Abstract
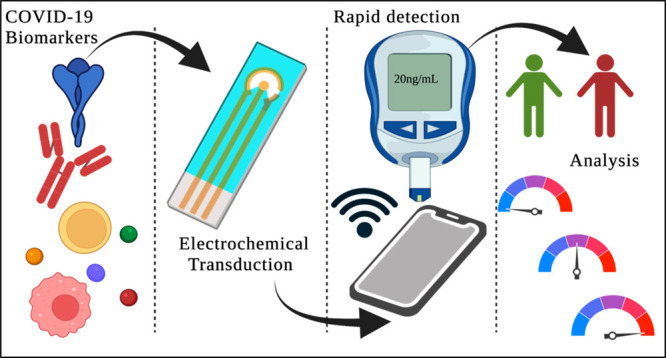
Rapid diagnosis is a critical aspect associated with controlling the spread of COVID-19. Electrochemical sensor platforms are ideally suited for rapid and highly sensitive detection of biomolecules. This review focuses on state-of-the-art of COVID-19 biomarker detection by utilizing electrochemical biosensing platforms. Point-of-care (POC) sensing is one of the most promising and emerging fields in detecting and quantifying health biomarkers. Electrochemical biosensors play a major role in the development of point-of-care devices because of their high sensitivity, specificity, and ability for rapid analysis. Integration of electrochemistry with point-of-care technologies in the context of COVID-19 diagnosis and screening has facilitated in convenient operation, miniaturization, and portability. Identification of potential biomarkers in disease diagnosis is crucial for patient monitoring concerning severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In this review, we will discuss the choice of biomarkers in addition to the various types of electrochemical sensors that have been developed to meet the needs of rapid detection and disease severity analysis.
1. Introduction
In the last two decades, infectious diseases caused by viruses have deteriorated human well-being. The current contemporary outbreak of coronavirus disease is caused by newly discovered infectious severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA virus and affects human health primarily through respiratory illness.1 An estimated 438 million cases with 5.9 million deaths were reported as of March 3, 2022.2 The catastrophic effect of the COVID-19 pandemic on the world economy and well-being is currently ongoing. It is imperative to use rapid diagnostic platforms with high sensitivity and selectivity toward COVID-19 to fight the battle effectively. Currently, diagnostic tests are based on nucleic acid amplification and immunological assays. The methods include real-time polymerase chain reaction (PCR), reverse transcription real-time PCR (rRT-PCR), real-time loop-mediated isothermal amplification (RT-LAMP), and reverse transcription–polymerase chain reaction (RTPCR). RTPCR is considered a gold standard method for detecting the presence of viruses. These methods are time-consuming, are expensive, and may result in false-negative results. It is preferable to conduct further serological analysis for false-negative patients, as this can help minimize disease spread and contact tracing. The emergence of SARS-CoV-2 has ushered the world to look for more robust and reliable detection strategies. Due to its rapid transmission and increasing mortality with infection severity, early detection of COVID-19 is of the utmost importance. Rapid detection within minutes or seconds, ideally within a few days, of infection is required. At-home detection methods with smartphone-based readouts are needed for early disease identification.3
A massive challenge for the current clinical scenario is the early onset of disease in a large population at an affordable cost. Finding alternatives to conventional laboratory diagnostic methods may be the most suitable way to address pandemic situations. Low sample volume requirement with no prior treatment of the samples is another big challenge. Point-of-care (POC) devices are such tools, which provide robust and sensitive results towards disease identification based on specific target biomarkers. There is a growing urgency to establish point-of-care devices for accurate and rapid diagnosis of disease onset as well as progression. Development of point-of-care devices needs to meet “ASSURED” criteria as per the recommendations from WHO (Figure 1). Biomarkers in the context of COVID-19 are either the spike protein, the RNA specific to the virus, or metabolites expressed by humans due to COVID-19 infections. In the scenario of COVID-19 testing, it is essential to focus on biomarker detection due to the rapid spread of COVID-19. In this context, biosensors are versatile tools for detecting early onset disease identification and progression with high sensitivity and selectivity. Biosensors are specific to the target analyte with low detection limits and rapid response within a few minutes.4
Figure 1.
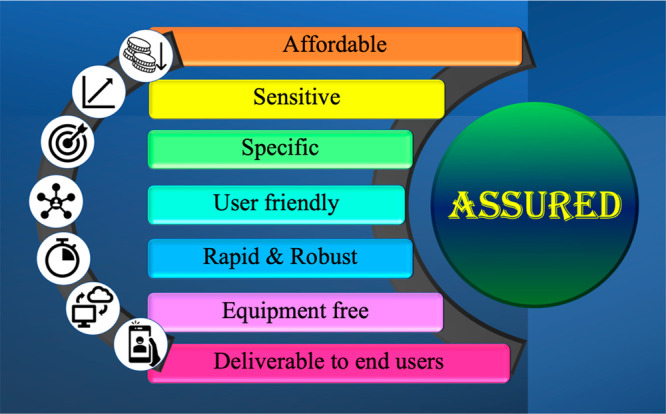
“ASSURED” guidelines indicating the features of point-of-care devices as suggested by the World Health Organization.
Electrochemical biosensors play a crucial role in the development of rapid and point-of-care diagnostic platforms, especially for the detection of biomarkers in the field of clinical diagnosis. POC devices utilizing electrochemical transduction mechanisms are well suited for biomarker analysis. These devices have a high affinity for biomarkers with enhanced sensitivity. Moreover, these methods reduce the cost of tests when compared to conventional laboratory diagnostic methods. Rapid and accurate analysis helps to increase the patient survival rate.
In this review article, we have chronicled the state of the art of electrochemical biosensors developed for the early diagnosis of COVID-19 and its related biomarkers. Moreover, this review critically discusses the role of each biomarker in disease onset and progression and electrochemical methods reported for the detection of these biomarkers.
2. Biomarkers of COVID-19 and Their Detection Methods with a Special Focus on Electrochemical Detection
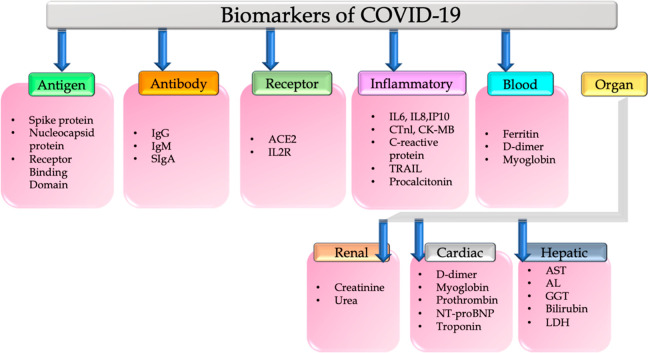
It is ideal to identify specific biomarkers that can indicate disease pathogenesis to effectively manage COVID-19. As shown in Figure 2, in this review we will discuss electrochemical biosensors detecting and quantifying antigens and antibodies associated with COVID-19 and the effect of the virus on inflammatory markers, cardiac biomarkers, hepatic biomarkers, renal biomarkers, hematological biomarkers, and biochemical biomarkers that are altered during disease progression. Additionally, the levels of biomarkers may be affected by the severity of the disease, which can be determined from other clinical perspectives, such as radiology studies, to determine the viral load, distribution, and severity.
Figure 2.
Classification of biomarkers of COVID-19.
2.1. Antigen- and Antibody-Based Biomarkers
The four major structural proteins of the coronavirus are the membrane (M), envelope (E), spike (S), and nucleocapsid (N). In addition to being a structural protein, the S protein is also a key component of fusion, entry, and antibody production within host cells. There is an N-terminal domain, an N-terminal S1 receptor-binding domain (RBD), and a C-terminal S2 subunit. SARS-CoV-2 invades the cell by interacting with the angiotensin-converting enzyme-2 (ACE2) protein. N-protein takes part in the viral replication process. The host system generates antibodies to fight it in response to the virus. Antibodies play a vital role in neutralizing the virus. The receptor-binding domain (RBD) of spike protein and nucleocapsid protein are the stereotypical antigens and are considered biomarkers of COVID-19 identification.5Figure 3 describes the structure of SARS-CoV-2 and its interaction with ACE2 followed by the host cell response. ACE2 is highly expressed in the endothelium of blood vessels. So, the higher expression of ACE2 by the endothelium leads to SARS-CoV-2 infection, which may lead to vascular dysfunction and then to a cytokine storm. Serological examination is an alternative method of COVID-19 identification by measuring antibodies. Through serological testing methods, identification of immunoglobulins generated in response to the antigens can be measured. Generation of these immunoglobulins such as IgG and IgM takes a few days to weeks after the person is infected with the virus. IgA is another predominant class of immunoglobulin in the respiratory tract with two subtypes IgA1 and IgA2, active in the airways and colon, respectively. It is produced by B-lymphocytes and expressed after 2 weeks of infection.6 Development of electrochemical methods for the selective measurement of these antigens and antibodies is the call for the scientific community. CR3022 is another interesting SARS-CoV-2 antibody against the RBD domain of spike protein. While published reports are available (as shown in Table 1) for the rapid and sensitive detection of spike protein, nucleocapsid protein, RBD domain, anti-spike protein antibody, antinucleocapsid protein antibody, anti-RBD protein antibody, IgG, IgM, and IgA using electrochemical methods, commercial availability of these sensors as point-of-care devices is still awaiting.
Figure 3.
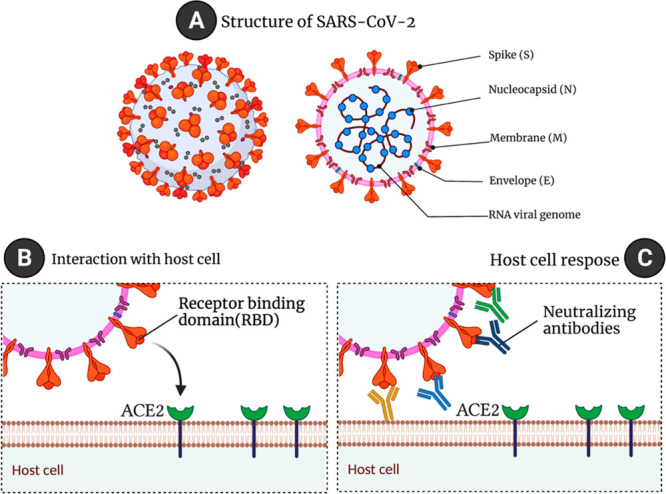
(A) An illustration of the structure of SARS-CoV-2 (left) and its cross-section (right). An illustration of (B) the interaction of the spike protein receptor-binding domain with ACE2 and of (C) neutralizing antibodies developed in response to being exposed to the virus (created using Biorender).
Table 1. Electrochemical Biosensors Reported for the Detection of COVID-19 Biomarkersa.
| description | sensing method | biomarkers | performance metrics | buffer/medium | reference |
|---|---|---|---|---|---|
| ECoVSens | electrochemical immunosensor | spike protein domain 1 | 10 fM | spiked saliva | (19) |
| SARS-CoV-2 RapidPlex | electrochemical immunosensor | nucleocapsid protein | 0.1–0.8 μg/mL | serum | (17) |
| 0.5–2μg/mL | saliva | ||||
| S1-IgG | 20–40 μg/mL | serum | |||
| 0.2–0.5 μg/mL | saliva | ||||
| S1-IgM | 20–50 μg/mL | serum | |||
| 0.6–5 μg/mL | saliva | ||||
| CRP | 10–20 μg/mL | serum | |||
| 0.1–0.5 μg/mL | saliva | ||||
| RGO-3D electrodes | electrochemical immunosensor | S1 antibody | 2.8 fM | fetal bovine serum and rabbit serum | (20) |
| RBD antibody | 16.9 fM | ||||
| portable, cell-based biosensor | electrochemical | spike protein | 10 fg to 1 μg/mL, 3 min | cell suspension | (21) |
| 1 fg/mL (LOD) | |||||
| AuNP–mAb | electrochemical combined with colorimetric | spike protein | 48 ng/mL (LOD) | saliva | (23) |
| 1 pg to 10 ng/mL | |||||
| MIL-53 with Au@Pt | electrochemical | nucleocapsid protein | 0.025–50 ng/mL | serum | (24) |
| 8.33 pg/mL (LOD) | |||||
| PET/Au/Ab | electrochemical immunosensor | RBD domain of spike protein | <5 min (96-well plate) | serum | (25) |
| 3D-printed COVID-19 immunosensor | electrochemical immunosensor | RBD domain of spike protein | 1–50 μg/mL | spiked serum | (26) |
| e-PAD:paper-based sensor | electrochemical | IgG and IgM | 1 ng/mL (LOD), 30 min | serum | (18) |
| spike protein | 1–1000 ng/mL | ||||
| 0.11 ng/mL (LOD) | |||||
| MIP-sensor | electrochemical | nucleocapsid protein | 2.22–111 fM | nasopharyngeal swab | (27) |
| 15 fM (LOD) | |||||
| OCETs | electrochemical | IgG | 10 fM to 100 nM, 5 min | serum, saliva | (28) |
| COVID-19 ROS system | electrochemical | reactive oxygen species | 30 s | sputum | (9) |
mAb, spike antibody; Au-NP, gold nanoparticles, MIL-53 with Au@Pt, metal–organic framework MIL-53(Al) decorated with gold@platinum; PET/Au/Ab, polyethylene terephthalate/gold/antibody; MIP, molecularly imprinted polymers; OCETs, organic electrochemical transistors.
2.2. Inflammatory Biomarkers
Inflammation is a crucial occurrence in a healthy immune system to fight against infections. An abnormal response in the inflammatory system may lead to several body dysfunctions such as severe cell damage, irregular functioning of multiple systems, organ dysfunction, or even death.
Uncontrolled inflammation plays a major part in several pathological events inside the body, for instance respiratory illness, aging, cancer, and sepsis. Innate and adaptive immunity systems differ from person to person with age. The immunosenescence process is responsible for making innate immunity more active toward the infection by increasing the number of pro-inflammatory cytokines, natural killer cells, etc. (Figure 4). These pro-inflammatory cytokines may serve as biomarkers to estimate COVID-19 disease severity. SARS-CoV-2 infection induces cytokine storm by increasing the levels of interleukins such as IL-6, IL-10, TNF-α, and IL-2R. An increase in these levels will eventually decrease the levels of CD4+ and CD8+ T cells which leads to the suppression of IFN-γ and correlates well with COVID-19 severity. IL-6 levels were strongly correlated with SARS-CoV-2 viral load in patients. A 10-fold increase in the IL-6 levels was reported in COVID-19 patients with more viral load than in patients with normal or less viral load. Hence, Il-6 is considered a promising biomarker of COVID-19. Interferon-gamma, also called IP-10, is a small protein responsible for the generation of natural killer cells and T-cells. IP-10 and MCP-1 were elevated in COVID-19 patients when compared to healthy patients. Several other cytokines, IL-1β, IL-8, IL-10, IL-21, and HMGB1, were also responsible for death caused by cytokine release syndrome or cytokine storm whereas procalcitonin (PCT), IP-10, and MCP-1 had a strong correlation with coagulation parameters.7 C-reactive protein (CRP) is another plasma protein induced by IL-6 and other types of cytokines. Significantly higher levels of CRP were reported in COVID-19 patients admitted to critical care units. CRP levels were elevated in the early stages of infection, making it an advisible clinical biomarker of COVID-19.8 Measurement of reactive oxygen species in COVID-19 patients was reported recently.9 Though many reports are available for electrochemical detection of the cytokines in several diseases or disorders, reports of testing for these inflammatory biomarkers in COVID-19 patients are scanty. Rapid analysis is strongly endorsed to enhance the quality of patient bedside monitoring.
Figure 4.
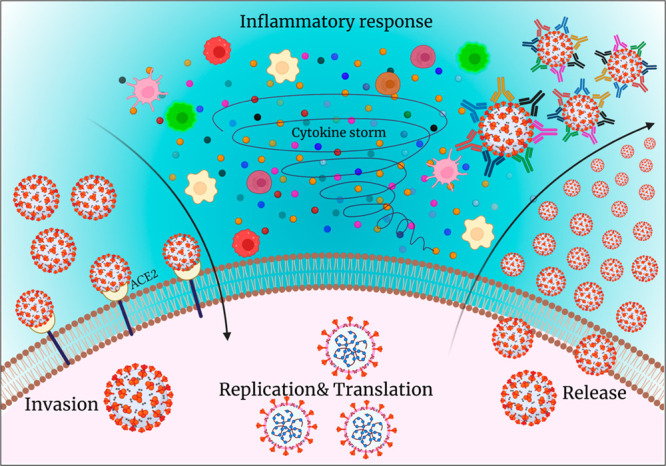
Stages of disease progression: invasion, replication, translation, and host cell response to virus by generating antibodies and cytokines (created using Biorender).
2.3. Biochemical and Hematological Biomarkers
A high mortality rate was reported in COVID-19 patients with underlying cardiovascular, kidney, and liver dysfunctions. The trigger for these cardiovascular comorbidities is the interaction between spike protein and ACE2 which is the initial step for the virus to enter the host cell.10 The ACE2 receptor is highly expressed in the tissues of the lungs, heart, blood vessels, and kidneys. D-dimer is considered as one of the crucial biomarkers of COVID-19 progression in infected patients, especially with metabolic disorders. D-dimer, a product of fibrin degradation, is a small protein fragment containing two D fragments of fibrin joined by a cross-link that presents in the blood. The rise in D-dimer levels is common in thrombosis and other conditions such as heart and liver diseases, pregnancy, and trauma. The concentration of D-dimer levels is helpful to diagnose thrombosis. An increase in the CRP and IL-6 levels promotes the rise in the levels of cardiac troponin. Myoglobin, fibrinogen, NT-pro BNP, and CK-MB are also considered as other potential markers of cardiac function. A rise in the levels of all these biomarker levels was reported in COVID-19 patients.11 Albumin, creatinine, blood urea nitrogen, and total bilirubin levels are significantly higher in nonsurviving patients of COVID-19. Other important biomarkers such as AST, ALT, GGT, and LDH are also highly expressed in COVID-19 patients. Total bilirubin and LDH are also significantly higher in nonsurvivors of COVID-19 when compared to survivors. In contrast, very low levels of albuminemia, hemoglobin, eosinophils, thrombocytes, and lymphocytes and high levels of white blood cells were reported in COVID-19 nonsurvivors. All these biochemical parameters are very important in monitoring COVID-19 (Figure 5) and not yet reported for COVID-19 risk analysis.12
Figure 5.
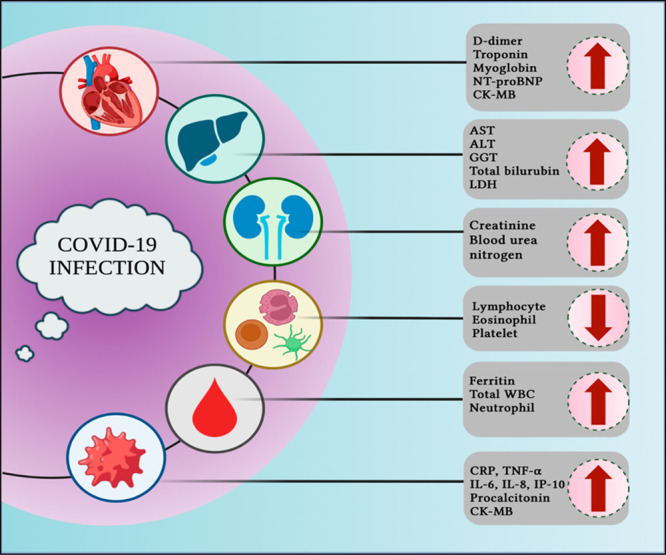
Organ-related and blood-based biomarkers of COVID-19 (created using Biorender).
2.4. Electrochemical Sensors Reported for COVID-19 Detection That Meet ASSURED Criteria
In order to identify the analyte of interest, various electrochemical techniques were employed such as cyclic voltammetry, differential pulse voltammetry, square wave voltammetry, amperometry, and electrochemical impedance spectroscopy. There are advantages and disadvantages to each of these techniques, and researchers prioritize the ones they use based on principles such as dynamic range, cross-reactivity, and specificity of the analyte they are trying to detect. Several research reports on the detection of COVID-19 biomarkers are available, and some of the reports that satisfy at least one of the ASSURED criteria are summarized in this section. Recently, Evair et al. reported an ultrasensitive disposable screen-printed electrochemical sensor using magnetic beads and gold nanoparticles. This assay detects and quantifies SARS-C0 V-2 spike protein in saliva by conjugating ACE2 peptides with magnetic beads (MBs) and gold nanoparticles (AuNPs). MBs-ACE2/SARS-CoV2 spike protein/ACE2 AuNPs bioconjugate was successfully prepared and distributed on the surface of a screen-printed working electrode while magnetic beads were externally positioned under the electrode. Differential pulse voltammetry was used for the reduction of AuCl4– to Au, and subsequent electrons generated were used for the quantification of the spike protein. As this screen-printed electrode is an eight-electrode array, this can be utilized for simultaneous analysis of different samples within the same duration. This technique also presented 100% sensitivity and 93.7% specificity towards spike protein, confirming the ability of the sensor as a point-of-care device with low cost and as a noninvasive technique with high specificity.13 A biosensor utilizing electrochemical capillary flow immunoassay that is rapid and requires low sample volume was reported to detect very low quantities of nucleocapsid protein. This electrochemical capillary-flow assay sensor also utilizes a screen-printed electrode, and the working electrode was conjugated with anti-N antibody labeled with HRP for specific detection of nucleocapsid protein. Chronoamperometry was used to quantify nucleocapsid protein from 1 ng/mL to 1000 ng/mL, and a proof-of-concept was provided for use as a point-of-care device by coupling this sensor with an NFC potentiostat operated by a smartphone for data visualization. This assay meets ASSURED criteria, as it utilizes very low-cost sensing equipment, requires low sample volumes of <10 μL, and allows rapid analysis with high sensitivity.3 A cotton-tipped electrode was successfully utilized for the detection of SARS-CoV-2 by quantifying nucleocapsid protein with a limit of detection of 0.8 pg/mL. A screen-printed electrode was modified with carbon nanofibers with diazonium by an electrografting method. Quantification of nucleocapsid protein was carried out by using square wave voltammetry in a potential window of −0.3 V to 0.5 V at a step potential of −5 mV with 20 mV amplitude and 25 Hz frequency. After successful development of the immunoassay, sensor performance was evaluated in nasal fluid collected from healthy volunteers, spiked with known concentrations of nucleocapsid protein, and measured using square wave voltammetry as described above. The sensor showed >90% of the recovery for low and high concentrations of nucleocapsid protein. Sensor performance was tested for cross-reactivity with other molecules such as FluA and HCoV antigens. Though it reacted to the nonspecific antigens, a significant increase in the response was observed only for SARS-CoV-2, confirming its selectivity. This sensor was successfully utilized for distinguishing COVID-19-infected patients from healthy ones. Because of its low cost and ability for rapid analysis, the sensor can be implemented for patient bedside monitoring.14 Another sensor was developed by Andrea et al. using square wave voltammetry for the successful detection of spike protein using an aptamer as a capture probe with high specificity in serum and saliva within 15 s. This rapid and reagentless measurement provides scope for the deployment of this sensor at clinical sites.15 Fabrication of an electrochemical biosensor based on a molecularly imprinted polymer was reported for successful detection of spike protein with a limit of detection of 64 fM in nasopharyngeal samples. This developed sensor was connected to a portable potentiostat and smartphone and assessed for its ability to analyze patient samples.16 An immunosensing platform was reported that measures nucleocapsid protein, IgG and IgM antibodies, and C-reactive protein in both serum and saliva samples to distinguish between COVID-19-positive and -negative. This device can send data to a mobile device, which is helpful for telemedicine approaches and personalized treatments.17 A paper-based wax screen-printed portable and hand-held sensor (ePAD) modified with graphene oxide was developed for detecting the RDB domain of spike protein, IgG, and IgM in serum samples. The low-cost, low-volume sensor showed 100% sensitivity and 90% specificity with a low detection limit of 1 ng/mL.18 In an eCoVSens portable device, screen-printed electrodes decorated with gold nanoparticles have been used to detect spike proteins within 10–30 s, allowing rapid analysis, but the device is expensive because it utilizes nanoparticles.19 S1 and RBD antigens were immobilized on reduced graphene oxide nanoflakes using EDC–NHS chemistry for the successful detection of S1 and RBD antibodies. Antibody detection was achieved by electrochemical transduction on a PDMS-based 3D-printed microfluidic chip. This method resulted in very high sensitivity and low cross-reactivity and was connected to a smartphone-based portable potentiostat to make it more reliable for on-site monitoring.20 An ultrarapid cell-based biosensor which can detect nucleocapsid protein within 3 s was developed using a screen-printed eight-electrode array. The PDMS layer was molded on a screen-printed electrode to separate the electrode channels. This sensor was able to detect nucleocapsid protein using low sample volumes (20 μL). The sensor surface was covered with a membrane-engineered Vero cell suspension, different concentrations of spike proteins were added, and electrochemical responses toward these concentrations were measured. A linear dynamic range of 10 fg −10 μg/mL was reported with a rapid detection time of 3 s.21 A nanocomposite of buffer-based zinc oxide and reduced graphene oxide was successfully used for the detection of nucleocapsid protein. This screen-printed immunosensor utilizes electrochemical impedance spectroscopy for detection and showed 82% sensitivity in clinical samples with a detection time of 15 min.22 The biosensors described in Table 1 use a variety of electrochemical methods and various materials. However, only a few of them meet the ASSURED criteria outlined in Introduction of this review article to qualify as a hand-held and portable device with high sensitivity and selectivity and affordable price according to WHO guidelines.
3. Conclusions
With the outbreak of the COVID-19 pandemic, the world has experienced that the threat caused by infectious diseases has become more evident. Pathogen and host interactions were more vulnerable and unpredicted in many cases. The scientific community has now opened doors to learn lessons from interdisciplinary aspects such as immunology, engineering, pathology, material science, nanoscience, electrochemistry, infectious diseases, virology, cellular and molecular biology, emergency medicine, environmental sciences, genetics, biomolecular chemistry, and proteomics. It is highly challenging to modulate the immune system toward the host response. Researchers face critical challenges in evaluating the host response to infection. We need rapid and robust technologies with efficient and coherent methodologies to tackle the situation effectively. The systematic and logical control of disease progression, medication treatments, and improvement of immune health can all be achieved through the spatial and temporal profiling of biomarkers. The development of point-of-care devices to enhance the sensitivity of the testing for biomarkers has long-term benefits, not just for COVID-19 but for all emerging infectious diseases in the future.
Biographies

Madhurantakam Sasya received her M.Sc degree in Nanomaterials and Technologies from Sri Venkateswara University, India, and was selected for a DST-INSPIRE fellowship to pursue her Ph.D. at SASTRA University. When she was a student in college, she found that interdisciplinary studies between engineering and life sciences were interesting to her as well as essential for advances in medicine. Her Ph.D. thesis focuses on the development of electrochemical biosensors that can quantify biomarkers of metabolic syndrome. She developed a few types of advanced electrochemical biosensors and published 12 research articles in peer-reviewed international journals. She was the recipient of the Karnataka DST Nano Fellowship Award in 2017 and the Metrohm India-IIT Bombay Young Chemist Award in 2019. She joined the Department of Bioengineering at the University of Texas at Dallas after completing her JSPS-Postdoctoral Fellowship at Osaka University, Japan. Her research focuses on the development of electrochemical point-of-care biosensing devices for health monitoring applications.

Dr. Sriram Muthukumar is the CEO and cofounder of EnLiSense LLC and also an Adjunct Associate Professor at the University of Texas at Dallas in the Department of Materials Science and Engineering. His research focuses on the development of ultrasensitive sensors for various applications, including biosensing and environmental use cases. He has over 20 years of experience in leading teams and developing scalable processes in electronic materials, device fabrication, and packaging technologies and high-volume manufacturing methods. He is an inventor on 17 granted patents and several patent applications pending and has authored and coauthored over 50 peer-reviewed publications to date. He received his Bachelor of Technology (BTech) degree from the Indian Institute of Technology in India in Metallurgical Engineering in 1997. He went on to complete his Ph.D. in Ceramics and Materials Engineering from Rutgers, The State University of New Jersey, in 2003. After his Ph.D., he worked at Intel, as a lead investigator in research and development programs. He also held positions at Maxim Integrated Products and Qorvo Inc., where he was instrumental in bringing new sensor technologies to market.

Shalini Prasad is a Professor in the Department of Bioengineering at The University of Texas at Dallas since 2011. She received her Bachelor’s degree in Electronics and Communication Engineering from the University of Madras, India, in 2001 and obtained her Ph.D. degree in Electrical Engineering from the University of California, Riverside, in 2004. Prior to UT Dallas, she worked as Research Assistant Professor at National Nanotechnology Infrastructure Network Node, Arizona State University, for 2 years. From 2005 to 2011 she worked as an Assistant Professor at Portland State University and Wichita State University and as an Adjunct Assistant Professor at Oregon Health Sciences University in the Department of Electrical Engineering and Biomedical Engineering, respectively. Her present research lab, Biomedical Microdevices and Nanotechnology Laboratory, focuses on “point-of-care” diagnostic devices and near-future therapeutic platforms, addressing the public health challenges of rapid and cost-effective medical care.
Author Contributions
S. Madhurantakam was responsible for conceptualization, original draft writing, review, and editing, S. Muthukumar was responsible for conceptualization, review, and editing, and S.P. was responsible for conceptualization, review, and editing. All authors have approved the final version of the manuscript.
The authors declare the following competing financial interest(s): S. Prasad and S. Muthukumar have a significant interest in EnLiSense LLC, a company that may have a commercial interest in the results of this research and technology. The potential individual conflict of interest has been reviewed and managed by The University of Texas at Dallas and played no role in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the report for publication.
References
- Ji T.; Liu Z.; Wang G. Q.; Guo X.; Akbar khan S.; Lai C.; Chen H.; Huang S.; Xia S.; Chen B.; Jia H.; Chen Y.; Zhou Q.. Detection of COVID-19: A Review of the Current Literature and Future Perspectives. Biosens. Bioelectron. 2020, 166, 112455. 10.1016/j.bios.2020.112455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO COVID-19 Dashboard. Covid19.Who.Int.
- Samper I. C.; Sánchez-Cano A.; Khamcharoen W.; Jang I.; Siangproh W.; Baldrich E.; Geiss B. J.; Dandy D. S.; Henry C. S. Electrochemical Capillary-Flow Immunoassay for Detecting Anti-SARS-CoV-2 Nucleocapsid Protein Antibodies at the Point of Care. ACS Sensors 2021, 6 (11), 4067–4075. 10.1021/acssensors.1c01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhurantakam S.; Karnam J. B.; Brabazon D.; Takai M.; Ahad I. U.; Balaguru Rayappan J. B.; Krishnan U. M. Nano”: An Emerging Avenue in Electrochemical Detection of Neurotransmitters. ACS Chemical Neuroscience 2020, 11, 4024–4047. [DOI] [PubMed] [Google Scholar]
- Ghaffari A.; Meurant R.; Ardakani A. COVID-19 Serological Tests: How Well Do They Actually Perform?. Diagnostics 2020, 10 (7), 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinti I.; Mortari E. P.; Fernandez Salinas A.; Milito C.; Carsetti R. IgA Antibodies and IgA Deficiency in SARS-CoV-2 Infection. Frontiers in Cellular and Infection Microbiology 2021, 11, 655896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rica R.; Borges M.; Gonzalez-Freire M. COVID-19: In the Eye of the Cytokine Storm. Frontiers in Immunology 2020, 11, 558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermali M.; Khalsa R. K.; Pillai K.; Ismail Z.; Harky A. The Role of Biomarkers in Diagnosis of COVID-19 – A Systematic Review. Life Sciences 2020, 254, 117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miripour Z. S.; Sarrami-Forooshani R.; Sanati H.; Makarem J.; Taheri M. S.; Shojaeian F.; Eskafi A. H.; Abbasvandi F.; Namdar N.; Ghafari H.; Aghaee P.; Zandi A.; Faramarzpour M.; Hoseinyazdi M.; Tayebi M.; Abdolahad M.. Real-Time Diagnosis of Reactive Oxygen Species (ROS) in Fresh Sputum by Electrochemical Tracing; Correlation between COVID-19 and Viral-Induced ROS in Lung/Respiratory Epithelium during This Pandemic. Biosens. Bioelectron. 2020, 165, 112435. 10.1016/j.bios.2020.112435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancio M.; Ciccocioppo R.; Rocco P. R. M.; Levine B. L.; Bronte V.; Bollard C. M.; Weiss D.; Boelens J. J.; Hanley P. J. Emerging Trends in COVID-19 Treatment: Learning from Inflammatory Conditions Associated with Cellular Therapies. Cytotherapy 2020, 22 (9), 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiga M.; Wang D. W.; Han Y.; Lewis D. B.; Wu J. C. COVID-19 and Cardiovascular Disease: From Basic Mechanisms to Clinical Perspectives. Nature Reviews Cardiology 2020, 17, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danwang C.; Endomba F. T.; Nkeck J. R.; Wouna D. L. A.; Robert A.; Noubiap J. J. A Meta-Analysis of Potential Biomarkers Associated with Severity of Coronavirus Disease 2019 (COVID-19). Biomarker Research 2020, 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento E. D.; Fonseca W. T.; de Oliveira T. R.; de Correia C. R. S. T. B.; Faça V. M.; de Morais B. P.; Silvestrini V. C.; Pott-Junior H.; Teixeira F. R.; Faria R. C.. COVID-19 Diagnosis by SARS-CoV-2 Spike Protein Detection in Saliva Using an Ultrasensitive Magneto-Assay Based on Disposable Electrochemical Sensor. Sens. Actuators, B 2022, 353,131128. 10.1016/j.snb.2021.131128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S.; Zourob M. Development of a Low-Cost Cotton-Tipped Electrochemical Immunosensor for the Detection of SARS-CoV-2. Anal. Chem. 2021, 93 (3), 1826–1833. 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- Idili A.; Parolo C.; Alvarez-Diduk R.; Merkoçi A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sensors 2021, 6 (8), 3093–3101. 10.1021/acssensors.1c01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayankojo A. G.; Boroznjak R.; Reut J.; Öpik A.; Syritski V.. Molecularly Imprinted Polymer Based Electrochemical Sensor for Quantitative Detection of SARS-CoV-2 Spike Protein. Sens. Actuators, B 2022, 353.131160. 10.1016/j.snb.2021.131160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente-Rodríguez R. M.; Lukas H.; Tu J.; Min J.; Yang Y.; Xu C.; Rossiter H. B.; Gao W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3 (6), 1981–1998. 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoh A.; Pimpitak U.; Rengpipat S.; Hirankarn N.; Chailapakul O.; Chaiyo S.. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176.112912. 10.1016/j.bios.2020.112912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S.; Roberts A.; Shahdeo D.; Gandhi S.. ECovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of NCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. bioRxiv 2020. 10.1101/2020.04.24.059204 [DOI] [Google Scholar]
- Ali M. A.; Hu C.; Jahan S.; Yuan B.; Saleh M. S.; Ju E.; Gao S. J.; Panat R.. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33 ( (7), ).2170046. 10.1002/adma.202170046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikou S.; Moschopoulou G.; Tsekouras V.; Kintzios S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-COV-2 S1 Spike Protein Antigen. Sensors (Switzerland) 2020, 20 (11), 3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghayegh F.; Salahandish R.; Hassani M.; Sanati-Nezhad A.. Highly Stable Buffer-Based Zinc Oxide/Reduced Graphene Oxide Nanosurface Chemistry for Rapid Immunosensing of SARS-CoV-2 Antigens. ACS Appl. Mater. Interfaces 2022.1410844. 10.1021/acsami.1c24475 [DOI] [PubMed] [Google Scholar]
- Karakuş E.; Erdemir E.; Demirbilek N.; Liv L.. Colorimetric and Electrochemical Detection of SARS-CoV-2 Spike Antigen with a Gold Nanoparticle-Based Biosensor. Anal. Chim. Acta 2021, 1182.338939. 10.1016/j.aca.2021.338939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.; Liang Z.; Hu O.; He Q.; Sun D.; Chen Z.. An Electrochemical Dual-Aptamer Biosensor Based on Metal-Organic Frameworks MIL-53 Decorated with Au@Pt Nanoparticles and Enzymes for Detection of COVID-19 Nucleocapsid Protein. Electrochim. Acta 2021, 387.138553. 10.1016/j.electacta.2021.138553 [DOI] [Google Scholar]
- Rashed M. Z.; Kopechek J. A.; Priddy M. C.; Hamorsky K. T.; Palmer K. E.; Mittal N.; Valdez J.; Flynn J.; Williams S. J.. Rapid Detection of SARS-CoV-2 Antibodies Using Electrochemical Impedance-Based Detector. Biosens. Bioelectron. 2021, 171.112709. 10.1016/j.bios.2020.112709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J.; Pumera M.. 3D-Printed COVID-19 Immunosensors with Electronic Readout. Chemical Engineering Journal 2021, 425.131433. 10.1016/j.cej.2021.131433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziq A.; Kidakova A.; Boroznjak R.; Reut J.; Öpik A.; Syritski V.. Development of a Portable MIP-Based Electrochemical Sensor for Detection of SARS-CoV-2 Antigen. Biosens. Bioelectron. 2021, 178.113029. 10.1016/j.bios.2021.113029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Yang A.; Song J.; Wang N.; Lam P.; Li Y.; Ka-Wai Law H.; Yan F.. Ultrafast, Sensitive, and Portable Detection of COVID-19 IgG Using Flexible Organic Electrochemical Transistors. Sci. Adv. 2021, 7 ( (38), ) 10.1126/sciadv.abg8387. [DOI] [PMC free article] [PubMed] [Google Scholar]