Abstract
Objective
Recent studies have detailed the genomic landscape of endometrial cancer (EC); however, no study has focused on genetic alterations in advanced EC. We performed genomic profiling of patients with advanced EC using targeted next-generation sequencing (NGS).
Methods
Archival tissue samples from 21 patients diagnosed with stage III and IV EC were obtained and subjected to NGS. Our data and the cancer genome atlas dataset were combined, and somatic mutation patterns were analyzed and compared according to the stage and histological type. Additionally, survival effects of specific mutated genes were analyzed.
Results
Somatic mutation patterns of 38 genes were identified in 263 EC samples, and the most commonly mutated genes were PTEN and PIK3CA. PTEN was the most common in endometrioid histology, while PPP2R1A was the most commonly mutated gene in serous histology. The mutation rates of PPP2R1A and TP53 were significantly higher in advanced EC sample than in stage I samples (22.5% vs. 4.3% [p<0.001] and 8.4% vs. 1.4% [p=0.021], respectively). Survival analysis of the total population and endometrioid subgroup revealed that patients with PPP2R1A mutations had significantly shorter survival than did those without mutations (p=0.005 and p<0.001, respectively).
Conclusion
PPP2R1A mutations might have a role in dismal prognosis of advanced EC.
Keywords: Endometrial Cancer, Survival Analysis, PPP2R1A, TP53
Synopsis
Both PPP2R1A and TP53 mutations were significantly higher in stage III/IV than in stage I EC samples. Mutated PPP2R1A and TP53 were associated with decreased survival rate. PPP2R1A and TP53 mutations might contribute to poor oncological outcomes in patients with stage III/IV EC.
INTRODUCTION
Endometrial cancer (EC) is the most common female genital tract cancer in Western countries. In Korea, the incidence of newly diagnosed ECs is progressively increasing [1]. Most cases are early stage endometrioid ECs that have favorable outcomes, but advanced ECs have a dismal prognosis. By definition, advanced ECs are classified as International Federation of Gynecology and Obstetrics (FIGO) stage III-IV ECs that comprise approximately 20% of all EC cases. Advanced EC is a substantially heterogeneous disease entity with multiple risk factors and highly diverse clinical outcomes. For example, within the same FIGO stage, patients with EC involving multiple metastatic sites have worse outcomes than those with EC involving a solitary site [2,3]. In addition to clinicopathological characteristics, molecular and genomic profiling might reveal the pathogenesis and natural history of advanced ECs.
Cancer spread, including distant metastasis, is considered to be deriven from the accumulation of somatic mutations followed by choosing the fittest clones, eventually resulting in metastatic spread [4]. Somatic mutations play a crucial role in cancer growth, invasion, and metastasis and thus, induce cancer progression. Most cancer treatment guidelines are based on somatic mutations in cancer-related genes. And identification of genomic changes and molecular markers in advanced EC could lead to more tailored therapies. Various molecular markers and pathways that may be potential therapeutic targets have been explored. These include, but are not limited to, the PTEN and PI3K/AKT/mTOR pathway [5,6,7,8], KRAS/BRAF/NRAS (Ras/Raf pathway) [9], homologous recombination deficiency (HRD) pathway [10,11], mismatch repair deficiency [12], ARID1A [13,14], FGFR2 [15], HER2/neu [16], and p53 [17,18]. However, data regarding molecular and genomic profiling, specifically focusing on advanced ECs, are lacking. In the Cancer Genome Atlas (TCGA) data published in 2013, almost three-fourths of the enrolled cases had early-stage EC [19]. Furthermore, in TCGA, no comparative analysis was performed between early stage and advanced ECs.
In this study, we examined the genomic landscape of patients with advanced EC using targeted next-generation sequencing (NGS). Additionally, we combined TCGA data and our data to compare mutational patterns between early stage and advanced EC cases, as well as within advanced EC cases. Furthermore, we assessed specific somatic mutations to determine the differential survival outcomes between early stage and advanced ECs.
MATERIALS AND METHODS
1. Study population and data collection
Formalin-fixed paraffin-embedded (FFPE) tumors from 21 patients diagnosed with advanced EC between 2010 and 2017 at our hospital (Korea University Medical Center; KUMC) were included in this study. Written informed consent was obtained from all patients. Patients who underwent hysterectomy with bilateral salpingo-oophorectomy and pelvic and/or para-aortic lymphadenectomy and were diagnosed with FIGO stage III and IV were considered eligible. Tumors with histological subtypes such as neuroendocrine, dedifferentiated, undifferentiated, or mesonephric-like EC were excluded. Clinicopathological data, including patient age, date of operation, last follow-up or date of death, and tumor stage, grade, and histological type, were collected from electronic medical records in our hospital. This study was approved by our Institutional Review Board (Korea University Guro Hospital Institutional Review Board [KUGH17282-001]). All methods and processes of this study were carried out in accordance with relevant guidelines and regulations.
2. Nucleic acid isolation and library preparation and sequencing
DNA was extracted from FFPE tumors using a total DNA extraction kit (RecoverALL, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. The Oncomine Comprehensive Assay™, which comprises the DNA Oncomine™ Focus Assay (Thermo Fisher Scientific) and RNA Oncomine™ Fusions assay (Thermo Fisher Scientific), designed to examine 143 oncogenes and tumor suppressor genes covering single nucleotide variants (SNVs), copy number alterations, insertions and deletions (INDELs), and fusions was used. DNA libraries were prepared using the Ion AmpliSeq™ Library Kit 2.0 (Thermo Fisher Scientific) following the manufacturer's instructions. The purified libraries were quantified using an Ion Universal Quantitation Kit. The kit contained Escherichia coli DH10B DNA as the standard for the quantification of libraries. The concentration of final library molecules was 50 pM, which is appropriate for downstream template preparation. Template was prepared using the Ion Chef System (Thermo Fisher Scientific). Sequencing was performed using the Ion Torrent S5XL Machine platform (Thermo Fisher Scientific) with an Ion 540 Chip Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
3. Data analysis
Analysis of raw sequencing data was performed using the Torrent Suite software version 5.10.1 (Thermo Fisher Scientific) with default analysis parameters. Variant Caller version 4.4.2.2 and Coverage Analysis version 4.4.3.3 plug-ins (Thermo Fisher Scientific) were used for variant calling and sequencing coverage analysis, respectively. The parameters applied to achieve adequate sequencing quality were as follows: mean sequencing depth, 1,200× and 250×; coverage, 90%; and total mapped fusion panel reads ≥20,000. Variant calling was performed using Ion Reporter software v5.4 with manufacturer-recommended settings. An allelic frequency of approximately 5% was used as the cutoff for variants (SNV, INDEL). Gene copy number estimation was performed for variants with a mean absolute percent difference score of >0.5. An average copy number of ≥4 indicated gain or amplification. Regarding translocations, ≥50,000 read counts per million were interpreted as positive results.
4. Statistical analysis and data visualization
Somatic mutation data were downloaded from TCGA, and all analyses were performed using R package version 4.0.3 [20]. Analyses were performed using R package TCGAbiolinks and GDCquery function with the following parameters: project, TCGA-UCEC; data category, simple nucleotide variation; data type, annotated somatic mutation; and workflow type, MuTect2 annotation. We processed the data in a mutation annotation format and merged our sequencing data by genomic location. Reference genome hg19 was used for the analysis. As TCGA-UCEC somatic mutation data were referenced to genome hg38, we used ‘liftOver’ function to convert hg38 to hg19. The mutation landscape, including SNVs, INDELs, and mutational burden, across our samples and TCGA were plotted using Genomic Visualization in R.
5. Variant interpretation
All the genetic alterations were interpreted using a four-tier system according to the standards and guidelines for the interpretation and reporting of sequence variants in cancer: tier I, variants of strong clinical significance such as biomarkers with FDA-approved therapies included in professional guidelines or those with proven diagnostic, prognostic or therapeutic effects in well-powered studies; tier II, variants of potential clinical significance such as biomarkers that predict response or resistance to FDA-approved therapies for a different tumor type or investigational therapies; tier III, variants of unknown clinical significance; and tier IV, benign or likely benign variants [21]. Variants corresponding to tier IV (benign or likely benign) have not been reported.
RESULTS
1. Mutation landscape of combined dataset
A total of 21 patients from the KUMC and 530 samples from TCGA were available for somatic mutation analysis. We selected 242 of 530 samples from TCGA dataset that were histologically classified as endometrioid or serous EC. Thereafter, we merged the two somatic mutation datasets and retrieved data of 86 alleles in 38 genes with mutations common between the KUMC and TCGA datasets (Fig. S1). 21 patients from KUMC and 242 samples from TCGA (total cases=263) were included in somatic mutation analysis. The clinical and pathological characteristics of 21 patients from the KUMC are summarized in Table S1.
Of the 242 samples from TCGA, 158 were stage I EC, 52 were stage III EC, and 10 were stage IV EC. Regarding the histological type, 206 samples were endometrioid EC and 36 were serous EC. Considering stage I samples, 144 were endometrioid EC and 14 were serous EC, and considering stages III/IV samples, 40 were endometrioid EC and 22 were serous EC.
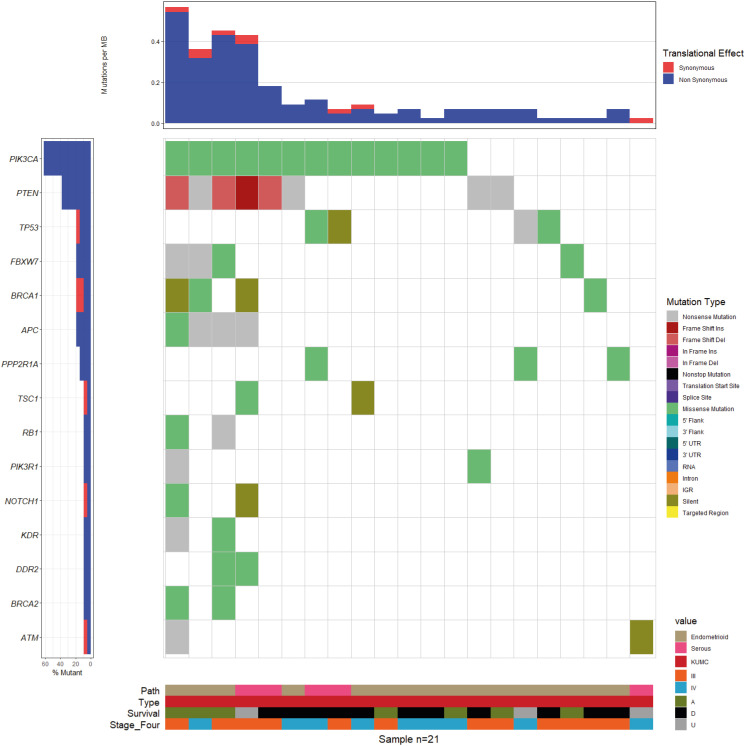
The distribution of somatic mutations in tumor samples of 21 patients from our institution is presented in Fig. 1. Targeted NGS analysis produced a mean of 9,503,261 reads, 97.84% of which were mapped on the reference genome hg19. The mean coverage depth per sample was 3,809×, and the percent base reads on target was 92.80%. After filtering out the processes, we identified 129 somatic variants (Table S2). Somatic variants were classified according to their effect on transcripts as missense (67.4%), nonsense (15.5%), silent (7.0%), frameshift deletion (7.0%), frameshift insertion (1.6%), 3′ untranslated region (0.8%), and in-frame deletion (0.8%). The amino acid changes reported for endometrioid or serous carcinoma of the endometrium primary site in the COSMIC (http://cancer.sanger.ac.uk/cosmic) database were compared with our results to confirm the pathogenicity of somatic mutations. In TCGA samples, more pathogenicity was observed in serous (92.31%; 72 of 78) than in endometrioid samples (81.82%; 459 of 561) (p=0.02281). Among the somatic mutations identified, 531 of 639 (83.10%) in TCGA samples and 93 of 129 (72.09%) in the KUMC samples were found to be pathogenic. In the 21 patients from KUMC, PTEN (36 of 129) was the most mutated gene followed by PIK3CA (25 of 129). INDEL and copy number variation (CNV) patterns were detected, and provided as Table S3.
Fig. 1. The somatic mutation landscape of 21 patients from Korea University Medical Center (KUMC) was depicted as waterfall plot. The upper panel demonstrates the mutational burden per megabase DNA (MB) for each patient. The left panel indicates the percentage of mutation of 21 patients in each 15 genes. The cut-off of indicated mutation was 0.05, so the mutation found in at least two of the 21 patients to be displayed in this plot. The lower panel separated into four sections, and each sections indicates pathological type (16 endometrioid and 5 serous) sample source (21 KUMC), survival status (A; 7 alive, D; 11 dead, U; 3 unknown), and two FIGO stages (12 stage III and 9 stage IV).
2. Somatic mutation patterns by stage
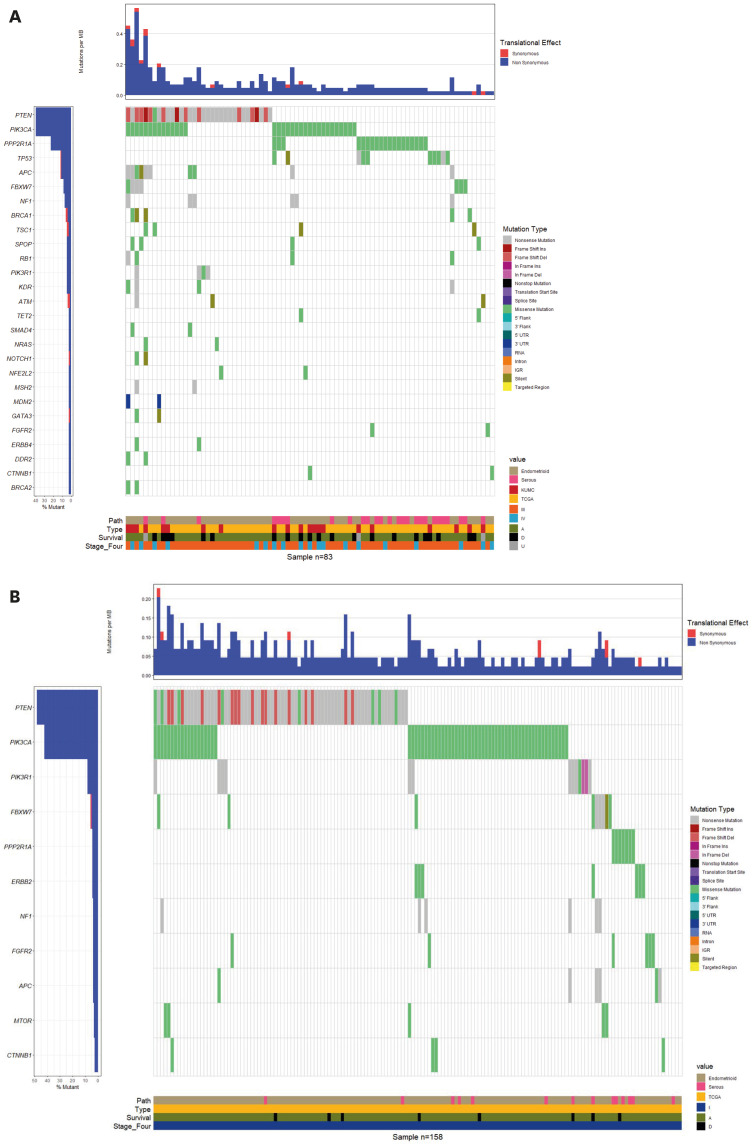
To investigate the differential somatic mutation patterns in advanced EC (FIGO stage III or IV), we categorized all samples into FIGO stages I and III/IV and visualized their distribution on a waterfall plot (Fig. 2). A total of 391 somatic mutations were found in 158 samples classified as FIGO stage I (Fig. 2B), and 313 somatic mutations in 83 samples classified as FIGO stage III/IV (Fig. 2A). After selecting only “Pathogenic” mutations by NGS, no significant difference was found in the number of somatic mutations between the FIGO stages I and III/IV groups (p=0.1761): 324 of 391 mutations in stage I (82.9%) versus 246 of 313 mutations in stage III/IV (78.6%). However, the number of serous EC samples in the FIGO stage III/IV groups was greater than that in stage I group (p<0.00001): stage I, 14 of 158 (9.0%) samples and stage III/IV, 27 of 83 (32.5%) samples. Regarding the frequency of somatic mutations in stage III/IV samples, the two most commonly mutated genes were PTEN and PIK3CA. The frequencies of PTEN mutations in stages I and III/IV samples were 46.3% (182 mutations) and 33.5% (105 mutations), respectively. The frequencies of PIK3CA mutations in stages I and III/IV were 27.6% (108 mutations) and 17.9% (56 mutations), respectively. The number of somatic mutations of PPP2R1A observed in stage III/IV samples were 19 (stage III: 14 and stage IV: 5, the third most common mutation) and in stage I samples were seven.
Fig. 2. The somatic mutation landscape of each patient was depicted as waterfall plot. (A) The left panel indicates the percentage of mutation of total 83 patients in each 27 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least two of the 83 patients to be displayed in this plot. The lower panel was separated into four sections, and each sections indicates that pathological type (61 endometrioid and 22 serous), sample source (21 KUMC and 62 TCGA), survival status (A; 59 alive, D; 21 dead, U; 3 unknown), and two FIGO stages (64 stage III and 19 stage IV). (B) The left panel indicates the percentage of mutation of total 158 patients in each 11 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least four of the 158 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (144 endometrioid and 14 serous), sample source (158 TCGA), survival status (A; 150 alive, D; 8 dead), and FIGO stage (158 stage I). The detailed gene mutation type was indicated to a unique color. The upper panel demonstrates the mutational burden per megabase DNA (MB) for each patient.
KUMC, Korea University Medical Center; TCGA, the cancer genome atlas.
3. Somatic mutation patterns by histological type
We investigated the differences in somatic mutation patterns between the endometroid and serous types. Hence, we compared stage III/IV ECs with stage I ECs according to the endometrioid (Fig. S2) and serous types (Fig. S3) on waterfall plots. In endometrioid type, 227 and 363 somatic mutations were identified in 56 patients with stage III/IV ECs (Fig. S2A), and in 144 with stage I ECs (Fig. S2B), respectively. In serous type, 86 and 28 somatic mutations were identified in 27 patients with stage III/IV ECs (Fig. S3A), and 14 with stage I ECs (Fig. S3B), respectively. In endometroid type, PTEN and PI3KCA were the two most commonly mutated genes, whereas PPP2R1A mutations were most common in the serous type. TP53 mutations were the second most frequently observed mutations in patients with serous histology and stage III/IV EC (Fig. S3A).
Specifically, the number of somatic mutations in PTEN and PIK3CA genes in the endometrioid type were 23 (24.7%) and 16 (17.2%) and in the serous type were 13 (36.1%) and 9 (25.0%), respectively. Thereafter, analysis of somatic mutations in TCGA samples (n=62) of stage III/IV EC revealed 182 somatic mutations. In the endometroid type, 132 somatic mutations in PTEN and PIK3CA, with frequency in the order mentioned, were found, which is identical to the KUMC results. In the serous type, the PPP2R1A gene had the highest mutation frequency (52.0%; 26 of 50), followed by PIK3CA (16.0%), TP53 (12.0%), and PTEN (10.0%) genes. The somatic mutation patterns within the same histological type were similar in the KUMC and TCGA datasets.
4. Confusion matrix by top 8 genes and stages
A Fisher’s exact test was used to calculate the odds ratios (ORs) and p-values of the eight frequently mutated genes (PTEN, PIK3CA, PPP2R1A, PIK3R1, FBXW7, FGFR2, TP53, and CTNNB1) (Table 1). PPP2R1A had a significantly higher mutation frequency in stage III/IV samples than in stage I samples (16 vs. 6, OR=6.05, p<0.001). In the endometrioid type, PPP2R1A mutation rate was marginally higher in stage III/IV samples than in stage I samples (4 vs. 2, OR=5.462, p=0.053). The mutation rate of TP53 was significantly higher in stage III/IV samples than in stage I samples (6 vs. 2, OR=6.078, p=0.021). The mutation frequency of PIK3CA was significantly higher in stage I samples than in stage III/IV samples (49 vs. 14, OR=0.451, p=0.020), unlike that of PPP2R1A.
Table 1. Comparison of mutation frequencies of 8 most commonly mutated genes between stage III/IV and stage I endometrial cancers.
| Gene | Total, stage I | Total, stage III/IV | p-value | Odds ratio | Endometrioid, stage I | Endometrioid, stage III/IV | p-value | Odds ratio | Serous, stage I | Serous, stage III/IV | p-value | Odds ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTEN | 63 | 27 | 0.327 | 0.727 | 61 | 24 | 1.000 | 1.020 | 2 | 3 | 1.000 | 0.783 |
| PIK3CA | 49 | 14 | 0.020 | 0.451 | 45 | 9 | 0.034 | 0.421 | 4 | 5 | 0.694 | 0.595 |
| PPP2R1A | 6 | 16 | 0.000 | 6.050 | 2 | 4 | 0.053 | 5.462 | 4 | 12 | 0.329 | 2.143 |
| PIK3R1 | 8 | 0 | 0.053 | 0.000 | 7 | 0 | 0.194 | 0.000 | 1 | 0 | 0.350 | 0.000 |
| FBXW7 | 3 | 5 | 0.128 | 3.312 | 2 | 3 | 0.135 | 4.019 | 1 | 2 | 1.000 | 1.083 |
| FGFR2 | 5 | 1 | 0.667 | 0.373 | 4 | 1 | 1.000 | 0.636 | 1 | 0 | 0.350 | 0.000 |
| TP53 | 2 | 6 | 0.021 | 6.078 | 1 | 2 | 0.190 | 5.296 | 1 | 4 | 0.640 | 2.364 |
| CTNNB1 | 1 | 2 | 0.273 | 3.877 | 1 | 2 | 0.190 | 5.296 | 0 | 0 | 1.000 | NA |
| Sum | 158 | 83 | 144 | 56 | 14 | 26 |
NA, not applicable.
5. Survival plot, univariate, and multivariate analysis by somatic mutations
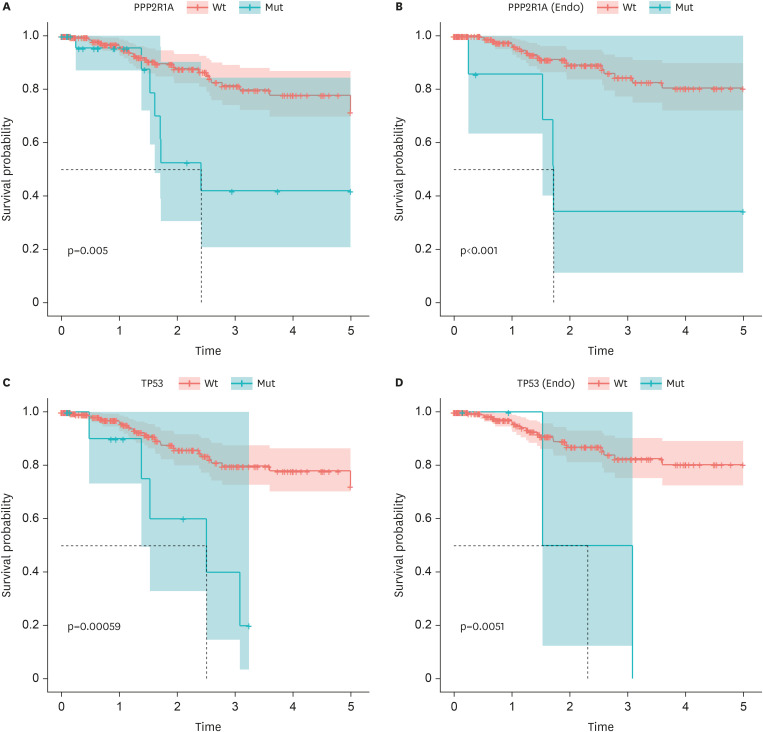
Thereafter, using Kaplan–Meier survival plots, we investigated which somatic mutations of the eight most frequently mutated genes affected survival outcome. Each survival analysis included all patients from the KUMC and TCGA dataset. Moreover, survival analyses according to the histological type (endometrioid) were performed (Fig. 3). First, survival analysis was performed according to PPP2R1A mutations, which showed a significantly higher rate in patients with stage III/IV EC than in patients with stage I EC. The survival proportion was significantly lower in patients with PPP2R1A mutations than in those with wild-type PPP2R1A (Fig. 3A; 73.1% vs. 88.6%, p=0.005). Notably, the negative prognostic effect of PPP2R1A mutations was more prominent with endometrioid EC than with serous EC (Fig. 3B; 41.9% vs. 91.2%, p<0.001). The survival proportion was significantly lower with TP53 mutations than with wild-type TP53 in all patients and those with endometrioid EC (Fig. 3C and D; 61.5% vs. 88.4%, 50.0% vs. 90.4%, p=0.00059 and 0.0051, respectively). However, because the proportion of patients with TP53 mutation was extremely low, it was difficult to analyze each histological type separately.
Fig. 3. Kaplan-Meier survival plots of PPP2R1A and TP53 mutation. (A) Of 263 patients, 26 patients had the PPP2R1A mutation. 27 patients out of 237 with PPP2R1A wild-types and 7 out of 26 with the PPP2R1A mutations were dead. (B) Among 222 patients with endometrioid types, 7 patients had the PPP2R1A mutations. 19 patients out of 215 with PPP2R1A wild-types and 4 out of 7 with PPP2R1A mutations were dead. (C) Of 263 patients, 13 patients had the TP53 mutations. 29 patients out of 250 with TP53 wild-types and 5 out of 13 with TP53 mutations were dead. (D) Out of 222 patients with endometrioid, 4 patients had the TP53 mutations. 21 patients out of 218 with TP53 wild-types and 2 out of 4 with TP53 mutations were dead.
We performed univariate and multivariate Cox regression analyses to determine the prognostic effect of clinicopathological factors (age, stage, grade, and histologic type) and the eight most frequently mutated genes on survival outcomes. In the univariate analysis, stage III/IV, grade 3, and the presence of PPP2R1A and TP53 mutations were significantly associated with decreased survival rate. However, only stage III/IV was found to be an independent prognostic factor for decreased survival proportion in the multivariate analysis (Table 2).
Table 2. Univariate and multivariate analysis of clinicopathological factors and 8 most frequently mutated genes for survival.
| Parameters | HR (95% CI) | p-value | |
|---|---|---|---|
| Univariate analysis | |||
| Age | 1.015 (0.984–1.047) | 0.351 | |
| STAGE III/IV | 4.727 (2.219–10.068) | <0.0001 | |
| Grade 3 | 2.397 (1.029–5.583) | 0.043 | |
| Histology Serous | 2.089 (0.929–4.694) | 0.075 | |
| PTEN | 0.574 (0.27–1.219) | 0.149 | |
| PIK3CA | 0.903 (0.442–1.845) | 0.779 | |
| PPP2R1A | 3.174 (1.289–7.818) | 0.012 | |
| PIK3R1 | 0.507 (0.118–2.179) | 0.362 | |
| FBXW7 | 0.449 (0.061–3.308) | 0.432 | |
| FGFR2 | 2.171 (0.291–16.189) | 0.449 | |
| TP53 | 3.983 (1.38–11.494) | 0.011 | |
| CTNNB1 | 1.077 (0.146–7.926) | 0.942 | |
| Multivariate analysis | |||
| STAGE III/IV | 3.303 (1.418–7.691) | 0.006 | |
| Grade 3 | 1.455 (0.583–3.63) | 0.422 | |
| PPP2R1A | 1.76 (0.661–4.688) | 0.257 | |
| TP53 | 2.017 (0.65–6.26) | 0.225 | |
CI, confidence interval; HR, hazard ratio.
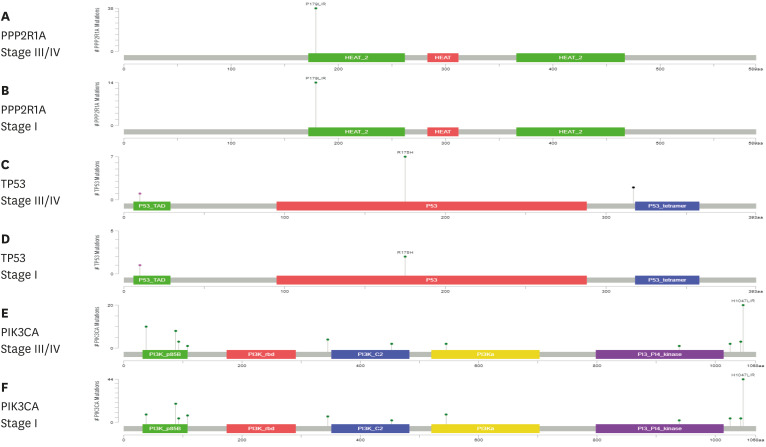
The lollipop plots of the PPP2R1A, TP53, and PIK3CA genes are presented in Fig. 4. The X-axis represents each domain located in the gene. The Y-axis represents the frequency of the mutations. In PPP2R1A, only one P179L/R mutation in the HEAT_2 domain was found. In TP53, the R175H mutation was found in stages I and III/IV samples, whereas the mutation located upstream of the P53 tetramer domain was found only in stage III/IV samples.
Fig. 4. The distribution and loci of mutations of 3 genes (PPP2R1A, TP53, and PIK3CA) in endometrial cancer (stage III/IV versus I) were depicted. Diagrams represent the protein domain of each genes. The presence of a mutation is shown on the x axis as lollipop, and the frequency of mutation is shown on the y axis. Missense mutations are presented as green circles, and other mutation types were presented as pink circles. Plots were generated using cBioPortal (www.cBioPortal.org).
DISCUSSION
In this study, we conducted genomic profiling in patients with FIGO stage III/IV EC using targeted NGS technology and compared the somatic mutation pattern between patients with FIGO stages I and III/IV EC. We merged 21 stage III/IV EC samples from KUMC with histological-type (either endometrioid or serous) labeled samples from TCGA. Consequently, PPP2R1A and TP53 were found to be more frequently mutated in stage III/IV EC samples than in early-stage EC samples. To the best of our knowledge, this is the first study to report the genomic landscape focused on stage III/IV EC and to compare somatic mutation patterns between stages I and III/IV ECs. In addition, we demonstrated differential mutation patterns by histological subtypes and survival data according to specific gene mutations.
PPP2R1A encodes the Aα subunit of PP2A, which is one of the four major serine/threonine phosphatases [22]. PP2A is involved in a variety of cellular pathways such as cell growth and survival and has been implicated as a tumor suppressor [23]. PPP2R1A mutations occur in type I EC, but at much lower frequencies (2.5%–6.9%) [24]. As expected, the PPP2R1A mutation rate was higher in serous type EC samples than in endometrioid EC samples in our study, which is consistent with findings of previous studies [25,26]. However, a significantly higher mutation rate of PPP2R1A was detected in stage III/IV EC samples than in stage I EC samples in our study, which is a novel finding. In line with this finding, Kaplan–Meier survival analyses revealed that the 5-year survival rate was significantly lower in patients with PPP2R1A mutations than in those without mutations. Consistent with our findings, the cBioportal reported a 5-year survival rate of 50% for patients with serous EC harboring PPP2R1A mutations compared with 80% for those without PPP2R1A mutations [27]. Our findings suggest that PPP2R1A mutations contribute to poor prognosis in patients with stage III/IV EC regardless of the histological subtype.
TP53 is a tumor suppressor gene. TP53 mutations are highly prevalent in so-called “high copy number” subtypes characterized by serous and high grade endometrioid histology. The cBioportal has indicated that TP53 mutations are related to poor prognosis in EC [19]. These data indicated a 5-year survival rate of 60% for patients with TP53 mutations compared with up to 90% for those without mutations. In our study, the mutation rate of TP53 was 12.2% in patients with serous EC. A previous study reported that the frequency of whole 133 TP53 mutation locations was >90% in serous and had a different frequency in the endometrioid subtype [19]. We confirmed TP53 mutations in three locations (chr17:7579883, chr17:7578406, and chr17:7576897 of hg19 assembly); hence, a lower frequency of mutations was found in our study than in the previous study.
In terms of the genomic landscape, PTEN and PIK3CA mutations were most common mutations in stage III/IV and I EC samples. Both PTEN and PIK3CA mutations had predominantly high frequency in TCGA data, ranging from 77% to 94% and from 53 to 71%, respectively, and were almost evenly distributed throughout the four molecular subgroups, except for the copy-number high group, in which the TP53 mutation rate was 92% [19]. Čerina et al. [28] conducted comprehensive genomic profiling in 32 metastatic uterine cancers. In their study, TP53 mutation was the most frequent genomic alteration, and PIK3CA, ARID1A, and PTEN were followed. Unfortunately, no stratified analysis focusing on advanced stage EC was done, which is different from our study.
Although statistically insignificant, the FBXW7 mutation rate was higher in stage III/IV EC samples than in stage I EC samples. FBXW7 mutations are most frequently reported in type II EC and are related to the PI3K pathway activation through mTOR stabilization [29]. Additionally, a similar mutation pattern was observed for a small number of CTNNB1 mutations. Generally, the mutation frequency of the CTNNB1 gene is lower in type II EC cases than in type I EC cases. β-catenin, a crucial component of the Wnt signaling pathway, stimulates the transcription of genes related to cell proliferation and survival [30]. Further, the role of FBXW7 and CTNNB1 mutations in the poor prognosis of stage III/IV ECs requires further investigation.
Lollipop plots of PPP2R1A, TP53, and PIK3CA showed different mutation loci according to the histological type and FIGO stage. In PPP2R1A gene, P179L/R mutation, located in the HEAT_2 domain was identified. In a previous study, the P179L mutation was detected in serous EC [31]. P179R is a recurrent factor that induces global changes in the A-subunit protein structure and loss of interaction with the catalytic subunit and reduces holoenzyme stability [32]. In TP53, the three mutations were located in the three different domains; however, the mutation located in the P53_tetramer was only detected in stage III/IV EC samples. In the P53 domain, R175H is an enriched somatic mutation in tumors with infiltrating immune cells [33,34].
The major strength of our study is that this is the first to investigate differential somatic mutation patterns in patients with stage III/IV EC presenting poor oncological outcomes. To overcome the small sample size, we combined our data with TCGA dataset for a comprehensive analysis. Given that we used TCGA dataset, the largest pool of genomic information of EC patients in the world, selection bias could be minimized.
This study had some limitations. First, because we used targeted NGS technology, a commercial multi-gene panel, the number of genes that could be investigated was limited. If whole genome or exome sequencing was applied, clinically significant genes other than PPP2R1A or TP53 could be identified. Second, although 548 samples with clinical information in TCGA dataset were identified, only 242 samples were retrieved by variant calling of somatic mutation. That’s because an overlap of variant calling results was seen in 21 KUMC samples (16 endometrioid and 5 serous) and 242 TCGA samples (206 endometrioid and 36 serous). If somatic mutation data of more patients with EC patients are analyzed by histological type, more accurate and statistically meaningful results can be obtained. Third, considering TP53, only three genomic locations of the 133 indicated in TCGA were analyzed. Although this study showed similar patterns to that in TCGA, more statistical significance could be obtained if more somatic mutation locations were analyzed. Lastly, we had only 21 patients with advanced EC in our institution who met inclusion criteria. For this reason, our results should be interpreted as preliminary data.
Herein, we found that both PPP2R1A and TP53 mutations were significantly higher in stage III/IV EC samples than in stage I EC samples. Compared with wild-type, mutated PPP2R1A and TP53 were associated with decreased survival rate. The negative effect of PPP2R1A mutation on survival outcome was more prominent in endometrioid EC population than in serous EC population. PPP2R1A and TP53 mutations might contribute to poor oncological outcomes in patients with stage III/IV EC. Because of small sample size, we could only show preliminary results at this stage. Larger-scaled, confirmatory study with prospectively collected samples is warranted.
Footnotes
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: H.J.H., C.H.W., L.J.K., G.J.A.
- Data curation: H.J.H., O.Y.T.
- Investigation: C.Y.
- Methodology: C.H.W.
- Software: G.J.A.
- Writing - original draft: H.J.H., C.H.W., O.Y.T., L.J.K., C.Y.
- Writing - review & editing: H.J.H., C.H.W., O.Y.T., L.J.K., C.Y.
SUPPLEMENTARY MATERIALS
Clinical and pathological features of 21 patients from Korea University Medical Center
Features of 129 somatic variants of 21 patients from Korea University Medical Center
Insertion and deletion (INDEL) and copy number variation (CNV) patterns of 21 patients from Korea University Medical Center
Top 16 frequently mutated genes are shown in the waterfall plot. (A) From combined KUMC and TCGA somatic mutation analysis, total somatic mutations were retrieved 86 locus, 38 genes in the total 263 patients. The cut-off of indicated mutation was 0.02, so the mutation found in at least six of the 263 patients to be displayed in this plot. A confusion matrix of the counts at each combination of factors, and factors were sample sources (KUMC and TCGA), endometroid, serous, stage, and sum of each sources. From endometroid (B), and serous (C) pathological type, waterfall plots were depicted.
(A) The left panel indicates the percentage of mutation of total 56 patients in each 23 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least two of the 56 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (56 endometrioid), sample source (16 KUMC and 40 TCGA), survival status (A, 41 alive; D, 14 dead; U, 1 unknown), and two FIGO stages (42 stage III and 14 stage IV). (B) The left panel indicates the percentage of mutation of total 144 patients in each 11 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least three of the 144 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (144 endometrioid), sample source (144 TCGA), survival status (A, 138 alive; D, 6 dead), and one FIGO stage (144 stage I).
(A) The left panel indicates the percentage of mutation of total 27 patients in each 16 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least one of the 27 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (27 serous), sample source (5 KUMC and 22 TCGA), survival status (A, 18 alive; D, 7 dead; U, 2 unknown), and two FIGO stages (22 stage III and 5 stage IV). The upper panel demonstrates the mutational burden per megabase DNA (MB) for each patient. (B) The left panel indicates the percentage of mutation of total 14 patients in each 9 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least one of the 14 patients to be displayed in this plot. The lower panel was separated into four sections, and each sections indicates that pathological type (14 serous), sample source (14 TCGA), survival status (A, 12 alive; D, 2 dead), and one FIGO stage (14 stage I).
References
- 1.Lim MC, Won YJ, Ko MJ, Kim M, Shim SH, Suh DH, et al. Incidence of cervical, endometrial, and ovarian cancer in Korea during 1999–2015. J Gynecol Oncol. 2019;30:e38. doi: 10.3802/jgo.2019.30.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greven KM, Corn B, Lanciano RM, Case D, Randall ME. Pathologic stage III endometrial carcinoma: significance of extrauterine sites. Radiat Oncol Investig. 1996;4:122–128. [Google Scholar]
- 3.Mariani A, Webb MJ, Keeney GL, Haddock MG, Aletti G, Podratz KC. Stage IIIC endometrioid corpus cancer includes distinct subgroups. Gynecol Oncol. 2002;87:112–117. doi: 10.1006/gyno.2002.6789. [DOI] [PubMed] [Google Scholar]
- 4.Lee WC, Reuben A, Hu X, McGranahan N, Chen R, Jalali A, et al. Multiomics profiling of primary lung cancers and distant metastases reveals immunosuppression as a common characteristic of tumor cells with metastatic plasticity. Genome Biol. 2020;21:271. doi: 10.1186/s13059-020-02175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philip CA, Laskov I, Beauchamp MC, Marques M, Amin O, Bitharas J, et al. Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial cancer cell lines to PARP inhibitors. BMC Cancer. 2017;17:638. doi: 10.1186/s12885-017-3639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oza AM, Elit L, Tsao MS, Kamel-Reid S, Biagi J, Provencher DM, et al. Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: a trial of the NCIC Clinical Trials Group. J Clin Oncol. 2011;29:3278–3285. doi: 10.1200/JCO.2010.34.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slomovitz BM, Jiang Y, Yates MS, Soliman PT, Johnston T, Nowakowski M, et al. Phase II study of everolimus and letrozole in patients with recurrent endometrial carcinoma. J Clin Oncol. 2015;33:930–936. doi: 10.1200/JCO.2014.58.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slomovitz BM, Lu KH, Johnston T, Coleman RL, Munsell M, Broaddus RR, et al. A phase 2 study of the oral mammalian target of rapamycin inhibitor, everolimus, in patients with recurrent endometrial carcinoma. Cancer. 2010;116:5415–5419. doi: 10.1002/cncr.25515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman RL, Sill MW, Thaker PH, Bender DP, Street D, McGuire WP, et al. A phase II evaluation of selumetinib (AZD6244, ARRY-142886), a selective MEK-1/2 inhibitor in the treatment of recurrent or persistent endometrial cancer: an NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2015;138:30–35. doi: 10.1016/j.ygyno.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 11.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38:1–10. doi: 10.1200/JCO.19.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda T, Banno K, Okawa R, Yanokura M, Iijima M, Irie-Kunitomi H, et al. ARID1A gene mutation in ovarian and endometrial cancers (Review) Oncol Rep. 2016;35:607–613. doi: 10.3892/or.2015.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winterhoff B, Konecny GE. Targeting fibroblast growth factor pathways in endometrial cancer. Curr Probl Cancer. 2017;41:37–47. doi: 10.1016/j.currproblcancer.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Fader AN, Roque DM, Siegel E, Buza N, Hui P, Abdelghany O, et al. Randomized phase II trial of carboplatin-paclitaxel versus carboplatin-paclitaxel-trastuzumab in uterine serous carcinomas that overexpress human epidermal growth factor receptor 2/neu. J Clin Oncol. 2018;36:2044–2051. doi: 10.1200/JCO.2017.76.5966. [DOI] [PubMed] [Google Scholar]
- 17.Samarnthai N, Hall K, Yeh IT. Molecular profiling of endometrial malignancies. Obstet Gynecol Int. 2010;2010:162363. doi: 10.1155/2010/162363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X, Bi J, Li Y, Yang S, Zhang Y, Li M, et al. AZD1775 increases sensitivity to olaparib and gemcitabine in cancer cells with p53 mutations. Cancers (Basel) 2018;10:10. doi: 10.3390/cancers10050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 23.Ruediger R, Ruiz J, Walter G. Human cancer-associated mutations in the Aα subunit of protein phosphatase 2A increase lung cancer incidence in Aα knock-in and knockout mice. Mol Cell Biol. 2011;31:3832–3844. doi: 10.1128/MCB.05744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remmerie M, Janssens V. Targeted Therapies in Type II Endometrial Cancers: Too Little, but Not Too Late. Int J Mol Sci. 2018;19:19. doi: 10.3390/ijms19082380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogani G, Ray-Coquard I, Concin N, Ngoi NY, Morice P, Enomoto T, et al. Uterine serous carcinoma. Gynecol Oncol. 2021;162:226–234. doi: 10.1016/j.ygyno.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haesen D, Abbasi Asbagh L, Derua R, Hubert A, Schrauwen S, Hoorne Y, et al. Recurrent PPP2R1A mutations in uterine cancer act through a dominant-negative mechanism to promote malignant cell growth. Cancer Res. 2016;76:5719–5731. doi: 10.1158/0008-5472.CAN-15-3342. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Čerina D, Matković V, Katić K, Lovasić IB, Šeparović R, Canjko I, et al. Precision oncology in metastatic uterine cancer; Croatian first-year experience of the comprehensive genomic profiling in everyday clinical practice. Pathol Oncol Res. 2021;27:1609963. doi: 10.3389/pore.2021.1609963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao J, Ge MH, Ling ZQ. Fbxw7 tumor suppressor: a vital regulator contributes to human tumorigenesis. Medicine (Baltimore) 2016;95:e2496. doi: 10.1097/MD.0000000000002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagendra DC, Burke J, 3rd, Maxwell GL, Risinger JI. PPP2R1A mutations are common in the serous type of endometrial cancer. Mol Carcinog. 2012;51:826–831. doi: 10.1002/mc.20850. [DOI] [PubMed] [Google Scholar]
- 32.Taylor SE, O’Connor CM, Wang Z, Shen G, Song H, Leonard D, et al. The highly recurrent PP2A Aα-subunit mutation P179R alters protein structure and impairs PP2A enzyme function to promote endometrial tumorigenesis. Cancer Res. 2019;79:4242–4257. doi: 10.1158/0008-5472.CAN-19-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behring M, Vazquez AI, Cui X, Irvin MR, Ojesina AI, Agarwal S, et al. Gain of function in somatic TP53 mutations is associated with immune-rich breast tumors and changes in tumor-associated macrophages. Mol Genet Genomic Med. 2019;7:e1001. doi: 10.1002/mgg3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo W, Parkhurst M, Robbins PF, Tran E, Lu YC, Jia L, et al. Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol Res. 2019;7:534–543. doi: 10.1158/2326-6066.CIR-18-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and pathological features of 21 patients from Korea University Medical Center
Features of 129 somatic variants of 21 patients from Korea University Medical Center
Insertion and deletion (INDEL) and copy number variation (CNV) patterns of 21 patients from Korea University Medical Center
Top 16 frequently mutated genes are shown in the waterfall plot. (A) From combined KUMC and TCGA somatic mutation analysis, total somatic mutations were retrieved 86 locus, 38 genes in the total 263 patients. The cut-off of indicated mutation was 0.02, so the mutation found in at least six of the 263 patients to be displayed in this plot. A confusion matrix of the counts at each combination of factors, and factors were sample sources (KUMC and TCGA), endometroid, serous, stage, and sum of each sources. From endometroid (B), and serous (C) pathological type, waterfall plots were depicted.
(A) The left panel indicates the percentage of mutation of total 56 patients in each 23 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least two of the 56 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (56 endometrioid), sample source (16 KUMC and 40 TCGA), survival status (A, 41 alive; D, 14 dead; U, 1 unknown), and two FIGO stages (42 stage III and 14 stage IV). (B) The left panel indicates the percentage of mutation of total 144 patients in each 11 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least three of the 144 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (144 endometrioid), sample source (144 TCGA), survival status (A, 138 alive; D, 6 dead), and one FIGO stage (144 stage I).
(A) The left panel indicates the percentage of mutation of total 27 patients in each 16 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least one of the 27 patients to be displayed in this plot. The lower panel was separated into four sections, and each section indicates that pathological type (27 serous), sample source (5 KUMC and 22 TCGA), survival status (A, 18 alive; D, 7 dead; U, 2 unknown), and two FIGO stages (22 stage III and 5 stage IV). The upper panel demonstrates the mutational burden per megabase DNA (MB) for each patient. (B) The left panel indicates the percentage of mutation of total 14 patients in each 9 genes. The cut-off of indicated mutation was 0.02, so the mutation found in at least one of the 14 patients to be displayed in this plot. The lower panel was separated into four sections, and each sections indicates that pathological type (14 serous), sample source (14 TCGA), survival status (A, 12 alive; D, 2 dead), and one FIGO stage (14 stage I).