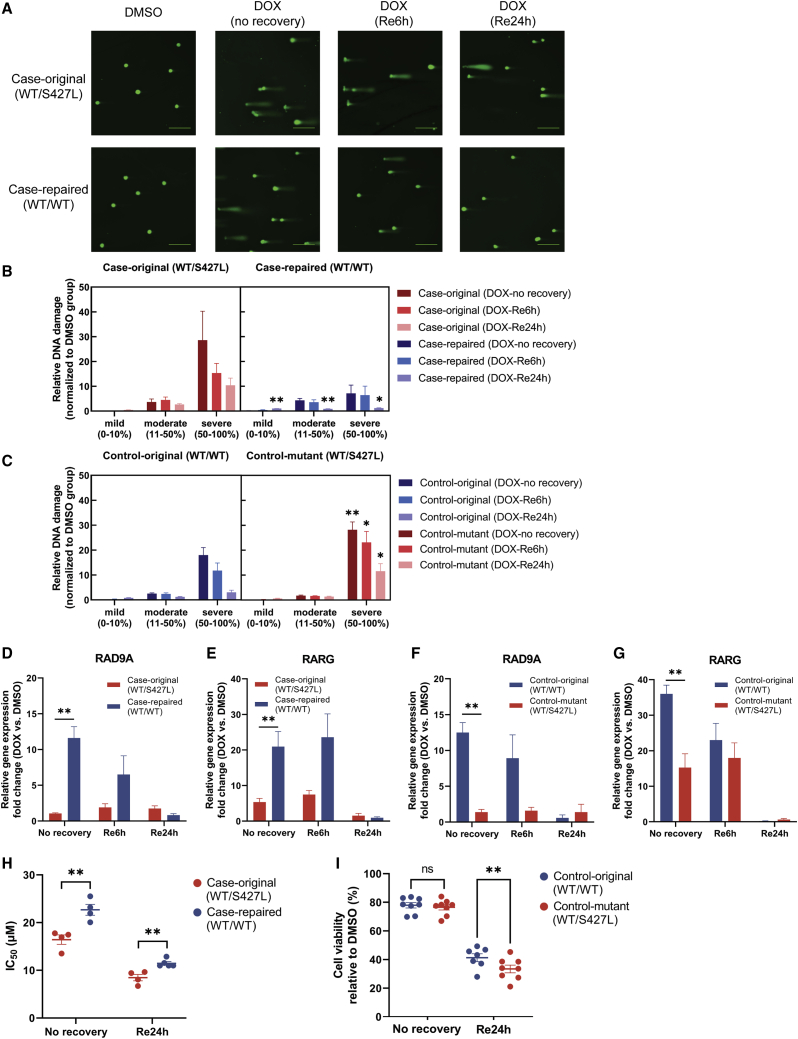
As our RNA-seq data implicated DNA repair pathways as being affected byimpaired target signaling due to RARG-S427L, we next set out to validate the functional significance of this observation by performing single-cell electrophoresis (comet assay) to investigate DNA breaks after doxorubicin treatment and recovery. DNA breaks were categorized as mild, moderate, or severe based on the tail DNA percentage (Figures 4A and 4B). After 24 h of 1 μM doxorubicin treatment in DIC-case iPSC-CMs, over 90% of the cells in RARG-WT/S427L and RARG-WT/WT showed a moderate or severe degree of DNA damage. With 6 and 24 h of recovery post-treatment, cell DNA damage levels were decreased in both genotypes. However, after 24 h of recovery, DNA damage levels remain significantly higher in RARG-WT/S427L compared with RARG-WT/WT: moderate (2.7-fold versus 0.86-fold; p = 0.009) and severe (10.4-fold versus 1.2-fold; p = 0.03). Similar results were seen in DIC-control isogenic lines with significantly higher severe level of DNA damage in RARG-WT/S427L in all time points (Figure 4C).
Figure 4.
Impact of RARG-S427L on doxorubicin-induced DNA damage and repair
(A) Comet assay results visualized under fluorescence microscope (scale bars represent 200 μm).
(B and C) Relative DNA damage fold change in RARG-WT/WT and RARG-WT/S427L assay (n = 3; 100–150 cells were quantified per sample in each independent experiment).
(D–G) Gene expression of DNA repair gene (D and F) RAD9A and (E and G) RARG with 6 and 24 h of recovery after doxorubicin treatment in DIC-case cell lines measured by qPCR (n = 3).
(H and I) IC50 and cell viability percentages after 24 h of doxorubicin treatment with no recovery time or with 24 h of recovery (n = 4–8). All experiments are biological, independent replicates and are shown as mean ± SEM; ∗p < 0.05; ∗∗p < 0.01; t test.
Finally, we assessed the expression of DNA repair gene RAD9A (RAD9 checkpoint clamp component A) with recovery post-treatment. RAD9A was identified in both GTRD and hTFtarget databases as having a high abundance of RARE-binding sites in its promoter region (Kolmykov et al., 2021; Zhang et al., 2020). Using qPCR, RAD9A showed 11.6-fold versus 1.1-fold (p = 0.003) increase in RARG-WT/WT versus RARG-WT/S427L after 24 h of 1 μM doxorubicin treatment in DIC case (Figure 4D). After 24 h of recovery, gene expression of RAD9A had decreased in RARG-WT/WT but maintained a similar expression level as 6-h recovery in RARG-WT/S427L. The expression pattern of RAD9A was similar to that of RARG, in that both transcripts remained elevated in RARG-WT/S427L compared with RARG-WT/WT after 24 h of recovery (Figure 4E). Similar expression trends in RAD9A and RARG were also seen in DIC-control isogenic lines (Figures 4F and 4G). In addition, cell viability analysis revealed that cardiomyocytes continued dying after removal of doxorubicin and half-maximal inhibitory concentration (IC50) dropped from 13–25 μM (no recovery) to 6–12 μM (24 h of recovery). Cell viability was higher in RARG-WT/WT after recovery for 24 h than RARG-WT/S427L in both DIC-case and control isogenic lines (Figures 4H and 4I). These results suggest that RARG-S427L leads to greater cell death as a result of an impaired ability to mediate in DNA repair.
Discussion
RARG-S427L is associated with an increased risk of DIC (Aminkeng et al., 2015). In our previous study, we identified that the presence of the RARG p.S427L variant is necessary and sufficient to increase susceptibility to doxorubicin-induced cardiotoxicity and that the presence of this variant was associated with an increase in double-stranded DNA breaks, reactive oxygen species production, and cell death in iPSC-derived cardiomyocytes (Christidi et al., 2020). Here, we used patient-specific iPSC-CMs to investigate the structural and functional effects of this genetic variant. The specific advances of the current study are (1) the identification of the predicted structural consequences of RARG-S427L using molecular dynamic simulations; (2) providing direct evidence that RARG-S427L decreases the binding affinity to target gene promotors; (3) performing global transcriptional analysis of RARG-S427L in response to doxorubicin treatment, which identified an inability to up-regulate genes involved in DNA repair in response to doxorubicin; and (4) providing evidence that RARG-S427L results in increased DNA damage in response to doxorubicin, which may underlie the increased susceptibility to doxorubicin-induced cell death. Collectively, these observations expand our understanding of the molecular mechanisms by which RARG protects from DIC by orchestrating a DNA repair response to doxorubicin and provides detailed insights into the pathogenesis of this adverse drug reaction.
RARs are transcription factors that can both activate or repress target genes in the presence or absence of ligands. However, RARG mainly plays an activation role of its target genes due to more stabilized contacts between helices 3 and 12 compared with isotype RARA (Farboud et al., 2003). Although deletion of the F domain of RARG was found to enhance co-activators binding in vitro (Farboud and Privalsky, 2004), we found the RAR activity was enhanced in RARG-WT/WT more than RARG-WT/S427L in response to doxorubicin treatment, which aligns with our prediction from MD results that the activation function of the protein is impaired in RARG-S427L.
We identified DNA damage and repair pathways as being among the most down-regulated pathways in RARG-WT/S427L cardiomyocytes, but these were not down-regulated in RARG-WT/WT cardiomyocytes (Figure 3D). Although not shown in the top regulated pathways, a subgroup of genes in the DNA repair pathway (R-hsa-73894) were significantly up-regulated in RARG-WT/WT (log p = −3.6), but not in RARG-WT/S427L. Among these genes, RAD9A is one of the top RARG-regulated genes that is activated in RARG-WT/WT and found to promote DNA damage repair through stimulating DNA polymerase beta binding at sites of DNA damage (Toueille et al., 2004). In addition to direct regulation effect of RARG on RAD9A, other key DNA repair genes, such as MSH2, ERCC1, and RAD51B, may also be affected via protein association networking with RAD9A (Figure 3F), indicating potential indirect effects of RARG on the DNA repair pathway through activating RAD9A expression. Combining these RNA-seq data with the results of lower RA singling activation and impaired ability to mediate DNA repair in RARG-WT/S427L, these results suggest that RARG-S427L attenuates the DNA repair response to doxorubicin in iPSC-CMs.
It is worth noting that, in addition to the DNA repair pathway we have highlighted, it is possible that RARG regulates other pathways that are also involved in DIC. We observed that mitochondrial number was reduced by doxorubicin to a greater extent in RARG-WT/S427L than RARG-WT/WT cardiomyocytes (Figures 3G and 3H), implicating this as another potential pathway by which RARG acts. We also specifically investigated the expression of genes involved in the apoptosis and p53 pathways and found that the degree of doxorubicin regulation was similar between genotypes (Figure 3D), which suggests that these are unlikely to be the major pathways by which the RARG variant acts. Similarly, although reactive oxygen species generation is an important component of DIC, we did not observe major changes in the regulation of genes in this process between genotypes. A recent study confirmed the important role of RARG-S427L in DIC (Magdy et al., 2021) and also reported an effect of RARG on DIC through suppression of topoisomerase 2β (TOP2B) expression and activation of the extracellular regulated kinase (ERK) pathway, highlighting that RARG may influence DIC through multiple pathways and mechanisms.
Our study has limitations that are worth noting. Firstly, iPSC-CMs are heterogeneous populations of cells with a mixture of atrial-, ventricular-, and nodal-like cardiac cells and express different transcriptional patterns across the differentiation stages (Karbassi et al., 2020). The heterogeneous cell population could introduce variability in the bulk RNA-seq and cellular assay we used. Also, our results were generated using two iPSC-CM lines, one from a DIC case and one from a DIC control, which both underwent genome editing to generate isogenic cell lines that differ only at the RARG-S427L variant. This approach has the advantage of allowing us to isolate the effect of the S427L variant by comparing each cell line with its genome-edited pair; however, it is also possible that RARG could have different effects in the context of different genetic backgrounds that may emerge when studying CMs from a larger number of iPSC lines.
In conclusion, we identify a mechanism by which RARG-S427L increases susceptibility to doxorubicin through reduced activation of the DNA repair pathway. These results expand our understanding of the molecular mechanisms of DIC and suggest specific cellular pathways that could be targeted for the prevention or treatment of DIC related to DNA repair. In addition, by uncovering a cellular mechanism linking RARG-S427L to DIC risk, our results highlight the opportunity to identify patients at greatest risk for this devastating adverse drug reaction by pharmacogenetic profiling of this variant prior to doxorubicin treatment.
Experimental procedures
A comprehensive description of the methodology is included in the supplemental information.
Cell culture and CRISPR-Cas9 genome editing
The iPSC lines used in this study were derived and described as part of a previous study (Christidi et al., 2020). In brief, we collected peripheral blood and performed re-programming to iPSCs from a patient who had experienced DIC (case) and a doxorubicin-treated individual who did not experience DIC (control). Then, we performed CRISPR-Cas9-mediated genome editing in the heterozygous case cell line (RARG-WT/S427L) to correct the genotype to homozygous WT. In the control cell line that was homozygous WT (RARG-WT/WT), we performed genome editing to introduce p.S427L in the heterozygous state (Figure S2A). All patient samples collection, process protocols, and experiments were approved by the University of British Columbia-Providence Health Care (UBC-PHC) Research Ethics Board.
RNA sequencing
iPSC-CMs were replated in 12-well plates and treated with 1 μM doxorubicin in the experiment group and vehicle (DMSO) in the control group for 24 h. Each condition has three replicates. Total RNA was extracted with illustra RNAspin Mini Isolation Kit (Cytiva) and used for library preparation following the standard protocol for the NEBnext Ultra ii Stranded mRNA (New England Biolabs) and sequencing that was performed on the NextSeq 500 (Illumina) with 30 million paired-end reads.
Statistics
All statistical analyses were conducted with GraphPad Prism 8.3. In graphs, values are expressed as mean ± SE of mean (SEM) and represent independent, biological repeats (at least three) unless otherwise stated. Statistical analyses were performed using Student’s t test when comparing two groups. p < 0.05 was considered statistically significant. Graphical abstract was created with BioRender.com.
Accession numbers
The accession number for the RNA sequencing datasets reported in this paper is GEO: GSE181517.
Author contributions
Conceptualization, H.H., E.C., S.S., M.K.D., G.F.T., and L.R.B.; methodology, H.H., E.C., S.S., and M.K.D.; investigation, H.H., E.C., and L.R.B.; formal analysis, H.H.; writing – original draft, H.H. and L.R.B.; funding acquisition, L.R.B.; supervision, L.R.B.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This study was funded by the Canadian Institute of Health Research (grant PJT153118 to L.R.B.). L.R.B. is a Michael Smith Foundation for Health Research Scholar and a Canada Research Chair in Precision Cardiovascular Disease Prevention.
Published: March 31, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.03.002.
Supplemental information
References
- Aminkeng F., Bhavsar A.P., Visscher H., Rassekh S.R., Li Y.L., Lee J.W., Brunham L.R., Caron H.N., van Dalen E.C., Kremer L.C., et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat. Genet. 2015;47:1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Li Y.F., Matsa E., Wu H.D., Ong S.G., Sharma A., Holmstrom A., Chang A.C., Coronado M.J., Ebert A.D., et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christidi E., Huang H.J., Shafaattalab S., Maillet A., Lin E., Huang K., Laksman Z., Davis M.K., Tibbits G.F., Brunham L.R. Variation in RARG increases susceptibility to doxorubicin-induced cardiotoxicity in patient specific induced pluripotent stem cell-derived cardiomyocytes. Sci. Rep. 2020;10:10363. doi: 10.1038/s41598-020-65979-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro T.N., Sibille N., Germain P., Barthe P., Boulahtouf A., Allemand F., Bailly R., Vivat V., Ebel C., Barducci A., et al. Interplay of protein disorder in retinoic acid receptor heterodimer and its corepressor regulates gene expression. Structure. 2019;27:1270–1285 e1276. doi: 10.1016/j.str.2019.05.001. [DOI] [PubMed] [Google Scholar]
- di Masi A., Leboffe L., De Marinis E., Pagano F., Cicconi L., Rochette-Egly C., Lo-Coco F., Ascenzi P., Nervi C. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Mol. Aspects Med. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Farboud B., Hauksdottir H., Wu Y., Privalsky M.L. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. Mol. Cell Biol. 2003;23:2844–2858. doi: 10.1128/Mcb.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B., Privalsky M.L. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol. Endocrinol. 2004;18:2839–2853. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- Karbassi E., Fenix A., Marchiano S., Muraoka N., Nakamura K., Yang X., Murry C.E. Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020;17:341–359. doi: 10.1038/s41569-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmykov S., Yevshin I., Kulyashov M., Sharipov R., Kondrakhin Y., Makeev V.J., Kulakovskiy I.V., Kel A., Kolpakov F. GTRD: an integrated view of transcription regulation. Nucleic Acids Res. 2021;49:D104–D111. doi: 10.1093/nar/gkaa1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linschoten M., Teske A.J., Cramer M.J., van der Wall E., Asselbergs F.W. Chemotherapy-related cardiac dysfunction A systematic review of genetic variants modulating individual risk. Circ. Genom. Precis. Med. 2018;11:e001753. doi: 10.1161/CIRCGEN.117.001753. [DOI] [PubMed] [Google Scholar]
- Magdy T., Jiang Z., Jouni M., Fonoudi H., Lyra-Leite D., Jung G., Romero-Tejeda M., Kuo H.H., Fetterman K.A., Gharib M., et al. RARG variant predictive of doxorubicin-induced cardiotoxicity identifies a cardioprotective therapy. Cell Stem Cell. 2021;28:2076–2089.e7. doi: 10.1016/j.stem.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J.P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl. Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.P., Shen F., Gardner L., Radovich M., Li L., Miller K.D., Jiang G., Lai D., O'Neill A., Sparano J.A., et al. Genome-wide association study for anthracycline-induced congestive heart failure. Clin. Cancer Res. 2017;23:43–51. doi: 10.1158/1078-0432.CCR-16-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal P.K., Iliskovic N., Li T., Kumar D. Adriamycin cardiomyopathy: pathophysiology and prevention. Faseb J. 1997;11:931–936. doi: 10.1096/fasebj.11.12.9337145. [DOI] [PubMed] [Google Scholar]
- Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M.O., Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova M., Russell R., 3rd Anthracycline cardiotoxicity: prevalence, pathogenesis and treatment. Curr. Cardiol. Rev. 2011;7:214–220. doi: 10.2174/157340311799960645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesinger A., Boink G.J.J., Christoffels V.M., Devalla H.D. Retinoic acid signaling in heart development: application in the differentiation of cardiovascular lineages from human pluripotent stem cells. Stem Cell Rep. 2021;16:2589–2606. doi: 10.1016/j.stemcr.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu W., Zhang H.M., Xie G.Y., Miao Y.R., Xia M.X., Guo A.Y. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genom. Proteom. Bioinf. 2020;18:120–128. doi: 10.1016/j.gpb.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.