Abstract
(−)-β-d-2,6-Diaminopurine dioxolane (DAPD), is a nucleoside reverse transcriptase (RT) inhibitor with activity against human immunodeficiency virus type 1 (HIV-1). DAPD, which was designed as a water-soluble prodrug, is deaminated by adenosine deaminase to give (−)-β-d-dioxolane guanine (DXG). By using calf adenosine deaminase a Km value of 15 ± 0.7 μM was determined for DAPD, which was similar to the Km value for adenosine. However, the kcat for DAPD was 540-fold slower than the kcat for adenosine. In CEM cells and peripheral blood mononuclear cells exposed to DAPD or DXG, only the 5′-triphosphate of DXG (DXG-TP) was detected. DXG-TP is a potent alternative substrate inhibitor of HIV-1 RT. Rapid transient kinetic studies show the efficiency of incorporation for DXG-TP to be lower than that measured for the natural substrate, 2′-deoxyguanosine 5′-triphosphate. DXG-TP is a weak inhibitor of human DNA polymerases α and β. Against the large subunit of human DNA polymerase γ a Ki value of 4.3 ± 0.4 μM was determined for DXG-TP. DXG showed little or no cytotoxicity and no mitochondrial toxicity at the concentrations tested.
Human immunodeficiency virus (HIV) type 1 (HIV-1) encodes an Mg2+-dependent reverse transcriptase (RT) which is required for virus replication and which is a proven target for the chemotherapy of HIV infection. Currently, strategies for the treatment of HIV infection involve the use of one or more inhibitors of HIV RT in combination with an HIV protease inhibitor. 2′,3′-Dideoxynucleoside or related nucleoside analogues that lack a free 3′-hydroxyl group, such as zidovudine (AZT), dideoxyinosine, stavudine, and lamivudine (3TC), are potent inhibitors of the RT and, when used in this combination regimen to treat HIV infection, strongly suppress virus replication and provide a durable antiviral response. However, drug-related adverse events and/or resistance eventually limits the usefulness of these compounds. Consequently, novel nucleoside analogues that are better tolerated and that have a different resistance profile are needed.
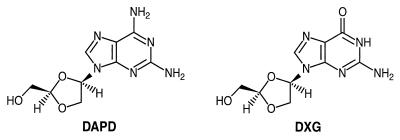
The 1-β-d enantiomer of the purine nucleoside analogue (−)-β-d-dioxolane guanosine (DXG; Fig. 1) has demonstrated both anti-HIV and anti-hepatitis B virus activities in vitro. However, because of the poor solubility and low oral bioavailability associated with DXG (2, 3; R. F. Schinazi, unpublished data), a more aqueous soluble and bioavailable prodrug, (−)-β-d-2,6-diaminopurine dioxolane (DAPD; Fig. 1), has been synthesized (10). Our results show that the in vitro anti-HIV activity seen upon treatment with DAPD is almost entirely due to the generation of DXG by the action of adenosine deaminase (ADA) on DAPD. DAPD is converted to DXG by the ADA-catalyzed hydrolysis of the 6-amino position of the molecule (Fig. 1). DXG is subsequently phosphorylated to the 5′-triphosphate (DXG-TP), which is a strong alternative substrate inhibitor of the HIV RT. Neither compound shows significant cytotoxicity. A compelling reason for the development of DAPD is that cross-resistance to DXG is not associated with mutations which confer resistance to AZT, 3TC, or adefovir and multidrug resistance (1, 8; J. Mewshaw, D. Wakefield, B. Hooper, L. Trost, B. McCreedy, and K. Borroto-Esoda, Program abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 924, 1999). This lack of cross-resistance offers the possibility of salvage therapy for those who have failed current nucleoside therapy. DAPD is currently being assessed for safety and efficacy in phase I/II clinical trials (D. Richman, H. Kessler, J. Eron, M. Thompson, F. Raffi, J. Jacobson, J. Harris, B. McCreedy, J. Bigley, and F. Rousseau, Program abstr. 7th Conf. Retrovir. Opportunistic Infect. abstr. G425p, 2000).
FIG. 1.
Structures of DAPD and DXG.
MATERIALS AND METHODS
Sources of nucleosides and nucleotides.
DAPD and DXG were synthesized by published procedures (10). [5′-3H]DAPD (specific activity, 9.4 Ci/mmol) and [8-3H]DXG (specific activity, 1.0 Ci/mmol) were obtained from Moravek Biochemicals, Inc., Brea, Calif. The 5′-triphosphate of DAPD (DAPD-TP) and DXG (DXG-TP) were synthesized by Inspire Pharmaceuticals, Research Triangle Park, N.C. The 5′-triphosphates of thymidine, cytidine, adenosine, and guanosine were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.). α-32P-labeled dGTP, dATP, and dTTP were from Amersham Pharmacia Biotech (Piscataway, N.J.). Adenosine was obtained from Calbiochem (San Diego, Calif.).
Antiviral activities of DAPD and DXG.
DAPD and DXG were evaluated for activity against the LAI strain of HIV-1 in MT2 cells and in human peripheral blood mononuclear cells (PBMCs) stimulated with phytohemagglutinin (PHA). MT2 cells were infected with virus at a multiplicity of infection (MOI) of 0.01 for 2 to 4 h at 37°C. The infected cells (3 × 104 cells/well) were seeded into 96-well cell culture plates containing fivefold serial dilutions of DAPD or DXG in complete medium (RPMI 1640 [Life Technologies, Rockville, Md.] containing 10% fetal bovine serum [Life Technologies] and 20 μg of gentamicin [Life Technologies] per ml). Following a 5-day incubation at 37°C in a humidified 5% CO2 atmosphere, the antiviral activity was determined by the 3,3′[1[(phenylamino) carbonyl]-3,4-tetrazolium-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate (XTT) assay of Weislow et al. (16). For evaluation of activity in PBMCs, cells were infected with virus for 4 h at 37°C at an MOI of 0.001. Infected cells (105 cells/well) were seeded into 96-well cell culture plates containing serial dilutions of test compound as described above. Cultures were incubated for 4 days at 37°C, and antiviral activity was determined by measuring the levels of p24 antigen in the culture supernatant by using the HIV-1 Antigen Microelisa System (Organon Teknika, Durham, N.C.).
Antiviral activity in the presence of inhibitors of ADA.
DAPD was tested in the presence of the ADA inhibitors erythro-3-(adenin-9-yl)-2-nonanol (EHNA; a generous gift from Glaxo Wellcome) and deoxycoformycin (DCF; Supergen, San Ramón, Calif.). Antiviral assays in the presence of EHNA were performed with MT2 cells infected with the LAI strain of HIV-1, as described above. Briefly, EHNA (100 μM) was added to the cells at the time of infection, and control cells were infected in the absence of EHNA. Following a 2-h infection, the cells were seeded into 96-well cell culture plates containing serial dilutions of DAPD and the appropriate concentration of EHNA. Cultures were incubated for 5 days, and antiviral activity was assessed by the XTT assay (16).
To determine the effect of DCF on the activity of DAPD, PHA-stimulated human PBMCs cultured in a 25-cm2 flask (5 × 106 cells per 10 ml) were infected with the LAI strain of HIV-1 at an MOI of 0.1 and were preincubated with DCF for 30 min prior to the addition of DAPD. Tenfold serial dilutions of DAPD (0.01 to 100 μM) were tested in combination with 10 μM DCF. AZT was used as a positive control. After 6 days of incubation, 1 ml of supernatant was harvested from each culture and was centrifuged at 11,750 × g for 2 h to pellet the virus. The pellet was solubilized by vortexing in 100 μl of solubilization buffer containing 0.5% Triton X-100, 0.8% NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 20% glycerol, and 0.1 M Tris (pH 7.8) and was assayed for RT activity. To assay for RT activity, 10 μl of each sample was added to 75 μl of an RT reaction mixture [0.06 M Tris (pH 7.8), 0.012 M NaCl, 0.006 M dithiothreitol, 0.006 mg of poly(rA) · oligo(dT)12–18 per ml, 96 μg of dATP per ml, 1 μM [3H]thymidine-5′-triphosphate], and the mixture was incubated at 37°C for 2 h. The reaction was stopped and the nucleic acid was precipitated by the addition of 100 μl of 10% trichloroacetic acid-containing 0.05% sodium pyrophosphate. The acid-insoluble product was harvested onto filter paper with a cell harvester (Packard Instrument Co., Meriden, Conn.), and the radioactivity was detected with a direct beta counter (Packard Instrument Co.).
Cytotoxicity assays.
MT2 cells and PHA-stimulated human PBMCs were seeded at densities of 3 × 104 and 1 × 105 cells/well, respectively, in 96-well cell culture plates containing twofold serial dilutions of DAPD or DXG. The concentrations of DAPD ranged from 0.48 to 1,000 μM, and the concentrations of DXG ranged from 0.24 to 500 μM. The cultures were incubated for 5 days at 37°C in a humidified 5% CO2 atmosphere and were then incubated with XTT for 3 h. Cytotoxicity was determined by comparing treated cultures with the untreated control (16).
Human bone marrow stem cell toxicity assay.
The method used to assess the toxicities of DAPD and DXG toward human bone marrow stem cells of the granulocyte-macrophage and erythroid lineages has been described previously (13, 14). Briefly, mononuclear cells were isolated from heparinized human bone marrow from donors by Ficoll-Hypaque gradient centrifugation. The cells were then washed twice with Hanks balanced salt solution and counted with a hemocytometer, and viability was assessed by trypan blue exclusion. The mononuclear cells (105/plate) were grown in the presence of various concentrations of test compound either in a double agar layer containing 75 to 100 U of granulocyte-macrophage colony-stimulating factor or in methylcellulose containing 1 U of erthropoietin.
Mitochondria toxicity assays.
The effect of DAPD and DXG on mitochondrial function in HepG2 cells was studied by measuring the concentrations of lactic acid in extracellular medium and the mitochondrial DNA content and by evaluation of structural effects by electron microscopy (5, 6, 11). Each experiment was performed three times.
(i) Effect on cell growth and lactic acid production.
The effect of DAPD or DXG on the growth of HepG2 cells was determined by incubating cells in the presence of various concentrations of DAPD or DXG (0, 0.1, 1, and 10 μM). Cells (5 × 104 per well) were incubated for 4 days at 37°C in 12-well culture dishes. At the end of the incubation period the cell number was determined with a hemocytometer. To measure the effects of DAPD and DXG on lactic acid production, HepG2 cells from a stock culture were diluted and plated in 12-well culture plates at 2.5 × 104 cells per well. Various concentrations (0, 0.1, 1, and 10 μM) of DAPD or DXG were added, and the cultures were incubated at 37°C in a humidified 5% CO2 atmosphere for 4 days. At day 4 the number of cells in each well was determined and the culture medium was collected. The culture medium was filtered with Arcodisc filters, and the lactic acid content in the medium was determined with a lactic acid assay kit (r-biopharm, Marshall, Mich.).
(ii) Effect on mitochondrial DNA synthesis.
HepG2 cells (2.5 × 104/sample) were plated into 12-well cell culture plates and treated with DAPD or DXG at concentrations of 0, 0.1, 1, and 10 μM, and the plates were incubated at 37°C in a humidified 5% CO2 atmosphere. After a 4-day incubation, the culture medium was changed every other day until termination of the experiment at 14 days, with fresh medium containing the appropriate dilution of compound added with each change of culture medium. At the end of the treatment period, approximately 5 × 105 cells were heated under alkaline conditions and blotted onto a Zeta-Probe GT nylon membrane for slot blot analysis. Mitochondrial DNA was detected by using a probe specific for human mitochondrial DNA that encompassed nucleotide positions 4212 to 4242. A 625-bp fragment of human beta-actin was used as a control probe. The DNA isolated from the treated cells was fixed to the membrane by heating for 10 min at 100°C and hybridized with the oligonucleotide probes for mitochondrial DNA or the beta-actin gene sequence.
(iii) Electron microscopic evaluation.
HepG2 cells (2.5 × 104 cells/ml) were seeded into tissue cultures dishes (35 by 10 mm) in the presence of 0, 0.1, 1, and 10 μM DAPD or DXG. Following a 4-day incubation period, the medium with and without compound was changed every other day. At day 8, the cells were fixed with 1% glutaraldehyde for 1 h, rinsed in sodium phosphate buffer, and fast fixed in 1% osmium tetroxide for 1 h. The cells were gradually dehydrated with increasing concentrations of ethanol (50 through 100%) and then with propylene oxide. The cells were infiltrated with Epon and embedded in Epon. Thin sections were prepared with a Reichter-Jung ultramicrotome, stained with uranyl acetate and lead citrate, and examined with a Hitachi 7000 electron microscope.
Intracellular metabolism of DAPD and DXG.
Intracellular metabolism studies were performed with PHA-stimulated human PBMCs and CEM cells with [5′-3H]DAPD (specific activity, 9.4 Ci/mmol) and [8-3H]DXG (specific activity, 1.0 Ci/mmol). Radiolabeled compounds were determined to be 98% pure by reverse-phase liquid chromatography. Approximately 2 × 106 cells/ml were incubated with 5 μM [3H]DAPD or 5 μM [3H]DXG at 37°C for 12, 24, 36, and 48 h in a humidified incubator with 5% CO2. Following the incubation, the cells were washed twice with ice-cold, serum-free medium. Cell extracts were prepared by the addition of 200 μl of cold 60% methanol–distilled water (vol/vol) and were stored at −70°C until analyzed by ion-exchange high-pressure liquid chromatography (HPLC) in which the chromatograph was coupled to a Flo-One radioactive flow detector (Packard Instrument Co.) equipped with a 5μ Hydropore SAX column (4.6 by 100 mm; Varian, Walnut Creek, Calif.). The elution was performed with 10% methanol and a gradient of ammonium dihydrogen phosphate buffer (pH 5.5) starting at 10 mM for 1 min and then increasing linearly to 125 mM ammonium dihydrogen phosphate over the next 24 min; the ammonium dihydrogen phosphate was maintained at this concentration for 5 min to allow elution of the 5′-triphosphate derivative. Elution was performed at a flow rate of 0.5 ml/min. The phosphorylated derivatives of DAPD and DXG were identified by using unlabeled phosphorylated standards of DAPD and DXG. Additionally, the identities of the phosphorylated derivatives of DAPD or DXG were confirmed by digesting the samples with bovine intestinal mucosa alkaline phosphatase (EC 3.1.3.1; Sigma Chemical Co., St. Louis, Mo.) and analyzing the digested samples by HPLC for conversion of the phosphorylated derivatives of DAPD and DXG to the corresponding nucleoside.
The susceptibility of DAPD to deamination was assessed with PHA-stimulated human PBMCs and CEM cells following a 24-h incubation with radiolabeled DAPD. The isolated nucleoside peak obtained by the ion-exchange HPLC method described above was analyzed by reverse-phase liquid chromatography with a Whatman Partispher C18 column. An isocratic elution was performed at a flow rate of 1 ml/min with 10 mM ammonium dihydrogen phosphate and 7% methanol. The amount of extracellular DAPD was also assessed by HPLC.
Enzyme assays. (i) ADA.
Calf ADA, obtained from Boehringer Mannheim (Indianapolis, Ind.), was used to determine the steady-state kinetic parameters (Km and kcat) for DAPD and adenosine by the method of initial rates (4). Enzyme reactions were performed at 25°C in 50 mM HEPES (pH 7.5). The concentrations of DAPD and adenosine, which was used as the control, ranged from 2.5 to 100 μM, and the enzyme concentration was 1.5 nM. Reaction progress was monitored by measurement of the UV absorbance with a Hewlett-Packard 8453 UV spectrometer at 265 and 245 nm for adenosine and DAPD, respectively (15). Kinetic parameters were calculated by nonlinear least-squared analysis curve fitting to the equation v = Vmax[S]/([S] + Km), where v is velocity, S is the substrate, and Vmax and Km correspond to standard definitions.
(ii) Steady-state HIV RT and DNA polymerase α, β, and γ assays.
The standard 50-μl reaction mixture for the HIV RT (ChimeriX, Madison, Wis.) contained 50 mM Tris-Cl (pH 7.8), 5 mM MgCl2, 0.025% Triton X-100, 0.012 U of poly(rC) · oligo(dG)12–18 or the heteropolymeric template primer r44 · d23mer at a concentration of 1.5 μM, and 50 μM (each) dATP, dCTP, and dTTP (Fig. 2). Enzyme reactions were performed at 37°C and were started by the addition of HIV RT (final concentration, 18 nM).
FIG. 2.
Sequences of oligonucleotide substrates. ∗, 5′-32P labeling of oligomer.
Reaction mixtures (50 μl) for human DNA polymerase α (provided by William C. Copeland) contained 50 mM Tris-Cl (pH 7.8), 5 mM MgCl2, 200 μg of bovine serum albumin fraction V per ml, 0.5 mg of activated calf thymus DNA per ml, and dATP, dCTP, and dTTP at 50 μM each. The reactions were started by the addition of enzyme to a final concentration of 1.7 nM, and the reaction mixtures were incubated at 37°C.
The reaction mixtures (50 μl) for human DNA polymerase β (ChimeriX) contained 50 mM Tris-Cl (pH 7.8), 5 mM MgCl2, 200 μg of bovine serum albumin fraction V per ml, 0.5 mg of activated calf thymus DNA per ml, 1 mM dithiothreitol, 4 mM N-ethylmaleimide, and 50 μM (each) dATP, dCTP, and dTTP. The reactions were started by the addition of 20 nM enzyme, and the reaction mixtures were incubated at 37°C.
For human DNA polymerase γ, the large subunit of the enzyme (provided by William C. Copeland) was used in the assay (12). The reaction mixtures (50 μl) contained 20 mM Tris-Cl (pH 8.0), 2 mM β-mercaptoethanol, 0.1 mg of bovine serum albumin fraction V per ml, 10 mM MgCl2, 0.4 mg of activated calf thymus DNA per ml, and deoxynucleoside triphosphates (dNTPs) at 50 μM. The reactions were started by the addition of enzyme to a final concentration of 5.2 nM, and the reaction mixtures were incubated at 37°C.
All polymerase reactions were terminated at various time intervals by spotting 5-μl aliquots of the reaction mixture onto DE81 paper. The paper was washed three times with 5% Na2HPO4, followed by a single wash with water and a single wash with ethanol. Kinetic constants were determined by the method of initial rates (4).
(iii) Pre-steady-state kinetic analysis.
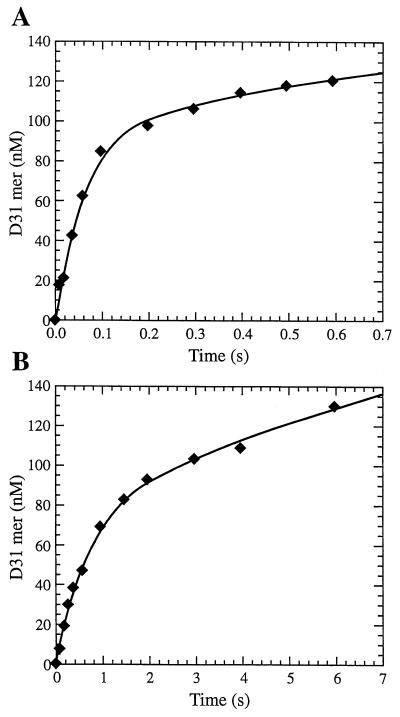
Transient kinetic experiments were performed by the rapid quench method as described previously (7, 9) with a KinTek Instruments model RQF-3 rapid-quench-flow apparatus (built at State College, Pa.). The reactions were carried out by mixing a solution containing the preincubated complex of HIV-1 RT (100 nM) and 5′-labeled DNA-DNA (D30/D45) or DNA-RNA (D30/R45) duplex (300 nM) (Fig. 2) with a solution of 10 mM MgCl2 and various concentrations of the dNTPs. The reactions were quenched with 0.3 M EDTA at time intervals ranging from 3 ms to 3 min. The products from the reaction were quantitated by sequencing gel analysis. The product formation occurred in a fast exponential phase, followed by a slower linear phase (Fig. 3). Data from burst experiments were fit to a burst equation, [product] = A[1 − e−kobsdt + ksst], where A represents the amplitude of the burst that correlates with the concentration of enzyme in the active form, kobsd is the observed first-order rate constant for dNTP incorporation, kss is the observed steady-state rate constant, and t is time. The dissociation constant, Kd, for dNTP binding to the complex of RT and primer-template was calculated by fitting the data to the hyperbolic equation kobsd = (kpol × [dNTP]/(Kd + [dNTP]), where kpol is the maximum rate of dNTP incorporation, [dNTP] is the corresponding concentration of dNTP, and Kd is the equilibrium dissociation constant for the interaction of dNTP with the enzyme · DNA complex.
FIG. 3.
Pre-steady-state burst kinetics showing incorporation of dGMP (A) or DXG-monophosphate (B) into a DNA-DNA 30/45-mer primer-template by HIV-1 RT. A preincubated mixture of DNA-DNA 30/45-mer primer-template (300 nM) and HIV-1 RT (active site concentration, 100 nM) was mixed with 75 μM dGTP (A) or 30 μM DXG-TP (B) in 10 mM MgCl2 containing buffer to start the reaction. The reactions were quenched at the indicated time interval with EDTA (0.3 M) and were analyzed by sequencing gel electrophoresis. The solid line represents the fit to the burst equation as described in the text. The curves in panel A represent fits with A equal to 101 ± 9 nM, kobsd equal to 15 ± 2 s−1, and kss equal to 0.36 ± 0.03 s−1 for dGTP, and the curves in panel B represent fits with A equal to 81 ± 4 nM, kobsd equal to 11.3 ± 0.1 s−1, and kss equal to 0.10 ± 0.01 s−1 for DXG-TP.
RESULTS
Anti-HIV activities and cytotoxicities of DAPD and DXG.
The activities of DAPD and DXG against the LAI strain of HIV-1 determined with MT2 cells and PHA-stimulated human PBMCs are given in Table 1. A median 50% effective concentration (EC50) of 12.5 ± 2.5 μM was determined for DAPD in MT2 cells, while DXG was found to be substantially more active, with a median EC50 of 3.4 ± 0.9 μM. When the activities were tested with PBMCs by a p24-based enzyme-linked immunosorbent assay, the EC50s were substantially lower than those determined with MT2 cells. DXG was again more active, with a median EC50 of 0.25 ± 0.17 μM.
TABLE 1.
Anti-HIV-1 activities of DAPD and DXG
| Compound | Median EC50a (μM)
|
|
|---|---|---|
| MT2 cells | Human PBMCs | |
| DAPD | 12.5 ± 2.5b | 4.0 ± 2.2b |
| DXG | 3.4 ± 0.9 | 0.25 ± 0.17 |
| DAPD + DCF | NDc | >1,000 |
| DAPD + EHNA | >150 | ND |
Assays were performed as described in Materials and Methods.
Data are the means ± standard deviations of six measurements.
ND, not determined.
To determine if DXG was responsible for the anti-HIV activity seen upon exposure of infected cultures to DAPD, assays were performed in the presence of the adenosine-adenylate deaminase inhibitor DCF and the specific ADA inhibitor EHNA. In the presence of DCF the EC50 of DAPD measured with PBMCs increased >250-fold, and in the presence of EHNA the EC50 of DAPD measured with MT2 cells increased >12-fold (Table 1). As a control, the antiviral activity of DXG was also assessed in the presence of EHNA and DCF. Neither DCF nor EHNA had any effect on the antiviral activity of DXG, and furthermore, neither DCF nor EHNA demonstrated any innate anti-HIV activity (data not shown).
The cytotoxicities of DAPD and DXG were determined in cell growth assays with PHA-stimulated human PBMCs and the laboratory-adapted MT2 and CEM cell lines (Table 2). DAPD and DXG showed little cytotoxicity toward PBMCs and MT2 and CEM cells, with the 50% cytotoxic concentrations (CC50s) of DAPD being >1 mM for all three cell types and the CC50s of DXG being >500 μM for all three cell types.
TABLE 2.
Evaluation of cytotoxicities of DAPD and DXG to various cells
| Compound | CC50 (μM)
|
||||
|---|---|---|---|---|---|
| MT2 cells | CEM cells | PBMCs | BFU-E | CFU-GM | |
| DAPD | >1,000 | >1,000 | >1000 | >100 | >100 |
| DXG | >500 | >500 | >500 | >100 | >100 |
| AZT | 20.0a | 14.3a | >100a | 0.1 | 2.0 |
Data are from reference 13.
Because of the apparent correlation between toxicity to bone marrow progenitor cells in vitro and bone marrow suppression in vivo seen with other antiviral agents, DAPD and DXG were tested for bone marrow toxicity by an in vitro human bone marrow progenitor stem cell assay. The results of those studies (Table 2) showed that neither DAPD nor DXG was toxic (CC50s, >100 μM) toward human bone marrow progenitor stem cells of the granulocyte-macrophage lineage (CFU-granulocyte macrophage [CFU-GM]) or the erythroid lineage (burst-forming unit–erythroid [BFU-E]).
Effects of DAPD and DXG on mitochondrial function in HepG2 cells.
DAPD and DXG were tested for potential toxicity toward human mitochondria by incubating HepG2 cells for 14 days in the presence of increasing concentrations of compound. At all concentrations tested, DAPD and DXG showed no adverse effects on cell growth, mitochondrial DNA synthesis, mitochondrial structure, lipid droplet formation, or lactic acid production (Table 3). Higher concentrations of DAPD and DXG (25 and 50 μM) were tested for their effects on cell growth, mitochondrial DNA synthesis, and lactic acid production. No effect on cell growth or lactic acid production was observed after exposure to DAPD and DXG at concentrations up to 50 μM. DXG at concentrations up to 50 μM had no effect on mitochondrial DNA synthesis. However, DAPD at concentrations of 25 and 50 μM caused 36% ± 4% and 64% ± 6% decreases in mitochondrial DNA synthesis, respectively. Dideoxycytidine and 2′-fluoro-5-methyl-β-d-arabinofuranosyluracil (d-FMAU) were included in these studies as positive controls for mitochondrial toxicity. At a concentration of 1 μM, ddC caused a 68% reduction in mitochondrial DNA synthesis. Exposure of HepG2 cells to 25 μM d-FMAU resulted in a 47% decrease in mitochondrial DNA synthesis and a 10-fold increase in lactic acid production compared with those for untreated control cells.
TABLE 3.
Effects of DAPD and DXG on cell proliferation and mitochondrial function and morphology in HepG2 cells
| Compound | Concn (μM) | Cell no. (104/ml) | l-Lactate density (mg/106 cells) | % of mitochondrial DNA content to that for control | Lipid droplet formation | Mitochondrial morphology |
|---|---|---|---|---|---|---|
| Control | 0 | 16.0 ± 0.9a | 2.4 ± 0.2a | 100 | Negative | Normal |
| DAPD | 0.1 | 16.2 ± 0.4 | 2.3 ± 0.1 | 104 ± 10a | NDb | ND |
| DAPD | 1.0 | 15.9 ± 0.8 | 2.3 ± 0.2 | 100 ± 11 | ND | ND |
| DAPD | 10.0 | 13.2 ± 0.8 | 3.0 ± 0.1 | 92 ± 5 | Negative | Normal |
| Control | 0 | 25.2 ± 0.8 | 2.35 ± 0.02 | 100 | Negative | Normal |
| DXG | 0.1 | 26.1 ± 0.8 | 2.28 ± 0.20 | 106 ± 5 | ND | ND |
| DXG | 1.0 | 25.2 ± 0.6 | 2.45 ± 0.08 | 107 ± 8 | ND | ND |
| DXG | 10.0 | 24.9 ± 0.8 | 2.36 ± 0.10 | 99 ± 9 | Negative | Normal |
| Controlc | 0 | 42.9 ± 0.2 | 1.52 ± 0.04 | 100 | ND | ND |
| DAPDc | 25.0 | ND | ND | 64 ± 4 | ND | ND |
| DAPD | 50.0 | 45.7 ± 0.1 | 1.40 ± 0.13 | 36 ± 6 | ND | ND |
| DXGc | 25.0 | ND | ND | 103 ± 11 | ND | ND |
| DXG | 50.0 | 45.5 ± 0.2 | 1.62 ± 0.06 | 98 ± 3 | ND | ND |
Data are the means ± standard deviations of three measurements.
ND, not determined.
The second experiment was performed with higher concentrations of DAPD and DXG.
Enzymatic conversion of DAPD to DXG.
The ability of ADA to utilize DAPD as a substrate was determined by using purified calf thymus ADA. The steady-state kinetic parameters for the deamination of DAPD were measured and compared with the values obtained for the natural substrate, adenosine (Table 4). The Km value of 11 ± 0.9 μM obtained for DAPD is similar to the Km value for adenosine (15 ± 0.7 μM). However, the kcat for DAPD is 540-fold slower than the kcat for adenosine. Comparison of the overall substrate efficiency (kcat/Km) of DAPD to that of adenosine showed that DAPD was 360-fold less efficiently used as a substrate for ADA. DAPD (100 μM) was also deaminated to DXG when DAPD was incubated with whole human blood. Under these conditions, in which excess enzyme was present, 50% of the total DAPD was converted to DXG in 2.4 h. When adenosine (100 μM) was incubated with human blood under identical conditions, 50% of the adenosine was converted to inosine in 0.6 min. In addition, the specific ADA inhibitor EHNA (10 μM) completely inhibited the conversion of DAPD to DXG by human blood (data not shown).
TABLE 4.
Catalytic constants for DAPD deamination by calf ADAa
| Compound | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| Adenosine | 15 ± 0.7b | 108 ± 9.0 | 7.2 |
| DAPD | 11 ± 0.9 | 0.2 ± 0.01 | 0.02 |
Assays were performed at 25°C and pH 7.5 by the method of initial rates as described in Materials and Methods.
Data are the means ± standard deviations of two measurements.
Metabolism of DAPD and DXG in cell culture.
The anabolism of DXG was detected in both PBMCs and CEM cells after incubation of the cells with 5 μM radiolabeled DXG. Both the parent nucleoside and the corresponding 5′-mono-, 5′-di-, and 5′-triphosphate forms were detected and identified as described in Materials and Methods. DXG-TP was detected after 12 h of exposure, and by 24 h a steady-state level appears to have been achieved (Table 5).
TABLE 5.
Anabolism of DXG in human PBMCs and CEM cells incubated with 5 μM DXGa
| Cell type | Incubation time (h) | Intracellular concn (pmol/106 cells)
|
|||
|---|---|---|---|---|---|
| DXG | DXG-MP | DXG-DP | DXG-TP | ||
| PBMCs | 12 | 1.88 | 3.51 | 3.37 | 0.63 |
| PBMCs | 24 | 2.76 | 1.88 | 2.98 | 1.79 |
| PBMCs | 36 | 2.93 | 2.34 | 2.52 | 1.61 |
| PBMCs | 48 | 3.55 | 2.22 | 2.43 | 1.20 |
| CEM cellsb | 24 | 0.22 | 0.13 | 0.53 | 4.40 |
| CEM cellsc | 24 | 3.69 | 0.69 | 1.25 | 1.10 |
Cells were incubated with 5 μM [8-3H]DXG (specific activity, 1.0 Ci/mmol). MP, monophosphate; DP, diphosphate.
Exponentially growing cells.
Resting cells.
The metabolism of DAPD in PHA-stimulated human PBMCs and CEM cells was also assessed (Table 6). As expected, DAPD was readily deaminated to DXG. While both DXG and DAPD were detected, DAPD levels in PBMCs were 27-fold higher than the level of DAPD determined in CEM cells; the level of DXG was roughly the same in both cell types. As a control for deamination by the cell culture medium, DAPD was incubated at 37°C for 24 h in RPMI 1640 containing 10% fetal bovine serum. At the end of the 24-h incubation, only 8% of the DAPD was converted to DXG.
TABLE 6.
Metabolism of DAPD in human PBMCs and CEM cells incubated with 5 μM DAPDa
| Cell type | Intracellular concn (pmol/106 cells)
|
||||
|---|---|---|---|---|---|
| DAPD | DXG | DXG-MP | DXG-DP | DXG-TP | |
| PBMCs | 0.54 | 3.93 | Blb | Bl | Bl |
| CEM cells | 0.02 | 2.30 | 0.07 | 0.04 | 0.46 |
Cells were incubated for 24 h with 5 μM [5′-3H]DAPD (specific activity, 9.4 Ci/mmol). MP, monophosphate; DP, diphosphate.
Bl, below the limit of detection of 0.01 pmol/106 cells.
The intracellular levels of DAPD and DXG and their phosphorylated derivatives were quantitated. No phosphorylation of DAPD to the corresponding mono-, di-, or triphosphate forms was detected in either cell type. However, DXG-TP was detected (0.46 pmol/106 cells) in rapidly dividing CEM cells but not in PBMCs following a 24-h incubation with 5 μM DAPD (Table 6). These results show that DAPD was deaminated to DXG and was subsequently phosphorylated to DXG-TP. To determine if the growth rate of cells can influence the anabolism of DXG, a rapidly growing culture of CEM cells and a slowly growing culture of CEM cells were incubated with 5 μM radiolabeled DAPD for 24 h. The level of DXG-TP was fourfold higher in rapidly dividing CEM cells compared with the levels of DXG-TP detected in the slowly growing cells (Table 5).
Determination of steady-state and pre-steady-state kinetic constants for DXG-TP.
The steady-state inhibition constant (Ki) for DXG-TP was measured for the HIV-1 RT and human DNA polymerases α and β and the large subunit of human DNA polymerase γ by using poly(rC) · oligo(dG)12–18 or activated calf thymus DNA as the template-primer (Table 7). With the HIV-1 RT and human DNA polymerases, DXG-TP functioned as a competitive inhibitor of the incorporation of dGTP into the template-primer. With HIV RT a Ki value of 0.019 ± 0.002 μM was determined for DXG-TP with the homopolymeric primer-template poly(rC) · oligo(dG)12–18 (Table 7). The Ki was also determined for DXG-TP with the 23/44 DNA-RNA primer-template. With this template-primer a Ki value of 0.7 ± 0.2 μM was calculated. In contrast to the results obtained with HIV-1 RT, DXG-TP was only a weak inhibitor of the human α and β DNA polymerases (Table 7). DXG-TP was an inhibitor of the large subunit of DNA polymerase γ, giving a Ki value of 4.0 ± 0.4 μM. The Ki/Km ratios for each of the polymerases tested show that DXG-TP is a better inhibitor of the HIV-1 RT than the human DNA polymerases α, β, and γ.
TABLE 7.
Steady-state kinetic constants for dGTP and DXG-TP for HIV-1 RT and human DNA polymerases
| Polymerase | Km (μM) for dGTPa | Ki (μM) for DXG-TPa | Ki/Km |
|---|---|---|---|
| HIV-1 RTb | 1.2 ± 0.4 | 0.019 ± 0.002 | 0.016 |
| HIV-1 RTc | 0.6 ± 0.2 | 0.73 ± 0.17 | 1.2 |
| DNA polymerase αd | 1.7 ± 0.7 | 78 ± 17 | 45.9 |
| DNA polymerase βd | 2.3 ± 0.5 | 32 ± 2 | 13.9 |
| DNA polymerase γd | 3.0 ± 0.4 | 4.3 ± 0.4 | 1.4 |
Data are the means ± standard deviations of four measurements.
Poly(dC) · oligo(dG)12–18 was used as the template-primer.
Heteropolymeric 23/44 primer-template used in the assay.
Activated calf thymus DNA was used as the template-primer.
To demonstrate that DXG functioned as a substrate for the HIV RT, enzyme reactions were performed by incubating 200 nM enzyme and the [32P]r44/d25-mer template-primer (D25/R44; Fig. 2) (100 nM) with 1.0 μM DXG-TP at 37°C for 20 min. The product of the reaction was separated on a 15% polyacrylamide gel, which can detect the addition of a single nucleotide to the template-primer. The results of these experiments showed that the length of the template-primer was extended only by a single nucleotide (data not shown), indicating that DXG-TP was a substrate for the HIV-RT and that incorporation resulted in chain termination.
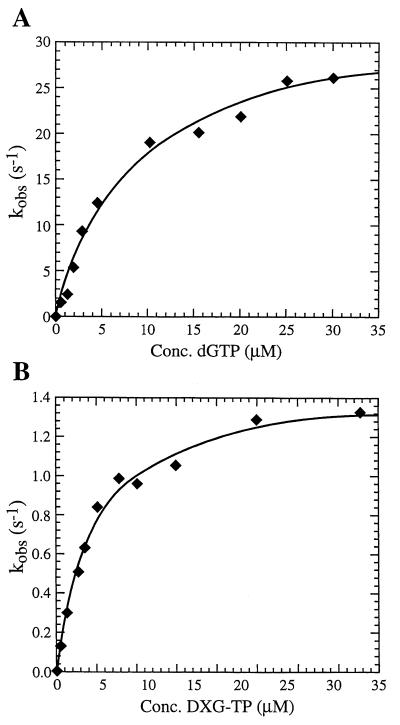
A pre-steady-state analysis of the alternative substrate inhibitor DXG-TP was run to directly observe the catalytic events occurring at the active site of the enzyme. Rapid transient kinetic experiments were performed to determine the rate of polymerization (kpol), the equilibrium dissociation constant (Kd), and the efficiency of incorporation (kpol/Kd). Pre-steady-state burst experiments were performed as described in the Materials and Methods section. The results from these experiments (Fig. 3) show a rapid exponential phase corresponding to an initial burst of dNTP incorporation and a slower linear phase of product formation (kss). The Kd for the interaction of dGTP and DXG-TP with the enzyme · primer-template complex as well as kpol were determined from the observed burst rates from a series of reaction time course experiments conducted with various concentrations of nucleotide substrate. Representative Kd curves for dGTP and DXG-TP are shown in Fig. 4. The Kd and kpol values, determined from these experiments with the D30/R45 and D30/D45 primer-templates, are summarized in Table 8. With both the D30/R45 and the D30/D45 primer-templates the Kd for DXG-TP was similar to that determined for the natural nucleotide substrate, dGTP. However, substantial differences exist between the kpol of dGTP and the kpol of DXG-TP with both the DNA-RNA and DNA-DNA primer-templates. Comparison of the kpol values in Table 8 shows that DXG-TP is incorporated much more slowly than the natural substrate, dGTP. Differences of 22.6- and 11-fold were observed with the D30/R45 and the D30/D45 primer-templates, respectively. The slower rate of incorporation for DXG-TP with both primer-templates compared with that for dGTP resulted in an approximately 17-fold lower incorporation efficiency (kpol/Kd) for DXG-TP compared to that for the natural substrate (Table 8).
FIG. 4.
Determination of Kd and kpol for dGTP incorporation (A) and DXG-TP incorporation (B) into a DNA-RNA 30/45-mer primer-template. The polymerase rates were plotted against dGTP or DXG-TP concentrations, and the data were fit to a hyperbola to give a Kd of 12 ± 2 μM and a kpol of 34 ± 2 s−1 for dGTP and a Kd of 8.8 ± 1.1 μM and a kpol of 1.5 ± 0.1 s−1 for DXG-TP.
TABLE 8.
Pre-steady-state kinetic constants for dGTP and DXG-TP incorporation into D30/D45 and D30/R45 primer-templates
| Primer-template | dGTP analog | kpol (s−1) | Kd (μM) | kss (s−1) | kpol/Kd (μM−1 s−1) |
|---|---|---|---|---|---|
| D30/D45 | dGTP | 16 ± 1a | 6.1 ± 0.9 | 0.36 ± 0.03 | 2.6 |
| DXG-TP | 1.46 ± 0.03 | 2.5 ± 0.2 | 0.28 ± 0.09 | 0.58 | |
| D30/R45 | dGTP | 34 ± 2 | 12 ± 2 | 0.15 ± 0.06 | 2.8 |
| DXG-TP | 1.5 ± 0.1 | 8.8 ± 1.1 | 0.14 ± 0.03 | 0.17 |
Values are means ± standard errors.
DISCUSSION
Since the discovery of AZT, a number of nucleoside analogues have been identified as potent and promising anti-HIV-1 agents. Among these, 1,3-dioxolane and 1,3-oxathiolane nucleosides are structurally unique in that an oxygen atom or a sulfur atom replaces the 3′ carbon of the carbohydrate moiety, respectively. Given the precedent set by the oxathiolane nucleosides, it would be anticipated that the dioxolane analogues would interact differently than other nucleoside analogues with the target enzyme, RT. This different interaction would be anticipated to manifest itself as a different resistance and cross-resistance profile. This has been shown to be true (1, 8, 13; Mewshaw et al., 39th ICAAC), in that recombinant viruses and clinical isolates of HIV-1 containing typical mutations for nucleoside and nonnucleoside RT inhibitors (M41L, D67N, T69D, K70R, K103N, M184V, G190A, T215Y, and K219Q), alone or in combination, remain sensitive to DXG. Furthermore, viruses with mutations associated with multinucleoside resistance due to SS or SG insertions between codons 68 and 69 were sensitive to DXG (Mewshaw et al., 39th ICAAC).
The lack of water solubility and the correspondingly poor bioavailability noted for DXG led to the synthesis of the more water soluble 2,6-diamino analogue DAPD (2, 3; Schinazi, unpublished data). DAPD is converted to DXG by the hydrolytic action of ADA at the 6-amino position (Fig. 1). Although the Km value measured for DAPD is comparable to that of the natural substrate adenosine, the overall substrate efficiency of DAPD is substantially less owing to a significantly slower kcat value. Nonetheless, DAPD is efficiently converted to DXG in whole human blood (L. L. Keifer, P. A. Furman, I. L. Liberman, K. Borroto-Esoda, E. L. Hill, C. K. Chu, G. R. Painter, and R. F. Schinazi, Program abstr. 12th World AIDS Conf., abstr. 12358, 1998). In this study 50% of the DAPD was converted to DXG in 2.4 h. The rate of conversion of DAPD in whole human blood is similar to the rate of conversion observed in a phase I clinical trial (half-life of ∼1 h) (L. H. Wang, J. W. Bigley, R. L. St. Claire, N. Sista, F. Rousseau, and the DAPD-101 Clinical Trial Group, Program abstr. 7th Conf. Retrovir. Opportunistic Infect., abstr. 103, 2000).
As can be seen from the data in Table 1, the anti-HIV activities observed in both MT2 cells and PBMCs are significantly higher for DXG. However, there is significant activity apparently associated with DAPD, particularly in PBMCs. This activity could be attributable to the action of ADA on DAPD in the cell cultures. To determine if this is the case, antiviral assays were performed in the presence of the ADA inhibitor EHNA or the adenosine-adenylate deaminase inhibitor DCF. Inclusion of EHNA or DCF resulted in a significant decrease in the antiviral activity of DAPD, suggesting that the activity seen for DAPD is due to conversion of some fraction of DAPD to DXG. Thus, it is clear that the vast majority of the antiviral activity associated with DAPD is attributable to conversion to DXG. These data do not rule out the possibility that adenylate deaminase is involved in the conversion of DAPD monophosphate into DXG monophosphate.
Anabolism studies in cell culture with radiolabeled DAPD show that DAPD is not phosphorylated to the corresponding 5′-triphosphate. Instead, DAPD is deaminated to DXG and is subsequently phosphorylated to DXG-TP. Interestingly, incubation of PBMCs and CEM cells with DAPD results in significantly higher intracellular levels of DAPD in PBMCs compared with the concentrations in CEM cells. However, the levels of DXG in both cell types are similar. That intracellular DXG most likely arises from the intracellular deamination of DAPD to DXG is suggested from the control study in which DAPD was incubated with cell culture medium containing 10% fetal bovine serum for 24 h. In that study 92% of the DAPD remained unchanged. In both CEM cells and PBMCs incubated with DXG, all three phosphorylated derivatives are detected. As might be anticipated, the level of the DXG-TP is higher in actively growing cells than in resting cells.
In this report we show that the DXG-TP is an alternative substrate inhibitor that competitively inhibits the HIV-catalyzed incorporation of dGTP into the template-primer. In addition to a steady-state kinetic evaluation of the inhibition of RT by DXG-TP, we have used rapid transient kinetics to study directly the events occurring at the active site of the RT and to determine the kinetic parameters kpol and Kd for DXG-TP. These parameters provide information regarding the rate of conformational change during the incorporation of DXG-TP and a direct measure of the affinity of DXG-TP for the enzyme · primer-template complex. Additionally, the efficiency of incorporation (kpol/Kd) can be calculated. This parameter takes into account compensatory changes in binding and catalysis, allowing comparisons to be made between a nucleoside 5′-triphosphate analog and the corresponding natural substrate. The characteristic burst curve of DXG monophosphate incorporation suggested that DXG-TP was used as a substrate by HIV-1 RT in a manner similar to that for dGTP (Fig. 3). However, the incorporation of DXG monophosphate happened at a slower rate than the incorporation of dGMP. When DNA served as the template, dGTP was incorporated approximately 11 times faster than DXG-TP. The Kd values show that the binding affinity for dGTP was similar to that determined for DXG-TP. When the template was RNA, dGTP was again incorporated faster than DXG-TP (approximately 23 times faster), and the Kd values for both substrates were similar. Comparison of the overall efficiency of incorporation of dGTP with that of DXG-TP showed that dGTP was incorporated approximately 17 times more efficiently than DXG-TP into both primer-templates. No differences were seen in the kpol/Kd determined for dGTP or for DXG-TP with either the D30/R45 or the D30/D45 primer-template.
In vitro cytotoxicity assays showed that DAPD and DXG are not significantly toxic toward either human lymphocyte cell lines or primary human cells such as PBMCs and human bone marrow progenitor cells. The therapeutic indices for DAPD with MT2 cells and PBMCs were >77 and >500, respectively, and the therapeutic indices for DXG with MT2 cells and PBMCs were >128 and >5,000, respectively. To give further insight into the origins of the selective anti-HIV activities exhibited by DAPD and DXG, the inhibition of human DNA polymerases α, β, and γ by the 5′-triphosphate of DXG was examined under steady-state conditions. DXG-TP was a relatively weak inhibitor of the α and β DNA polymerases. On the other hand, DNA polymerase γ proved to be somewhat sensitive to inhibition by DXG-TP. The lack of correlation between the enzymatic and cellular data may be due to the fact that DXG-TP may not be efficiently taken up by mitochondria or that DXG is not taken up and is subsequently phosphorylated by mitochondria. Alternatively, DAPD, which does affect mitochondrial DNA synthesis in cell culture, may be efficiently taken up by mitochondria, where it could be deaminated to DXG by mitochondrial ADA and subsequently phosphorylated to DXG-TP. Additional studies will be required to differentiate between these and other possible causes of the differential effect on mitochondrial DNA synthesis seen with DAPD and DXG. Because the levels of DAPD in vivo are low due to rapid deamination to DXG and because the concentrations of DAPD required to inhibit mitochondrial DNA synthesis are high, the likelihood that administration of DAPD will affect mitochondrial DNA synthesis in vivo is very low. To date, no toxicities associated with mitochondrial toxicity have been observed in animals exposed to high doses of DAPD in chronic toxicology studies.
In summary, DAPD possesses all of the desirable preclinical attributes that a nucleoside analogue requires to be considered a viable clinical candidate. The compound is water soluble, is a potent inhibitor of HIV replication, and shows little or no cytotoxicity. These desirable properties are translating in the phase I/II clinical trials currently under way (Richman et al., 7th Conf. Retrovir. Opportunistic Infect.). First, DAPD is bioavailable and is efficiently converted in the peripheral circulation to DXG. Second, DXG is proving to be a potent inhibitor of viral replication in vivo, inducing up to a 1.45-log drop in viral load in drug-naïve patients at a dose of 300 mg. With the caveat that the time of exposure of patients to DAPD is relatively short, the adverse effect profile of the drug is extremely clean, in accordance with the cytotoxicity studies reported in this paper. Finally, as anticipated from the unique structure, DAPD is showing some activity in patients who have failed multiple-nucleoside therapy.
ACKNOWLEDGMENTS
R.F.S. and C.K.C. are supported in part by grant AI-25899 from the National Institutes of Health (NIH), the U.S. Department of Veterans Affairs, and a grant from Triangle Pharmaceuticals. P.A.F., L.L.K., and R.F.S. were supported in part by grant AI-40775 from NIH, and K.S.A. was supported by grant GM49551 from NIH.
REFERENCES
- 1.Bazmi H Z, Hammond J L, Cavalcanti S C H, Chu C K, Schinazi R F, Mellors J W. In vitro selection of mutations in human immunodeficiency virus type 1 reverse transcriptase that decrease susceptibility to (−)-β-d-dioxolane-guanosine and suppress resistance to 3′-azido-3′deoxythymidine. Antimicrob Agents Chemother. 2000;44:1783–1788. doi: 10.1128/aac.44.7.1783-1788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Boudinot F D, Chu C K, McClure H M, Schinazi R F. Pharmacokinetics of (−)-β-d-2-aminopurine dioxolane and (−)-β-d-2-amino-6-chloropurine dioxolane and their antiviral metabolite (−)-β-d-dioxolane guanine in rhesus monkeys. Antimicrob Agents Chemother. 1996;40:2332–2336. doi: 10.1128/aac.40.10.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H, Schinazi R F, Rajagopalan P, Gao Z, Chu C K, McClure H M, Boudinot F D. Pharmacokinetics of (−)-β-d-dioxolane guanine and prodrug (−)-β-d-diaminopurine dioxolane in rats and monkeys. AIDS Res Hum Retrovir. 1999;15:1625–1630. doi: 10.1089/088922299309667. [DOI] [PubMed] [Google Scholar]
- 4.Cleland W W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63A:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Schinazi R F, Gosselin G, Imbach J-L, Chu C K, Rando R F, Revankar G R, Sommadossi J-P. Effect of β-enantiomeric and racemic nucleoside analogues on mitochondrial functions in HepG2 cells. Biochem Pharmacol. 1996;52:1577–1584. doi: 10.1016/s0006-2952(96)00562-x. [DOI] [PubMed] [Google Scholar]
- 6.Faraj A, Fowler D A, Bridges E G, Sommadossi J-P. Effects of 2′,3′-dideoxynucleosides on proliferation and differentiation of human pluripotent progenitors in liquid culture and their effects on mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38:924–930. doi: 10.1128/aac.38.5.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng J Y, Anderson K S. Mechanistic studies comparing the incorporation of (+) and (−) isomers of 3TCTP by HIV-1 reverse transcriptase. Biochemistry. 1999;38:55–63. doi: 10.1021/bi982340r. [DOI] [PubMed] [Google Scholar]
- 8.Gu Z, Wainberg M A, Nguyen-Ba N, L'Heureux L, DeMuys J-M, Bowlin T L, Rando R F. Mechanism of action and in vitro activity of 1′,3′-dioxolanylpurine nucleoside analogues against sensitive and drug-resistant human immunodeficiency virus type 1 variants. Antimicrob Agents Chemother. 1999;43:2376–2382. doi: 10.1128/aac.43.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr S G, Anderson K S. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;46:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 10.Kim H O, Schinazi R F, Shanmuganathan K, Jeong L S, Beach J W, Nampalli S, Cannon D L, Chu C K. l-β-(2S,4S)- and l-α-(2S, 4R)-Dioxolanyl nucleosides as potential anti-HIV agents: asymmetric synthesis and structure-activity relationships. J Med Chem. 1993;36:519–528. doi: 10.1021/jm00057a001. [DOI] [PubMed] [Google Scholar]
- 11.Lewis W, Levine E S, Griniuviene B, Tankersley K O, Coacino J M, Sommadossi J-P, Wantanbe K A, Perrino F W. Fialuridine and its metabolites inhibit DNA polymerase γ at sites of multiple adjacent analog incorporation, decrease mtDNA abundance, and cause mitochondrial defects in cultured hepatoblasts. Proc Natl Acad Sci USA. 1996;93:3592–3597. doi: 10.1073/pnas.93.8.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longley M J, Ropp P A, Lim S E, Copeland W C. Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 13.Schinazi R F, McMillan A, Cannon D, Mathis R, Lloyd R M, Peck A, Sommadossi J-P, St. Clair M, Wilson J, Furman P A, Painter G, Choi W-B, Liotta D C. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine. Antimicrob Agents Chemother. 1992;36:2423–2431. doi: 10.1128/aac.36.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommadossi J-P, Carlisle R, Schinazi R F, Zhou Z. Uridine reverses the toxicity of 3′-azido-3′-deoxythymidine in normal human granulocyte-macrophage progenitor cells in vitro without impairment of the antiretroviral activity. Antimicrob Agents Chemother. 1988;7:997–1001. doi: 10.1128/aac.32.7.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spector T. Progress curve analysis of adenosine deaminase-catalyzed reactions. Anal Biochem. 1984;138:242–245. doi: 10.1016/0003-2697(84)90796-6. [DOI] [PubMed] [Google Scholar]
- 16.Weislow O S, Kiser R, Fine D L, Bader J, Shoemaker R H, Boyd M R. New soluble formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J Natl Cancer Inst. 1989;81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]