Abstract
Chemical mutagenesis of Staphylococcus aureus RN450 generated two strains that displayed a stable reduction (30- to 60-fold) in susceptibility to evernimicin. Cell-free translation reactions demonstrated that the resistance determinant was located in the ribosomal fraction. Compared to ribosomes isolated from a wild-type strain, ribosomes from the mutant strains displayed an 8- to 10-fold reduction in affinity for [14C]evernimicin. In contrast, the mutants displayed no alteration in either binding affinity or in vitro susceptibility to erythromycin. Exponential cultures of the mutant strains accumulated significantly less [14C]evernimicin than the wild-type strain, suggesting that accumulation is dependent on the high affinity that evernimicin displays for its binding site. Sequencing rplP (encodes ribosomal protein L16) in the mutant strains revealed a single base change in each strain, which resulted in a substitution of either cysteine or histidine for arginine at residue 51. Introduction of a multicopy plasmid carrying wild-type rplP into the mutant strains restored sensitivity to evernimicin, confirming that the alterations in rplP were responsible for the change in susceptibility. Overexpression of the mutant alleles in S. aureus RN450 had no effect on susceptibility to evernimicin, demonstrating that susceptibility is dominant over resistance.
Evernimicin (SCH 27899) is a novel oligosaccharide antibiotic (8, 22) with activity against a broad range of gram-positive pathogenic bacteria including glycopeptide-resistant enterococci, methicillin-resistant staphylococci, and penicillin-resistant streptococci (11). We previously demonstrated that evernimicin inhibits protein synthesis in Staphylococcus aureus and in a susceptible Escherichia coli strain (13). The antibiotic binds with high affinity to a single site on the 50S subunit. This binding site appears to be unique to evernimicin and a structurally similar compound, avilamycin, since an assay designed to identify compounds that blocked the binding of evernimicin to 70S ribosomes identified only avilamycin from among a collection of antibiotics known to bind the 50S subunit (13). Mutations that confer reduced susceptibility to evernimicin in Streptococcus pneumoniae have been mapped to both rplP (3), which encodes 50S-associated protein L16, and the operons encoding 23S rRNA (4). The mutations in the 23S rRNA are located in two separate stem-loops that are associated with the peptidyl transferase center; one of these loops has been cross-linked to L16 (15). In this study we characterize in detail two S. aureus strains with reduced susceptibility to evernimicin. We demonstrate that single amino acid substitutions in ribosomal protein L16 reduce the binding of evernimicin to 70S ribosomes. We also show that accumulation of the antibiotic is reduced in the mutant strains. These data further support the contention that evernimicin acts by binding the ribosome and inhibiting some aspect of translation.
(Portions of this work were presented at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., 1999.)
MATERIALS AND METHODS
Antibiotics.
Evernimicin was isolated at Schering Plough Research Institute (SPRI) and was solubilized as the clinical formulation at 16 μg/ml. Linezolid was synthesized at SPRI. Quinupristin-dalfopristin and avilamycin were gifts from Rhone Poulenc-Rorer and Elanco Animal Health, respectively. Chloramphenicol, clindamycin, erythromycin, and lincomycin were purchased from Sigma Chemical Co. MIC determinations were performed in microtiter plates using tryptic soy broth (TSB). Antibiotics were added in doubling dilutions, and the MIC was recorded as the lowest concentration of antibiotic that completely inhibited bacterial growth after 16 h of incubation at 37°C.
Bacterial strains and plasmids.
S. aureus strains RN450 and a pyrF derivative of S. aureus RN4220, WZ4220, were obtained from the SPRI culture collection. Strains were grown in TSB or on tryptic soy agar (TSA). pYR7 is an E. coli/S. aureus shuttle vector with a gram-positive temperature-sensitive replicon (unpublished data).
Mutant selection.
Strain RN450 was treated with ethyl methanesulfonate as described previously (14). Under the conditions employed here there was no loss in cell viability. After being washed the cells were plated on TSA containing evernimicin at 8 μg/ml and incubated for 24 h at 37°C.
Preparation and fractionation of ribosomes.
Ribosomes and S100 extracts, prepared from S. aureus as described previously (12), were resuspended and dialyzed, respectively, against B3 (10 mM MgCl2, 20 mM Tris [pH-7.8], 30 mM NH4Cl, 0.1 mM EDTA, 6 mM β-mercaptoethanol).
Cell-free translation reactions.
Reactions using a poly(A) template with [14C]lysine (specific activity of 310 mCi/mmol; purchased from NEN) were performed as described previously (21). Radiolabeled polypeptides were precipitated with 5% trichloroacetic acid containing 0.05% tungstic acid (17), heated to 90°C for 15 min, and then applied to 96-well filter plates (Millipore). Radioactivity was determined by liquid scintillation counting.
Binding assays.
[14C]evernimicin (specific activity of 8 mCi/mmol) was prepared as described previously (10), and [14C]erythromycin (specific activity of 55 mCi/mmol) was purchased from NEN. Binding reactions were performed as described previously (13).
Accumulation assays.
[14C]evernimicin was added to exponentially growing cultures in TSB, and at timed intervals triplicate 6-ml samples were withdrawn, filtered through 0.2-μM-pore-size filters, and washed twice with 6 ml of 3% NaCl. After the filters were dried, the amount of drug was determined by liquid scintillation counting. A correction was made to account for nonspecific binding of [14C]evernimicin to filters (the value was determined by filtering a broth control containing only the labeled drug). Dinitrophenol (DNP) and N,N′-dicyclohexylcarbodiimide (DCCD) were added at 2 mM and 20 μM, respectively, immediately before addition of [14C]evernimicin.
Cloning of rplP alleles and allele replacement.
The penP promoter from Bacillus licheniformis (9) was amplified by PCR with oligonucleotides 2949 (5′-GGATCCGGTGGAAACGAGGTCA-3′) and 2950 (5′-CGACGATATTTACACGTTTTGGTAGTAACA-3′) and fused to rplP from RN450 by amplification with oligonucleotides 2949 and 0797 (5′-GAATTCTTAGCTTTCATTTGTTTCACC-3′). The fusion product was cloned into pSK265 generating pPAM14. The cloning was repeated for rplP alleles from strains RN450-70 and RN450-77 generating pPAM15 and pPAM16. Allele replacement was performed by amplifying rplP along with 600 bp of flanking DNA from strain RN450-70 with oligonucleotides 58 (5′-TAGGAGCTCCCGTGCTTCATTCCGTCGTGT-3′) and 59 (5′-TAGGGTACCGAGTATTTTACTCGTTTA-3′). The PCR fragment was cloned into pYR7 generating pPAM27. WZ4220(pPAM27) was grown overnight in TSB at 30°C, plated on TSA with evernimicin at 0.5 μg/ml, and incubated for 24 h at 42°C. Evernimicin-resistant colonies were tested for plasmid loss by screening for loss of plasmid-mediated chloramphenicol resistance at 30°C. Both the allele exchange and the sequences of all inserts were confirmed by DNA sequencing.
RESULTS
Isolation of strains displaying reduced susceptibility to evernimicin.
Following exposure of S. aureus RN450 to ethyl methanesulfonate, evernimicin-resistant colonies arose at a frequency of 1.9 × 10−7 per CFU. From the same batch of cells, streptomycin-resistant isolates arose at a frequency of 1.7 × 10−7 per CFU. Out of 18 isolates analyzed, 16 exhibited temperature growth defects. Isolates RN450-70 and RN450-77 grew at all temperatures and were chosen for further study. Compared to RN450, RN450-70 and RN450-77 exhibited 60- and 30-fold reductions in evernimicin susceptibility, respectively (Table 1). With the exception of susceptibilities to avilamycin, the susceptibilities of RN450-70 and RN450-77 to protein synthesis inhibitors were unchanged (Table 1). The mutant strains grew more slowly in TSB at 37°C than the wild-type progenitor; the doubling times of RN450-70 and RN450-77 were 37 min, and RN450 had a doubling time of 22 min (data not shown).
TABLE 1.
MICs of common protein synthesis inhibitors
| Drug | MIC (μg/ml) for:
|
||
|---|---|---|---|
| RN450 (wild-type L16) | RN450-77 (Arg 51 → Cys) | RN450-77 (Arg 51 → His) | |
| Evernimicin | 0.02 | 1.25 | 0.625 |
| Avilamycin | 0.4 | >25 | >25 |
| Chloramphenicol | 3 | 3 | 3 |
| Clindamycin | 0.1 | 0.1 | 0.1 |
| Erythromycin | 0.2 | 0.2 | 0.2 |
| Gentamicin | 1.6 | 1.6 | 1.6 |
| Kanamycin | 3.2 | 1.6 | 1.6 |
| Lincomycin | 1 | 1 | 1 |
| Linezolid | 1 | 1 | 1 |
| Quinupristin-dalfopristin | 1 | 1 | 1 |
| Tetracycline | 0.08 | 0.16 | 0.16 |
Ribosomes are less susceptible to inhibition by evernimicin in vitro.
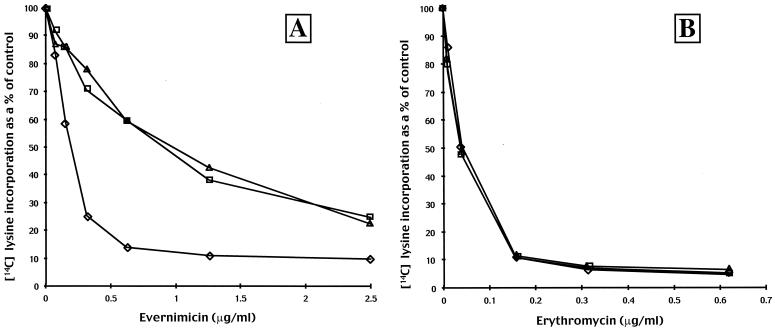
We performed cell-free translation assays using an S100 extract isolated from the wild-type strain (RN450) and ribosomes isolated from RN450, RN450-70, and RN450-77. In agreement with previous data (13), ribosomes from RN450 were inhibited in a dose-dependent manner with the maximal level of inhibition, 90% that of the antibiotic-negative control, occurring at an evernimicin concentration of approximately 1 μg/ml (600 nM; Fig. 1A). At the same drug concentration ribosomes isolated from strains RN450-70 and RN450-77 were inhibited approximately 50%. To achieve 90% inhibition required 10-fold more evernimicin (6 μM; data not shown). Under the same conditions erythromycin was equally effective in inhibiting translation irrespective of the ribosome source (Fig. 1B).
FIG. 1.
Effect of evernimicin (A) and erythromycin (B) on in vitro translation reactions. Reactions were performed using an S100 extract from S. aureus RN450 and ribosomes isolated from strains RN450 (diamonds), RN450-70 (squares), and RN450-77 (triangles). Incorporation of [14C]lysine into hot trichloracetic acid-precipitable material is expressed as a percentage of that for the control (no antibiotic) reaction.
Ribosomes display reduced binding of [14C]evernimicin.
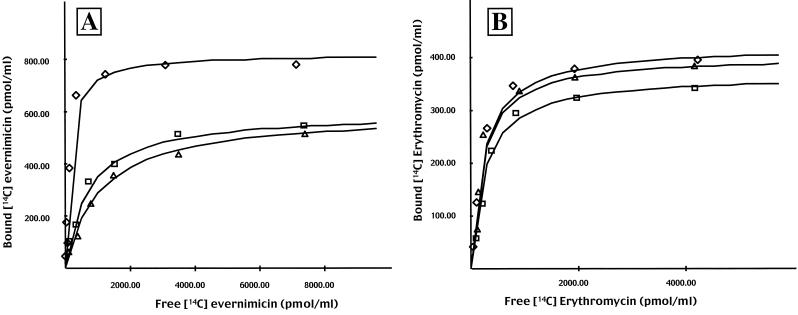
Binding reactions were performed using [14C]evernimicin and ribosomes purified from RN450, RN450-70, and RN450-77. At low levels of [14C]evernimicin, all three sets of ribosomes exhibited a dose-dependent linear increase in antibiotic binding (Fig. 2A). At higher levels binding reached a plateau and no further binding was observed. Nonlinear regression plots yielded a dissociation constant (Kd) of 135 nM for ribosomes isolated from RN450, a value similar to that reported previously (13). At saturation the stoichiometry of [14C]evernimicin to ribosomes was approximately 1:1 (960 pmol of ribosomes bound 800 pmol of [14C]evernimicin). In contrast, ribosomes isolated from strains RN450-70 and RN450-77 displayed a lower affinity for [14C]evernimicin; Kds were 700 and 1,050 nM, respectively. Ribosomes from strains RN450-70 and RN450-77 also bound less antibiotic; 960 pmol of both sets of ribosomes bound 580 pmol of [14C]evernimicin. Increasing the amount of labeled antibiotic beyond those levels shown in Fig. 2A did not result in additional drug binding (data not shown). On repeating these assays with [14C]-labeled erythromycin we observed a similar initial dose-dependent increase followed by a plateau (Fig. 2B). However, in this instance there were no major differences between the three sets of ribosomes.
FIG. 2.
Nonlinear regression analysis of [14C]evernimicin (A) and [14C]erythromycin (B) binding to 70S ribosomes. Binding was performed with ribosomes isolated from S. aureus strains RN450 (diamonds), RN450-70 (squares), and RN450-77 (triangles). The amounts of bound [14C]evernimicin and [14C]erythromycin were calculated from standard curves.
rplP mutations are recessive in a merodiploid background.
The sequencing of the rDNA operons and rplP in RN450, RN450-70, and RN450-77 revealed that the latter two strains each contained single nucleotide substitutions in rplP resulting in a conversion of residue 51, arginine, to cysteine or histidine, respectively. To ascertain if these mutations were responsible for the reduction in susceptibility, we introduced multicopy plasmid pMAN14, which carries a wild-type copy of rplP under the control of the penP promoter, into RN450, RN450-70, and RN450-77. The presence of either pMAN14 or the empty vector, pSK265, in RN450 had no effect on susceptibility to evernimicin (Table 2). In contrast, pMAN14 completely restored evernimicin susceptibility to RN450-77 and lowered the MIC approximately eightfold in RN450-70. The rplP alleles from RN450-70 and RN450-77 were also cloned under the control of the penP promoter, and the resultant plasmids were introduced into RN450. Neither plasmid caused an alteration in the evernimicin MIC (Table 2). To determine if the inability to fully revert strain RN450-70 to evernimicin sensitivity was due to an unlinked mutation, we introduced the rplP mutation from RN450-70 into the chromosome of wild-type strain WZ4220. The MIC for the resultant strain, WZ4220-70, was comparable to that for RN450-70, and introduction of pMAN14 into WZ4220-70 again only partially restored evernimicin susceptibility (Table 2).
TABLE 2.
MICs for S. aureus strains carrying plasmids that contain rplP alleles
| Complementing plasmid | Evernimicin MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| RN450 | RN450-70 | RN450-77 | WZ4220 | WZ4220-70 | |
| pSK265 (vector alone) | 0.02 | 1.25 | 0.625 | 0.02 | 0.625 |
| pPAM14 (contains wild-type rplP) | 0.02 | 0.16 | 0.02 | 0.02 | 0.16 |
| pPAM15 (contains rplP from RN450-70) | 0.02 | NTa | NT | NT | NT |
| pPAM16 (contains rplP from RN450-77) | 0.02 | NT | NT | NT | NT |
NT, not tested.
Strains with a reduced susceptibility to evernimicin exhibit reduced accumulation of evernimicin.
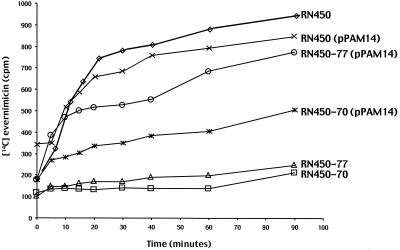
When added to an exponential culture of S. aureus RN450 at 0.02 μg/ml (i.e., at the MIC), [14C]evernimicin was rapidly accumulated over the first 20 min, after which accumulation slowed (Fig. 3). Addition of a 500-fold excess of unlabeled evernimicin at the 20-min time point led to a linear decrease in cell-associated label such that after 20 min there was no cell-associated [14C]evernimicin (data not shown). We repeated the accumulation assay in the presence of either protonophore DNP or inhibitor of ATP synthesis DCCD. After correcting for changes in the growth rate, resulting from the addition of DNP and DCCD, we found that there was no change in either the rate at which [14C]evernimicin accumulated or the amount of it that accumulated (data not shown). In contrast to RN450, strains RN450-70 and RN450-77 showed a greatly reduced accumulation of [14C] evernimicin (Fig. 3); it should be noted that over the time course of these experiments there was no significant differences in the optical densities of the cultures (data not shown). Introduction of pPAM14 (contains wild-type rplP) into strains RN450-70 and RN450-77 either fully (RN450-77) or partially (RN450-70) restored the accumulation of [14C]evernimicin.
FIG. 3.
Accumulation of [14C]evernimicin by various strains of S. aureus. [14C]evernimicin was added at 0.02 μg/ml to exponentially growing cultures of the indicated strains at time zero. Triplicate samples were taken at the indicated time points, and the amounts of cell-associated radiolabel were determined as described in Materials and Methods.
DISCUSSION
Evernimicin binds the bacterial 50S ribosomal subunit and inhibits some aspect of the elongation process (13). In this study we demonstrate for the first time that mutations in rplP affect evernimicin binding to ribosomes, drug accumulation, and the susceptibility of the ribosomes in cell-free assays. We analyzed two strains of S. aureus that displayed a stable reduction in susceptibility to evernimicin. Cell-free translation assays localized the resistance determinant to the ribosomal fraction, and we subsequently determined that both strains contained missense mutations in residue 51 of ribosomal protein L16. Three other recent studies highlight the importance of this region of L16 in mediating evernimicin resistance. In S. pneumoniae substitutions in either residue 51 or 52 conferred evernimicin resistance (3, 4). In a second study 22 enterococcal isolates were analyzed (2). The bacteria, Enterococcus faecium and Enterococcus faecalis, were isolated from animals on the basis of their avilamycin resistance and were subsequently shown to have reduced susceptibility to evernimicin (coresistance to both avilamycin and evernimicin has been noted previously [1]). Each isolate carried a single amino acid change in either residue 52 or 56 of L16. An in-house analysis of a further 13 enterococcal isolates from the same source as above found substitutions in the same residues of L16 (unpublished data). Thus, it would appear that mutations that confer resistance to evernimicin are confined to a small region of L16. Furthermore, the degree of conservation of this region (Fig. 4) suggests that this domain is critical for ribosomal activities.
FIG. 4.
Alignment of rplP from S. pneumoniae (Sp), E. faecium (Efm), E. faecalis (Efc), and S. aureus (Sa). Shown are the locations of amino acid substitutions that result in reduced susceptibility to evernimicin in S. aureus and S. pneumoniae (3). Stars, identical residues in all four proteins.
It is unclear if evernimicin binds L16 directly or whether the amino acid changes described above influence drug binding in an allosteric manner. Drugs to which the latter process applies are streptomycin and spectinomycin; mutations in S12 and S5, respectively, confer resistance, but ribosomes lacking either protein showed no alteration in streptomycin or spectinomycin binding (19, 20). One finding that argues against direct binding is that overproduction of L16 did not increase cellular accumulation of evernimicin. L16 has been implicated in the binding a variety of antibiotics; chloramphenicol and derivative monoiodoamphenicol preferentially bound L16 (16, 18) and ribosomes stripped of L16 no longer bound virginiamycin S (6). However, we demonstrated that these drugs (instead of virginiamycin S we used semisynthetic derivative quinupristin-dalfopristin) did not block the binding of labeled evernimicin to 70S ribosomes, suggesting that the binding sites of the three drugs do not overlap (13). In addition, evernimicin-resistant strains do not exhibit cross-resistance to either chloramphenicol or quinupristin-dalfopristin. However, we cannot rule out the possibility that evernimicin, along with chloramphenicol and virginiamycin S, bind L16 at different sites.
Accumulation of evernimicin by exponentially growing cells appears to be driven primarily by the drugs' high affinity for the ribosome. This is evidenced by the finding that cells with alterations in L16, which diminish evernimicin binding to ribosomes in vitro, accumulate significantly less antibiotic than wild-type cells. Introduction of a plasmid encoding wild-type L16 effectively restored accumulation. In addition, exposure of cells to either a protonophore or an inhibitor of ATP synthesis did not inhibit accumulation, suggesting that the drug enters the cells by passive diffusion. Incubation of the cells with an excess of unlabeled drug resulted in a complete loss of cell-associated labeled evernimicin; the half-life of this reaction was 10 min. Utilizing purified ribosomes we previously estimated the half-lives of antibiotic-ribosome complexes to be 20 min (13). The twofold discrepancy may be due to differences in temperature; the in vivo and in vitro reactions were performed at 37°C and room temperature, respectively.
Expression of rplP+ in a mutant background resulted in a decrease in the evernimicin MIC. However, the magnitude of the decrease was allele specific; strain RN450-77 was converted to full sensitivity, while RN450-70 remained partially resistant. We ruled out the possibility that the residual resistance in RN450-70 was due to a second unlinked mutation by introducing the rplP mutation into a wild-type strain and repeating the complementation assay. In the reverse experiment, expression of the mutant rplP alleles from the same regulatory elements did not change the susceptibility of a wild-type strain. Presumably in the merodiploid strain the mutated form of L16 is at a competitive disadvantage compared to wild-type L16. Together these data argue that mutations in rplP, like those in the genes encoding ribosomal proteins L4 and L22, which confer resistance to erythromycin (7), are recessive in a merodiploid strain. The recessive nature of evernimicin-resistant alleles may explain why resistance mutations in rDNA operons occur far less frequently than rplP mutations. Most organisms have multiple rDNA operons, and presumably the cell must ensure, by a process of allelic exchange, that all (or the majority) of the rrn operons carry the mutation. To date such mutations have only been found in S. pneumoniae and only after prolonged incubation at low evernimicin concentrations (4).
In summary, substitutions in L16 that result in reduced susceptibility to evernimicin are confined to a small highly conserved region. This finding, plus the observation that alterations in L16 often slow cell growth, may limit the spread of such strains in a clinical setting.
ACKNOWLEDGMENTS
Paul McNicholas, Paul Mann, and David Najarian contributed equally to this work.
We thank Scott Walker for helpful discussions and Bruce Malcolm for help with binding studies.
REFERENCES
- 1.Aarestrup F M. Association between decreased susceptibility to a new antibiotic for treatment of human diseases, everninomycin (evernimicin), and resistance to an antibiotic used for growth promotion in animals, avilamycin. Microb Drug Resist. 1998;4:137–141. doi: 10.1089/mdr.1998.4.137. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup F M, Jensen L B. Presence of variations in ribosomal protein L16 corresponding to susceptibility of enterococci to oligosaccharides (avilamycin and evernimicin) Antimicrob Agents Chemother. 2000;44:3425–3427. doi: 10.1128/aac.44.12.3425-3427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adrian P V, Zhao W, Black T A, Shaw K J, Hare R S, Klugman K P. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin ( SCH27899): implications for mechanism of action. Antimicrob Agents Chemother. 2000;44:732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adrian P V, Mendrick C, Loebenberg D, McNicholas P, Shaw K J, Klugman K P, Hare R S, Black T A. Evernimicin ( SCH27899) inhibits a novel ribosome target site: an analysis of 23S ribosomal DNA mutants. Antimicrob Agents Chemother. 2000;44:3101–3106. doi: 10.1128/aac.44.11.3101-3106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernabeu C, Vázquez D, Ballesta J P G. The involvement of protein L16 on ribosomal peptidyl transferase activity. Eur J Biochem. 1977;79:469–472. doi: 10.1111/j.1432-1033.1977.tb11829.x. [DOI] [PubMed] [Google Scholar]
- 6.Bethune M-P, Nierhaus K H. Characterization of the binding of virginiamycin S to Escherichia coli ribosomes. Eur J Biochem. 1978;86:187–191. doi: 10.1111/j.1432-1033.1978.tb12298.x. [DOI] [PubMed] [Google Scholar]
- 7.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganguly A K, Pramanik B, Chan T M, Sarre O, Liu Y-T, Morton J, Girijavallabhan V. The structure of new oligosaccharide antibiotics, 13–384 components 1 and 5. Heterocycles. 1989;28:83–88. [Google Scholar]
- 9.Gray O, Chang S. Molecular cloning and expression of Bacillus licheniformis β-lactamase gene in Escherichia coli and Bacillus subtilis. J Bacteriol. 1981;145:422–428. doi: 10.1128/jb.145.1.422-428.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesk D, Gunnarsson I, Koharski D, McNamara P, Schwartz J L, Thonoor M, Wirth M. Synthesis of [3H]- and [14C]- SCH27899 by fermentation and evaluation of in vivo label stability. J Label Compd Radiopharm. 1999;42:159–167. [Google Scholar]
- 11.Jones R N, Barrett M S. Antimicrobial activity of SCH27899, oligosaccharide member of the everninomicin class with a wide gram-positive spectrum. J Clin Microbiol Infect. 1995;1:35–43. doi: 10.1111/j.1469-0691.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 12.Mahmood R, Compagnone-Post P, Khan S A. An in vitro coupled transcription-translation system from Staphylococcus aureus. Gene. 1991;106:29–34. doi: 10.1016/0378-1119(91)90562-p. [DOI] [PubMed] [Google Scholar]
- 13.McNicholas P M, Najarian D J, Mann P A, Hesk D, Hare R S, Shaw K J, Black T A. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell free systems derived from both gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 15.Muralikrishna P, Cooperman B S. Ribosomal components neighboring the 2475 loop in Escherichia coli 50S subunits. Biochemistry. 1995;34:115–121. doi: 10.1021/bi00001a014. [DOI] [PubMed] [Google Scholar]
- 16.Nierhaus D, Nierhaus K N. Identification of the chloramphenicol-binding protein in Escherichia coli ribosomes by partial reconstitution. Proc Natl Acad Sci USA. 1973;70:2224–2228. doi: 10.1073/pnas.70.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pezzuto J M, Hecht S M. Amino acid substitutions in protein biosynthesis. Poly(A)-directed polyphenylalanine synthesis. J Biol Chem. 1980;255:865–869. [PubMed] [Google Scholar]
- 18.Pongs O, Bald R, Erdmann V A. Identification of chloramphenicol-binding protein in Escherichia coli ribosomes by affinity labeling. Proc Natl Acad Sci USA. 1973;70:2229–2233. doi: 10.1073/pnas.70.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiner G, Nierhaus K H. Protein involved in the binding of dihydrostreptomycin to ribosomes of Escherichia coli. J Mol Biol. 1973;81:71–82. doi: 10.1016/0022-2836(73)90248-9. [DOI] [PubMed] [Google Scholar]
- 20.Spahn C M, Prescott C D. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 21.Staehlin T, Marglott D R. Preparation of Escherichia coli ribosomal subunits active in polypeptide synthesis. Methods Enzymol. 1971;20:449–455. [Google Scholar]
- 22.Weinstein M J, Lundemann G M, Oden E M, Wagman G H. Everninomicin, a new antibiotic complex from Micromonospora carbonaceae. Antimicrob Agents Chemother. 1965;1964:24–32. [PubMed] [Google Scholar]