Abstract

A two-pronged computational approach was taken to study the promiscuity of the SAM+-dependent methyl transferase AtHTMT1 from thale cress with several nucleophiles (Cl–, Br–, I–, NCO–, NCS–). First, enzyme-free methyl transfer reactions were studied with M05/6-311+G(2d,p) DFT calculations and electrostatic continuum models (PCM/SMD) for various chemical environments. Second, QM/MM MD simulations with semiempirical Hamiltonians (PM7, PM6-D3, AM1, PM6-D3H4) and the AMBER 14SB force field were used to study the enzyme catalyzed reaction in silico. The combination of the DFT and MD results shows that reactant desolvation generally accelerates the reaction, but it cannot explain the selectivity of the enzyme. The critical position of H2O molecules at the reactive site favors the reaction of NCS– over Cl– and Br– in agreement with experiments, but not observed in the quantum calculations for the cytosol. The addition of selected H2O molecules to the N terminus of NCS– greatly increases its reactivity, while H2O molecules attached to Cl– slow the reaction. The partial solvation of the nucleophiles in the reactive pouch holds the key to understanding the reactivity of AtHTMT1.
1. Introduction
S-adenosyl-l-methionine (SAM)-dependent halide/thiocyanate methyl transferases (HTMTs) catalyze the transfer of a methyl group from the sulfonium cation in SAM+ to a nucleophile X– yielding CH3X and S-adenosyl-l-homocysteine (SAH) as summarized in eq 1.1−3
 |
1 |
The promiscuity of HTMTs, which accept a wide range of nucleophiles, has been successfully exploited to replace the CH3 group at the sulfonium cation with other residues R to produce SAM analogues R–SAH+,
| 2 |
which then can be used to selectively mark targeted biomolecules with other methyl transferases (MTs) as possible medical diagnostics.4−7
This versatile promiscuity is not only a useful lab-property of HTMTs. It can also be exploited for the green production of methyl halides from nonfood biomass as valuable industrial and agricultural chemicals.8 The production of CH3Cl can be observed on a large scale in vivo. Methyl chloride is a significant source of atmospheric chlorine (17%), but its emission is not controlled, as about 55% of it is produced by plants.9−11
The HTMTs linked to the plantal emission of CH3Cl typically accept a wide range of nucleophiles, such as SH–, CN–, SCN–, Cl–, Br−, and I–.12−17 Methyl thiocyanate has been found to be part of the plantal defense against bacterial infections,18 but no such biological benefit has been found for CH3Cl so far.
The analysis of the promiscuity of HTMTs and their catalytic mechanism hold the key to understanding plantal CH3Cl production. It has been speculated that the emission of CH3Cl can be used to remove excess Cl– from the plant, but SAM is an energetically expensive commodity for the cell.13,19 The quest of Cl– removal consuming SAM appears to be energetically prohibitively expensive and can be suppressed by feeding NaSCN to the plant.20 The study of the enzymatic mechanism provides the answer to the question of whether the production of CH3Cl is a simple, though expensive, byproduct of the NCS– metabolism.
The kinetic and structural data for the HTMT from thale cress (Arabidopsis thalina, AtHTMT1) lay the foundation for the computational study of HTMT promiscuity with kinetic and structural data from a single source.14 It has been reported that AtHTMT1 works most efficiently with the NCS– anion in the following order: NCS– > I– > Br– > Cl–. (The minus sign “–” in ambidentate ligands marks the ligand atom later binding directly to the CH3 group from the SAM molecule.) The reproduction of this order will be used to judge the quality of the computational model. Simulations agreeing with the experiment then can be used to deduce the physical principles controlling the promiscuity of AtHTMT1.
There are theoretical studies on the chemistry of SAM,21−32 but none of them focuses on AtHTMT1 and its reactivity with small nucleophiles. In this work, starting from the PDB structure, the free energy changes in the rate-determining step (rds) of the formation of CH3Cl, CH3Br, CH3I, CH3OCN, CH3NCO, CH3NCS, and CH3SCN are studied with QM/MM MD simulations to explore the details of the underlying mechanism.33 To quantify which nucleophile benefits the most from the catalytic environment, the MD simulations including AtHTMT1 are compared with enzyme-free QM calculations of the methyl transfer with an electrostatic continuum model using water as solvent to mimic the uncatalyzed reaction in the cytosol.
Comparing results obtained from QM/MM MD simulations with those from DFT calculations with an SMD model for solvent appears to be counterintuitive as the two methods differ dramatically in the number of atoms involved. Nevertheless, both methods strive to reproduce chemical reality starting from different assumptions of the solvation process. The heterogeneous environment is better described by the atomistically detailed MD simulations, while the homogeneous environment of a dilute aqueous solution can be more efficiently handled with a continuum model. The large size of the atomistic model limits the range of computationally affordable electronic structure methods. The principally lower computational costs of a continuum model allow us to use high level quantum models and thereby to analyze individual bonds at a much more sophisticated level. As both methods result in a macroscopically correct picture of the process, their direct comparison yields complementing information for each other of the chemical process studied.
The dielectric constant (ϵ) of the interior of enzymes has been estimated to lie between 4 and 20,34 and model studies with (CH3)3S+ mimicking SAM show that the methyl transfer is greatly accelerated in environments with low values for ϵ.35 A change of the value of ϵ in the electrostatic continuum model from 78 (water) to 4.24 (diethyl ether) in the QM calculations provides an estimate of the magnitude of the electrostatic effect on the reaction rate caused by the desolvation of the reactants in the enzyme. The X-ray structure of AtHTMT1 shows the presence of additional H2O molecules at the reactive site, indicating chemical interactions of the nucleophile with these molecules. The interaction between a small solute ion and its environment is addressed in continuum models by the addition of explicit solvent molecules to the quantum calculation.36 The number and position of these solvent molecules are an integral part of the continuum model and its parametrization. The addition of an explicit H2O molecule to the small nucleophile in the continuum solvation models can then be used to identify the effect of an individual hydrogen bond at the reactive site on the catalytic process.
The QM/MM MD and the ab initio QM calculations with an electrostatic continuum handle the effect of the chemical environment differently. Therefore, it is necessary to be prudent comparing their results. The main focus of this work lies on the relative reactivities of various nucleophiles X– and chemical trends presuming that both approaches describe the macroscopic reality correctly. Details of the computational and technical setup will be discussed in the Methods section. The Results section is split into subsections for the results from the computational blocks as laid-out above, and all observed effects will be condensed into one model in the final subsection, which leads to the conclusion about the catalytic mechanism and the promiscuity of AtHTMT1.
2. Methods
SAM has an isoelectric point (pHI) of 7.2437 and the pKa values of the COOH and NH3+ groups in the methionine branch are 1.80 and 7.80, respectively. Hence, three possible protonation states of the methione chain in the SAM molecule (anionic aSAM+, bipolar bSAM+, neutral nSAM+; Scheme 1) were considered for the continuum QM calculations assuming temperate pH values for the cytosol. The acronyms SAM and SAH are extended by a superscript on the left (‘a’, ‘b’ or ‘n’) to indicate the protonation state of the methionine chain. A superscript on the right indicates the positive charge on the sulfur atom in SAM.
Scheme 1. Protonation States of Methionine in SAM: (a) Anionic, aSAM+; (b) Bipolar, bSAM+; (n) Neutral, nSAM+.
The electric charges on the methionine branch of SAM are involved with the methyl transfer reaction. To study this effect, the QM calculations were carried out with all three protonation states. The QM/MM MD simulations, on the other hand, were done exclusively with bSAM+, as a preliminary analysis indicates its dominance in an enzymic environment, vide infra.
2.1. Standard QM Calculations without the Enzyme
The comparatively small number of atoms involved in the reaction between SAM+ and a nucleophile X– without the enzyme and an electrostatic continuum model for the chemical environment facilitates the application of midlevel quantum models to the problem. The properties of the sulfonium cation in SAM+ (marked red in eq 1) dominate the SN2 reaction of the methyl transfer. Extensive method testing on the analogue methyl transfer in trimethylsulfonium chloride [(CH3)3SCl] shows that the M05/6-311+G(2d,p) computational level can describe the internal SN2 reaction adequately in various solvents.35,38 The initial geometry of the SAM+ molecule for the geometry optimization is the equilibrated structure of SAM+ in aqueous solution taken from our previous QM/MM MD study of the hydrolysis of SAM+.22 All quantum calculations using continuum models were done with the Gaussian 09 software packages.39 All results from the geometry optimizations were verified with additional frequency calculations. Minima showed no imaginary frequencies, and the results for transition states showed only one aligned with the SN2 reaction path. Changes in Gibbs free energy (ΔG°) were calculated for 298.15 K, 1 atm and 1 mol/L. The correction to changes in Gibbs free energy caused by the transfer of the quantum results from data of the gas phase [(1 mol)/(24.5 L)] to solution [1 mol/L] is denoted as δG°,
| 3 |
where Δξ is the difference in the sums of stoichiometric coefficients of the reaction.35,40
The X-ray structure of AtHTMT1 indicates that H2O molecules in the reactive cavity near the nucleophile are likely to influence the enzymatic reaction.14 Hence, it is necessary to add one explicit H2O molecule into the continuum calculations.36 The calculations for Cl– and Br– in water and diethyl ether with and without explicit H2O molecules were implemented with the PCM and SMD models.36,41,42 Details (geometries, energies, frequencies) of these calculations can be found in section S.4 in the Supporting Information (SI).
The two solvation models yield different results for the relative reactivity of Cl– and Br–. This difference is briefly discussed here to justify the choice of solvation model for the uncatalyzed methyl transfer reaction in the cytosol. The barrier height ΔTSGenv°
| 4 |
is defined as the difference in free Gibbs energy between the reactive complex (RC) and the transition state (TS) in a given environment (env) defined by the solvent model and the number of explicit H2O molecules. Closely related to ΔTSGenv° is ΔHalGenv
| 5 |
defined as the difference in barrier height for the Br– and Cl– ions. It quantifies the change in reaction speed of the methyl transfer as the nucleophile changes.
Tables S.1 to S.3 in the SI list the data for ΔTSGenv° from the benchmark calculations, and Table 1 lists the results for ΔHalGenv. The addition of an explicit H2O molecule to the continuum solvation model raises the barrier obtained from both PCM and SMD calculations (Tables S.1, S.2, S.3, SI). Without any explicit H2O molecule, both continuum models predict Br– to be a better nucleophile having a lower barrier (Table 1). However, the addition of one H2O molecule changes the relative nucleophilicity of the halogenides in two cases. This effect is even more remarkable if the solvent separated reactants are used as reference (Tables S.1 to S.3). The PCM model yields [Cl·H2O]− to react faster than [Br·H2O]−, while the SMD model preserves [Br·H2O]− as the faster one regardless of the protonation state of the SAM molecule. Kinetic experiments with sulfonium cations in ethanol as solvent show that Br– is the better nucleophile of the two in agreement with general textbook chemistry.43−46
Table 1. ΔHalG° for Different Chemical Environments Mimicked by Various Continuum Modelsa.
The H2O molecules in the reactive cavity of the enzyme as revealed by X-ray crystallographic analysis14 can interact with the nucleophile; hence, the correct description of the interactions among the nucleophile and surrounding H2O molecules is critical for the computational model. The data in Table 1 indicate that only the SMD model describes this situation adequately. Therefore, we use the SMD model for the continuum calculations.
The calculations for Cl– and Br– also highlight the effect of the SAM+ protonation state on the methyl transfer. The absolute barrier heights (Tables S.1 to S.3) show that nSAM+ and bSAM+ have similar values of ΔTSGenv° with nSAM+ always being the smaller one. The Coulomb repulsion between the halide ion and the overall negative charge on the methionine branch of aSAM+ can be seen in the markedly higher barriers for aSAM+. The SMD calculations for the noncatalyzed methyl transfer reaction in the cytosol with all nucleophiles were done with nSAM+ to compare the results with the fastest possible reaction in aqueous solution.
2.2. QM/MM MD Simulations with the Enzyme
The structure of AtHTMT1 with SAH at the reactive site was obtained from the Protein Data Bank.14,47 The protonation state of the enzyme was determined by GROMACS and that of the SAH molecule with CHIMERA.48,49 The bSAH+ molecule was turned into bSAM+ by replacing a H2O molecule in the cavity with the new methyl group. The influence of the environment on the methyl transfer reaction can be deduced from the comparison of the QM/MM and the QM calculations, as nSAM+ and bSAM+ yield similar barriers for the methyl transfer to Cl– and Br–.
The AMBER 14SB force field, the general amber force field (GAFF), and the TIP3P force field were used to describe AtHTMT1, bSAM+, and the 94 560 water molecules, respectively.50−52 Periodic boundary conditions were used on a cubic water-box of (10 nm)3. The Coulomb and van der Waals cutoff radii were 0.9 nm, and long-range electrostatic interactions were calculated with the particle mesh Ewald (PME) method.53 To preserve electrostatic neutrality and to keep the salt concentration close to 0.150 mol/L, Na+ and Cl– ions were added to the system.
Figure 1 shows a snapshot of the methyl transfer from bSAM+ to Cl– in AtHTMT1, where the MM environment and the quantum H2O molecules in the solvation shell of bSAM+ and Cl– are omitted for clarity sake. Quantum atoms were described using the semiempirical methods PM7,54 PM6-D3,55,56 AM1,57 and PM6-D3H458 as defined in the Gaussian 09/1639,59 and MOPAC2011660 program suites. The collective variable λ (PLUMED),61 defined as the difference between the C–S (d1) and C–Cl (d2) bond lengths
| 6 |
was followed to calculate free energy changes using the umbrella sampling/weighted-histogram analysis method (US/WHAM).62,63
Figure 1.

Methyl transfer from bSAM+ to Cl–. d1 and d2 are used to span the collective variable λ. The quantum region is depicted using CPK model and licorice for the MM region. White, H; turquoise, C; blue, N; red, O; yellow, S; green, Cl. Link atoms are colored pink.
The MD simulations started with a steepest descent geometry optimization of the whole simulation cell followed by a set of MM MD simulations beginning with a 2 ns NVT simulation [TVR = 300 K (new velocity rescaling),64 τ = 2 fs (time steps)] with a restraint on the movement of the heavy backbone atoms and d2 (40 000 kJ/mol/nm2). The restraints on the backbone atoms were then lifted stepwise (1000, 100, 10, 0 kJ/mol/nm2) in a series of 2, 2, 2, 4, and 60 ns NpT simulations [TVR = 300 K, pBer = 1 bar (Berendsen barostat), τ = 2 fs], which allows the density of the system to relax to its natural state. The last step of the equilibration phase was a 180 ns NpT simulation [TVR = 300 K, pBer = 1 bar, τ = 2 fs] without any constraints, which yielded a constant rmd (root mean displacement) value for the heavy atoms in the backbone of the enzyme, while the running average of the total energy and its standard deviation have become constant. The equilibrated structure was then used to construct the QM/MM model for the methyl transfer reaction and to determine the number of QM H2O molecules for the solvation model by promoting all H2O molecules within 500 pm of the nucleophile to the quantum level.
The collective variable λ and the methyl transfer were driven in steps of 0.015 nm (40 ps each) from −0.18 to 0.225 nm or −0.195 to 0.225 nm depending on the nucleophile with a length constraint of 40 000 kJ/mol/nm2. The angle of attack of the nucleophile (4000 kJ/mol/rad2) and position of the QM H2O molecules in their solvation shells (2000 kJ/mol) was constrained with functions provided by PLUMED to avoid their diffusion away from the nucleophile. The transition region was resampled with smaller λ steps and/or larger constraints, if individual windows in this region sampled poorly. The overall ΔG° curve was then constructed from the individual segments.
3. Results and Discussion
3.1. Standard QM Calculations without the Enzyme
Table 2 lists the results of ΔTSGenv° (eq 4) from the SMD calculations with the neutral nSAM+ molecule. The reactive complex was used as the energetic reference due to its conceptual similarity to the enzyme–substrate complex in the Michaelis–Menten model of enzymatic reactivity. It should be noted that the reactions for Cl– and Br– without explicit H2O molecules in diethyl ether and that for Br– with an explicit H2O molecule appear to be barrier-less if the solvent separated reactants are taken as the energetic reference, because the formation of the contact ion pair in the reactive complex in ether is associated with a huge gain in Gibbs free energy (Table S.1), if the solvent separated reactants are chosen as reference.
Table 2. ΔTSG° (eq 4) and Δ+1G° (eq 10) for the Methyl Transfer from the Neutral nSAM+ Molecule to Various Nucleophiles Obtained from SMD Calculations with and without an Explicit H2O Moleculea.
| X–b | envb | ΔTSG° | envb | ΔTSG° | ΔDSG° | Δ+1Gwat° | Δ+1Geth° |
|---|---|---|---|---|---|---|---|
| Cl– | water | 104.1 | ether | 95.8 | –8.3 | ||
| Br– | 88.2 | 82.2 | –5.9 | ||||
| NCO– | 138.1 | 132.5 | –5.6 | ||||
| OCN– | 106.6 | 94.2 | –12.4 | ||||
| NCS– | 105.4 | 106.4 | 1.0 | ||||
| SCN– | 112.7 | 101.9 | –10.8 | ||||
| Cl– | water + 1c | 109.5 | ether + 1c | 99.6 | –9.9 | 5.4 | 3.8 |
| Br– | 94 | 94.4 | 0.4 | 5.8 | 12.2 | ||
| NCO– | 145.8 | 136.3 | –9.6 | 7.8 | 3.8 | ||
| NCO– | 142.7 | 139.6 | –3.1 | 4.6 | 7.2 | ||
| OCN– | 104 | 87.8 | –16.2 | –2.6 | –6.4 | ||
| OCN– | 111.4 | 97.7 | –13.7 | 4.8 | 3.4 | ||
| NCS– | 111.3 | 114.4 | 3.0 | 5.9 | 7.9 | ||
| NCS– | 112.5 | 103.6 | –8.8 | 7.1 | –2.8 | ||
| SCN– | 112.8 | 109.3 | –3.5 | 0.1 | 7.4 | ||
| SCN– | 113.3 | 106.4 | –7.0 | 0.6 | 4.4 |
All energies in kJ mol–1.
env, Chemical environment; X–, nucleophile.
One explicit H2O molecule included. The atom connecting to the H2O molecule is marked in bold-italic text.
The results for ΔTSGwat° from the SMD calculations with water as solvent and no explicit H2O molecule show that the X– with the more nucleophilic atom attacking the methyl group has lower reaction barriers and reacts faster with nSAM+ (eq 7)
| 7 |
where the ambidentate ions SCN– and OCN– engage in faster reactions with the softer end of them attacking the methyl group favoring S over N and N over O.45,46 Kinetic experiments with various trimethylsulfonium salts [(CH3)3S+·X–] in methanol (ϵ = 32.6) and ethanol (ϵ = 24.9) show that (CH3)3S+·Br– decomposes at a similar rate to (CH3)3S+·NCS–. Hence, the position of the NCS– ion in eq 7 suggests that more types of interaction between the nSAM+ and X– than just the nucleophilic attack have to be considered regarding the reactivity of SAM+.
The desolvation of the reactants has been found to be important for the catalysis of the SN2 methyl transfer.33,35 The difference ΔDSG° between the ΔTSGenv° values (eq 4) for water and ether
| 8 |
is used to quantify the desolvation effect (Table 2) with ether marking the lower end of the effective permittivity range for the interior of an enzyme.34
The data for ΔDSG° from SMD calculations with no explicit H2O molecule show that the methyl transfer from the nSAM+ donor to the nucleophile is generally enhanced with the exception of the NCS– ion. Sulfur atoms typically do not form strong hydrogen-bond networks; hence, a small positive value for ΔDSG° is not surprising. The interaction strength of each ion with the environment leads to the following rearrangement of the reactivity order by barrier height
| 9 |
The relative softness of the nucleophile appears to be less dominant in an apolar environment, but the halides seem to be generally more reactive.
The effect of H2O molecules on the methyl transfer can be quantified similarly. The difference between the values for ΔTSG° in the same continuum model solvent (sol) with (sol+1) and without (sol) an explicit H2O molecule is defined as Δ+1Gsol°
| 10 |
[sol: solvent continuum model, water (wat) or ether (eth)]. The explicit H2O molecule can be attached to either end of the ambidentate OCN– and SCN– ions.
The attachment of one H2O molecule to Br– effectively removes the desolvation effect (ΔDSG° = 0.4 kJ mol–1) while the changes in ΔDSG° for Cl– are small. Indeed, the values of Δ+1Geth° for Cl– and Br– are at opposite ends of the range for positive values of Δ+1Geth.
The data of Δ+1Gsol° (Table 2) indicate that H2O molecules in the vicinity of the nucleophile generally increases the barrier of the methyl transfer except notably OCN– and NCS– with the H2O molecule attached to the more electronegative atom that does not interact with the methyl group. Shifting the H2O molecule from the outer end of the nucleophile to the attacking end raises the barrier height by 10.3 kJ mol–1 on average. This effect is more prominent for NCS– than for OCN–, where the attachment of a H2O molecule to the sulfur atom removes the benefits of reactant desolvation (ΔDSG° = 3.0 kJ mol–1).
Table 3 lists the Michaelis-Menten parameters of four methyl transferases for Cl–, Br–, NCS– (the experimental trio) and I– obtained from experiment14−17 and their interpretation by us (section S.2). The kinetic data for the same kind of enzyme in the marine diatom microalgae (Phaeodactylum tricornutum), daikon radish (Raphanus sativus), and rice (Oryza sativa) are given for comparison. The enzymatic efficiency kcat/KM varies between 9.9 × 1 0–4 and 2.2 × 1 03 s–1 M–1, while the barrier height varies in a much smaller numeric range between 80 and 90 kJ mol–1. The variability in kcat/KM can be traced back to the enzyme’s ability to bind the substrate X– visible in KM. The highest values for KM are typically observed with Cl– and the lowest with NCS–.
Table 3. Experimental Data and Their Interpretation.
| enzyme | X– | KMa [mM] | vmaxa [nmol/(min mg)] | kcatb,d (10–3/s) | kcat/KMb,d [1/(s M)] | ΔTSG°c,d (kJ/mol) | ref |
|---|---|---|---|---|---|---|---|
| AtHTMT1 | NCS– | 0.099 ± 0.020 | 43.6 ± 2.52 | 19.2 ± 1.11 | 193.9 ± 40.85 | 82.82 ± 0.14 | (14) |
| AtHTMT1 | Br– | 24.87 ± 2.785 | 11.4 ± 0.31 | 5.02 ± 0.14 | 0.202 ± 0.023 | 86.15 ± 0.07 | (14) |
| AtHTMT1 | Cl– | 145.2 ± 26.56 | 2.43 ± 0.12 | 1.07 ± 0.05 | 0.007 ± 0.001 | 89.98 ± 0.12 | (14) |
| PtHTMT | NCS– | 7.9 | — | 10.2 | 1.29 | 84.39 | (15) |
| PtHTMT | I– | 8.6 | — | 51.6 | 6.00 | 80.37 | (15) |
| PtHTMT | Br– | 72.8 | — | 1.04 | 0.0143 | 90.05 | (15) |
| PtHTMT | Cl– | 637.9 | — | 0.31 | 0.00048 | 93.05 | (15) |
| RsHTMT | NCS– | 0.04 | 185.185 | 89.5 | 2237.65 | 80.37 | (16) |
| RsHTMT | I– | 4.47 | 139.286 | 67.3 | 15.0608 | 81.09 | (16) |
| RsHTMT | Br– | 177.34 | 34.965 | 16.9 | 0.09530 | 84.58 | (16) |
| RsHTMT | Cl– | 1657.40 | 3.381 | 1.63 | 0.00099 | 90.46 | (16) |
| OsHol1 | NCS– | 0.15 | 0.966 | 0.42 | 2.91 | 92.29 | (17) |
| OsHol1 | I– | 0.07 | 5.190 | 2.27 | 34.13 | 88.12 | (17) |
| OsHol1 | Br– | 44.6 | 3.390 | 1.48 | 0.03 | 89.18 | (17) |
| OsHol1 | Cl– | no activity observed | — | — | — | (17) | |
Copied from the reference.
Calculated from published kinetic data when vmax is available; otherwise copied from the reference.
This work, calculated from experimental kinetic data.
Error range calculated directly from the error range stated for the data obtained from experiment.
The fastest reaction, which has the largest values for vmax and kcat, is observed with AtHTMT1 and NCS– ions having the lowest barrier of 82.8 kJ mol–1. The highest AtHTMT1 barrier of 90.0 kJ mol–1 is observed in the reaction with Cl–. By contrast, the quantum calculation with a continuum solvation model (Table 2) shows that NCS– is the slowest of the trio and Br– is the fastest. Although the quantum calculation with a continuum solvation model indicates that the desolvation of the reactant will increase the rate of the reaction (typically ΔDSG° < 0 and Δ+1G° > 0), it fails to reproduce the relative order of reactivity of the nucleophiles observed in the experiments with enzymes. Experiments show that the enzymatic environment not only enhances the methyl transfer, but also favors NCS– over other nucleophiles. Therefore, it is necessary to include the enzyme explicitly in the calculations.
3.2. QM/MM MD Simulations with the Enzyme
The CHIMERA analysis of SAH in AtHTM1 indicates that bSAH+ is the dominating protonation state,48,49 while the lowest barriers for the methyl transfer in an aqueous environment are observed with nSAH. The initial analysis with Cl– and Br– as nucleophiles indicates that both protonation states lead to similar barrier height (Table 1). The docking of SAM+ to AtHTMT1 locks its protonation state to bSAM+ while the fast, unhindered proton movement in the aqueous environment enables the methyl transfer reaction to pass through the energetically more favorable transition state with nSAM+. The difference in protonation state depending on the chemical environment ensures that the methyl transfer reaction always passes through its lowest transition state.
The current study of methyl transfer in the solution and in the enzyme follows different mechanisms as summarized in eq 11,
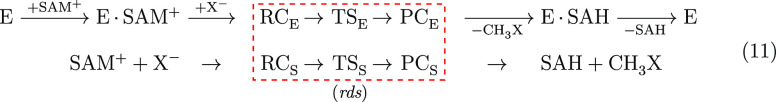 |
11 |
where E is the enzyme, S is the solution, RC is the reactive complex, TS is the transition state, PC is the product complex, E·SAM+ represents SAM+ docked to the enzyme, and E·SAH+ represents SAH docked to the enzyme. The rate determining step (rds) in which the reaction passes from RC to PC is unimolecular in both cases, and is governed only by the concentration of RC. Hence, the rate determining steps can be compared directly.
The change in free energy in the enzymatic environment (Genz°) related to the methyl transfer as it proceeds along the reaction coordinate λ (eq 6) obtained from QM/MM MD simulations with the PM6-D3H4 Hamiltonian for Cl–, Br–, and NCS– is shown in Figure 2. The first minimum in the Genz curves marks the [X–···(E·bSAM+)] complex with the bSAM+ molecule being bound to the enzyme. The structure associated with this minimum in free energy can be compared directly with the reactive complex (RC) in the simple SN2 reactions with an electrostatic continuum model and the second minimum with the product complex (PC). By focusing solely on the reaction series RC → TS → PC, it is possible to compare the DFT results directly with those of the MD simulations.
Figure 2.
ΔRCGenz° curves obtained with the PM6-D3H4 Hamiltonian for Cl–, Br–, and NCS–.
The changes in Gibbs free energy Genz° are plotted relative to the energy of the first minimum Gmin, which has been identified as the reactive complex (Figure 2).
| 12 |
where Gmin° is the energetic reference, and the maximum in ΔRCGenz curve becomes equivalent to ΔTSGenv° (eq 4). ΔTSGenz is also the rate determining barrier for the methyl transfer reaction in the enzyme as shown in Figure 2, and the barrier of the second step in the Michaelis-Menten model of enzyme kinetics.
Table 4 summarizes the methyl transfer barriers ΔTSGenz° for all nucleophiles X– obtained with four semiempirical Hamiltonians. No barrier was observed for the methyl transfer with the AM1 Hamiltonian for I– and NCS–. However, experiments on the enzyme kinetics of AtHTMT1 (Table 3) indicate a barrier of 82.8 kJ mol–1 for NCS–. The barrier in the reaction of I– with the other enzymes varies between 81.1 and 88.1 kJ mol–1. Hence, the results obtained with the AM1 Hamiltonian are dismissed from further discussions. The experiments with AtHTMT1 indicate that I– reacts more rapidly than Br– and Cl–.14 This observation is not reproduced by the series of MD simulations using the PM7 Hamiltonian, and the PM7 calculations are dismissed from further discussions, too.
Table 4. Barrier Heights ΔTSGenz° for the Methyl Transfer from bSAM+ to X– and the Number of H2O Molecules nH2O Solvating X– in AtHTMT1 Obtained from a Single QM/MM MD Simulation with Four Different Semi-empirical Methods and the AMBER 14SB Force Fielda.
| PM7 |
PM6-D3 |
AM1 |
PM6-D3H4 |
|||||
|---|---|---|---|---|---|---|---|---|
| X– | ΔTSGenz° | nH2O | ΔTSGenz° | nH2O | ΔTSGenz° | nH2O | ΔTSGenz° | nH2O |
| Cl– | 75.7 | 7 | 89.5 | 7 | 19.0 | 7 | 71.1c | 7 |
| Br– | 70.4 | 7 | 83.0 | 8 | 5.9 | 6 | 60.9 | 7 |
| I– | 87.2 | 5 | 54.3 | 5 | —b | 5 | 42.4 | 5 |
| NCO– | 119.1 | 7 | 87.8 | 7 | 45.1 | 7 | 76.8 | 7 |
| OCN– | 86.1 | 5 | 69.6 | 5 | 72.4 | 5 | 69.5 | 5 |
| NCS– | 60.6 | 3 | 59.8 | 5 | —b | 3 | 51.7 | 5 |
| SCN– | 100.6 | 3 | 93.1 | 3 | 69.4 | 3 | 87.6 | 3 |
All energies are in kJ mol–1.
No apparent maximum in the free energy curve.
Average of six simulations, std. dev. 1.2 kJ mol–1.
The AM1 Hamiltonian has been used successfully in other MD simulations30−32 of the enzyme catalyzed methyl transfer, but semiempirical Hamiltonians cannot universally transfer due to their empirical nature. The data in Table 4 indicate the necessity to validate their application when changes in the chemical problem studied cannot be ignored.
The MD simulations with the PM6-D3 and PM6-D3H4 Hamiltonians yield the same order of barrier heights ΔTSGenz° of the experimental trio (Cl– > Br– > NCS–). The terminal reaction speed vmax observed in experiment14 and the barrier heights deduced from them are given in Table 3. The barriers obtained with the PM6-D3 Hamiltonian are generally closer to barrier heights calculated from the experimental data and hence are more credible. The disagreement between experiment and MD simulation increases with the reaction speed of the nucleophile. It is the largest (23.0 kJ mol–1) for NCS– and the smallest (0.5 kJ mol–1) for Cl–. The data in Table 3 show that NCS– is generally the more reactive nucleophile than I– with the exception of RsHTMT (daikon radish, Raphanus sativus), where the barrier for I– is similar to that for NCS– (ΔTSGI– ≳ ΔTSGNCS–°. Schmidberger et al. claimed I– to be more efficient than NCS–, but no vmax value for I– is given.14 In agreement with the experiment, the MD simulations with the two PM6 Hamiltonians yield larger barrier heights for NCS– then for I–.
The reverse methyl transfer reaction
| 13 |
has been studied in detail.5 The rate constant for the catalyzed step kcat was found to be 364.4 ± 12.7 min–1. The barrier of the inverse reaction ΔTSinvGenz is estimated to be 69.0 kJ mol–1 by transition state theory.65 ΔTSinvGenz can be calculated from MD results by taking the second minimum (products) as the energetic reference (Tables S6 to S9). The PM6-D3H4 and PM6-D3 Hamiltonians yield similar barrier heights of 72.9 and 75.3 kJ mol–1. Both values are in reasonably good agreement with the experimental values close to the threshold of chemical accuracy of 1 kcal mol–1. The PM6 Hamiltonians describe the reverse reaction from the uncharged products CH3X and SAH to the transition state well, but the calculated barriers for the forward reaction with NCS– and I– appear to be too low compared to the experiment (Table 3), which suggests that the ionic reactive complex RC with these nucleophiles is not well described by these semiempirical Hamiltonians. The simulations with the PM6 Hamiltonians describe the acceleration of the methyl group transfer to NCS– and I– in agreement with experiment, but are likely to overestimate the effect.
The MD simulations for Cl– with the PM6-D3H4 Hamiltonian were done six times. The six values for ΔTSGenz° are found in the range of 71.6 ± 1.8 kJ mol–1 with an average value of 71.1 kJ mol–1 and a standard deviation of 1.2 kJ mol–1. The barriers for the studied nucleophiles are separated on average by 7.5 kJ mol–1 so that the relative reactivity orders obtained from a single simulation can be trusted provided that the errors in the barriers for all nucleophiles are similar. The exceptions are Cl– and OCN–, the barriers of which are separated by merely 1.6 kJ mol–1, and can be regarded as approximately equal. The reactivity order of the seven nucleophiles in the AtHTMT1 catalyzed reaction based on ΔTSGenz by QM/MM MD simulations with the PM6 Hamiltonians are as follows
| 14 |
with I– being the most reactive and SCN– being the least. The positions of the halides Br– and Cl– changes with the Hamiltonians so that the pair OCN–/NCO– becomes more reactive with the PM6-D3 Hamiltonian. The rows in eq 14 become equal excluding the pair OCN–/NCO–. The high regio-selectivity of AtHTMT1 is clearly visible as NCS– and SCN– are located at opposite ends of the reactivity scale, and CH3SCN is the favored product.
The number of H2O molecules nH2O in the solvation shell of the nucleophile (Table 4) is defined as the number of H2O molecules within 500 pm from X– in the enzymatic environment. The nucleophile in the enzymatic environment is always surrounded by H2O molecules. The enzymatic environment does not desolvate the nucleophile completely, but the X– ions keep parts of their solvation shell from the aqueous environment. Hence, the nucleophiles can be regarded as partially solvated. The most H2O molecules can be found around the halides (Cl–, Br–) and the least around NCS– and SCN–. This fact is independent of the semiempirical Hamiltonians, and suggests that NCS– and SCN– ions fit the best into the reactive cavity. They are the ions with the smallest number of solvent molecules and hence the most activated nucleophiles, which partially explains why NCS– becomes the most reactive (highest value for vmax, Table 3) of the experimental trio.
Both Cl– and Br– have KM values significantly larger than 1 (Table 3), which indicates that the equilibrium for the dissociation of the enzyme–substrate complex lies on the side of the unbound enzyme following the definition of KM by Michaelis and Menten66 or equivalently that KM is dominated by the dissociation of the enzyme–substrate complex in the steady state interpretation by Briggs and Haldane.67 The dissociation can be seen in MD simulations for Cl– without constraints on the position of the nucleophile where the Cl– ion quickly escapes from the reactive pouch of AtHTMT1.
Schmidberger et al. reported a site-directed mutagenesis (Tyr172/Phe), which changes Tyrosine 172 to phenylalanine, yielded an enzyme showing a reduced “efficiency with chloride ions..., but otherwise a similar efficiency for bromide and thiocyanate”.14 The MD simulations provide a possible explanation for this observation. Figure 3 panels a and b display snapshots from the trajectories for the methyl transfer with Cl– and NCS– in the reactive complex, which is the first minimum in Figure 2. The orientation of the tyrosine OH group changes over time. When it points into the general direction of the SAM methyl group, it can form a chain of hydrogen bonds to the attacking Cl– ion (Figure 3a). These transitory hydrogen-bond chains can stabilize the position of the wandering Cl– ion in the SN2 transition state and then facilitate the reaction. The directing property of Tyr172 is visible in the decrease of vmax from 2.43 nmol min–1 (mg protein)−1 for the native enzyme to 0.91 for the mutant.14 In the case of the NCS– ion (Figure 3b), this chain points to the S atom of the NCS– ion. Sulfur atoms are generally weak hydrogen-bond acceptors so that a possible hydrogen-bond chain is weak and has only a small influence on the transition state. A chain to the N atom of the NCS– ion perpendicular to the S···HO axis would be longer and less effective. Therefore, the changes in vmax caused by the mutation are within the margin of error of the reported experiment.14
Figure 3.
Snapshots for the position of the nucleophile X– relative to the bSAM+ molecule and its coordination. (a) QM/MM MD simulation reactive complex with Cl–. (b) QM/MM MD simulation reactive complex with NCS–. (c) SMD transition state calculation for ether as solvent. Atom colors: see Figure 1. Cartoons a,b: gray ribbons, enzyme backbone; blue dotted line, reaction path element d2; magenta dotted line, hydrogen bonds.
In summary, ΔTSGenz° obtained from MD simulation correctly describes the relative reactivities of the nucleophiles observed in experiment by the value of vmax, the regio-selectivity of AtHTMT1, and the simulations provide clues for the understanding of the value of KM. The simulations correctly describe the essentials of methyl transfer in thale cress.
3.3. The Role of the Environment in the Catalytic Process
The combination of the results from the standard quantum calculations and the MD simulations provides insight into the mechanism of the enzymatic reaction with AtHTMT1. The number nH2O of H2O molecules in the vicinity of the nucleophile X– differs greatly in the two approaches. It is very small and fixed in the standard quantum calculation, while it is large and fluctuating in the MD simulations. Hence, the observed solvation shell of X– in the MD simulations is a simulation result in contrast to that in the standard quantum calculations, where it is a rigid, user-defined part of the calculation. The comparison of both can be used to determine the role of solute–solvent interactions in the enzymatic environment.
The MD barriers (Table 4) calculated with the PM6 Hamiltonians are smaller than their SMD counterparts (Table 2) both in water and ether with or without explicit H2O molecules with the exception of the PM6-D3 result for Br– and Br– in ether. The lower MD barriers provide strong evidence for the catalytic efficiency of AtHTMT1. The PM6-D3 barriers are higher, though closer to their experimental counterparts, than those obtained with the PM6-D3H4 Hamiltonian, which suggests a smaller acceleration of the SN2 reaction by the PM6-D3 Hamiltonian.
The enzymatic gain ΔgainG° is defined as the difference in barrier height between the PM6-D3 MD simulations and the M05 quantum calculations. The NCS– ion has the second largest ΔgainG° value being −35.8 kJ mol–1 (PM6-D3H4, −53.7 kJ mol–1). This gain is accompanied by a boost in regio-selectivity |ΔregioGwat°| = |ΔTSGwat°–ΔTSGwat°| = 7.3 kJ mol–1 in water to 33.3 kJ mol–1 (PM6-D3H4, 36.0 kJ mol–1) in the enzyme. The NCO– ion has the largest activity boost with ΔgainG° being −50.3 kJ mol–1 (PM6-D3H4: −61.3 kJ mol–1), but with a severely reduced enzymatic regio-selectivity |ΔregioGenz| = |ΔTSOCN–Genz – ΔTSNCO–Genz| = 18.2 kJ mol–1 (PM6-D3H4, 7.3 kJ mol–1). The DFT calculations with the SMD solvation model for water without explicit H2O molecules are less regioselective than the MD simulations with the PM6-D3 Hamiltonian.
The enzyme’s preference for the pseudohalides attacking the methyl group with their softer atoms is observed in the change of relative nucleophilic activities. The SMD water calculations with no explicit H2O molecules (reactivity order in eq 7) for the cytosol indicate that NCS– would be the least reactive nucleophile of the experimental trio (Cl–, Br–, NCS–), while the MD simulations for the same reaction catalyzed by AtHTMT1 (reactivity order in eq 14) suggest that NCS– is the most reactive. This change in reactivity order cannot be attributed to a simple desolvation effect because the same order can be observed with ether as the SMD solvent (eq 9). The addition of an explicit H2O molecule to the nucleophile does not change the reactivity order either; therefore, the change in the reactivity order has to be credited to the enzymatic environment.
Figure 3c shows the transition state of the methyl transfer to NCS– with an explicit H2O molecule attached to the S atom in ether as the SMD solvent. The orthogonality of the C–S and S–H bonds caused by lack of overlap between the 3s and 3p orbitals is well preserved in the enzymatic environment (Figure 3b). The orthogonality of the C–S and S–H bonds directs the N atom of NCS– into the void in the lower half of the figure, where it can interact with the solvent H2O molecules in the reactive cavity. The data of Δ+1Geth° for the energetic costs of adding a H2O molecule to the nucleophile in Table 2 show that moving a H2O molecule from the S to the N atom in NCS– lowers the barrier of the methyl transfer by 10.7 kJ mol–1. Therefore, AtHTMT1 not only catalyzes the methyl transfer by desolvating the nucleophile to support the underlying SN2 reaction, but also facilitates the reaction by placing H2O molecules near the N atom of the NCS– ion. Such an advantageous arrangement of H2O molecules follows directly from the ability of the atoms in the nucleophile to accept hydrogen bonds. SMD calculations with ether as solvent show that the formation of the S···H hydrogen bond (244 pm) in NCS–···H2O is endergonic (ΔG° = +4.9 kJ mol–1) and the correction for particle densities (eq 3) makes it slightly exergonic in solution (−3.0 kJ mol–1). The corresponding N···H hydrogen bond in OH2···NCS– (189 pm) is slightly exergonic in the gas phase (−1.4 kJ mol–1) and strongly in solution (−9.3 kJ mol–1). Hence, not much chemical work is required to desolvate the attacking S atom of NCS– in the enzymatic environment while keeping the N atom solvated to accelerate the reaction. The NCS– ion holds parts of his solvation shell in the catalytic process.
The ability of AtHTMT1 to selectively desolvate the attacking atom in the nucleophile is not very strong for Cl–. The longer Cl···H bond (221 pm) is stronger in solution (−11.5 kJ mol–1), and the Cl– ion keeps parts of its solvation shell on entering the enzyme (Table 4). The H2O molecules in the vicinity of Cl– slow the methyl transfer as the quantum calculations indicate (Δ+1G°, Table 2).
The data for Cl– and NCS– reveal the remarkable fine-tuning of the desolvation of the nucleophile in AtHTMT1. Nucleophile desolvation generally accelerates the SN2 reaction of the methyl transfer. The partial preservation of the nucleophiles solvation shell boosts the reaction of NCS–, meanwhile it reduces the catalytic effect for the reaction with Cl–. It propels NCS– from the least to the most reactive nucleophile of the experimental trio and hence limits the loss of SAM+ to the accidental production of CH3Cl.
4. Conclusions
A set of quantum SMD calculations and QM/MM MD simulations was done to understand the promiscuity of AtHTMT1 and the production of methyl chloride by thale cress and other plants. Published structural data were used as starting point for the computational analysis, and the companion kinetic data14 for Cl–, Br–, and NCS– (experimental trio) to validate the results. The combination of the results from both computational methods shows that neither technique alone is sufficient to explain the mechanism of the observed promiscuity.
The uncatalyzed reaction in the cytosol was studied with M05/6-311+G(2d,p) DFT calculations and water as SMD solvent, while the QM/MM (PM6-D3H4, AMBER 14SB, TIP3P) MD simulations were used to study the enzyme catalyzed reaction. The direct comparison of the absolute barrier heights obtained from the DFT calculations with those from the MD simulations indicate that NCS– has the largest gain in reaction speed for a nucleophile of the experimental trio, which supports the hypothesis of AtHTMT1 being part of the plantal defense against bacterial infections.18 The preference for NCS– is apparent in the order of reaction speeds for the experimental trio. NCS– is the slowest reacting nucleophile in water (Table 2) in contrast to the simulations for the enzyme catalyzed one, which predicts NCS– to be the fastest of the trio (Table 4) in agreement with experiments (Table 3).
Reactant desolvation has been suggested to be the acting principle in enzyme catalysis.33 Water-free calculations with ether as SMD solvent support this idea as barrier heights generally decrease in ether solution, but the relative barrier heights for the experimental trio and the order of reaction rates do not change. Only the MD simulations with AtHTMT1 reproduce the reactivity order correctly. Furthermore, the calculated absolute barrier heights for the rate determining step in the enzymatic reaction are found to be in much better agreement with those obtained from the experimental data (Table 3). The average difference in barrier height between the MD simulations with the PM6-D3 Hamiltonian and the experimental value is 8.9 kJ mol–1 (PM6-D3H4, 25.1 kJ mol–1) with a standard deviation of 12.3 kJ mol–1 (PM6-D3H4, 6.1 kJ mol–1) for the experimental trio.
The MD simulations provide the atomistic details necessary to understand the methyl transfer in vivo. The NCS– ion fits best into the reactive site of enzyme since it is surrounded by the least H2O molecules. The NCS– and SCN– ions are the most desolvated and most activated ions. The regio-selectivity appears to be controlled by the abilities of the S and N atoms to be engaged in SN2 reactions.
The H2O molecules hover around the N atom in NCS– in the enzymatic process, which reflects the ability of an individual atom in NCS– to engage in hydrogen bonding. The concept of partial desolvation not only focuses on the number of H2O molecules close to the nucleophile, but also comprises the hydrogen bonding partners of the H2O molecules. No H2O molecules can be found close to the S atom of NCS–, but the H2O molecules of the partial solvation shell of NCS– can be found close to the N atom. Such a configuration of H2O molecules improves the reactivity of the nucleophile as shown by the quantum calculations. The partial solvation shell of the nucleophile in the reactive pouch of the enzyme favors the reactivity of NCS– and consequently the formation of CH3SCN. Direct interactions between AtHTMT1 and reactants controlling the methyl transfer are not observed in the MD simulation.
The Cl– and Br– ions amass the most H2O molecules around them. The enzyme cannot effectively desolvate the Cl– ion, because the Cl···H hydrogen bond is much stronger than its S···H counterpart. The partial preservation of the solvation shell of the Cl– ion effectively deactivates the nucleophile. It seems that the fine-tuning of the solvation shell around the nucleophile by the enzyme disfavors the small halides.
The quantum chemical analysis35 of the methyl transfer between a trimethyl sulfonium ion [(CH3)3S+] as stand-in for nSAM+ and Cl– shows that the formation of intermediate [(CH3)3S+·Cl–] ion pairs depends strongly on the permeability of the solvent. Therefore, the partial solvation of the nucleophile in the enzymatic environment also prevents Cl– from binding to the reactive site as observed in MD simulations without spatial constraints. The Cl– ion prefers to move out of the reactive cavity. The tendency of Cl– to leave the reactive pouch is also observed in the experimental KM value being much larger than 1, while it can still engage in the methyl transfer reaction. Hence, the formation of CH3Cl by AtHTMT1 may be regarded as an unfortunate byproduct of the general acceleration of SN2 reactions in the enzymatic environment.
The model of a partial solvation shell for the nucleophile in AtHTMT1 can be verified in experiments with cyanate ions. The model concludes that the softer N atom attacks the methyl group while the H2O molecules of its partial solvation shell gather around the O atom. The simulations with the PM6-D3 Hamiltonian yield barrier heights in better agreement with those calculated from experimental data than the simulations with the PM6-D3H4 Hamiltonian. The calculations with the PM6-D3 Hamiltonian show that CH3NCO will be formed in such an experiment with a vmax value larger than that for the formation of CH3Br (eq 14) while the PM6-D3H4 Hamiltonian indicates a lower value. In contrast, the DFT calculations with a H2O molecule attached to the nucleophile indicate that the reaction between nSAM+ and the halides should be faster than that of OCN– in the uncatalyzed reaction with water as solvent. The proposed model of partial solvation to explain the basic selectivity of AtHTMT1 can assist the rational design of new, more complex biotechnological techniques.
Acknowledgments
This work has been supported by the Ministry of Science and Technology, Taiwan (Grant Nos. MOST 107-2113-M-007-012-MY3 and MOST 110-2113-M-007-017).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c07327.
Additional tables; evaluation of published Michaelis–Menten data; geometries, energies, frequencies from QM calculations; geometry data for the QM region in MD simulations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Manley S. L. Phytogenesis of halomethanes: A product of selection or a metabolic accident?. Biogeochemistry 2002, 60, 163–180. 10.1023/A:1019859922489. [DOI] [Google Scholar]
- Cantoni G. L. S-Adenosylmethionine; A New Intermediate Formed Enzymatically from L-Methionine and Adenosinetriphosphate. J. Biol. Chem. 1953, 204, 403–416. 10.1016/S0021-9258(18)66148-4. [DOI] [PubMed] [Google Scholar]
- Roje S. S-Adenosyl-l-methionine: Beyond the universal methyl group donor. Phytochemistry (Elsevier) 2006, 67, 1686–1698. 10.1016/j.phytochem.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Struck A.-W.; Thompson M. L.; Wong L. S.; Micklefield J. S -Adenosyl-Methionine-Dependent Methyltransferases: Highly Versatile Enzymes in Biocatalysis, Biosynthesis and Other Biotechnological Applications. ChemBioChem. 2012, 13, 2642–2655. 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- Tang Q.; Grathwol C. W.; Aslan-Üzel A. S.; Wu S.; Link A.; Pavlidis I. V.; Badenhorst C. P. S.; Bornscheuer U. T. Directed Evolution of a Halide Methyltransferase Enables Biocatalytic Synthesis of Diverse SAM Analogs. Angew. Chem., Int. Ed. 2021, 60, 1524–1527. 10.1002/anie.202013871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.; Pavlidis I. V.; Badenhorst C. P. S.; Bornscheuer U. T. From Natural Methylation to Versatile Alkylations Using Halide Methyltransferases. ChemBioChem. 2021, 22, 2584–2590. 10.1002/cbic.202100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold E.; Dalhoff C.; Klimasauskas S.; Lukinavicius G.. New S-Adenosyl-L-Methionine Analogs with Extended Activated Groups for Transfer by Methyltransferases. European Patent, EP 1 874 790 B1, 18.08.2010.
- Bayer T. S.; Widmaier D. M.; Temme K.; Mirsky E. A.; Santi D. V.; Voigt C. A. Synthesis of Methyl Halides from Biomass Using Engineered Microbes. J. Am. Chem. Soc. 2009, 131, 6508–6515. 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- Nohende Ajavon A.-L.; Newman P. A.; Pyle J. A.; Ravishankara A.. Scientific Assessment of Ozone Depletion: 2014; World Meteorological Organization: Geneva, Switzerland, 2014.
- Montzka S. A.; Reimann S.; Engel A.; Krüger K.; O’Doherty S.; Sturges W. T.; Blake D.; Dorf M.; Fraser P.; Froidevaux L.; Jucks K.; Kreher K.; Kurylo M. J.; Mellouki A.; Miller J.; Nielsen O.-J.; Orkin V. L.; Prinn R. G.; Rhew R.; Santee M. L.; Stohl A.; Verdonik D.. Chapter 1: Ozone-Depleting Substances (ODSs) and Related Chemicals. In Scientific Assessment of Ozone Depletion: 2010, Global Ozone Research and Monitoring Project-Report No.52; Nohende Ajavon A.-L., Newman P. A., Pyle J. A., Ravishankara A. R., Eds.; World Meteorological Organization: Geneva, Switzerland, 2011.
- Clerbaux C.; Cunnold D. M.; Anderson J.; Engel A.; Fraser P. J.; Mahieu E.; Manning A.; Miller J.; Montzka S. A.; Nassar R.; Prinn R.; Reimann S.; Rinsland C. P.; Simmonds P.; Verdonik D.; Weiss R.; Wuebbles D.; Yokouchi Y.. Chapter 1: Long-Lived Compounds. In Scientific Assessment of Ozone Depletion 2006, Global Ozone Research and Monitoring Project-Report No. 50; Nohende Ajavon A.-L., Albritton D. L., Watson R. T., Eds.; World Meteorological Organization: Geneva, Switzerland, 2007.
- Ohsawa N.; Tsujita M.; Morikawa S.; Itoh N. Purification and Characterization of a Monohalomethane-producing Enzyme S-adenosyl-L-methionine: Halide Ion Methyltransferase from a Marine Microalga, Pavlova pinguis. Biosci., Biotechnol., Biochem. 2001, 65, 2397–2404. 10.1271/bbb.65.2397. [DOI] [PubMed] [Google Scholar]
- Itoh N.; Tsujita M.; Ando T.; Hisatomi G.; Higashi T. Formation and Emission of Monohalomethanes from Marine Algae. Phytochemistry (Elsevier) 1997, 45, 67–73. 10.1016/S0031-9422(96)00786-8. [DOI] [Google Scholar]
- Schmidberger J.; James A.; Edwards R.; Naismith J.; O’Hagan D. Halomethane Biosynthesis: Structure of a SAM-Dependent Halide Methyltransferase from Arabidopsis thaliana. Angew. Chem., Int. Ed. 2010, 49, 3646–3648. 10.1002/anie.201000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda H.; Itoh N. Isolation and characterization of a gene encoding a S-adenosyl-L-methionine-dependent halide/thiol methyltransferase (HTMT) from the marine diatom Phaeodactylum tricornutum: Biogenic mechanism of CH3I emissions in oceans. Phytochemistry (Elsvier) 2011, 72, 337–343. 10.1016/j.phytochem.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Itoh N.; Toda H.; Matsuda M.; Negishi T.; Taniguchi T.; Ohsawa N. Involvement of S-Adenosylmethionine-dependent Halide/Thiol Methyltransferase (HTMT) in Methyl Halide Emissions from Agricultural Plants: Isolation and Characterization of an HTMT-Coding Gene from Raphanus sativus (Daikon Radish). BMC Plant Biol. 2009, 9, 116–116. 10.1186/1471-2229-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa Y.; Nakamura T. Rice OsHOL1 and OsHOL2 proteins have S-adenosyl-L-methionine-dependent methyltransferase activities toward iodide ions. Plant Biotechnol. (Tokyo, Jpn.) 2012, 29, 103–108. 10.5511/plantbiotechnology.12.0207a. [DOI] [Google Scholar]
- Nagatoshi Y.; Nakamura T. Arabidopsis HARMLESS TO OZONE LAYER Protein Methylates a Glucosinolate Breakdown Product and Functions in Resistance to Pseudomonas Syringae PV. Maculicola. J. Biol. Chem. 2009, 284, 19301–19309. 10.1074/jbc.M109.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham G. D.S-Adenosylmethionine. https://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0000662.pub2 (accessed 2021-10-29).
- Rhew R. C.; Østergaard L.; Saltzman E. S.; Yanofsky M. F. Genetic Control of Methyl Halide Production in Arabidopsis. Curr. Biol. 2003, 13, 1809–1813. 10.1016/j.cub.2003.09.055. [DOI] [PubMed] [Google Scholar]
- Schmidt T.; Schwede T.; Meuwly M. Computational Analysis of Methyl Transfer Reactions in Dengue Virus Methyltransferase. J. Phys. Chem. B 2014, 118, 5882–5890. 10.1021/jp5028564. [DOI] [PubMed] [Google Scholar]
- Lankau T.; Kuo T. N.; Yu C. H. Computational Study of the Degradation of S-Adenosyl Methionine in Water. J. Phys. Chem. A 2017, 121, 505–514. 10.1021/acs.jpca.6b09639. [DOI] [PubMed] [Google Scholar]
- Yue Y.; Chu Y.; Guo H. Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations. Molecules 2015, 20, 10032–10046. 10.3390/molecules200610032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P. R. M.; Araújo J. d. O.; Silva J. R. A.; Alves C. N.; Lameira J.; Lima A. H. Exploring Chloride Selectivity and Halogenase Regioselectivity of the SalL Enzyme through Quantum Mechanical/Molecular Mechanical Modeling. J. Chem. Inf. Model. 2020, 60, 738–746. 10.1021/acs.jcim.9b01079. [DOI] [PubMed] [Google Scholar]
- Qian P.; Guo H.-B.; Yue Y.; Wang L.; Yang X.; Guo H. Understanding the Catalytic Mechanism of Xanthosine Methyltransferase in Caffeine Biosynthesis from QM/MM Molecular Dynamics and Free Energy Simulations. J. Chem. Inf. Model. 2016, 56, 1755–1761. 10.1021/acs.jcim.6b00153. [DOI] [PubMed] [Google Scholar]
- Araújo E.; Lima A. H.; Lameira J. Catalysis by solvation rather than the desolvation effect: exploring the catalytic efficiency of SAM-dependent chlorinase. Phys. Chem. Chem. Phys. 2017, 19, 21350–21356. 10.1039/C7CP02811C. [DOI] [PubMed] [Google Scholar]
- Lima A. H.; Alves C. N.; Prasad R.; Lameira J. Exploring the origin of the catalytic power and product specificity of SET domain protein methyltransferase. Mol. BioSyst. 2016, 12, 2980–2983. 10.1039/C6MB00414H. [DOI] [PubMed] [Google Scholar]
- Zhao C.; Li Y.; Wang C.; Chen H. Mechanistic Dichotomy in the Activation of SAM by Radical SAM Enzymes: QM/MM Modeling Deciphers the Determinant. ACS Catal. 2020, 10, 13245–13250. 10.1021/acscatal.0c03384. [DOI] [Google Scholar]
- Jindal G.; Warshel A. Exploring the Dependence of QM/MM Calculations of Enzyme Catalysis on the Size of the QM Region. J. Phys. Chem. B 2016, 120, 9913–9921. 10.1021/acs.jpcb.6b07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M.; Martí S.; Andrés J.; Moliner V.; Tuñón I.; Bertrán J.; Williams I. H. Theoretical Modeling of Enzyme Catalytic Power: Analysis of ”Cratic” and Electrostatic Factors in Catechol O -Methyltransferase. J. Am. Chem. Soc. 2003, 125, 7726–7737. 10.1021/ja0299497. [DOI] [PubMed] [Google Scholar]
- Ruggiero G. D.; Williams I. H.; Roca M.; Moliner V.; Tuñón I. QM/MM Determination of Kinetic Isotope Effects for COMT-Catalyzed Methyl Transfer Does Not Support Compression Hypothesis. J. Am. Chem. Soc. 2004, 126, 8634–8635. 10.1021/ja048055e. [DOI] [PubMed] [Google Scholar]
- Martí S.; Roca M.; Andrés J.; Moliner V.; Silla E.; Tuñón I.; Bertrán J. Theoretical insights in enzyme catalysis. Chem. Soc. Rev. 2004, 33, 98–107. 10.1039/B301875J. [DOI] [PubMed] [Google Scholar]
- Liscombe D. K.; Louie G. V.; Noel J. P. Architectures, Mechanisms and Molecular Evolution of Natural Product Methyltransferases. Nat. Prod. Rep. 2012, 29, 1238–1250. 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- Schutz C. N.; Warshel A. What are the dielectric ”constants” of proteins and how to validate electrostatic models?. Proteins: Struct., Funct., Bioinf. 2001, 44, 400–417. 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- Lankau T.; Yu C.-H. Solvent Effects on the Intramolecular Conversion of Trimethylsulfonium Chloride to Dimethyl Sulfide and Methyl Chloride. Phys. Chem. Chem. Phys. 2014, 16, 26658–26671. 10.1039/C4CP03965C. [DOI] [PubMed] [Google Scholar]
- Marenich A. V.; Cramer C. J.; Truhlar D. G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. 10.1021/jp810292n. [DOI] [PubMed] [Google Scholar]
- Farooqui J.; Kim S.; Paik W. K. Measurement of Isoelectric Point of S-Adenosyl-L-methionine and its Metabolic Products by an Isoelectric Focusing Technique. Electrophoresis 1983, 4, 261–265. 10.1002/elps.1150040402. [DOI] [Google Scholar]
- Zhao Y.; Schultz N. E.; Truhlar D. G. Exchange-Correlation Functional with Broad Accuracy for Metallic and Nonmetallic Compounds, Kinetics, and Noncovalent Interactions. J. Chem. Phys. 2005, 123, 161103. 10.1063/1.2126975. [DOI] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam J. M.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C.; Ochterski J. W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas O.; Foresman J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 09, rev. B.01; Gaussian Inc.: Wallingford, CT, 2009.
- Truhlar D. G.; Cramer C. J.; Lewis A.; Bumpus J. A. Molecular modeling of environmentally important processes: Reduction potentials. J. Chem. Educ. 2004, 81, 596–604. 10.1021/ed081p596. [DOI] [Google Scholar]
- Tomasi J.; Mennucci B.; Cammi R. Quantum Mechanical Continuum Solvation Models. Chem. Rev. 2005, 105, 2999–3094. 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- Scalmani G.; Frisch M. J. Continuous Surface Charge Polarizable Continuum Models of Solvation. I. General Formalism. J. Chem. Phys. 2010, 132, 114110. 10.1063/1.3359469. [DOI] [PubMed] [Google Scholar]
- Gleave J. L.; Hughes E. D.; Ingold C. K. 54. Mechanism of substitution at a saturated carbon atom. Part III. Kinetics of the degradations of sulphonium compounds. J. Chem. Soc. (Resumed) 1935, 236. 10.1039/jr9350000236. [DOI] [Google Scholar]
- Pocker Y.; Parker A. J. Kinetics and Mechanism of Decomposition of the Trimethylsulfonium Cation. J. Org. Chem. 1966, 31, 1526–1531. 10.1021/jo01343a051. [DOI] [Google Scholar]
- Pearson R. G.Chemical Hardness, 1st ed.; VCH Verlagsgesellschaft: Weinheim, Germany, 1977. [Google Scholar]
- Sykes P.Reaktionsmechanismen der Organischen Chemie, 9th ed.; VCH Verlagsgesellschaft: Weinheim, Germany, 1988. [Google Scholar]
- Bernstein F. C.; Koetzle T. F.; Williams G. J. B.; Meyer E. F. Jr; Brice M. D.; Rodgers J. R.; Kennard O.; Shimanouchi T.; Tasumi M. The Protein Data Bank. Eur. J. Biochem. 1977, 80, 319–324. 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle Mesh Ewald: An N · log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Stewart J. J. P. Optimization of Parameters for Semiempirical Methods VI: More modifications to the NDDO Approximations and Re-Optimization of Parameters. J. Mol. Model. 2013, 19, 1–32. 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. J. P. Optimization of Parameters for Semiempirical Methods V: Modification of NDDO Approximations and Application to 70 Elements. J. Mol. Model. 2007, 13, 1173–1213. 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme S.; Antony J.; Ehrlich S.; Krieg H. A Consistent and Accurate ab initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys. 2010, 132, 154104. 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- Dewar M. J. S.; Zoebisch E. G.; Healy E. F.; Stewart J. J. P. Development and Use of Quantum Mechanical Molecular Models. 76. AM1: A New General Purpose Quantum Mechanical Molecular Model. J. Am. Chem. Soc. 1985, 107, 3902–3909. 10.1021/ja00299a024. [DOI] [Google Scholar]
- Řezáč J.; Hobza P. Advanced Corrections of Hydrogen Bonding and Dispersion for Semiempirical Quantum Mechanical Methods. J. Chem. Theory Comput. 2012, 8, 141–151. 10.1021/ct200751e. [DOI] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Keith T. A.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16, rev. D.01; Gaussian Inc.: Wallingford, CT, 2016.
- Stewart J. J. P.MOPAC2016; Stewart Computational Chemistry, 2016.
- Tribello G. A.; Bonomi M.; Branduardi D.; Camilloni C.; Bussi G. PLUMED 2: New Feathers for an Old Bird. Comput. Phys. Commun. 2014, 185, 604–613. 10.1016/j.cpc.2013.09.018. [DOI] [Google Scholar]
- Torrie G.; Valleau J. Nonphysical Sampling Distributions in Monte Carlo Free-Energy Estimation: Umbrella Sampling. J. Comput. Phys. 1977, 23, 187–199. 10.1016/0021-9991(77)90121-8. [DOI] [Google Scholar]
- Kumar S.; Bouzida D.; Swendsen R. H.; Kollman P. A.; Rosenberg J. M. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I: The Method. J. Comput. Chem. 1992, 13, 1011–1021. 10.1002/jcc.540130812. [DOI] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical Sampling Through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Lienhard G. E. Enzymatic Catalysis and Transition-State Theory. Science 1973, 180, 149–154. 10.1126/science.180.4082.149. [DOI] [PubMed] [Google Scholar]
- Johnson K. A.; Goody R. S. The Original Michaelis Constant: Translation of the 1913 Michaelis-Menten Paper. Biochemistry 2011, 50, 8264–8269. 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs G. E.; Haldane J. B. S. Note on the Kinetics of Enzyme Action. Biochem. J. 1925, 19, 338–339. 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





