Abstract
The tumor microenvironment (TME) is a well-recognized system that plays an essential role in tumor initiation, development, and progression. Intense intercellular communication between tumor cells and other cells (especially macrophages) occurs in the TME and is mediated by cell-to-cell contact and/or soluble messengers. Emerging evidence indicates that noncoding RNAs (ncRNAs) are critical regulators of the relationship between cells within the TME. In this review, we provide an update on the regulation of ncRNAs (primarily micro RNAs [miRNAs], long ncRNAs [lncRNAs], and circular RNAs [circRNAs]) in the crosstalk between macrophages and tumor cells in hepatocellular carcinoma (HCC). These ncRNAs are derived from macrophages or tumor cells and act as oncogenes or tumor suppressors, contributing to tumor progression not only by regulating the physiological and pathological processes of tumor cells but also by controlling macrophage infiltration, activation, polarization, and function. Herein, we also explore the options available for clinical therapeutic strategies targeting crosstalk-related ncRNAs to treat HCC. A better understanding of the relationship between macrophages and tumor cells mediated by ncRNAs will uncover new diagnostic biomarkers and pharmacological targets in cancer.
Keywords: noncoding RNA (ncRNA), tumor microenvironment (TME), microRNA (miRNA), long noncoding RNA (lncRNA), circular RNA (circRNA), tumor-associated macrophage (TAM), hepatocellular carcinoma (HCC)
Graphical abstract
Tumor-related macrophages (TAMs) and tumor-cell-derived ncRNAs (mainly miRNAs, lncRNAs and circRNAs) in hepatocellular carcinoma act as oncogenes or tumor suppressors to influence tumor progression, not only by regulating almost all physiological and pathological processes of tumor cells but also by modulating the infiltration, activation, polarization, or function of TAMs.
Introduction
Each year, 900,000 people are diagnosed with hepatocellular carcinoma (HCC), with 800,000 people dying of it.1 HCC has become the most common primary malignancy in the adult population, but the rate of early detection is very low, as most HCC cases are not diagnosed until reaching advanced tumor stages.2 At present, surgical resection is still the primary treatment for HCC, but as many as 70% of HCC patients who undergo resection relapse after 5 years.3 Moreover, for most patients with advanced HCC, sorafenib appears to be the most clinically effective chemotherapy drug, but its use and function are limited by many uncertain factors.4 Therefore, the overall 5-year survival rate of HCC is less than 18%, and new therapies are urgently needed to improve diagnostic efficiency and prolong the survival period of patients.5
Increasing evidence suggests that targeting the tumor microenvironment (TME) represents a potential novel therapeutic approach for cancer treatment, as the TME is responsible for the continued uncontrolled growth, metastasis, immunosuppression, and drug resistance of cancer.6, 7, 8 Depending on the type of tumor, the TME comprises many different elements, but it generally includes stromal cells, fibroblasts, endothelial cells, and immune cells as well as various cytokines, chemokines, and other factors secreted by the host and tumor cells.9 Intercellular communication among these cells mediated by various cytokines, chemokines, and other factors from the host or tumor cells determines the direction of tumor development, particularly the communication between tumor cells and macrophages.10 Macrophages in the TME, which are referred to as tumor-associated macrophages (TAMs), are the primary infiltrating immune cell population in most tumor types.11,12 TAMs are becoming a promising target for cancer immunotherapeutic strategies and are being used in many paraclinical and clinical attempts to treat tumors, including HCC.13,14
Recently, the potential of noncoding RNAs (ncRNAs) as biomarkers and targets for HCC diagnosis and treatment has been evaluated in multiple reports.15, 16, 17 ncRNA is a functional RNA molecule that is transcribed from DNA but not translated into protein.18 ncRNAs can be divided into two categories based on their length: long ncRNAs (lncRNAs) and short ncRNAs. lncRNAs are transcribed RNA molecules longer than 200 nucleotides (nt) in length and include two subclasses: pseudogenes and circular RNAs (circRNAs).19 In contrast, short ncRNAs are less than 200 nt and usually <30 nt. The short ncRNAs include at least four types: microRNAs (miRNAs), transfer RNA-derived small RNAs (tsRNAs), small nucleolar RNA (snoRNAs), and piwi interacting RNAs (piRNAs).19, 20, 21 Both short and lncRNAs have been shown to play roles in gene expression at multiple levels, such as gene silencing, chromatin remodeling, DNA methylation, and histone modification.22 Dysregulation of these ncRNAs plays important roles in the development of many human diseases, notably tumorigenesis.19,22, 23, 24 In HCC, ncRNAs have been proved to be key contributors to various biological and pathologic processes for tumor development and progression, making them potential therapeutic targets for HCC treatment.25,26
In this review, we describe the biology, function, and therapeutic strategies of macrophages and ncRNAs in tumors. Moreover, we focus on the crosstalk between macrophages and tumor cells mediated by ncRNAs and their functions and mechanisms in HCC. We primarily summarize the regulation of miRNAs, lncRNAs, and circRNAs derived from macrophages or tumor cells, as oncogenes or tumor suppressors, on the biological activity and function of their source cells and target cells. In addition, we explore the options available for clinical therapeutic strategies targeting crosstalk-related ncRNAs for HCC treatment.
Macrophages in tumors
According to their origins, TAMs are primarily divided into two subgroups: tissue-resident or monocyte-derived TAMs. Tissue-resident TAMs originate from fetal/embryonic hematopoietic progenitors and express high levels of F4/80, CX3CR1, and major histocompatibility complex (MHC) class II.27,28 Monocyte-derived TAMs originate from adult bone marrow hematopoietic stem cells with high expression of CD11b, CCR2, and MHC class II27. Tissue-resident TAMs often settle in specific tissues and organs and have many historical names, such as Langerhans cells (epidermis), Kupffer cells (liver), and microglia (brain).29, 30, 31, 32 Obviously, these macrophages are one of the earliest cells interacting with transformed cells in the process of tumor formation.33 Monocyte-derived TAMs originate from circulating monocytes in the peripheral circulation. In response to the internal environment after tumor formation, these cells are recruited to the TME and rapidly differentiate into macrophages.34 A variety of regulatory factors within the TME, including growth factors, chemokines, cytokines, peptides, and adhesion molecules, are involved in the recruitment and differentiation of monocyte-derived TAMs, and they are secreted or expressed by host or tumor cells.35
Both tissue-resident TAMs and monocyte-derived TAMs are associated with tumor progression. Tissue-resident TAMs appear to participate in at least the occurrence of tumors based on their specificity of tissue residency. Currently, the roles and molecular mechanisms of tissue-resident TAMs in tumorigenesis remain to be further investigated, which may be due to the lack of a deeper understanding of the origin and ontology of these macrophages.36,37 Recently, tissue-resident TAMs have been demonstrated to play important roles in cancer development by enhancing tumor growth and metastasis,38,39 while depletion of these cells significantly inhibits tumor progression.36 Moreover, tissue-resident macrophages can further promote cancer progression through their enhanced proliferation and expansion mediated by interleukin (IL)-4 within the TME.40 In general, tissue-resident TAMs are more related to the generation of tumor extracellular matrix and tumor vascular signals.27 However, monocyte-derived TAMs are more often induced to an immunosuppressive state in the body with the inhibition of immune cells and the activation of immunosuppressive cells.35,41,42 In addition, monocyte-derived TAMs are traditionally regarded as classic TAMs and have been well characterized to play important roles in tumor development by stimulating the proliferation and survival of tumor cells through various signaling pathways.27,35
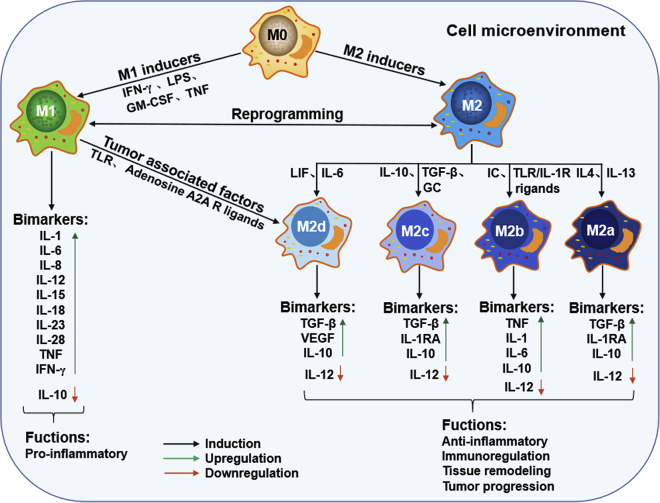
Macrophages are highly plastic, with polarized and nonpolarized states. Nonpolarized macrophages are known as the M0 type and can be reprogrammed into polarized M1 (classical activated macrophages) and M2 (alternatively activated macrophages) types, as shown in Figure 1. Moreover, the M1 or M2 type can also be reprogrammed into each other.27 Macrophage polarization is induced by microbial agents and cytokines in the local cell microenvironment.27,35 The inducers of M1 mainly include lipopolysaccharide (LPS), granulocyte macrophage colony-stimulating factor (GM-CSF), interferon gamma (IFN-γ), and tumor necrosis factor alpha (TNF-α), whereas M2 polarizes in response to Th2 cytokines, such as IL-4, IL-6, IL-10, IL-13, transforming growth factor β (TGF-β), glucocorticoid (GC), immune complex (IC), Toll-like receptor (TLR) or IL-1R ligands, and leukemia inhibitory factor (LIF) hormones.42,43 Moreover, based on the different stimulants, the M2 type can be divided into four subgroups (M2a, M2b, M2c, and M2d) (Figure 1). Differential cytokine production is a key biomarker of polarized macrophages; the M1 feature is typically IL-12, while the M2 feature is IL-10. M1 macrophages are highly potent effector cells that have anti-inflammatory and immunoregulatory roles, while M2 cells are involved in proinflammatory processes, adaptive Th1 immunity, tissue remodeling and repair, and tumor progression.27
Figure 1.
The polarization of macrophages and their characteristics
In response to different inducers in cell microenvironment, macrophages polarize into two extreme phenotypes (M1 and M2) and display distinct functions. M2 macrophages can be divided into four advanced subtypes, M2a, M2b, M2c, and M2d, based on their different stimuli and roles.
In the tumor environment, polarization of TAMs is regulated by the type of cancer, the stage of the tumor, clinical interventions, and the TME in particular.44 Some cytokines and molecules in the TME, such as IFN-γ, TNF-α, monoacylglycerol lipase (MGLL), and CD68, can induce TAM M1 polarity with corresponding antitumor activity.42,44, 45, 46, 47, 48, 49, 50 Hypoxia-inducible factors (HIFs), peripheral cannabinoid CB2 receptors (CB2Rs), CD163, signal transducer and activators of transcription (STATs), and arginase (Arg) are primarily responsible for TAM M2 polarity and tumor progression.42,44,47,51 M1 TAMs function as the main killer cells for suppressing tumors, while the role of M2 TAMs is to promote tumorigenesis and development. Moreover, in the special tumor microenvironment, polarized TAMs usually interact with other immune cells, such as natural killer (NK) cells, B cells, CD8+ cells (CTLs), γδ T cells, or CD4+ CD25+ Foxp3+ regulatory T (Treg) cells, to regulate each other, further resulting in tumor suppression or progression (Figure 2).27,35 Recently, many clinical studies have also observed that high levels of M1 TAMs in tumor tissue predict better survival outcomes in patients with cancer,52,53 while M2 TAM enrichment predicts poor prognosis.54 Moreover, TAMs in many types of cancer usually exhibit M2 polarity, making TAMs an attractive target for cancer therapy.55 Emerging therapeutic strategies targeting TAMs, especially M2 TAMs, have been investigated to remove them and/or re-educate them by inhibiting monocyte/macrophage recruitment or activation, directly eliminating macrophages, or modulating macrophage polarization from the M2 to M1 state.12,56,57
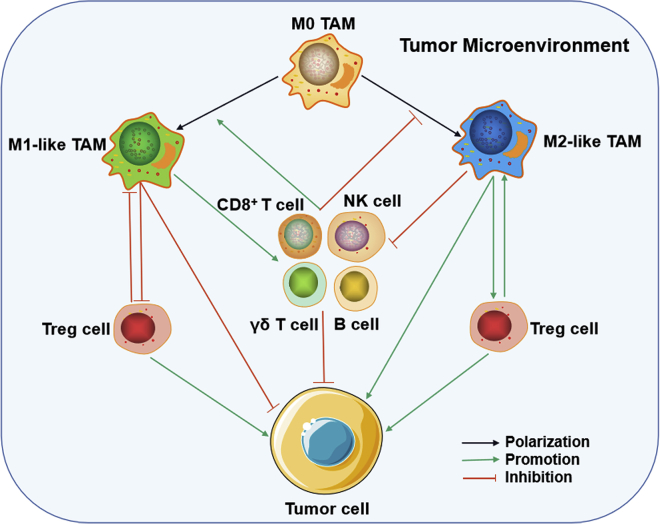
Figure 2.
TAM-involved tumor immune microenvironment
Various tumor-infiltrating cells, mainly including TAMs (i.e. M0, M1- and M2-like type), NK cells, CD8+ cells, γδ T cells, and regulatory T (Treg) (CD4+ CD25+ Foxp3+ Treg) cells, form a niche in the tumor microenvironment. Within the niche, intense communication occurs between macrophages and other immune cells through cell-to-cell contact and/or soluble messengers, resulting in tumor suppression or progression.
NcRNAs in tumors
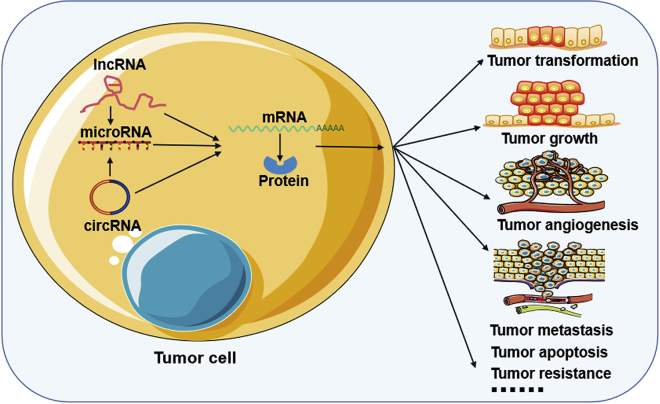
A growing number of studies have shown that ncRNAs, especially miRNAs, lncRNAs, and circRNAs, play important roles in cancer progression, including cancer transformation, growth, angiogenesis, metastasis, and drug resistance (Figure 3). Among ncRNA species, miRNAs are the most well-characterized group in cancer thus far.19,58 miRNAs are small endogenous ncRNAs with a length of 21–24 nt that are encoded in introns, exons, or intergenic regions of protein-coding genes.59 miRNAs recognize and bind to other RNAs, especially messenger RNAs (mRNAs) involved in fundamental cellular processes, in a base-complementation-dependent manner and regulate the expression of their target RNAs by inducing RNA cleavage or degradation or blocking RNA translation.19,60 Deregulation or genetic changes in miRNAs have been observed in all cancer types studied and have been demonstrated to act as tumor suppressors or oncogenes that play crucial roles in the pathogenesis of cancer in transformation, proliferation, survival, apoptosis, and several steps of the metastatic cascade.24,61, 62, 63 In addition, variation in the levels of some miRNAs is helpful for diagnosing early-stage cancer or defining the prognosis of late-stage cancer patients, which makes miRNAs useful as biomarkers and therapeutic targets.19,64 Currently, the application of biotechnologies targeting miRNAs in cancer includes eliminating oncogenic miRNAs using anti-miRNA oligonucleotides or small-molecule inhibitors and restoring suppressor miRNAs using viral vector-based gene restoration or miRNA mimics.65
Figure 3.
ncRNA functions in tumor progression
miRNAs regulate gene expression by inducing RNA cleavage or degradation or blocking RNA translation. lncRNAs function as molecular signals, decoys, guides, or scaffolds to mediate gene expression by interacting with DNAs, mRNAs, proteins, or other ncRNAs. circRNAs mediate gene expression by acting as miRNA sponges and protein scaffolds. Both of those ncRNAs play important roles in cancer progression, such as tumor transformation, growth, and angiogenesis.
Compared with miRNAs, lncRNAs can be either cis acting or trans acting to regulate gene expression at all levels: epigenetic, transcriptional, and posttranscriptional.66 lncRNAs were discovered a long time ago, although the term “lncRNA” was not coined until recently.19 Similar to mRNAs, lncRNA transcripts also consist of multiple exons and are enriched for H3K4 trimethylation at transcription start sites and H3K36 trimethylation across the genome.19,67 There are at least four groups of lncRNAs: intergenic, intronic, overlapping, and antisense.67,68 These lncRNAs function as molecular signals, decoys, guides, or scaffolds to mediate gene expression, which is usually achieved by interacting with DNAs, mRNAs, other ncRNAs, each other, or proteins.69, 70, 71 To date, many lncRNAs are often overexpressed in cancer and have been associated with poor outcomes.72,73 These lncRNAs have been elucidated as oncogenes that promote tumor progression by enhancing tumor growth, metastasis, and invasiveness.19 However, some lncRNAs acting as suppressors of cancer development are expressed at low levels in cancer and are related to good prognosis.72,73 These lncRNAs can be targeted using multiple approaches in cancer treatment, including knockdown of pathogenic RNAs by RNA interference (RNAi), antisense morpholino oligonucleotides, nucleic acid limitation, CRISPR-Cas9, transcriptional upregulation, and steric inhibition of lncRNA function using small molecules or therapeutic manipulation of lncRNA promoters.19,74
circRNAs are a newly described type of ncRNA with a covalently closed circular structure without 3′ or 5′ tails that is produced by back-splicing of premRNA. circRNA is widely expressed in eukaryotes and primarily located in the cytoplasm with highly stable and conservative properties that protect themselves from degradation by ribonuclease R (RNase R).75,76 According to their different compositions, circRNAs are classified into three categories: exonic circRNAs (ecircRNAs), intronic circRNAs (ciRNAs), and exon-intron circRNAs (eIciRNAs).77 Although the detailed mechanism of the biogenesis of cyclic RNAs is still being elucidated and confirmed, most circRNAs are formed by exon cyclization, and some cyclic RNAs are lasso structures formed by intron cyclization.78,79 However, an understanding of the biogenesis mechanisms, complexity, and functionality of these circRNAs remains elusive and needs further study. At present, some circRNAs have been found to be involved in gene expression by acting as miRNA sponges or decoys, as protein scaffolds, as insulators of RNA-binding protein (RBP), or as regulators of nuclear splicing and transcription.75,80 Regarding tumors, an increasing number of circRNAs have been discovered to be dysregulated in several types of tumors and are involved in the development of cancer with respect to tumor growth, invasion, metastasis, apoptosis, drug resistance, and angiogenesis.78,81,82 In addition, circRNAs can be detected in body fluids, such as blood, exosomes, and urine, indicating that circRNAs may represent potential diagnostic and prognostic biomarkers and therapeutic targets in cancer.77,83 To date, no effective approach has been reported regarding circRNA clinical applications, which requires further investigation.
Regulation of ncRNAs in the crosstalk between macrophages and cancer cells in hepatocellular carcinoma
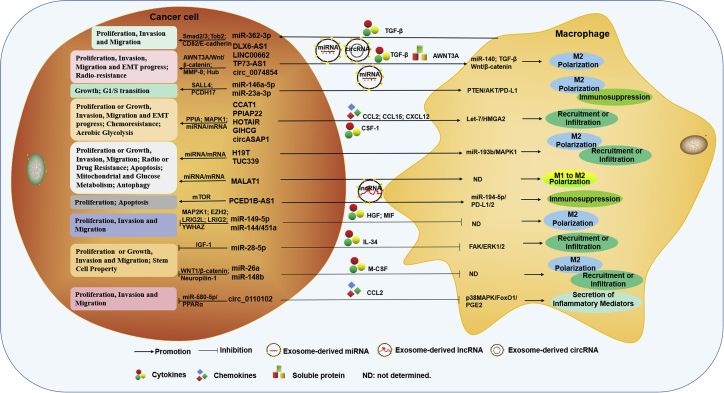
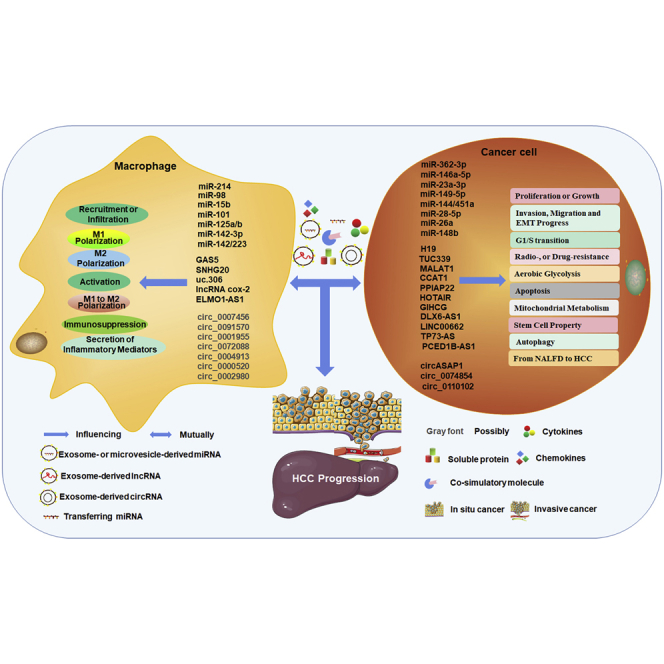
As described above, both macrophages and ncRNAs have been proved to play very important roles in tumor development and progression. As shown in Figures 4 and 5, crosstalk exists between macrophages and tumor cells, as both macrophages and tumor cells have been proved to directly transfer cellular components to other cells through cell-to-cell contact mediated by costimulation, secreted cytokines, or secreted extracellular vesicles (EVs), such as exosomes and microvesicles (MVs).84, 85, 86, 87, 88, 89 Among them, exosomes are the smallest vesicles (30–150 nm in diameter) and are produced as components of the plasma membrane of multivesicular bodies (MVBs) and released by the fusion of MVBs.90 They can mediate communication between tumor cells and their microenvironment through the transfer of information via their cargo and protect information molecules, including proteins, DNA, and ncRNAs, from degradation.91, 92, 93 In addition, these costimulatory molecules, cytokines, and EVs can influence each other within the TME and mediate the crosstalk between macrophages and tumor cells, resulting in the deterioration or slowing down of tumors.86,93
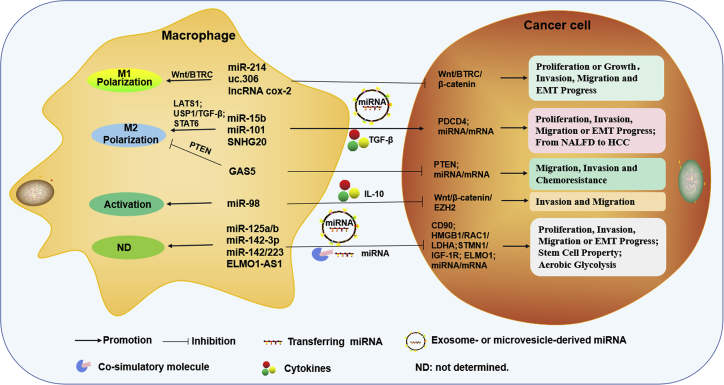
Figure 4.
Macrophage-derived ncRNA-mediated crosstalk between macrophages and HCC cells
Macrophage-derived ncRNAs not only regulate the activation or polarization of macrophages but also are involved in cancer cell proliferation, migration, invasion, EMT progress, stem cell properties, aerobic glycolysis, chemoresistance, or the progression from NAFLD to HCC, by cell-to-cell contact, cytokines, exosomes, or microvesicles.
Figure 5.
Tumor-derived ncRNA-mediated crosstalk between macrophages and HCC cells
Tumor-cell-derived ncRNAs not only regulate cancer cell proliferation, cell cycle, migration, invasion, EMT progress, stem cell property, mitochondrial metabolism, glucose metabolism, apoptosis, autophagy, and radio- or drug resistance but also are involved in the polarization or reprogramming, immunosuppression, secretion of inflammatory mediators, recruitment, or infiltration of macrophages by exosomes, cytokines, soluble protein, or chemokines.
In recent years, an increasing number of studies have demonstrated that ncRNAs can be expressed in both macrophages and tumor cells and are secreted in exosomes or microvesicles from these cells.94,95 These ncRNAs that are expressed extracellularly or secreted within EVs are well characterized to be involved in the biology of tumor cells and macrophages, making them important regulators of the crosstalk that occurs between these cell types. On the one hand, ncRNAs act as oncogenes or tumor suppressors in the proliferation, invasion, migration, apoptosis, or drug resistance of tumor cells, as mentioned above. On the other hand, they also mediate the infiltration, recruitment, activation, or polarization of macrophages.96 For example, miRNA-34-a (miR-34-a), miR-155, and lncRNA CCAT1 were found to play important roles in macrophage activation and M1 polarization.97, 98, 99, 100 Conversely, high levels of miR-21, miR-Let-7, and lncRNA MALAT1 are beneficial to macrophage M2 polarization.101, 102, 103, 104 Moreover, the regulation of ncRNAs in tumor cells has been demonstrated to be mediated by controlling macrophage phenotype and function.94,105,106 That is, ncRNAs, whether they arise from tumor cells or macrophages or whether they are self-expressed or derived from cell-derived EVs, can mediate the crosstalk between tumor cells or macrophages, hence affecting the development and progression of cancer, as shown in Tables 1, 2, and 3. We summarize the regulation of ncRNAs in the crosstalk between these two types of cells and their primary functions and mechanisms in HCC.
Table 1.
miRNAs associated with the crosstalk between macrophages and HCC cells
| miRNAs | Origin cells | Expression in HCC | Role in HCC | Ways for crosstalk | Function in macrophages and/or HCC cells | Reference |
|---|---|---|---|---|---|---|
| miR-15b | macrophages | upregulated | oncogene | exosomes | induce macrophage M2 polarization by LATS1; enhance HCC cell proliferation, invasion, and EMT progress by TGF-β or PDCD4 | 107, 108, 109, 110 |
| miR-101 | macrophages | downregulated | oncogene or suppressor | cytokines | induce macrophage M2 polarization by USP1/TGF-β axis; inhibit HCC cell proliferation, migration, and invasion | 111, 112, 113 |
| miR-125a/b | macrophages | downregulated | suppressor | exosomes | inhibit the biology of HCC cells or stem cells by CD90 | 114 |
| miR-142-3p | macrophages | downregulated | suppressor | microvesicles | inhibit HCC cell aerobic glycolysis, proliferation, migration, and invasion by HMGB1, RAC1, or LDHA | 115, 116, 117, 118 |
| miR-142/miR-223 | macrophages | downregulated | suppressor | cell-to-cell contact | inhibit HCC cell proliferation by STMN1 or IGF-1R | 110,119,120 |
| miR-98 | macrophages | downregulated | suppressor | cytokines | induce IL-10 secreting of macrophages; inhibiting HCC cell migration and invasion by Wnt/β-catenin/EZH2 axis | 64,121 |
| miR-214 | macrophages | downregulated | suppressor | ND | promote macrophage M1 polarization; inhibit HCC cell growth and invasion by β-catenin | 122, 123, 124, 125 |
| miR-146a-5p | HCC cells | upregulated or downregulated | oncogene (in mice) or suppressor | exosomes | induce macrophage M2 polarization and immunosuppression; promote HCC progression by SALL4 (in mice) | 126, 127, 128, 129, 130 |
| miR-23a-3p | HCC cells | upregulated | oncogene | exosomes | induce macrophage M2 polarization and immunosuppression by PTEN/AKT/PD-L1 axis; promote HCC cell G1/S transition by PCDH17 | 63,131 |
| let-7 | HCC cells | upregulated | oncogene | stimulating molecules | inhibit immune surveillance of macrophages by CD47-SIRPα axis; promote HCC cell growth and proliferation | 132, 133, 134 |
| miR-362-3p | HCC cells | upregulated | oncogene | cytokines | macrophages increase miR-362-3p expression in HCC cells by TGF-β/Smad2/3 axis; promote HCC cell proliferation, invasion, and metastasis by Tob2, CD82, or E-cadherin | 135,136 |
| miR-149-5p | HCC cells | downregulated | suppressor | cytokines | reduce macrophage M2 polarization by M-CSF; inhibit HCC cell invasion and migration by MAP2K1, LRIG2L, LRIG2, or MMP9 | 4,137,138 |
| miR-144/miR-451a | HCC cells | downregulated | suppressor | cytokines | re-educate macrophages from M2 to M1 polarization by HGF and MIF; inhibit HCC cell proliferation, invasion, and EMT progress by EZH2, YWHAZ, or MMP9 | 139, 140, 141, 142 |
| miR-26a | HCC cells | downregulated | suppressor | cytokines | suppress macrophage recruitment and M2 polarization by M-CSF; inhibit HCC cell growth and metastasis | 113,143, 144, 145, 146 |
| miR-148b | HCC cells | downregulated | suppressor | cytokines | Reduce macrophage infiltration and M2 polarization by M-CSF; suppress the biology of HCC cells or stem cells by WNT1/β-catenin or neuropilin-1 | 147, 148, 149 |
| miR-28-5p | HCC cells | downregulated | suppressor | cytokines | reduce macrophage infiltration by FAK/ERK1/2/IL-34; suppress the biology of HCC cells or stem cells by IGF-1 | 150, 151, 152, 153 |
Table 2.
lncRNAs associated with the crosstalk between macrophages and HCC cells
| lncRNAs | Origin cells | Expression in HCC | Role in HCC | Ways for crosstalk | Function in macrophages and/or HCC cells | Reference |
|---|---|---|---|---|---|---|
| SNHG20 | macrophages | upregulated | oncogene | ND | induce macrophage M2 polarization by STAT6; promote HCC cell invasion and EMT progress, enhance the progression from NAFLD to HCC | 154, 155, 156, 157 |
| uc.306 | macrophages | downregulated | suppressor | ND | induce macrophage M1 polarization by the Wnt/BTRC signaling | 158 |
| lncRNA cox-2 | macrophages | ND | suppressor | ND | induce macrophage M1 polarization; inhibit HCC cell proliferation, invasion, and migration | 99,159 |
| GAS5 | macrophages | downregulated | suppressor | ND | reduce macrophage M2 polarization by PTEN; inhibit HCC cell migration, invasion, and chemoresistance by miRNA/mRNA | 160, 161, 162, 163, 164 |
| ELMO1-AS1 | macrophages | downregulated | suppressor | ND | inhibit HCC cell proliferation, migration, and invasion by ELMO1 | 158,165 |
| CCAT1 | HCC cells | upregulated | oncogene | ND | induce macrophage infiltration by the Let-7/HMGA2 axis; promote HCC cell proliferation, migration, and invasion by many miRNA/mRNA axis | 166, 167, 168 |
| H19 | HCC cells | upregulated | oncogene | ND | induce macrophage infiltration and M2 polarization by the miR-193b/MAPK1 axis; promote HCC cell growth, migration, invasion, radio-, or drug resistance by many miRNA/mRNA axis | 169, 170, 171 |
| MALAT1 | HCC cells | upregulated | oncogene | ND | re-educate macrophages from M1 to M2 polarization; promote HCC cell proliferation, migration, chemotherapy, resistance, mitochondrial and glucose metabolism, inhibit cell apoptosis and autophagy by many miRNA/mRNA axis | 172, 173, 174, 175, 176 |
| PCED1B-AS1 | HCC cells | upregulated | oncogene | exosomes | induce macrophage immunosuppression by the miR-194-5p/PD-L1/2 axis; promote HCC cell proliferation, decrease cell apoptosis by mTOR signaling | 177 |
| TUC339 | HCC cells | upregulated | oncogene | exosomes | induce macrophage activation and M2 polarization; promote HCC cell cycle, proliferation, and adhesion | 105,178 |
| DLX6-AS1 | HCC cells | upregulated | oncogene | exosomes | induce macrophage M2 polarization by miR140; promote HCC cell viability, invasion, migration, and EMT progress by many miRNA/mRNA axis | 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135;136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170;171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212 |
| LINC00662 | HCC cells | upregulated | oncogene | soluble protein | induce macrophage M2 polarization by the AWNT3A/Wnt/β-catenin signaling; promote HCC cell proliferation and invasion by miR-15a/16/107/AWNT3A/Wnt/β-catenin signaling | 179,180 |
| TP73-AS1 | HCC cells | upregulated | oncogene | cytokines | induce macrophage M2 polarization by TGF-β; promote HCC cell proliferation, survival, and radioresistance by many miRNA/mRNA axis, including the miR-539-MMP-8 axis | 181, 182, 183 |
| PPIAP22 | HCC cells | upregulated | oncogene | chemokines | induce macrophage infiltration by CCL15 or CXCL12; promote HCC cell growth by the miR-197-3p/PPIA axis | 184 |
| HOTAIR | HCC cells | upregulated | oncogene | chemokines | induce macrophage recruitment by CCL2; promote HCC cell aerobic glycolysis, drug resistance, and EMT progress by many miRNA/mRNA axis | 185, 186, 187, 188 |
| GIHCG | HCC cells | upregulated | oncogene | ND | induce macrophage infiltration; promote HCC cell proliferation and metastasis by miR-200b/a/429 | 189,190 |
ND, not determined.
Table 3.
circRNAs associated with the crosstalk between macrophages and HCC cells
| circRNAs | Origin cells | Expression in HCC | Role in HCC | Ways for crosstalk | Function in macrophages and/or HCC cells | Reference |
|---|---|---|---|---|---|---|
| hsa_circ_0007456 hsa_circ_0091570hsa_circ_0001955hsa_circ_0072088hsa_circ_0004913hsa_circ_0000520hsa_circ_0002980 | macrophages Or HCC cells |
upregulated or downregulated or ND | oncogene or suppressor Or ND |
ND | induce or reduce macrophage infiltration, or ND; promote or inhibit the biology of HCC cells by the miRNA-mRNA axis, or ND | 191, 192, 193, 194, 195, 196, 197, 198, 199 |
| circASAP1 | HCC cells | upregulated | oncogene | cytokines | induce macrophage infiltration by regulating CSF-1; promote HCC cell proliferation, migration, and invasion by miR-326/miR-532-5p/MAPK1 signaling | 200,201 |
| hsa_circ_0074854 | HCC cells | upregulated | oncogene | exosomes | induce macrophage M2 polarization; promote HCC cell migration, invasion, and EMT progress by regulating HuR | 202 |
| hsa_circ_0110102 | HCC cells | downregulated | suppressor | chemokines | inhibit COX-2/PGE2 production of macrophages by the CCL2/CCR2/p38MAPK/FoxO1 signaling; inhibit HCC cell proliferation, migration, and invasion on the miR-580-5p-PPARα axis | 203,204 |
Regulatory functions mediated by macrophage-derived ncRNAs
Macrophage-derived miRNAs
Function as oncogenes by modulating macrophage polarization and promoting tumor cell proliferation, migration, and invasion
Leveraging the high stability and specificity in the body, exosome-derived ncRNAs are emerging as mediators of intercellular communication between cells of the same species or between cells of different species, as shown in Table 1. Li et al. showed that both the expression level and secretion level of miR-15b within exosomes were significantly increased in THP-1-derived macrophages treated with arsenite, an environmental toxin that is consequently present in air, food, and water.205,206 miR-15b is commonly upregulated in a variety of tumor tissues, including HCC, and it can be considered an indicator of poor prognosis when found in the serum of cancer-bearing patients.107,108 Downregulation of miR-15b suppresses cell proliferation, invasion, and epithelial–mesenchymal transition (EMT) in HCC cells by inhibiting TGF-β or programmed cell death 4 (PDCD4).107,109 Overexpression of miR-15 in macrophages treated with arsenite induced macrophage M2 phenotype polarization. When these miR-15b-enriched macrophages were cocultured with HCC cells, miR-15b was transferred from macrophages to HCC cells via exosomes. This transfer was demonstrated to promote cell proliferation, migration, and invasion by targeting LATS1 to inhibit activation of the Hippo pathway. In addition, the transfer was involved in the promotion of HCC formation in xenografts of nude mice.110 These results suggest that miR-15b not only favors the protumor ability of M2 TAMs on HCC cells but can also be transferred from macrophages to HCC cells via exosomes, leading to the deterioration of tumors.108,110
Cultured medium from macrophages, which includes many cytokines and chemokines, is commonly used to investigate the relationship between macrophages and HCC cells mediated by ncRNAs. Wei et al. found that, when cultured with medium derived from M2 macrophages treated with sorafenib, HCC cells exhibited decreased migration and invasion compared with cells treated with medium from untreated M2 macrophages.111 However, the decreased migration and invasion of HCC cells was reduced when cultured in the medium of M2 macrophages treated with sorafenib plus TGF-β.111 TGF-β is an essential factor for the protumor effects of TAMs on HCC cells.207 Moreover, the expression of miR-101 in M2-polarized macrophages was inhibited by sorafenib. Furthermore, inhibition of miR-101 enhanced the expression of its direct target DUSP1 to decrease the activation of ERK1/2, p38, and JNK as well as the production of TGF-β. Clinical data have also revealed that TGF-β levels in HCC tissues and serum are increased and positively correlated with the M2 macrophage marker CD206.111 This suggests that the miR-101/DUSP1/TGF-β axis is involved in the influences exerted by M2 TAMs on HCC progression. However, miR-101 was reported to be frequently downregulated in HCC tissues and plays suppressive roles in cell proliferation, migration, and invasion.112,113 Therefore, the expression and role of miR-101 in HCC tissues and TAMs need further investigation and verification.
Function as tumor suppressors by modulating macrophage polarization and inhibiting the biology of tumor cells or stem cells
Compared with miR-15b, which has protumor effects, miR-125a/b in exosomes from TAMs has been shown to exhibit antitumor potential. TAMs were isolated from human HCC tissues, and then the exosomes of TAMs were extracted and cocultured with human HCC cells. TAM-derived exosomes significantly promoted HCC cell proliferation, metastasis, and stem cell properties.114 miRNA profile assays determined that the expression of miR-125a/b was decreased in both the exosomes and cell lysates of TAMs isolated from HCC tissues. Not surprisingly, the expression of miR-125a/b was also downregulated in HCC cells. Overexpression of miR-125a/b suppressed HCC cell growth and stem cell properties (sphere formation). In addition, when HCC cells were transfected with miR-125a or miR-125b combined with TAM-derived exosomes, they exhibited a low proliferative state. That is, restoration of miR-125a or miR-125b in HCC cells antagonized the effect of TAM-derived exosomes. Furthermore, the suppressive effects of miR-125a/b on the proliferation of HCC cells were revealed to be mediated by targeting CD90.114 These data indicate that these TAMs tend to be the M2 type, and the promoting effect of these M2 TAM-derived exosomes on HCC cells is mediated by decreased secretion of miR-125a/b.114
Similar to exosome-derived miRNAs, microvesicle-derived miRNAs, including miR-142-3p, can also be transferred from TAMs to HCC cells. miR-142-3p, which is decreased in human HCC tissues and cell lines, was found to act as a tumor suppressor in aerobic glycolysis, proliferation, migration, and invasion of HCC cells by regulating HMGB1, RAC1, or LDHA.115, 116, 117 Interestingly, plasma levels of miR-142-3p within microvesicles were increased in a murine HCC model treated with propofol (2,6-diisopropylphenol, an intravenous antitumor agent).118,208 However, the expression of miR-142-3p was not upregulated in HCC cells treated with propofol, while HCC cells cocultured with TAMs or even microvesicles isolated from TAMs of propofol-treated HCC tissues exhibited significantly increased miR-142-3p, indicating that these TAMs tend to be the M1 type. In addition, miR-142-3p was demonstrated to be involved in the antitumor effects of propofol on HCC, suggesting that miR-142-3p in the MVs of TAMs is taken up by HCC cells, leading to tumor suppression.118
Aucher et al. demonstrated that macrophages transfer miRNAs to HCC cells primarily by delivering miRNAs to HCCs across intercellular contacts.110 Interestingly, not all miRNAs but only some specific miRNAs, such as miR-142 and miR-223, can be transferred from macrophages to HCC cells. Both miR-142 and miR-223 are downregulated in HCC tissues and considered good prognostic biomarkers of HCC.119,120 The two transferred miRNAs were found to be involved in the antitumor ability of primary human monocyte-derived macrophages against HCC cells. When cocultured with macrophages, the proliferation of HCC cells was significantly inhibited, and it could be restored by inhibitors of gap junctions, such as 18-alpha-GA, 2-octanol, and oleamide, suggesting that these TAMs tend to be the M1 type. In addition, the inhibitory effect of miR-142 and miR-223 on HCC cell proliferation was mediated by decreasing stathmin-1 (STMN1) and insulin-like growth factor-1 receptor (IGF-1R) in HCC cells.119,120 These data indicate that the transfer of miR-142 and miR-223 from macrophages to HCC cells occurs in a contact-dependent manner and plays an important role in the antitumor effects of primary human monocyte-derived macrophages on HCC cells.119,120
In addition to TGF-β, IL-10 is also an important anti-inflammatory molecule in the TME.209 Recently, IL-10 was shown to be involved in the miR-98-mediated relationship between macrophages and HCC cells.121 miR-98 is significantly decreased in HCC tissues and inhibits tumors by reducing the Wnt/β-catenin signaling pathway targeting EZH2.64 Similarly, the expression of miR-98 was also downregulated in HCC-associated TAMs. When cultured with medium from TAMs transferred with miR-98 mimics, the migration and invasion of HCC cells was significantly inhibited, while that of HCC cells was increased in miR-98 inhibitor-treated TAM medium, indicating that these TAMs tend to be the M2 type. Furthermore, IL-10, which was highly upregulated in HCC tissues and HCC-conditioned TAMs, was demonstrated to be a direct target of miR-98.121 Overexpression of IL-10 in HCC-conditioned TAMs reversed the inhibitory effects of TAMs treated with miR-98 mimics on HepG2 cells, indicating that the enhancing effect of TAM medium on HCC cells is mediated by IL-10 due to the low levels of miR-98 in TAMs.121
In addition, miR-214 was shown to be involved in the antitumor effects of M1-type TAMs against HCC cells in a murine HCC model.122 Transfecting TAMs isolated from HCC-bearing mice treated with norcantharidin (NCTD) with premiR-214 significantly increased the expression of miR-214 in TAMs. These miR-214-enriched TAMs displayed an M1 phenotype with higher levels of NOS2 but lower levels of Arg-1.122 The expression of miR-214 is downregulated in human HCC tissues and associated with tumor progression and poor prognosis. Ectopic expression of miR-214 suppressed tumor growth and angiogenesis in a murine xenograft model of human HCC.123 Further study found that overexpression of miR-214 in HCC cells inhibited tumor cell growth and invasion through suppression of β-catenin.124,125 Similarly, when TAMs isolated from an NCTD-treated murine HCC model were cocultured with H22 murine HCC cells, they significantly decreased the survival and inhibited the invasion of H22 cells.122 These data suggest that overexpression of miR-214 not only directly inhibits tumor cells but also induces M1 polarization of TAMs to further inhibit tumor cells.122, 123, 124, 125
Macrophage-derived lncRNAs
Function as oncogenes by inducing macrophage M2 polarization and promoting tumor cell invasion and EMT progression
Small nucleolar RNA host gene 20 (SNHG20), an lncRNA upregulated in many cancer types, has been frequently identified as an oncogene that contributes to cancer development and progression.210 Similar to other cancers, SNHG20 expression is upregulated in HCC tissues, including in nonalcoholic fatty liver disease (NAFLD)-related HCC, and predicts poor prognosis of patients. SNHG20 overexpression has been reported to play a promoting role in the invasion of HCC cells by targeting hepatocyte growth factor receptor (MET).154, 155, 156 In liver macrophage Kupffer cells (KCs) with the M2 phenotype isolated from HCC or NAFLD-HCC, SNHG20 was also markedly upregulated compared with that in KCs isolated from NAFLD and NAFLD-HCC adjacent tissues in both humans and mice. Knockdown of SNHG20 induced M1 polarization of macrophages, while SNHG20 overexpression caused M2 polarization.154 In addition, the promoting effect of SNHG20 on macrophage M2 polarization was shown to be mediated by STAT6 signaling, which is a key factor in the activation of M2 macrophages.154,157 Moreover, injection of macrophages infected with shRNA lentivirus targeting SNHG20 delayed the progression from NAFLD to HCC in mice.154 This finding indicates that the protumor effects of SNHG20 on HCC development and progression are further enhanced by SNHG20-induced M2 polarization. Knockdown of SNHG20 may represent a promising strategy for treating HCC and at least may delay malignant progression of NAFLD to HCC by regulating macrophage polarization.154,157
Function as tumor suppressors by modulating macrophage polarization and inhibiting tumor cell proliferation, invasion, migration, or chemoresistance
Transcribed ultraconserved regions (T-UCRs) are a special class of lncRNAs that are highly evolutionarily conserved across many species. Increasing results have shown that T-UCRs are involved in the pathogenesis and diagnosis of many human diseases, including cancer. In hepatobiliary cancers, compared with the normal liver and biliary epithelia, T-UCR uc.158 was increased in cholangiocarcinoma and exerted a promoting function in tumor growth. Mechanistically, the Wnt signaling pathway, which plays promoting roles in malignant tumor progression, and miR-193b, which functions as a tumor suppressor, were identified upstream and downstream, respectively.211 Unlike T-UCR uc.158, T-UCR uc.306 was found to be downregulated in HCC tissues and positively associated with poor survival outcomes in patients. Interestingly, uc.306 expression was significantly increased in M1-type TAMs but decreased in M2-type TAMs. In addition, BTRC, a key molecule in Wnt signaling, was predicted to be a target of uc.306, indicating that uc.306 may be a tumor suppressor involved in the antitumor effects of M1-type TAMs by targeting the Wnt signaling pathway in HCC cells.158
The lncRNA cox-2 is an lncRNA originating close to the COX-2 gene and has been demonstrated to regulate the transcription of immune and inflammatory genes.212,213 Recently, it was reported that cox-2 expression was also upregulated in M1 macrophages compared with Mo and M2 macrophages. In addition, suppression of the lncRNA cox-2 decreased macrophage polarization to the M1 type and depressed the antitumor ability of M1 macrophages to suppress proliferation, invasion, and migration of HCC cells. However, lncRNA cox-2 silencing in M2 macrophages strengthened tumor progression.95 The lncRNA cox-2 has been shown to regulate the progression of liver fibrogenesis.214 However, there are few reports concerning the lncRNA cox-2 in HCC at present. However, the lncRNA cox-2 is upregulated in HCC tissues and acts as a regulator of differentiation grade in HCC. Silencing of the lncRNA cox-2 in HCC cells significantly inhibits cell growth and induces apoptosis.159 These studies demonstrate that the lncRNA cox-2 is responsible for the M1 polarization of macrophages that suppresses tumor development.159,214 Nonetheless, the expression and function of the lncRNA cox-2 in HCC tissues and cells still need to be further elucidated.
Similarly, lncRNA growth arrest-specific transcript 5 (GAS5) is also upregulated in M1-type macrophages but downregulated in M2-type macrophages and TAMs.160 However, the expression levels of GAS5 are reduced in HCC tissues, and GAS5 functions as a tumor suppressor.161,162 Overexpression of GAS5 in HCC cells suppressed migration and invasion and overcame tumor cell chemoresistance by upregulating tumor suppressor genes, including phosphatase and tensin homolog deleted from chromosome 10 (PTEN).163,164 In TAMs, GAS5 overexpression enhanced M1-like polarization of TAMs but inhibited TAM M2-like polarization by upregulating PTEN.160 Taken together, these data indicate that the GAS5-PTEN axis plays important inhibitory roles in TAM M2 polarization and the tumor cell phenotype, leading to tumor suppression.160,163,164
In addition, lncRNA engulfment and cell motility 1 (ELMO1) antisense RNA 1 (ELMO1-AS1) was also discovered to be markedly increased in M1-type TAMs and decreased in M2-type TAMs from HCC tissues.158 In contrast, the expression of ELMO1-AS1 is downregulated in HCC tissues and negatively correlated with good patient outcomes. ELMO1-AS1 overexpression in HCC cells significantly inhibited cell proliferation, migration, and invasion by targeting engulfment and cell motility 1 (ELMO1).165 However, there is little research on the influence of ELMO1-AS1, discovered as a macrophage-related lncRNA in HCC, on the polarization and function of TAMs.158,165 The ELMO1-AS1-associated relationship between macrophages and tumor cells and its mechanism need to be further explored.
Macrophage-derived circRNAs
To date, there have been few studies on macrophage-derived circRNAs in tumors. Even in HCC, no relevant reports have been retrieved, as shown in Table 3. Zhou et al. constructed a circRNA-miRNA-mRNA regulatory network based on macrophage-related differentially expressed mRNAs (DEmRNAs) in HCC by calculating the immune cell fractions of HCC from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases.191,192 In a circRNA-miRNA-mRNA regulatory network, the hsa_circ_0007456-hsa-miR-139-5p axis was identified. hsa_circ_0007456 has some binding sites for miR-139-5p and may act as a miRNA sponge for hsa-miR-139-5p. Low expression of hsa-miR-139-5p is associated with a low infiltration level of macrophages, malignant clinicopathological features of tumors, and poor patient prognosis.191 miR-139-5p has been reported to be decreased in HCC samples and acts as a tumor suppressor to regulate cell invasion and proliferation.193 In addition, four DEmRNAs were predicted as targets of hsa-miR-139-5p: CDCA8, KPNA2, PRC1, and TOP2A, most of which are oncogenes that are increased in HCC tissues.191,192,194 These data indicate that hsa_circ_0007456 may act as an oncogene in the progression of HCC by sponging miR-139-5p.193,194
In addition, six other macrophage-related DEcircRNAs in HCC were included in the circRNA-miRNA-mRNA regulatory network: hsa_circ_0091570, hsa_circ_0001955, hsa_circ_0072088, hsa_circ_0004913, hsa_circ_0000520, and hsa_circ_0002980.192 Among them, hsa_circ_0001955 and hsa_circ_0072088 have been demonstrated to be upregulated in HCC and to function as protumor agents.195,196 In contrast, hsa_circ_0091570 and hsa_circ_0005075 have been shown to be downregulated in HCC and to function as antitumor agents.197, 198, 199 However, none of them have been found to be involved in the biology, polarization type, or function of macrophages related to HCC. Therefore, the expression relationships among these circRNAs and their downstream miRNAs and mRNAs need more detailed research.
Taken together, these studies focused on macrophages and identified the relationship between macrophages and HCC cells mediated by circRNAs. Unfortunately, as the results were all obtained from whole HCC tissue, including macrophages, tumor cells, and other immune cells, the source of the circRNAs in this network could not be confirmed (Table 3). Therefore, future studies should focus on the expression and function of circRNAs in both macrophages and tumor cells as well as the relationship between these two cell types and their mechanisms.
The regulatory effects mediated by tumor-cell-derived ncRNAs
Similar to the ncRNAs derived from macrophages described above (Figure 4), many tumor-derived miRNAs, lncRNAs, or circRNAs are also involved in the relationship between macrophages and HCC cells (Figure 5), acting as oncogenes or tumor suppressors through cell-to-cell contact, costimulatory molecules, exosomes, microvesicles, soluble proteins, cytokines, or chemokines (Tables 1, 2, and 3).
Tumor-cell-derived miRNAs
Function as oncogenes by modulating macrophage polarization and immunosuppression and promoting tumor cell proliferation, the cell cycle, invasion, and metastasis
Increasing reports have shown that tumor-derived exosomes can be taken up by macrophages, leading to their infiltration, activation, M2 polarization, and immunosuppressive activity.215 Some miRNAs, such as miR-146a-5p, are also selectively packaged into the exosomes of HCC cells and play important roles in macrophage features and functions. The present research on the expression level and function of miR-146a-5p in human HCC is contradictory. Some studies have shown that miR-146a-5p is highly expressed in HCC and plays an antitumor role, while other studies have reported the opposite views.126, 127, 128, 129 Recently, miR-146a-5p was shown to be significantly increased in both human and murine HCC cell lines and to be abundantly contained within exosomes. When cultured with miR-146a-5p-enriched exosomes from the murine HCC cell line Hepa1–6, the generation of CD206+ macrophages was increased, while the expression levels of TNF-α were downregulated. In addition, HCC exosome-treated macrophages exhibited enhanced immunosuppressive activity by inhibiting the T cell response and inducing T cell exhaustion.215 Mechanistically, unlike miR-146a-5p, which is regulated by STAT3 in human HCC cells, miR-146a-5p in murine HCC cells was shown to be regulated by Sal-like protein-4 (SALL4).130,215 In a murine model, blocking SALL4/miR-146a-5p signaling induced a T cell response, reversed T cell exhaustion, and inhibited HCC progression.215 These results suggest that transferring miR-146a-5p from HCC exosomes promotes TAM M2 polarization and immunosuppression by targeting SALL4, resulting in the promotion of HCC progression.130,215
Interestingly, the release of exosomes by HCC cells was recently shown to be increased by endoplasmic reticulum (ER) stress.131 ER stress plays an important role in tumor progression not only by regulating the intracellular biology of tumor cells but also by influencing the tumor microenvironment and immune responses.216 In human HCC, ER stress is upregulated and positively correlated with a decreased patient survival rate. Moreover, ER stress activation is also associated with increased programmed death ligand 1 (PD-L1) expression by macrophages in HCC cells through secretion of a higher number of exosomes.131 The programmed cell death protein 1 (PD-1)/PD-L1 axis is an important immune checkpoint that is responsible for tumor immune escape by suppressing the host antitumor immune response.217 PD-1 is expressed on immune cells, such as macrophages, T cells, and B cells, while PD-L1 is expressed on tumor cells.218,219 Therefore, the expression of PD-L1 in macrophages is unusual. These PD-L1high macrophages treated with exosomes derived from ER-stressed HCC cells were shown to significantly decrease the ratio of CD8+ T cells and induce the apoptosis of T cells. Mechanistically, miR-23a-3p in exosomes derived from ER-stressed HCC cells was shown to be involved in the upregulation of PD-L1 expression in macrophages by targeting the PTEN/protein kinase B (AKT) pathway.131 miR-23a-3p is upregulated in HCC tissues and cells and promotes G1/S cell cycle transition by regulating PCDH17.63 That is, the transfer of miR-23a-3p from ER-stressed tumor cells to macrophages induces PD-L1 expression in macrophages to polarize M2 type, resulting in the inhibition of T cell function and the promotion of HCC progression.63,131
Lethal-7 (let-7) was the first discovered human miRNA and generally functions as a tumor suppressor in human cancer.220 However, let-7i-5p was recently found to be significantly overexpressed in HCC tissues and shown to be an oncogenic molecule related to tumor immune escape.132 Although let-7i-5p expression is low in HCC cell lines, its silencing by antisense let-7i-5p (AS-let-7i-5p) significantly suppressed tumor cell growth and proliferation. In particular, let-7i-5p is involved in restoring the antitumor effects of histone deacetylase 6 (HDAC6) in HCC.132 HDAC6 is a tumor suppressor in HCC that functions by decreasing cell proliferation, invasion, and migration and increasing apoptosis.132,133 When HDAC6-treated HCC cells were cotransfected with a let-7i-5p mimic, all of the tumor-suppressing effects of HDAC6 were rescued. Further study indicated that the tumor-promoting effect of let-7i-5p occurs through inhibition of the thrombospondin-1 (TSP1)/CD47 signaling pathway.132 CD47 also serves as a ligand for signal regulatory protein α (SIRPα) expression on macrophages. The CD47-SIRPα axis inhibits the phagocytosis of tumor cells by M1 macrophages.134 Furthermore, let-7i-5p blocks the TSP1/CD47 pathway by targeting TSP1 to inhibit the antitumor roles of TSP1. Meanwhile, let-7i-5p indirectly releases more CD47 to bind SIRPα, thus escaping the immune monitoring function of M1 macrophages to further favor HCC development.132,134
miR-362-3p is upregulated in HCC tissues and cell lines and acts as an oncogene by promoting cell proliferation, invasion, and metastasis by targeting Tob2, CD82, or E-cadherin.135,136 TGF-β is one of the major cytokines in the TME and can be secreted by M2 TAMs.207 Interestingly, the expression of miR-362-3p can be increased by TGF-β, which indicates that TAMs are involved in the upregulation of miR-362-3p in HCC.136 When human HCC cells were cocultured with M2 TAMs derived from THP-1 cells, the expression of miR-362-3p in HCC cells was significantly increased. Moreover, TGF-β also positively regulated the miR-362-3p-mediated EMT process of HCC cells by increasing the expression of CD82 and E-cadherin, especially the phosphorylation of Smad2/3. Furthermore, Smad2/3 positively regulates the transcription of miR-362-3p by binding to its promoter.136 These results suggest that the protumor effects of M2 TAMs are also manifested by upregulation of the oncogene miR-362-3p through the TGF-β/Smad2/3 pathway.136
Function as tumor suppressors by modulating macrophage polarization and inhibiting the biology of tumor cells or stem cells
In contrast to miR-362-3p, miR-149-5p expression levels in HCC cells were decreased by M2 TAMs.4 miR-149-5p is decreased in HCC tissues and has been widely demonstrated to be a tumor suppressor gene by targeting MAP2K1, LRIG2 L, or LRIG2.137,138 When HCC cells were cocultured with M2 TAMs, the expression levels of miR-149-5p decreased in HCC cells with enhanced invasion and migration. Overexpression of miR-149-5p in HCC cells reversed the promoting function of M2 macrophages.4 In addition, MMP9 was shown to be a direct target of miR-149-5p, indicating that miR-149-5p/MMP9 signaling in HCC cells is involved in the pro-invasion and pro-migration effects of M2 TAMs on HCC.4,137,138
The expression levels of the miR-144/miR-451a cluster are decreased in HCC tissues and positively correlated with increased patient survival.139,140 As a tumor suppressor, the tumor-suppressive roles of the miR-144/451a cluster include inhibiting proliferation, EMT, and invasion of HCC cells by targeting EZH2 or YWHAZ.139,140 Recently, the tumor-suppressive roles of these miRNAs were found to be also mediated by regulating the tumor immune microenvironment. In a murine HCC model, TAMs isolated from miR-144/451a-overexpressing tumors exerted an increase in antitumor cytokines or molecules with the M1 phenotype but a decrease in M2-type cytokine production.141 Similarly, clinical data showed that the expression levels of miR-144 and miR-451a were positively correlated with the M1 marker HLA-DR but negatively associated with the M2 marker CD163.141 Mechanistically, miR-144/451a-induced remodeling of TAM polarization was mediated by targeting hepatocyte growth factor (HGF) and macrophage migration inhibitory factor (MIF) in HCC cells. MIF and HGF are essential cytokines that mediate the recruitment, differentiation, and M2 polarization of macrophages.142 In addition, the miR-144/miR-451a cluster is involved in the feedback circuit with EZH2 and the catalytic subunit of the polycomb repressive complex (PRC2) to repress its expression in HCC.141 These data indicate that restoring the expression of the miR-144/miR-451a cluster in HCC cells could not only directly inhibit tumor biology but also reprogram TAMs from the protumor M2 phenotype into the antitumor M1 phenotype.141,142
miR-26a in different tumor types has been reported to function as either an oncogene or tumor suppressor gene. In HCC, although the expression levels and roles of miR-26a are still controversial, miR-26a is generally identified as a tumor suppressor that is downregulated in HCC tissues and cell lines.113,143,144 Overexpression of miRNA-26a was reported to not only directly inhibit tumor growth and metastasis but also suppress the recruitment of macrophages to the TME.145,146 When macrophages derived from THP-1 cells were cultured in medium from miR-26a-enriched HCC cells, the migration of THP-1 cells was markedly decreased, and the expression of M1-type cytokines, such as IL-12b and IL-23, was upregulated. Conversely, those derived from THP-1 cells cultured with the medium of miR-26a-silenced HCC cells were increased. Comparing the medium from different HCC cells, the secretion of macrophage colony-stimulating factor 1 (M-CSF or CFS1) was found to be significantly decreased in the medium from miR-26a-enriched HCC cells compared with that in control or miR-26a-silenced HCC cells.146 M-CSF (CFS1) plays a pivotal role in the regulation of monocyte/macrophage characteristics in tumors.221 In addition, a clinical investigation showed that HCC patients displayed higher miR-26a expression and lower M-CSF expression and CD68+ macrophage (M0 type) numbers.146
Similar to miR-26a, miR-148b expression is downregulated in HCC tissues and negatively associated with the metastasis and prognosis of patients.147, 148, 149 miRNA-148b overexpression suppresses the biological characteristics of HCC cells or liver cancer stem cells (CSCs) by regulating the WNT1/β-catenin pathway or neuropilin-1.148 Moreover, miRNA-148b silencing increased the infiltration and M2 polarization of macrophages by targeting M-CSF (CSF1) to promote HCC growth and metastasis. The relationship among miR-148b, M-CSF expression, and TAM infiltration was also confirmed in HCC patients.149 These data suggest that overexpression of miR-26a or miR-148b in HCC cells inhibits the infiltration of macrophages by inhibiting the expression and secretion of M-CSF and by inducing M1 polarization of infiltrated macrophages, ultimately inhibiting HCC progression.148,149
In addition to CSF-1, IL-34 is another essential cytokine involved in monocyte/macrophage infiltration, differentiation, and function.150 CSF-1 and IL-34 have been described as "twin" cytokines sharing the common receptor CSF-1R and common signaling pathways, indicating that IL-34 might also be a target of miRNA in the crosstalk between cancer cells and macrophages.221 IL-34 has been shown to be a direct target of miR-28-5p in HCC cells. Similar to miR-148b, miR-28-5p was decreased in human HCC tissues, HCC cell lines, and liver CSCs.151 miR-28-5p overexpression markedly inhibits the proliferation and migration of HCC cells, and miR-28-5p overexpression inhibits the self-renewal and tumorigenesis of CSCs by targeting insulin-like growth factor-1 (IGF-1).152,153 In addition, miR-28-5p-silenced HCC cells express and secrete high levels of IL-34. When macrophages were cultured with IL-34-enriched medium from HCC cells, the chemotaxis and proliferation of macrophages were significantly increased, which could be rescued by anti-IL-34.151 Moreover, the promoting effects of miR-28-5p-IL-34 on TAM infiltration were also confirmed. Mechanistically, the focal adhesion kinase (FAK) and extracellular signal-related kinase 1 and 2 (ERK1/2) signaling pathways were revealed to be involved in IL-34-mediated anti-macrophage activity. Interestingly, TGF-β derived from TAMs suppressed the expression of miR-28-5p in HCC cells. The relationship between miR-28-5p-IL-34 expression and TAM infiltration was confirmed in HCC samples.151 Taken together, these data identified a miR-28-5p-IL-34 HCC-macrophage-positive feedback signaling loop that mediates TAM (M0 type) infiltration and HCC progression and metastasis.151, 152, 153
In addition, several other macrophage-related miRNAs have been identified in HCC, such as mir-194-5p, miR-424-5p, and miR-139-5p.177,191,222 Most of these miRNAs are involved in other ncRNA regulatory networks and play important roles in the antitumor or protumor functions of other ncRNAs, particularly lncRNAs and circRNAs, which will be discussed in the subsequent sections.
Tumor-cell-derived lncRNAs
Interestingly, all tumor-derived lncRNAs that can mediate the relationship between tumor cells and macrophages promote tumorigenesis and development. As shown in Figure 6, these oncogenic tumor-cell-derived lncRNAs not only can regulate their own biological functions but also can modulate macrophage infiltration, activation, polarization, or immunosuppression to facilitate tumor progression via costimulatory molecules, exosomes, soluble proteins, or cytokines.
Figure 6.
Promoting function of tumor-derived lncRNAs by mediating the crosstalk between macrophages and HCC cells
On the one hand, tumor-derived lncRNA acts as an oncogene in the biological function of the cells from which it originated, and on the other hand, it induces the M2-like polarization of TAMs while inhibiting its M1-like polarization cells through cytokines, chemokines, exosomes, or soluble proteins, thereby prompting macrophages to secrete a large amount of anti-inflammatory cytokines, further enhancing tumor progression.
The lncRNA CCAT1 is markedly increased in HCC tissues and is associated with poor prognosis. Overexpression of CCAT1 promotes the proliferation and migration of HCC cells, while silencing CCAT1 inhibits these phenotypes. Mechanistically, CCAT1 can act as a miRNA sponge of let-7, miR-30c-2-3p, or miR-181 to antagonize their functions, resulting in the derepression of miRNA targets, such as HMGA2, c-Myc, and ATG7.166, 167, 168 Recently, it was reported that, when cocultured either with TAMs in a Transwell system or with TAM supernatant, the expression levels of CCAT1 and HMGA2 were increased, while the expression level of let-7 was decreased. In addition, HMGA2 expression in HCC tissues was confirmed to be positively associated with TAM markers (CD68, CD163, and CD204). Interestingly, in a Transwell coculture system, HMGA2 overexpression in HCC cells promoted TAM migration and IL-10 expression, while IL-12 was inhibited.166 This indicates that a positive feedback signaling loop between TAMs enhances HCC progression by targeting CCAT1-Let-7-HMGA2 HCC macrophages, leading to TAM (M0 type) infiltration and HCC progression.166,168
Similarly, the lncRNA H19 and its competitive endogenous RNA (ceRNA) network are also enhanced by TAM.169 H19 is also upregulated in HCC tissues, correlates with worse patient outcomes, and acts as an oncogene to promote tumor growth, metastasis, and radioresistance or chemotherapeutic resistance. Mechanistically, many signaling pathways have been demonstrated to participate in the protumor effects of H19, which involves several miRNAs and their targets, such as miR-520a-3p-LIMK1, miR-193b-MAPK1, and miR-200b-3p-Zeb1.169, 170, 171 Moreover, H19 upregulation was recently revealed to be positively correlated with TAM infiltration, particularly M2-type TAMs. In the Transwell coculture system, H19 expression in HCC cells was significantly increased in response to TAM stimulation. In addition, the migration and invasion of HCC cells mediated by the H19-miR-193b-MAPK1-EMT axis was enhanced. Knockdown of H19 dramatically reduced the enhanced HCC aggressiveness induced by M2-like TAMs.169
In addition, MALAT1 is also upregulated in HCC tissues and is closely related to tumor progression, metastasis, and recurrence in patients. Knockdown of MALAT1 in HCC cells inhibits cell proliferation, migration, chemotherapy resistance, and mitochondrial and glucose metabolism while promoting apoptosis and autophagy.172, 173, 174, 175 When cultured in medium from MALAT1-silenced HCC cells, the migration and proliferation of human umbilical vein endothelial cells (HUVECs) are inhibited, leading to inhibition of tumor angiogenesis. MALAT1 downregulation significantly suppresses the M2-like polarization of macrophages but promotes their M1-like polarization. Furthermore, the reducing effects of MALAT1 silencing in HUVECs and macrophages were demonstrated to be due to the restoration of miR-140 expression in HCC cells.176 These results demonstrated that M2-like TAMs have the ability to reprogram HCC cells by targeting the lncRNA-centered signaling network, resulting in more aggressive tumors.176
PD-L1 is frequently expressed on many types of tumors to escape immune surveillance.219 In HCC tissues, both PD-L1 and PD-L2 were upregulated and positively associated with tumor size and tumor-node-metastasis (TNM) stage.177 In addition, the expression of lncRNA PCED1B-AS1 was increased and correlated with the clinical characteristics of HCC. It was further confirmed that PCED1B-AS1 promotes the expression of PD-L1 and PD-L2 by sponging miR-194-5p. In addition, PCED1B-AS1 exerted significant promoting roles in cell proliferation and colony formation through mammalian target of rapamycin (mTOR) signaling in HCC cells.177 Moreover, PCED1B-AS1 could be released from HCC exosomes and then taken up by M1 macrophages and T cells, resulting in an increase in apoptosis and inhibition of proliferation.177 These findings indicate that PCED1B-AS1 not only promotes tumor immune escape through the PD-1/PD-L1 axis but also enhances the malignant process of tumors through other pathways.177
The lncRNA TUC339 and distal-less homeobox 6 antisense 1 (DLX6-AS1) are also likely overexpressed in HCC tissues and HCC-derived exosomes.223,224 TUC339 promotes cycle progression, proliferation, and adhesion of HCC cells, and TUC339 transferred in exosomes from HCC cells to HCC cells further promotes tumor growth and progression.223 Meanwhile, HCC exosome-derived TUC339 can also be taken up by macrophages to regulate their activation and M2-like polarization.178 Downregulation of DLX6-AS1 in HCC cells was found to dramatically inhibit the biological functions of HCC cells, including cell viability, invasion, and migration, by targeting miRNA-involved signaling pathways.225, 226, 227, 228, 229 The density of TAMs in HCC tissues was positively correlated with the expression level of HCC exosome-derived DLX6-AS1.224 Further study demonstrated that, when treated with HCC-derived exosomes, TAMs could take up the transferred DLX6-AS1 and induce M2 polarization, leading to their antitumor ability to inhibit HCC cell migration, invasion, and EMT. Then, the transfer of DLX6-AS1 was confirmed to increase the expression of CXCL17 by sponging mmiR-15a-5p to mediate the polarization and function of TAMs.224 These results indicated that, in addition to directly affecting tumor cell characteristics, lncRNAs could also be secreted in tumor-derived exosomes and regulate immunological surveillance or M2-like polarization of macrophages, further accelerating the progression and metastasis of HCC.224
LINC00662 is upregulated in HCC tissues and associated with the clinical characteristics and prognosis of HCC patients.179,180 LINC00662 promotes HCC cell proliferation and invasion and suppresses apoptosis in HCC cells. In addition, LINC00662 induces M2-like macrophage polarization of macrophages cultured with conditioned medium from LINC00662-enriched HCC cells. Mechanistically, LINC00662 in HCC cells increased the expression and secretion of wingless-type MMTV integration site family member 3 (AWNT3A) by sponging miR-15a/16/107 to activate Wnt/b-catenin signaling, resulting in tumor cell proliferation and invasion.180 Moreover, secreted WNT3A was taken up by macrophages to trigger cell M2 polarization through Wnt/b-catenin signaling, suggesting that soluble proteins are involved in the LINC00662-mediated relationship between HCC cells and macrophages.179,180
As mentioned above, TGF-β can be increased by both macrophages and HCC cells and participates in miRNA-related intercellular communication.107,111,136,207 Recently, it was also reported that lncRNAs improve the expression of HCC-derived TGF-β, leading to M2 polarization of macrophages.181 The lncRNA TP73-AS1 was upregulated in HCC tumor tissues and was positively associated with patient survival. TP73-AS1 overexpression plays important promoting roles in proliferation, survival, and radioresistance through multiple signaling pathways by sponging certain miRNAs.182,183 The lncRNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. In particular, activation of the TP73-AS1-miR-539-MMP-8 axis was revealed to not only participate in tumor cell growth but also augment the M2 polarization of macrophages in response to HCC-secreted TGF-β, enhancing HCC tumor progression.181
Pseudogenes are a special type of lncRNA that have been less investigated in HCC. Recently, pseudogene peptidylprolyl isomerase A pseudogene 22 (PPIAP22) was found to be significantly upregulated in HCC tissues and negatively associated with good patient outcomes. Further study found that PPIAP22 may act as a ceRNA by sponging miR-197-3p to relieve repression of its parental gene peptidylprolyl isomerase A (PPIA).184 Similar to PPIAP22, upregulation of PPIA is also associated with tumor size and poor patient survival and has been shown to promote tumor growth.184,230 The expression levels of PPIAP22 or PPIA were correlated with the infiltration levels of tumor-infiltrating immune cells (TIICs), such as T cells, dendritic cells, and macrophages, and with the expression of several chemokines, including C-C motif chemokine ligand 15 (CCL15) and C-X-C motif chemokine ligand 12 (CXCL12).184 Chemokines are likely also involved in the lncRNA HOTAIR-mediated relationship in HCC. HOTAIR was upregulated in HCC tissues and associated with poor prognosis of patients. Many reports have shown that HOTAIR contributes to HCC progression primarily through the lncRNA-miRNA-mRNA axis.185, 186, 187 In addition, HOTAIR plays an important role in the recruitment of macrophages and myeloid-derived suppressor cells (MDSCs) into the TME by upregulating CCL2 expression in HCC cells.188 These results suggest that activation of PPIAP22 or HOTAIR promotes a malignant phenotype in cancer cells and regulates the infiltration of TIICs through the CCL-CCR or CXCL-CXCR pathway to enhance tumor progression.188
In addition, lncRNA GIHCG levels are highly expressed in HCC tissues compared with nontumor liver tissues and correlated with tumor clinicopathological characteristics and patient survival.189,190 GIHCG has been reported to be an oncogene that promotes the proliferation and metastasis of HCC by silencing the miR-200b/a/429 axis 224. In addition, upregulation of GIHCG is positively or negatively correlated with the proportions of TIICs, including macrophages.190 However, the expression and function of GIHCG in macrophages still need to be studied further.
Tumor-cell-derived circRNAs
Function as oncogenes by modulating macrophage infiltration and polarization and promoting tumor cell proliferation, migration, and invasion
circASAP1 is a newly identified circRNA derived from the ASAP1 gene. It is frequently upregulated in HCC tissues and is positively correlated with HCC metastasis and prognosis. Overexpression of circASAP1 in HCC cells significantly promotes cell proliferation, migration, and invasion in vitro as well as lung metastasis in vivo.200 Additionally, the promoting effects of circASAP1 on tumor progression were mediated by sponging miR-326 and 532-5p to activate p-MAPK1 signaling. In addition, the circRNA-miR-326/miR-532-5p axis increased the production and secretion of CSF-1 and promoted the infiltration levels of M2-like TAMs through CSF-1-CSF-L1 signaling.200,201 These results suggest that circASAP1 is involved in tumor progression not only by promoting HCC metastasis through miR-326/miR-532-5p-MAPK1 signaling but also by enhancing M2-like TAM infiltration through miR-326/miR-532-5p-CSF-1 signaling.200,201
Similarly, hsa_circ_0074854 was also found to be upregulated in HCC tissues and cells.202 Knockdown of hsa_circ_0074854 in HCC cells significantly reduced cell migration and invasion by inhibiting the stability of human antigen R (HuR) to suppress EMT. In addition, hsa_circ_0074854 in cells can be released via exosomes, and its levels in HCC cell-derived exosomes were upregulated compared with those in exosomes from noncancerous human hepatic cells. Knockdown of hsa_circ_0074854 in exosomes suppressed the M2-like polarization of macrophages, further inhibiting the migration, invasion, and EMT of HCC cells.202 These results indicate that hsa_circ_0074854 acts as an oncogene to promote tumor progression by inhibiting cell invasion and migration through regulating HuR-MET signaling and by suppressing the M2-like polarization of macrophages mediated by exosomes.202
Function as tumor suppressors by decreasing macrophage infiltration and inhibiting HCC cell proliferation, migration, and invasion
Unlike the low expression of circASAP1 and hsa_circ_0074854, hsa_circ_0110102 is downregulated in HCC cell lines and tissues. hsa_circ_0074854 is a newly identified circRNA derived from the ZNF562 gene, and its low expression is associated with poor prognosis.203 Knockdown of hsa_circ_0110102 in HCC cells markedly enhanced cell proliferation, migration, invasion, and metastasis in vitro and in vivo. Furthermore, hsa_circ_0110102 functions as a sponge for miR-580-5p, which is overexpressed in HCC with protumor ability. miR-580-5p significantly upregulates CCL2 expression and secretion by reducing the expression of the CCL2 inhibitor PPARα.203,204 The CCL2-CCR2 axis plays an important role in the chemotaxis of M1 TAMs to contribute to cancer progression.204 When cocultured with miR-580-5p-enriched HCC cells treated with miR-580-5p, the expression levels of CCR2 and secretion levels of the protumor cytokines COX-2 and PGE2 were decreased in macrophages.203,204 The inhibitory effects of miR-580-5p-enriched HCC cells on macrophages can be recovered by hsa_circ_0110102. Moreover, CCL2 is involved in the production of COX-2/PGE2 in macrophages by regulating FoxO1 expression in a p38MAPK-dependent manner.203,204 These results indicate that hsa_circ_0110102 acts as a tumor suppressor to decrease CCL2 expression and secretion in HCC cells by targeting the miR-580-5p-PPARα axis, suppressing the secretion of COX-2/PGE2 from M1 macrophages by inhibiting the CCL2-CCR2-p38MAPK-FoxO1 signaling pathway and attenuating tumor progression.204
Outlook on HCC diagnostics and therapeutics of ncRNAs
With the development of chemistry, biology, and medical science, an increasing number of miRNAs, lncRNAs, and circRNAs that could mediate the crosstalk between macrophages and tumor cells will be discovered in HCC. Their functions in the development, progression, drug resistance, recurrence, and metastasis of HCC will also become clearer. This suggests that crosstalk-related ncRNAs are potential diagnostic and prognostic biomarkers and molecular therapeutic targets for HCC. Therefore, upregulation of macrophage-derived ncRNAs or elimination of ncRNAs from tumor cells among these crosstalk-related ncRNAs is one of the important options for promising biomarkers for HCC diagnosis and prognosis or as capacity targets for HCC treatment.
Focusing on macrophage-derived ncRNAs, one strategy in preclinical or clinical trials for cancer treatment is RNA delivery-based reprogramming of TAMs. Good examples include charge-altering releasable transporters (CARTs) and nanoparticle (NP)-mediated RNA delivery. These two delivery systems can carry specific RNAs and deliver them into specific target cells. Targeting ncRNAs may be encapsulated in CARTs or NPs for subsequent delivery into TAMs, resulting in the overexpression of specific ncRNAs in TAMs. Consequently, these ncRNA-enriched TAMs could remodel their phenotype and function to inhibit tumor growth or metastasis.
Similarly, in tumor-derived ncRNA-based therapeutics, siRNAs can also be encapsulated within the two delivery systems to silence targeted ncRNA in tumor cells. Then, the specific ncRNA-silenced tumor cells will subsequently lose the functions of proliferation, invasion, and immune escape as well as the immunosuppressive function of promoting macrophage infiltration, activation, or M2 polarization, leading to tumor elimination and phenotype and function remodeling to inhibit tumor growth or metastasis. In addition, many other effective strategies, such as antisense morpholino oligonucleotides, small-molecule inhibitors, nucleic acid limitation, CRISPR-Cas9, or steric inhibition, as mentioned above about miRNA-, lncRNA-, or circRNA-based therapeutics, can be used to deplete the selected oncogenic RNA.19,65,74
In addition, miRNAs, lncRNAs, and circRNAs can all be detected in many human body fluids, such as blood, saliva, urine, breast milk, and serum exosomes in particular. Exosomes can protect these ncRNAs from degradation to reflect their true levels in the body. Thus, exosomal crosstalk-related ncRNAs may be stable tumor diagnosis and treatment markers that could easily be detected in HCC. However, uncertain factors in these ncRNA-based diagnoses and therapeutics may limit HCC treatment. Therefore, additional preclinical and clinical trials are required to verify the efficacy of existing methods and identify new strategies.
Conclusions and discussion
The tumor microenvironment is a well-characterized system that plays a key role in the initiation and progression of HCC. Tumor-related macrophages within the microenvironment form a niche for cancer development. Within the niche, intense communication occurs between macrophages and tumor cells through cell-to-cell contact and/or soluble messengers. The increasing studies, based on next-generation sequencing technologies, have shed light on the implication of ncRNAs in the process of communication.231,232 Unlocking the link between ncRNAs and cell communication may open up new fields for the discovery of new diagnosis and treatment strategies in HCC. This review focused on the regulation of ncRNAs in the crosstalk between macrophages and tumor cells and its function and mechanism in HCC, as shown in Figures 4 and 5. These ncRNAs are derived from macrophages or tumor cells and act as oncogenes or tumor suppressors not only to regulate almost all physiological and pathological processes of tumor cells but also to control macrophage infiltration, activation, polarization, and function, resulting in aggravation or delay of HCC progression (Tables 1, 2, and 3). In addition, the regulatory networks among different types of ncRNAs, such as lncRNA-miRNA or lncRNA-miRNA axes, indicate the complexity of ncRNA regulation in the communication among ncRNAs, macrophages, and tumor cells.233 Meanwhile, miRNAs have also been suggested to be checkpoints in regulatory networks, and this should be given more attention with respect to HCC diagnosis and treatment. Moreover, we found that ncRNAs derived from tumor cells almost all exhibit tumor-promoting functions, especially tumor-derived lncRNAs, while macrophage-derived ncRNAs exert antitumor functions (such as miRNAs and lncRNAs). In addition, the dynamic expression of these tumor-derived lncRNAs that were upregulated and positively associated with tumor size and TNM stage usually facilitates the progression of HCC, indicating that these lncRNAs may act as potential novel prognostic biomarkers and therapeutic targets in HCC treatment. These findings also indicate that although TAMs are generally considered to have characteristics that promote tumor development, as important natural immune cells, they are still trying to restore their immune monitoring function in some way; for example, through the expression of antitumor ncRNAs. This also suggests that, in the clinical treatment strategy of tumors, the method of reprogramming tumor-related macrophages has greater development potential than that of removing them.
In view of the regulatory roles of the aforementioned three types of ncRNAs in the relationship between tumor cells and immune cells, this suggests that other types of ncRNAs, such as enhancer RNAs (eRNAs), may also be involved in the relationship in HCC. eRNA is a type of ncRNA transcribed from enhancers that does not encode a protein but retains important functions in enhancer activation and gene regulation.234,235 Increasing evidence suggests that some eRNAs, such as eRNA-TAOK1e and eRNA-CELF2e, are abnormally expressed in tumors and may be key factors in tumorigenesis.236,237 In HCC, eRNA-APH1A is highly expressed in advanced tumor stages with larger tumor sizes and higher histological grades.238 Furthermore, many immune-related eRNAs (IReRNAs) were identified in HCC, and the prognostic role of IReRNAs was explored. Five IReRNAs (AC073257.2, AL445524.1, FAM120AOS, LINC00513, and STK3) were identified as an IReRNA signature (IReRS), and they are positively correlated with poor survival but inversely correlated with CD8+ T cells and M0 macrophage infiltration in HCC, especially AL445524.1.239 These data indicate that these eRNAs may play important roles in HCC immunity mediated by T cells and TAMs.
In addition to macrophages, many types of immune cells exist in the tumor microenvironment, and they play an important regulatory role in tumor immunity (Figure 3). Among them, tumor infiltrating lymphocytes (TILs) have the largest number of infiltrating cells. TILs include many subtypes of cells, such as CTLs, γδ T cells, Treg cells, and CD4+ IL-17+ (Th17) cells.35 Among them, CTL and γδ T cell subsets play a role in eliminating tumor cells, while Treg and Th17-cell subsets can trigger severe chronic inflammation and play an auxiliary role in tumorigenesis, invasion, and metastasis.240,241 Increasing evidence has shown that some novel ncRNAs are involved in the reciprocal regulatory relationship between these T cells and tumor cells in HCC. For example, Treg-derived lnc-EGFR was proved to stimulate Treg cell differentiation and CTL inhibition, leading to HCC progression by the lnc-EGFR-EGFR-NF-AT1/AP1-lnc-EGFR forward-feedback loop.242 CTL-derived lnc-NEAT1 was also proved to be involved in CTL cell apoptosis, and repression of NEAT1 could restrain cell apoptosis while enhancing the cytolysis activity against HCC by the miR-155/Tim-3 pathway.243 On the other hand, tumor-cell-derived lnc-FENDRR acted as a suppressor to inhibit the immune escape of tumor cells mediated by Treg cells by sponging miR-423-5p.244 These data indicated that ncRNA-mediated crosstalk between TILs and tumor cells plays an important role in mediating tumor immune escape during HCC development, and cell-to-cell contact and TIL- or tumor-secreted EVs may also act as a bridge in the crosstalk mediated by ncRNAs. Similar to T cells, B cells, as a core component of adaptive immunity, also play an important regulatory role in tumor immunity, although their infiltration in the tumor microenvironment is relatively small. B cells were found to not only secrete immunoglobulin (Ig) and express costimulatory molecules to participate in tumor immunity but also provide high levels of IL-10 secreted by the regulatory B (Breg) cell subset to suppress host immune responses and play a tumor-promoting role in cancer, including HCC.245,246 In addition, PD-1hi B cells, another newly identified subtype, were discovered in advanced-stage HCC patients. When activated by PD-1, these cells can produce large amounts of IL-10 to suppress tumor-specific T cell immunity.247 Interestingly, B cells were recently proved to be regulated by tumor cells. The expansion of the TIM-1+ Breg cell subset can be promoted by HCC-derived exosomes, resulting in CTL inhibition and HCC progression via the HMGB1-TLR2/4-MAPK pathway.248 This indicates that B cells and tumor cells in the tumor microenvironment also have a mutual regulatory relationship, and cytokines and exosomes play a mediating role in it. However, whether ncRNAs are involved in the relationship needs clarification.
Therefore, more research is needed to uncover more critical tumor-infiltrating cells and tumor cell crosstalk-related ncRNAs within the microenvironment, especially circRNAs and eRNAs, which have rarely been identified, to clarify their regulation, function, and mechanism in HCC.
Acknowledgments
This study was supported by the Natural Science Foundation of Shandong Province of China (ZR2020MH250 and ZR2020MH202), the National Natural Science Foundation of China (91849209, 81803016, and 32000495), and a project of Shandong Province Higher Educational Science and Technology Program (J18KA290).
Author contributions
Z.Z. and P.L. designed the review and contributed to manuscript preparation. Z.Z. and Z.W. wrote the manuscript. J.G., Z.L., and Y.W. provided technical and administrative support. Z.Z., T.Z., and M.L. prepared the figures. J.G. and Z.L. revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Zhixia Zhou, Email: zhou_zhixia@qdu.edu.cn.
Peifeng Li, Email: peifli@hotmail.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J., Reig M., Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 3.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 4.Liu G., Yin L., Ouyang X., Zeng K., Xiao Y., Li Y. M2 macrophages promote HCC cells invasion and migration via miR-149-5p/MMP9 signalin. J. Cancer. 2020;11:1277–1287. doi: 10.7150/jca.35444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 6.Dougan M., Dougan S.K. Targeting immunotherapy to the tumor microenvironment. J. Cell Biochem. 2017;118:3049–3054. doi: 10.1002/jcb.26005. [DOI] [PubMed] [Google Scholar]
- 7.Li H., Yu J., Wu Y., Shao B., Wei X. In situ antitumor vaccination: targeting the tumor microenvironment. J. Cell Physiol. 2020;235:5490–5500. doi: 10.1002/jcp.29551. [DOI] [PubMed] [Google Scholar]
- 8.Gong B.S., Wang R., Xu H.X., Miao M.Y., Yao Z.Z. Nanotherapy targeting the tumor microenvironment. Curr. Cancer Drug Targets. 2019;19:525–533. doi: 10.2174/1568009619666181220103714. [DOI] [PubMed] [Google Scholar]
- 9.Arneth B. Tumor microenvironment. Medicina (Kaunas) 2019;56:15. doi: 10.3390/medicina56010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinshaw D.C., Shevd L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu Y., Cheng P. Targeting tumor-associated macrophages for cancer immunotherapy. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188434. doi: 10.1016/j.bbcan.2020.188434. [DOI] [PubMed] [Google Scholar]
- 12.Ngambenjawong C., Gustafson H.H., Pun S.H. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X., Li L., Thorpe P.E., Yopp A.C., Brekken R.A., Huang X. Antibody-mediated blockade of phosphatidylserine enhances the antitumor effect of sorafenib in hepatocellular carcinomas xenografts. Ann. Surg. Oncol. 2016;23:583–591. doi: 10.1245/s10434-016-5107-5. [DOI] [PubMed] [Google Scholar]
- 14.Li X., Yao W., Yuan Y., Chen P., Li B., Li J., Chu R., Song H., Xie D., Jiang X., et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 15.Beg M.S., Brenner A.J., Sachdev J., Borad M., Kang Y.K., Stoudemire J., Smith S., Bader A.G., Kim S., Hong D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rashad N.M., El-Shal A.S., Shalaby S.M., Mohamed S.Y. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinom. Mol. Cell Biochem. 2018;447:125–136. doi: 10.1007/s11010-018-3298-8. [DOI] [PubMed] [Google Scholar]
- 17.Ma J., Zhang L., Bian H.R., Lu Z.G., Zhu L., Yang P., Zeng Z.C., Xiang Z.L. A noninvasive prediction nomogram for lymph node metastasis of hepatocellular carcinoma based on serum long noncoding RNAs. Biomed. Res. Int. 2019;2019:1710670. doi: 10.1155/2019/1710670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander R.P., Fang G., Rozowsky J., Snyder M., Gerstein M.B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 19.Slack F.J., Chinnaiyan A.M. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romano G., Veneziano D., Acunzo M., Croce C.M. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnusamy M., Yan K.W., Liu C.Y., Li P.F., Wang K. PIWI family emerging as a decisive factor of cell fate: an overview. Eur. J. Cell Biol. 2017;96:746–757. doi: 10.1016/j.ejcb.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z., Lin Z., Pang X., Tariq M.A., Ao X., Li P., Wang J. Epigenetic regulation of long non-coding RNAs in gastric cancer. Oncotarget. 2018;9:19443–19458. doi: 10.18632/oncotarget.23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J., Chen X., Wei P., Wang Y., Li P., Shao K. Regulation of pyroptosis in cardiovascular pathologies: role of noncoding RNAs. Mol. Ther. Nucleic Acids. 2021;25:220–236. doi: 10.1016/j.omtn.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan C., Zhang Y., Hao X., Gao J., Chen X., Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo M.J., Yun J., Kim S.G. Role of non-coding RNAs in liver disease progression to hepatocellular carcinoma. Arch. Pharm. Res. 2019;42:48–62. doi: 10.1007/s12272-018-01104-x. [DOI] [PubMed] [Google Scholar]
- 26.Klingenberg M., Matsuda A., Diederichs S., Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Ham S., Lima L.G., Lek E., Moller A. The impact of the cancer microenvironment on macrophage phenotypes. Front. Immunol. 2020;11:1308. doi: 10.3389/fimmu.2020.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadi A., Sharifi A., Pourpaknia R., Mohammadian S., Sahebkar A. Manipulating macrophage polarization and function using classical HDAC inhibitors: implications for autoimmunity and inflammation. Crit. Rev. Oncol. Hematol. 2018;128:1–18. doi: 10.1016/j.critrevonc.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott C.L., Zheng F., De Baetselier P., Martens L., Saeys Y., De Prijck S., Lippens S., Abels C., Schoonooghe S., Raes G., et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016;7:10321. doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molawi K., Wolf Y., Kandalla P.K., Favret J., Hagemeyer N., Frenzel K., Pinto A.R., Klapproth K., Henri S., Malissen B., et al. Progressive replacement of embryo-derived cardiac macrophages with age. J. Exp. Med. 2014;211:2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Hove H., Martens L., Scheyltjens I., De Vlaminck K., Pombo Antunes A.R., De Prijck S., Vandamme N., De Schepper S., Van Isterdael G., Scott C., et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 2019;22:1021–1035. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 33.Cotechini T., Atallah A., Grossman A. Tissue-resident and recruited macrophages in primary tumor and metastatic microenvironments: potential targets in cancer therapy. Cells. 2021;10:960. doi: 10.3390/cells10040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madsen D.H., Jurgensen H.J., Siersbaek M.S., Kuczek D.E., Grey Cloud L., Liu S., Behrendt N., Grontved L., Weigert R., Bugge T.H. Tumor-associated macrophages derived from circulating inflammatory monocytes degrade collagen through cellular uptake. Cell Rep. 2017;21:3662–3671. doi: 10.1016/j.celrep.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goswami K.K., Ghosh T., Ghosh S., Sarkar M., Bose A., Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y., Herndon J.M., Sojka D.K., Kim K.W., Knolhoff B.L., Zuo C., Cullinan D.R., Luo J., Bearden A.R., Lavine K.J., et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity. 2017;47:597. doi: 10.1016/j.immuni.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perdiguero E.G., Geissmann F. The development and maintenance of resident macrophages. Nat. Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loyher P.L., Hamon P., Laviron M., Meghraoui-Kheddar A., Goncalves E., Deng Z., Torstensson S., Bercovici N., Baudesson de Chanville C., Combadiere B., et al. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med. 2018;215:2536–2553. doi: 10.1084/jem.20180534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Etzerodt A., Moulin M., Doktor T.K., Delfini M., Mossadegh-Keller N., Bajenoff M., Sieweke M.H., Moestrup S.K., Auphan-Anezin N., Lawrence T. Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. J. Exp. Med. 2020;217:e20191869. doi: 10.1084/jem.20191869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins S.J., Ruckerl D., Cook P.C., Jones L.H., Finkelman F.D., van Rooijen N., MacDonald A.S., Allen J.E. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daurkin I., Eruslanov E., Stoffs T., Perrin G.Q., Algood C., Gilbert S.M., Rosser C.J., Su L.M., Vieweg J., Kusmartsev S. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71:6400–6409. doi: 10.1158/0008-5472.CAN-11-1261. [DOI] [PubMed] [Google Scholar]
- 42.Najafi M., Hashemi Goradel N., Farhood B., Salehi E., Nashtaei M.S., Khanlarkhani N., Khezri Z., Majidpoor J., Abouzaripour M., Habibi M., et al. Macrophage polarity in cancer: a review. J. Cell Biochem. 2019;120:2756–2765. doi: 10.1002/jcb.27646. [DOI] [PubMed] [Google Scholar]
- 43.Albini A., Bruno A., Noonan D.M., Mortara L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment, implications for immunotherapy. Front. Immunol. 2018;9:527. doi: 10.3389/fimmu.2018.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reichel D., Tripathi M., Perez J.M. Biological effects of nanoparticles on macrophage polarization in the tumor microenvironment. Nanotheranostics. 2019;3:66–88. doi: 10.7150/ntno.30052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D., Xie J., Fiskesund R., Dong W., Liang X., Lv J., Jin X., Liu J., Mo S., Zhang T., et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018;9:873. doi: 10.1038/s41467-018-03225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genard G., Wera A.C., Huart C., Le Calve B., Penninckx S., Fattaccioli A., Tabarrant T., Demazy C., Ninane N., Heuskin A.C., et al. Proton irradiation orchestrates macrophage reprogramming through NFkappaB signalin. Cell Death Dis. 2018;9:728. doi: 10.1038/s41419-018-0757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang W., Shi R., Kang X., Zhang X., Chen P., Zhang L., Hou A., Wang R., Zhao Y., Zhao K., et al. Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat. Commun. 2018;9:2574. doi: 10.1038/s41467-018-04999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Q., Xia J., Wang L., Wang X., Ma X., Deng Q., Lu Y., Kumar M., Zhou Z., Li L., et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J. Hematol. Oncol. 2018;11:58. doi: 10.1186/s13045-018-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cully M. Cancer, Re-educating tumour-associated macrophages with nanoparticles. Nat. Rev. Drug Discov. 2018;17:468. doi: 10.1038/nrd.2018.102. [DOI] [PubMed] [Google Scholar]
- 50.Flemming A. Cancer: Re-educating tumour-associated macrophages. Nat. Rev. Drug Discov. 2011;10:177. doi: 10.1038/nrd3399. [DOI] [PubMed] [Google Scholar]
- 51.Tan B., Shi X., Zhang J., Qin J., Zhang N., Ren H., Qian M., Siwko S., Carmon K., Liu Q., et al. Inhibition of Rspo-lgr4 facilitates checkpoint blockade therapy by switching macrophage polarization. Cancer Res. 2018;78:4929–4942. doi: 10.1158/0008-5472.CAN-18-0152. [DOI] [PubMed] [Google Scholar]
- 52.Ma J., Liu L., Che G., Yu N., Dai F., You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer. 2010;10:112. doi: 10.1186/1471-2407-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mei J., Xiao Z., Guo C., Pu Q., Ma L., Liu C., Lin F., Liao H., You Z., Liu L. Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: a systemic review and meta-analysis. Oncotarget. 2016;7:34217–34228. doi: 10.18632/oncotarget.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Z.Y., Luo R.Z., Peng R.J., Wang S.S., Xue C. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;7:1475–1480. doi: 10.2147/OTT.S61838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y., Song Y., Du W., Gong L., Chang H., Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 2019;26:78. doi: 10.1186/s12929-019-0568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belgiovine C., D'Incalci M., Allavena P., Frapolli R. Tumor-associated macrophages and anti-tumor therapies: complex links. Cell Mol. Life Sci. 2016;73:2411–2424. doi: 10.1007/s00018-016-2166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anfray C., Ummarino A., Andon F.T., Allavena P. Current strategies to target tumor-associated-macrophages to improve anti-tumor immune responses. Cells. 2019;9:46. doi: 10.3390/cells9010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan B.M., Robles A.I., Harris C.C. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y.F., Xu H.M., Yu F., Wang M., Li M.Y., Xu T., Gao Y.Y., Wang J.X., Li P.F. Crosstalk between MicroRNAs and peroxisome proliferator-activated receptors and their emerging regulatory roles in cardiovascular pathophysiology. PPAR Res. 2018;2018:8530371. doi: 10.1155/2018/8530371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carleton M., Cleary M.A., Linsley P.S. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 62.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiang Y., Yang Y., Lin C., Wu J., Zhang X. MiR-23a-3p promoted G1/S cell cycle transition by targeting protocadherin17 in hepatocellular carcinoma. J. Physiol. Biochem. 2020;76:123–134. doi: 10.1007/s13105-020-00726-4. [DOI] [PubMed] [Google Scholar]
- 64.Zhang J.J., Chen J.T., Hua L., Yao K.H., Wang C.Y. miR-98 inhibits hepatocellular carcinoma cell proliferation via targeting EZH2 and suppressing Wnt/beta-catenin signaling pathway. Biomed. Pharmacother. 2017;85:472–478. doi: 10.1016/j.biopha.2016.11.053. [DOI] [PubMed] [Google Scholar]
- 65.Corsini L.R., Bronte G., Terrasi M., Amodeo V., Fanale D., Fiorentino E., Cicero G., Bazan V., Russo A. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin. Ther. Targets. 2012;16:S103–S109. doi: 10.1517/14728222.2011.650632. [DOI] [PubMed] [Google Scholar]
- 66.Peng W.X., Koirala P., Mo Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattick J.S., Rinn J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 68.Marques A.C., Ponting C.P. Intergenic lncRNAs and the evolution of gene expression. Curr. Opin. Genet. Dev. 2014;27:48–53. doi: 10.1016/j.gde.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 69.Chen J., Wang R., Zhang K., Chen L.B. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic target. J. Cell Mol. Med. 2014;18:2425–2436. doi: 10.1111/jcmm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clark M.B., Mattick J.S. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 71.Gao J., Chen X., Shan C., Wang Y., Li P., Shao K. Autophagy in cardiovascular diseases: role of noncoding RNAs. Mol. Ther. Nucleic Acids. 2021;23:101–118. doi: 10.1016/j.omtn.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z., Lin Z., He Y., Pang X., Wang Y., Ponnusamy M., Ao X., Shan P., Li P., Wang J. The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Mol. Ther. Nucl. Acids. 2018;12:405–419. doi: 10.1016/j.omtn.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arun G., Diermeier S.D., Spector D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilusz J.E. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA. 2018;9:e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hentze M.W., Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang Q., Hann S.S. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–2092. doi: 10.2147/OTT.S233672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kristensen L.S., Hansen T.B., Veno M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen S., Zhao Y. Circular RNAs: characteristics, function, and role in human cancer. Histol. Histopathol. 2018;33:887–893. doi: 10.14670/HH-11-969. [DOI] [PubMed] [Google Scholar]
- 80.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lei B., Tian Z., Fan W., Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int. J. Med. Sci. 2019;16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y., Jia D.D., Zhang Y.F., Cheng M.D., Zhu W.X., Li P.F., Zhang Y.F. The emerging function and clinical significance of circRNAs in Thyroid Cancer and Autoimmune Thyroid Diseases. Int. J. Biol. Sci. 2021;17:1731–1741. doi: 10.7150/ijbs.55381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y., Liu J., Ma J., Sun T., Zhou Q., Wang W., Wang G., Wu P., Wang H., Jiang L., et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capece D., Fischietti M., Verzella D., Gaggiano A., Cicciarelli G., Tessitore A., Zazzeroni F., Alesse E. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophage. Biomed. Res. Int. 2013;2013:187204. doi: 10.1155/2013/187204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murray P.J., Wyn T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis D.M. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat. Rev. Immunol. 2007;7:238–243. doi: 10.1038/nri2020. [DOI] [PubMed] [Google Scholar]
- 87.Onfelt B., Nedvetzki S., Benninger R.K., Purbhoo M.A., Sowinski S., Hume A.N., MSeabra C., Neil M.A., French P.M., Davis D.M. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J. Immunol. 2006;177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- 88.Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T.D., Tridandapani S., et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Latifkar A., Hur Y.H., Sanchez J.C., Cerione R.A., Antonyak M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019;132:jcs222406. doi: 10.1242/jcs.222406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han C., Zhang C., Wang H., Zhao L. Exosome-mediated communication between tumor cells and tumor-associated macrophages: implications for tumor microenvironment. Oncoimmunology. 2021;10:1887552. doi: 10.1080/2162402X.2021.1887552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Q., Zhou L., Lv D., Zhu X., Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019;12:53. doi: 10.1186/s13045-019-0739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Milane L., Singh A., Mattheolabakis G., Suresh M., Amiji M.M. Exosome mediated communication within the tumor microenvironment. J. Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 94.Kwon Y., Kim M., Kim Y., Jung H.S., Jeoung D. Exosomal MicroRNAs as mediators of cellular interactions between cancer cells and macrophages. Front. Immunol. 2020;11:1167. doi: 10.3389/fimmu.2020.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beeraka N.M., Doreswamy S.H., Sadhu S.P., Srinivasan A., Pragada R.R., Madhunapantula S.V., Aliev G. The role of exosomes in stemness and neurodegenerative diseases-chemoresistant-cancer therapeutics and phytochemicals. Int. J. Mol. Sci. 2020;21:6818. doi: 10.3390/ijms21186818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mohapatra S., Pioppini C., Ozpolat B., Calin G.A. Non-coding RNAs regulation of macrophage polarization in cancer. Mol. Cancer. 2021;20:24. doi: 10.1186/s12943-021-01313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang L., Liao Y., Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J., Ding D., Jiang Z., Du T., Liu J., Kong Z. Long non-coding RNA CCAT1/miR-148a/PKCzeta prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79:105–112. doi: 10.1002/pros.23716. [DOI] [PubMed] [Google Scholar]
- 99.Ye Y., Xu Y., Lai Y., He W., Li Y., Wang R., Luo X., Chen R., Chen T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J. Cell Biochem. 2018;119:2951–2963. doi: 10.1002/jcb.26509. [DOI] [PubMed] [Google Scholar]
- 100.Cai X., Yin Y., Li N., Zhu D., Zhang J., Zhang C.Y., Zen K. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J. Mol. Cell Biol. 2012;4:341–343. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 101.Wang Z., Xu L., Hu Y., Huang Y., Zhang Y., Zheng X., Wang S., Wang Y., Yu Y., Zhang M., et al. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci. Rep. 2016;6:25602. doi: 10.1038/srep25602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Z.Z., Han Z.D., Liang Y.K., Chen J.X., Wan S., Zhuo Y.J., Cai Z.D., Deng Y.L., Lin Z.Y., Mo R.J., et al. TRIB1 induces macrophages to M2 phenotype by inhibiting IKB-zeta in prostate cancer. Cell Signal. 2019;59:152–162. doi: 10.1016/j.cellsig.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 103.Hsieh C.H., Tai S.K., Yang M.H. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering MiR-21-abundant exosomes. Neoplasia. 2018;20:775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang J.K., Ma L., Song W.H., Lu B.Y., Huang Y.B., Dong H.M., Ma X.K., Zhu Z.Z., Zhou R. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J. Cell Biochem. 2017;118:4821–4830. doi: 10.1002/jcb.26153. [DOI] [PubMed] [Google Scholar]
- 105.Weng Y.S., Tseng H.Y., Chen Y.A., Shen P.C., Al Haq A.T., Chen L.M., Tung Y.C., Hsu H.L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer. 2019;18:42. doi: 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tao S., Chen Q., Lin C., Dong H. Linc00514 promotes breast cancer metastasis and M2 polarization of tumor-associated macrophages via Jagged1-mediated notch signaling pathway. J. Exp. Clin. Cancer Res. 2020;39:191. doi: 10.1186/s13046-020-01676-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ji W.B., Liu X., Luo Y., Zhang W.Z. High expression of miR-15b predicts poor prognosis for hepatocellular carcinoma after curative hepatectomy. Oncol. Rep. 2016;36:1901–1908. doi: 10.3892/or.2016.4982. [DOI] [PubMed] [Google Scholar]
- 108.Liu A.M., Yao T.J., Wang W., Wong K.F., Lee N.P., Fan S.T., Poon R.T., Gao C., Luk J.M. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song S., Qiu X. LncRNA miR503HG inhibits epithelial-mesenchymal transition and angiogenesis in hepatocellular carcinoma by enhancing PDCD4 via regulation of miR-15b. Dig. Liver Dis. 2021;53:107–116. doi: 10.1016/j.dld.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 110.Aucher A., Rudnicka D., Davis D.M. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013;191:6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei X., Tang C., Lu X., Liu R., Zhou M., He D., Zheng D., Sun C., Wu Z. MiR-101 targets DUSP1 to regulate the TGF-beta secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget. 2015;6:18389–18405. doi: 10.18632/oncotarget.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao K., Li J., Zhao Y., Wang Q., Zeng Q., He S., Yu L., Zhou J., Cao P. miR-101 inhibiting cell proliferation, migration and invasion in hepatocellular carcinoma through downregulating girdin. Mol. Cells. 2016;39:96–102. doi: 10.14348/molcells.2016.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhuang C., Jiang W., Huang D., Xu L., Yang Q., Zheng L., Wang X., Hu L. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2016;40:386–396. doi: 10.1016/j.clinre.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y., Wang B., Xiao S., Li Y., Chen Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell Biochem. 2019;120:3046–3055. doi: 10.1002/jcb.27436. [DOI] [PubMed] [Google Scholar]
- 115.Hua S., Liu C., Liu L., Wu D. miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochem. Biophys. Res. Commun. 2018;496:947–954. doi: 10.1016/j.bbrc.2018.01.112. [DOI] [PubMed] [Google Scholar]
- 116.Fu Y., Sun L.Q., Huang Y., Quan J., Hu X., Tang D., Kang R., Li N., Fan X.G. miR-142-3p inhibits the metastasis of hepatocellular carcinoma cells by regulating HMGB1 gene expression. Curr. Mol. Med. 2018;18:135–141. doi: 10.2174/1566524018666180907161124. [DOI] [PubMed] [Google Scholar]
- 117.Wu L., Cai C., Wang X., Liu M., Li X., Tang H. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 2011;585:1322–1330. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 118.Zhang J., Shan W.F., Jin T.T., Wu G.Q., Xiong X.X., Jin H.Y., Zhu S.M. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J. Transl. Med. 2014;12:279. doi: 10.1186/s12967-014-0279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu Q., Xiang L., Yin L., Liu X., Yang D., Zhou J. Loss-of-function of miR-142 by hypermethylation promotes TGF-beta-mediated tumour growth and metastasis in hepatocellular carcinoma. Cell Prolif. 2017;50:e12384. doi: 10.1111/cpr.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bhattacharya S., Steele R., Shrivastava S., Chakraborty S., Di Bisceglie A.M., Ray R.B. Serum miR-30e and miR-223 as Novel Noninvasive Biomarkers for Hepatocellular Carcinoma. Am. J. Pathol. 2016;186:242–247. doi: 10.1016/j.ajpath.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li L., Sun P., Zhang C., Li Z., Zhou W. MiR-98 suppresses the effects of tumor-associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL-10. Biochimie. 2018;150:23–30. doi: 10.1016/j.biochi.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 122.Lu S., Gao Y., Huang X., Wang X. Cantharidin exerts anti-hepatocellular carcinoma by miR-214 modulating macrophage polarization. Int. J. Biol. Sci. 2014;10:415–425. doi: 10.7150/ijbs.8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shih T.C., Tien Y.J., Wen C.J., Yeh T.S., Yu M.C., Huang C.H., Lee Y.S., Yen T.C., Hsieh S.Y. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J. Hepatol. 2012;57:584–591. doi: 10.1016/j.jhep.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 124.Wang J., Li J., Wang X., Zheng C., Ma W. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem. Biophys. Res. Commun. 2013;439:47–53. doi: 10.1016/j.bbrc.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 125.Wang X., Chen J., Li F., Lin Y., Zhang X., Lv Z., Jiang J. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of beta-catenin. Biochem. Biophys. Res. Commun. 2012;428:525–531. doi: 10.1016/j.bbrc.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 126.Bandiera S., Pernot S., El Saghire H., Durand S.C., Thumann C., Crouchet E., Ye T., Fofana I., Oudot M.A., Barths J., et al. Hepatitis C virus-induced upregulation of MicroRNA miR-146a-5p in hepatocytes promotes viral infection and deregulates metabolic pathways associated with liver disease pathogenesis. J. Virol. 2016;90:6387–6400. doi: 10.1128/JVI.00619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vergadi, E., Vaporidi, K., Theodorakis, E.E., Doxaki, C., agoudaki, E., Ieronymaki, LE., Alexaki, V.I., Helms, M., Kondili, E., B. Soennichsen, et al. J. Immunol. 192, 394-406. [DOI] [PubMed]
- 128.Zhang X., Ye Z.H., Liang H.W., Ren F.H., Li P., Dang Y.W., Chen G. Down-regulation of miR-146a-5p and its potential targets in hepatocellular carcinoma validated by a TCGA- and GEO-based study. FEBS Open Bio. 2017;7:504–521. doi: 10.1002/2211-5463.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo J., Si Z.Z., Li T., Li J.Q., Zhang Z.Q., Chen G.S., Qi H.Z., Yao H.L. MicroRNA-146a-5p enhances radiosensitivity in hepatocellular carcinoma through replication protein A3-induced activation of the DNA repair pathway. Am. J. Physiol. Cell Physiol. 2019;316:C299–C311. doi: 10.1152/ajpcell.00189.2018. [DOI] [PubMed] [Google Scholar]
- 130.Sun X., Zhang J., Hou Z., Han Q., Zhang C., Tian Z. miR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression. Cell Cycle. 2015;14:243–252. doi: 10.4161/15384101.2014.977112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu J., Fan L., Yu H., Zhang J., He Y., Feng D., Wang F., Li X., Liu Q., Li Y., et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. 2019;70:241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang H.D., Kim H.S., Kim S.Y., Na M.J., Yang G., Eun J.W., Wang H.J., Cheong J.Y., Park W.S., Nam S.W. HDAC6 suppresses let-7i-5p to Elicit TSP1/CD47-mediated anti-tumorigenesis and phagocytosis of hepatocellular carcinoma. Hepatology. 2019;70:1262–1279. doi: 10.1002/hep.30657. [DOI] [PubMed] [Google Scholar]
- 133.Lv Z., Weng X., Du C., Zhang C., Xiao H., Cai X., Ye S., Cheng J., Ding C., Xie H., et al. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol. Carcinog. 2016;55:1024–1033. doi: 10.1002/mc.22345. [DOI] [PubMed] [Google Scholar]
- 134.Matlung H.L., Szilagyi K., Barclay N.A., van den Berg T.K. The CD47-SIRPalpha signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 2017;276:145–164. doi: 10.1111/imr.12527. [DOI] [PubMed] [Google Scholar]
- 135.Shen H., Li W., Tian Y., Xu P., Wang H., Zhang J., Li Y. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting Tob2 in hepatocellular carcinoma. J. Cell Biochem. 2015;116:1563–1573. doi: 10.1002/jcb.25110. [DOI] [PubMed] [Google Scholar]
- 136.Zhang Q., Huang F., Yao Y., Wang J., Wei J., Wu Q., Xiang S., Xu L. Interaction of transforming growth factor-beta-Smads/microRNA-362-3p/CD82 mediated by M2 macrophages promotes the process of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Sci. 2019;110:2507–2519. doi: 10.1111/cas.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou C., Wang P., Tu M., Huang Y., Xiong F., Wu Y. Long non-coding RNA PART1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells via miR-149-5p/MAP2K1 Axis. Cancer Manag. Res. 2020;12:3771–3782. doi: 10.2147/CMAR.S246311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ji D., Wang Y., Li H., Sun B., Luo X. Long non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates hepatocellular carcinoma progression. Biochem. Biophys. Res. Commun. 2019;512:176–181. doi: 10.1016/j.bbrc.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 139.Yao Y., Liu Y., Jin F., Meng Z. LINC00662 promotes oral squamous cell carcinoma cell growth and metastasis through miR-144-3p/EZH2 Axis. Yonsei Med. J. 2021;62:640–649. doi: 10.3349/ymj.2021.62.7.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wei G.Y., Hu M., Zhao L., Guo W.S. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019;23:5158–5167. doi: 10.26355/eurrev_201906_18180. [DOI] [PubMed] [Google Scholar]
- 141.Zhao J., Li H., Zhao S., Wang E., Zhu J., Feng D., Zhu Y., Dou W., Fan Q., Hu J., et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer. 2021;20:46. doi: 10.1186/s12943-021-01343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dong N., Shi X., Wang S., Gao Y., Kuang Z., Xie Q., Li Y., Deng H., Wu Y., Li M., et al. M2 macrophages mediate sorafenib resistance by secreting HGF in a feed-forward manner in hepatocellular carcinoma. Br. J. Cancer. 2019;121:22–33. doi: 10.1038/s41416-019-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang X., Zhang X.F., Lu X., Jia H.L., Liang L., Dong Q.Z., Ye Q.H., Qin L.X. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology. 2014;59:1874–1885. doi: 10.1002/hep.26941. [DOI] [PubMed] [Google Scholar]
- 144.Zhao W.T., Lin X.L., Liu Y., Han L.X., Li J., Lin T.Y., Shi J.W., Wang S.C., Lian M., Chen H.W., et al. miR-26a promotes hepatocellular carcinoma invasion and metastasis by inhibiting PTEN and inhibits cell growth by repressing EZH2. Lab. Invest. 2019;99:1484–1500. doi: 10.1038/s41374-019-0270-5. [DOI] [PubMed] [Google Scholar]
- 145.Yang X., Liang L., Zhang X.F., Jia H.L., Qin Y., Zhu X.C., Gao X.M., Qiao P., Zheng Y., Sheng Y.Y., et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 146.Chai Z.T., Zhu X.D., Ao J.Y., Wang W.Q., Gao D.M., Kong J., Zhang N., Zhang Y.Y., Ye B.G., Ma D.N., et al. microRNA-26a suppresses recruitment of macrophages by down-regulating macrophage colony-stimulating factor expression through the PI3K/Akt pathway in hepatocellular carcinoma. J. Hematol. Oncol. 2015;8:56. doi: 10.1186/s13045-015-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Z., Zheng W., Hai J. MicroRNA-148b expression is decreased in hepatocellular carcinoma and associated with prognosis. Med. Oncol. 2014;31:984. doi: 10.1007/s12032-014-0984-6. [DOI] [PubMed] [Google Scholar]
- 148.Zhang J.G., Shi Y., Hong D.F., Song M., Huang D., Wang C.Y., Zhao G. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/beta-catenin pathway. Sci. Rep. 2015;5:8087. doi: 10.1038/srep08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ke M., Zhang Z., Cong L., Zhao S., Li Y., Wang X., Lv Y., Zhu Y., Dong J. MicroRNA-148b-colony-stimulating factor-1 signaling-induced tumor-associated macrophage infiltration promotes hepatocellular carcinoma metastasis. Biomed. Pharmacother. 2019;120:109523. doi: 10.1016/j.biopha.2019.109523. [DOI] [PubMed] [Google Scholar]
- 150.Barve R.A., Zack M.D., Weiss D., Song R.H., Beidler D., Head R.D. Transcriptional profiling and pathway analysis of CSF-1 and IL-34 effects on human monocyte differentiation. Cytokine. 2013;63:10–17. doi: 10.1016/j.cyto.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 151.Zhou S.L., Hu Z.Q., Zhou Z.J., Dai Z., Wang Z., Cao Y., Fan J., Huang X.W., Zhou J. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 152.Shi X., Teng F. Down-regulated miR-28-5p in human hepatocellular carcinoma correlated with tumor proliferation and migration by targeting insulin-like growth factor-1 (IGF-1) Mol. Cell Biochem. 2015;408:283–293. doi: 10.1007/s11010-015-2506-z. [DOI] [PubMed] [Google Scholar]
- 153.Xia Q., Han T., Yang P., Wang R., Li H., Zhang J., Zhou X. MicroRNA-28-5p regulates liver cancer stem cell expansion via IGF-1 pathway. Stem Cells Int. 2019;2019:8734362. doi: 10.1155/2019/8734362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu J., Lu C., Xiao M., Jiang F., Qu L., Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed. Pharmacother. 2017;89:857–863. doi: 10.1016/j.biopha.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 155.Zhang D., Cao C., Liu L., Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J. Cancer. 2016;7:608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang B., Li X., Hu W., Zhou Y., Din Y. Silencing of lncRNA SNHG20 delays the progression of nonalcoholic fatty liver disease to hepatocellular carcinoma via regulating liver Kupffer cells polarization. IUBMB Life. 2019;71:1952–1961. doi: 10.1002/iub.2137. [DOI] [PubMed] [Google Scholar]
- 157.Begitt A., Cavey J., Droescher M., Vinkemeier U. On the role of STAT1 and STAT6 ADP-ribosylation in the regulation of macrophage activation. Nat. Commun. 2018;9:2144. doi: 10.1038/s41467-018-04522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Luo H.L., Chen J., Luo T., Wu F.X., Liu J.J., Wang H.F., Chen M., Li L.Q., Li H. Downregulation of macrophage-derived T-UCR uc.306 associates with poor prognosis in hepatocellular carcinoma. Cell Physiol. Biochem. 2017;42:1526–1539. doi: 10.1159/000479269. [DOI] [PubMed] [Google Scholar]
- 159.Bae S.H., Jung E.S., Park Y.M., Kim B.S., Kim B.K., Kim D.G., Ryu W.S. Expression of cyclooxygenase-2 (COX-2) in hepatocellular carcinoma and growth inhibition of hepatoma cell lines by a COX-2 inhibitor, NS-398. Clin. Cancer Res. 2001;7:1410–1418. [PubMed] [Google Scholar]
- 160.Wang X., Li F.Y., Zhao W., Gao Z.K., Shen B., Xu H., Cui Y.F. Long non-coding RNA GAS5 overexpression inhibits M2-like polarization of tumour-associated macrophages in SMCC-7721 cells by promoting PTEN expression. Int. J. Exp. Pathol. 2020;101:215–222. doi: 10.1111/iep.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen F., Li Y., Li M., Wang L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Am. J. Cancer Res. 2019;9:108–121. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162.Zhang F., Yang C., Xing Z., Liu P., Zhang B., Ma X., Huang L., Zhuang L. LncRNA GAS5-mediated miR-1323 promotes tumor progression by targeting TP53INP1 in hepatocellular carcinoma. Onco Targets Ther. 2019;12:4013–4023. doi: 10.2147/OTT.S209439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang L., Jiang J. GAS5 regulates RECK expression and inhibits invasion potential of HCC cells by sponging miR-135b. Biomed. Res. Int. 2019;2019:2973289. doi: 10.1155/2019/2973289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao P., Cui X., Zhao L., Liu L., Wang D. Overexpression of growth-arrest-specific transcript 5 improved cisplatin sensitivity in hepatocellular carcinoma through sponging miR-222. DNA Cell Biol. 2020;39:724–732. doi: 10.1089/dna.2019.5282. [DOI] [PubMed] [Google Scholar]
- 165.Luo T., Chen M., Zhao Y., Wang D., Liu J., Chen J., Luo H., Li L. Macrophage-associated lncRNA ELMO1-AS1: a novel therapeutic target and prognostic biomarker for hepatocellular carcinoma. Onco Targets Ther. 2019;12:6203–6216. doi: 10.2147/OTT.S213833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deng L., Huang S., Chen B., Tang Y., Huang F., Li D., Tang D. Tumor-linked macrophages promote HCC development by mediating the CCAT1/Let-7b/HMGA2 signaling pathway. Onco Targets Ther. 2020;13:12829–12843. doi: 10.2147/OTT.S283786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang J., Cai M., Jiang D., Xu L. Upregulated LncRNA-CCAT1 promotes hepatocellular carcinoma progression by functioning as miR-30c-2-3p sponge. Cell Biochem. Funct. 2019;37:84–92. doi: 10.1002/cbf.3375. [DOI] [PubMed] [Google Scholar]
- 168.Guo J., Ma Y., Peng X., Jin H., Liu J. LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J. Cell Biochem. 2019;120:17975–17983. doi: 10.1002/jcb.29064. [DOI] [PubMed] [Google Scholar]
- 169.Ye Y., Guo J., Xiao P., Ning J., Zhang R., Liu P., Yu W., Xu L., Zhao Y., Yu J. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310–322. doi: 10.1016/j.canlet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 170.Huang Z., Chu L., Liang J., Tan X., Wang Y., Wen J., Chen J., Wu Y., Liu S., Liao J., et al. H19 promotes HCC bone metastasis through reducing osteoprotegerin expression in a protein phosphatase 1 catalytic subunit alpha/p38 mitogen-activated protein kinase-dependent manner and sponging microRNA 200b-3p. Hepatology. 2021;74:214–232. doi: 10.1002/hep.31673. [DOI] [PubMed] [Google Scholar]
- 171.Wang D., Xing N., Yang T., Liu J., Zhao H., He J., Ai Y., Yang J. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218–7230. doi: 10.1002/cam4.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao Y., Zhou L., Li H., Sun T., Wen X., Li X., Meng Y., Li Y., Liu M., Liu S., et al. Nuclear-encoded lncRNA MALAT1 epigenetically controls metabolic reprogramming in HCC cells through the mitophagy pathway. Mol. Ther. Nucleic Acids. 2021;23:264–276. doi: 10.1016/j.omtn.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Malakar P., Stein I., Saragovi A., Winkler R., Stern-Ginossar N., Berger M., Pikarsky E., Karni R. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR-mediated translation of TCF7L2. Cancer Res. 2019;79:2480–2493. doi: 10.1158/0008-5472.CAN-18-1432. [DOI] [PubMed] [Google Scholar]
- 174.Feng C., Zhao Y., Li Y., Zhang T., Ma Y., Liu Y. LncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge. Arch. Bronconeumol. (Engl Ed. 2019;55:627–633. doi: 10.1016/j.arbres.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 175.Peng N., He J., Li J., Huang H., Huang W., Liao Y., Zhu S. Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer Cell Int. 2020;20:165. doi: 10.1186/s12935-020-01231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hou Z.H., Xu X.W., Fu X.Y., Zhou L.D., Liu S.P., Tan D.M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am. J. Physiol. Cell Physiol. 2020;318:C649–C663. doi: 10.1152/ajpcell.00510.2018. [DOI] [PubMed] [Google Scholar]
- 177.Fan F., Chen K., Lu X., Li A., Liu C., Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021;15:444–458. doi: 10.1007/s12072-020-10101-6. [DOI] [PubMed] [Google Scholar]
- 178.Li X., Lei Y., Wu M., Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int. J. Mol. Sci. 2018;19:2958. doi: 10.3390/ijms19102958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tian X., Wu Y., Yang Y., Wang J., Niu M., Gao S., Qin T., Bao D. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/beta-catenin signaling. Mol. Oncol. 2020;14:462–483. doi: 10.1002/1878-0261.12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guo T., Gong C., Wu P., Battaglia-Hsu S.F., Feng J., Liu P., Wang H., Guo D., Yao Y., Chen B., et al. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 2020;27:2191–2205. doi: 10.1038/s41418-020-0494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen J., Huang Z.B., Liao C.J., Hu X.W., Li S.L., Qi M., Fan X.G., Huang Y. LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-beta1 signaling. Cell Signal. 2020;75:109738. doi: 10.1016/j.cellsig.2020.109738. [DOI] [PubMed] [Google Scholar]
- 182.Song W., Zhang J., Xia Q., Sun M. Down-regulated lncRNA TP73-AS1 reduces radioresistance in hepatocellular carcinoma via the PTEN/Akt signaling pathway. Cell Cycle. 2019;18:3177–3188. doi: 10.1080/15384101.2019.1671089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 183.Ma C.X., Gao W.C., Tian L. LncRNA TP73-AS1 promotes malignant progression of hepatoma by regulating microRNA-103. Eur. Rev. Med. Pharmacol. Sci. 2019;23:4713–4722. doi: 10.26355/eurrev_201906_18052. [DOI] [PubMed] [Google Scholar]
- 184.Gu Y., Wang C., Chen S., Tang J., Guo X., Hu W., Cui A., Zhang D., Yu K., Chen M. A critical role of peptidylprolyl isomerase A pseudogene 22/microRNA-197-3p/Peptidylprolyl isomerase A Axis in hepatocellular carcinoma. Front. Genet. 2021;12:604461. doi: 10.3389/fgene.2021.604461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hu M., Fu Q., Jing C., Zhang X., Qin T., Pan Y. LncRNA HOTAIR knockdown inhibits glycolysis by regulating miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. Biomed. Pharmacother. 2020;125:109703. doi: 10.1016/j.biopha.2019.109703. [DOI] [PubMed] [Google Scholar]
- 186.Tang X., Zhang W., Ye Y., Li H., Cheng L., Zhang M., Zheng S., Yu J. LncRNA HOTAIR contributes to sorafenib resistance through suppressing miR-217 in hepatic carcinoma. Biomed. Res. Int. 2020;2020:9515071. doi: 10.1155/2020/9515071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Topel H., Bagirsakci E., Comez D., Bagci G., Cakan-Akdogan G., Atabey N. lncRNA HOTAIR overexpression induced downregulation of c-Met signaling promotes hybrid epithelial/mesenchymal phenotype in hepatocellular carcinoma cells. Cell Commun. Signal. 2020;18:110. doi: 10.1186/s12964-020-00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fujisaka Y., Iwata T., Tamai K., Nakamura M., Mochizuki M., Shibuya R., Yamaguchi K., Shimosegawa T., Satoh K. Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell line. Oncol. Lett. 2018;15:509–514. doi: 10.3892/ol.2017.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sui C.J., Zhou Y.M., Shen W.F., Dai B.H., Lu J.J., Zhang M.F., Yang J.M. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J. Mol. Med. (Berl) 2016;94:1281–1296. doi: 10.1007/s00109-016-1442-z. [DOI] [PubMed] [Google Scholar]
- 190.Xiao S., Huang S., Yang J. Overexpression of GIHCG is associated with a poor prognosis and immune infiltration in hepatocellular carcinoma. Onco Targets Ther. 2020;13:11607–11619. doi: 10.2147/OTT.S271966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen Y., Li Y., Zheng G., Zhou P. Construction and analysis of macrophage infiltration related circRNA-miRNA-mRNA regulatory networks in hepatocellular carcinoma. PeerJ. 2020;8:e10198. doi: 10.7717/peerj.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhou P., Zheng G., Li Y., Wu D., Chen Y. Construction of a circRNA-miRNA-mRNA network related to macrophage infiltration in hepatocellular carcinoma. Front. Genet. 2020;11:1026. doi: 10.3389/fgene.2020.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu J., Zhang T., Chen Y., Ha S. MiR-139-5p influences hepatocellular carcinoma cell invasion and proliferation capacities via decreasing SLITRK4 expression. Biosci. Rep. 2020;40 doi: 10.1042/BSR20193295. BSR20193295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu T., Zhang H., Yi S., Gu L., Zhou M. Mutual regulation of MDM4 and TOP2A in cancer cell proliferation. Mol. Oncol. 2019;13:1047–1058. doi: 10.1002/1878-0261.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xu R., Yin S., Zheng M., Pei X., Ji X. Circular RNA circZFR promotes hepatocellular carcinoma progression by regulating miR-375/HMGA2 Axis. Dig. Dis. Sci. 2021;66:4361–4373. doi: 10.1007/s10620-020-06805-2. [DOI] [PubMed] [Google Scholar]
- 196.Ding B., Fan W., Lou W. hsa_circ_0001955 enhances in vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS Axis. Mol. Ther. Nucleic Acids. 2020;22:445–455. doi: 10.1016/j.omtn.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang Y.G., Wang T., Ding M., Xiang S.H., Shi M., Zhai B. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–138. doi: 10.1016/j.canlet.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 198.Cheng B., Tian J., Chen Y. Identification of RNA binding protein interacting with circular RNA and hub candidate network for hepatocellular carcinoma. Aging (Albany NY) 2021;13:16124–16143. doi: 10.18632/aging.203139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hu Z.Q., Zhou S.L., Li J., Zhou Z.J., Wang P.C., Xin H.Y., Mao L., Luo C.B., Yu S.Y., Huang X.W., et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2020;72:906–922. doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 201.Laoui D., Van Overmeire E., De Baetselier P., Van Ginderachter J.A., Raes G. Functional relationship between tumor-associated macrophages and macrophage colony-stimulating factor as contributors to cancer progression. Front. Immunol. 2014;5:489. doi: 10.3389/fimmu.2014.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang Y., Gao R., Li J., Tang S., Li S., Tong Q., Li S. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int. J. Nanomedicine. 2021;16:2803–2818. doi: 10.2147/IJN.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang X., Sheng W., Xu T., Xu J., Gao R., Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARalpha/CCL2 pathway. Aging (Albany NY) 2021;13:11969–11987. doi: 10.18632/aging.202900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Teng K.Y., Han J., Zhang X., Hsu S.H., He S., Wani N.A., Barajas J.M., Snyder L.A., Frankel W.L., Caligiuri M.A., et al. Blocking the CCL2-CCR2 Axis using CCL2-neutralizing antibody is an effective therapy for hepatocellular cancer in a mouse model. Mol. Cancer Ther. 2017;16:312–322. doi: 10.1158/1535-7163.MCT-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li J., Xue J., Ling M., Sun J., Xiao T., Dai X., Sun Q., Cheng C., Xia H., Wei Y., et al. MicroRNA-15b in extracellular vesicles from arsenite-treated macrophages promotes the progression of hepatocellular carcinomas by blocking the LATS1-mediated Hippo pathway. Cancer Lett. 2021;497:137–153. doi: 10.1016/j.canlet.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 206.Zhang Y.F., Shan C., Wang Y., Qian L.L., Jia D.D., Zhang Y.F., Hao X.D., Xu H.M. Cardiovascular toxicity and mechanism of bisphenol A and emerging risk of bisphenol S. Sci. Total Environ. 2020;723:137952. doi: 10.1016/j.scitotenv.2020.137952. [DOI] [PubMed] [Google Scholar]
- 207.Batlle E., Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xu Y., Pan S., Jiang W., Xue F., Zhu X. Effects of propofol on the development of cancer in humans. Cell Prolif. 2020;53:e12867. doi: 10.1111/cpr.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Saraiva M., Vieira P., O'Garra A. Biology and therapeutic potential of interleukin-10. J. Exp. Med. 2020;217:e20190418. doi: 10.1084/jem.20190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhao W., Ma X., Liu L., Chen Q., Liu Z., Zhang Z., et al. SNHG20: a vital lncRNA in multiple human cancers. J. Cell Physiol. 2019:1–7. doi: 10.1002/jcp.28143. [DOI] [PubMed] [Google Scholar]
- 211.Carotenuto P., Fassan M., Pandolfo R., Lampis A., Vicentini C., Cascione L., Paulus-Hock V., Boulter L., Guest R., Quagliata L., et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268–1277. doi: 10.1136/gutjnl-2016-312278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tong Q., Gong A.Y., Zhang X.T., Lin C., Ma S., Chen J., Hu G., Chen X.M. LincRNA-Cox2 modulates TNF-alpha-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2016;30:1187–1197. doi: 10.1096/fj.15-279166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B., et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tang S.H., Gao J.H., Wen S.L., Tong H., Yan Z.P., Liu R., Tang C.W. Expression of cyclooxygenase-2 is correlated with lncRNA-COX-2 in cirrhotic mice induced by carbon tetrachloride. Mol. Med. Rep. 2017;15:1507–1512. doi: 10.3892/mmr.2017.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yin C., Han Q., Xu D., Zheng B., Zhao X., Zhang J. SALL4-mediated upregulation of exosomal miR-146a-5p drives T-cell exhaustion by M2 tumor-associated macrophages in HCC. Oncoimmunology. 2019;8:1601479. doi: 10.1080/2162402X.2019.1601479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lin Y., Jiang M., Chen W., Zhao T., Wei Y. Cancer and ER stress: mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed. Pharmacother. 2019;118:109249. doi: 10.1016/j.biopha.2019.109249. [DOI] [PubMed] [Google Scholar]
- 217.Han Y., Liu D., Li L. PD-1/PD-L1 pathway: current researches in cancer. Am. J. Cancer Res. 2020;10:727–742. [PMC free article] [PubMed] [Google Scholar]
- 218.Ai L., Xu A., Xu J. Roles of PD-1/PD-L1 pathway: signaling, cancer, and beyond. Adv. Exp. Med. Biol. 2020;1248:33–59. doi: 10.1007/978-981-15-3266-5_3. [DOI] [PubMed] [Google Scholar]
- 219.Gordon S.R., Maute R.L., Dulken B.W., Hutter G., George B.M., McCracken M.N., Gupta R., Tsai J.M., Sinha R., Corey D., et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 221.Munoz-Garcia J., Cochonneau D., Teletchea S., Moranton E., Lanoe D., Brion R., Lezot F., Heymann M.F., Heymann D. The twin cytokines interleukin-34 and CSF-1: masterful conductors of macrophage homeostasis. Theranostics. 2021;11:1568–1593. doi: 10.7150/thno.50683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yang Z., Zi Q., Xu K., Wang C., Chi Q. Development of a macrophages-related 4-gene signature and nomogram for the overall survival prediction of hepatocellular carcinoma based on WGCNA and LASSO algorithm. Int. Immunopharmacol. 2021;90:107238. doi: 10.1016/j.intimp.2020.107238. [DOI] [PubMed] [Google Scholar]
- 223.Braconi C., Valeri N., Kogure T., Gasparini P., Huang N., Nuovo G.J., Terracciano L., Croce C.M., Patel T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U S A. 2011;108:786–791. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang L.P., Lin J., Ma X.Q., Xu D.Y., Shi C.F., Wang W., Jiang X.J. Exosomal DLX6-AS1 from hepatocellular carcinoma cells induces M2 macrophage polarization to promote migration and invasion in hepatocellular carcinoma through microRNA-15a-5p/CXCL17 axis. J. Exp. Clin. Cancer Res. 2021;40:177. doi: 10.1186/s13046-021-01973-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 225.Long J., Bai Y., Yang X., Lin J., Yang X., Wang D., He L., Zheng Y., Zhao H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019;19:90. doi: 10.1186/s12935-019-0817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wu D.M., Zheng Z.H., Zhang Y.B., Fan S.H., Zhang Z.F., Wang Y.J., Zheng Y.L., Lu J. Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J. Exp. Clin. Cancer Res. 2019;38:237. doi: 10.1186/s13046-019-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 227.Zhang L., He X., Jin T., Gang L., Jin Z. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed. Pharmacother. 2017;96:884–891. doi: 10.1016/j.biopha.2017.10.056. [DOI] [PubMed] [Google Scholar]
- 228.Liu X., Peng D., Cao Y., Zhu Y., Yin J., Zhang G., Peng X., Meng Y. Upregulated lncRNA DLX6-AS1 underpins hepatocellular carcinoma progression via the miR-513c/Cul4A/ANXA10 axis. Cancer Gene Ther. 2021;28:486–501. doi: 10.1038/s41417-020-00233-0. [DOI] [PubMed] [Google Scholar]
- 229.Li D., Tang X., Li M., Zheng Y. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J. Cell Biochem. 2019;120:12290–12299. doi: 10.1002/jcb.28493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Cheng S., Luo M., Ding C., Peng C., Lv Z., Tong R., Xiao H., Xie H., Zhou L., et al. Downregulation of Peptidylprolyl isomerase A promotes cell death and enhances doxorubicin-induced apoptosis in hepatocellular carcinoma. Gene. 2016;591:236–244. doi: 10.1016/j.gene.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 231.Hong M., Tao S., Zhang L., Diao L.T., Huang X., Huang S., Xie S.J., Xiao Z.D., Zhang H. RNA sequencing, new technologies and applications in cancer research. J. Hematol. Oncol. 2020;13:166. doi: 10.1186/s13045-020-01005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ling Y., Zhang W.Y., Wang P.Y., Xie W.H., Yang W.Y., Wang D.A., Fan C.J. Three-dimensional (3D) hydrogel serves as a platform to identify potential markers of chondrocyte dedifferentiation by combining RNA sequencing. Bioact Mater. 2021;6:2914–2926. doi: 10.1016/j.bioactmat.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hsu C.K., Lin C.C., Hsiao L.D., Yang C.M. Mevastatin ameliorates sphingosine 1-phosphate-induced COX-2/PGE2-dependent cell migration via FoxO1 and CREB phosphorylation and translocation. Br. J. Pharmacol. 2015;172:5360–5376. doi: 10.1111/bph.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sartorelli V., Lauberth S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020;27:521–528. doi: 10.1038/s41594-020-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hsieh C.L., Fei T., Chen Y., Li T., Gao Y., Wang X., Sun T., Sweeney C.J., Lee G.-S.M., Chen S., et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U S A. 2014:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Li W., Hu Y., O S., Ma Q., Merkurjev D., Song X., Zhou X., Liu Z., Tanasa B., He X., et al. Condensin I and II complexes license full estrogen receptor alpha-dependent enhancer activation. Mol. Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lee J.H., Ruan H., Ye Y., Krakowiak J., Hu Q., Xiang Y., Gong J., Zhou B., Wang L., Lin C., et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNAtargeted therapy in cancer. Nat. Commun. 2019;10:4562. doi: 10.1038/s41467-019-12543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wu H., Zhang J., Bai Y., Zhang S., Zhang Z., Tong W., Han P., Fu B., Zhang Y., Shen Z. DCP1A is an unfavorable prognostic-related enhancer RNA in hepatocellular carcinoma. Aging (Albany NY) 2021;13:23020–23035. doi: 10.18632/aging.203593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cai S., Hu X., Chen R., Zhang Y. Identification and validation of an immune-related eRNA prognostic signature for hepatocellular carcinoma. Front. Genet. 2021;12:657051. doi: 10.3389/fgene.2021.657051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rei M., Pennington D.J., Silva-Santos B. The emerging protumor role of gammadelta T lymphocytes: implications for cancer immunotherapy. Cancer Res. 2015;75:798–802. doi: 10.1158/0008-5472.CAN-14-3228. [DOI] [PubMed] [Google Scholar]
- 241.Tanchot C., Terme M., Pere H., Tran T., Benhamouda N., Strioga M., Banissi C., Galluzzi L., Kroemer G., Tartour E. Tumour-infiltrating regulatory T cells: phenotype, role, mechanism of expansion in situ and clinical significance. Cancer Microenviron. 2013;6:147–157.216. doi: 10.1007/s12307-012-0122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Jiang, R., Tang, J., Chen, Y., Deng, L., Ji, J., Xie, Y., Wang, K., Jia, W., Chu, W.M., Sun, B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat. Commun. 8, 15129. [DOI] [PMC free article] [PubMed]
- 243.Yan K., Fu Y., Zhu N., Wang Z., Hong J.L., Li Y., Li W.J., Zhang H.B., Song J.H. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8 + T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019;110:1–8. doi: 10.1016/j.biocel.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 244.Yu Z.Y., Zhao H.Z., Feng X., Li H.B., Qiu C.H., Yi X.M., Tang H., Zhang J.W. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the treg-mediated immune escape of hepatocellular carcinoma cells. Mol. Ther. Nucleic Acids. 2019;17:516–529. doi: 10.1016/j.omtn.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Han Q.J., Zhao H.J., Jiang Y., Yin C.L., Zhang J. HCC-derived exosomes, critical player and target for cancer immune escape. Cells. 2019;8:558. doi: 10.3390/cells8060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Shao Y., Lo C.M., Ling C.C., Liu X.B., Ng K.T.-P., Chu A.C.Y., Ma Y.Y., Li C.X., Fan S.T., Man K. Regulatory B-cells accelerate hepatocellular carcinoma progression via CD40/CD154 signaling pathway. Cancer Lett. 2014;355:264–272. doi: 10.1016/j.canlet.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 247.Xiao X., Lao X.M., Chen M.M., Liu R.X., Wei Y., Ouyang F.Z., Chen D.P., Zhao X.Y., Zhao Q., Li X.F., et al. PD-1hi identifies a novel regulatory B-cell population in human hepatoma that promotes disease progression. Cancer Discov. 2016;6:546–559. doi: 10.1158/2159-8290.CD-15-1408. [DOI] [PubMed] [Google Scholar]
- 248.Ye L., Zhang Q., Cheng Y., Chen X., Wang G., Shi M., Zhang T., Cao Y., Pan H., Zhang L., et al. Tumor-derived exosomal HMGB1 fosters hepatocellular carcinoma immune evasion by promoting TIM-1(+) regulatory B-cell expansion. J. Immunother. Cancer. 2018;6:145. doi: 10.1186/s40425-018-0451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]