Abstract
Diabetic foot ulceration (DFU) is a serious complication of diabetes, and a major challenge for healthcare systems around the world. Further infection and ischemia in DFU can significantly prolong treatment and often result in limb amputation, with more severe cases resulting in terminal illness. Thus, early identification and regular monitoring is necessary to improve care, and reduce the burden on healthcare systems. With that in mind, this study attempts to address the problem of infection and ischemia classification in diabetic food ulcers, in four distinct classes. We have evaluated a series of VGG architectures with different layers, following numerous training strategies, including k-fold cross validation, data pre-processing options, augmentation techniques, and weighted loss calculations. In favor of transparency and reproducibility, we make all the implementations available through the Generally Nuanced Deep Learning Framework (GaNDLF, github.com/CBICA/GaNDLF. Our best model was evaluated during the DFU Challenge 2021, and was ranked 2nd, 5th, and 7th based on the macro-averaged AUC (area under the curve), macro-averaged F1 score, and macro-averaged recall metrics, respectively. Our findings support that current state-of-the-art architectures provide good results for the DFU image classification task, and further experimentation is required to study the effects of pre-processing and augmentation strategies.
Keywords: Diabetic foot, Classification, Deep learning, Convolutional neural network, DFUC2021, DFU, Ischemia, VGG, GaNDLF
1. Introduction
Diabetic foot ulcers (DFUs) are the most common complications of diabetes mellitus that usually take long time to heal and are among the leading causes of hospitalization and morbidity of patients with diabetes [1,2]. According to published estimates, DFU accounts for roughly 20% of hospital admissions of diabetic patients [3]. In addition, DFU leads to substantial emotional, physical, and financial distress that deteriorates the quality of life of patients and their caregivers [4]. If not managed properly, DFU combined with ischemia and infection can cause gangrene, lower limb amputation, and even death [1,2].
For healthcare systems with limited resources, DFU diagnosis can impose a substantial economic burden. As such, the clinical translation of computational methods for the automated assessment of DFU could be beneficial for all stakeholders in the healthcare system, namely, the clinical sites, patients, and caregivers. Such translation could specifically contribute in the early detection of DFU, as well as in the regular monitoring of patients’ condition by themselves or by their caregivers. For this purpose, several mobile device applications have been designed and developed for standardization and collection of DFU images, and for promoting self-care of DFU [5-8].
Recent advances in the fields of computer vision and machine learning have had a growing impact on medical imaging, including radiology, histopathology, and dermatology [9-25]. Of special note is the proliferation of several deep learning (DL) methods, which have shown superior performance in numerous computer vision and medical image computing tasks [26-31]. Lately, it is also shown that DL models succeed in classification, detection, and segmentation tasks on DFU images [32-38]. Specifically, previous work from Goyal et al. [33] focuses on the recognition of ischemia and infection on DFU images, but their work tries to solve binary classification problem of ischemia (ischemia vs. non-ischemia) and infection (infection vs. non-infection) separately.
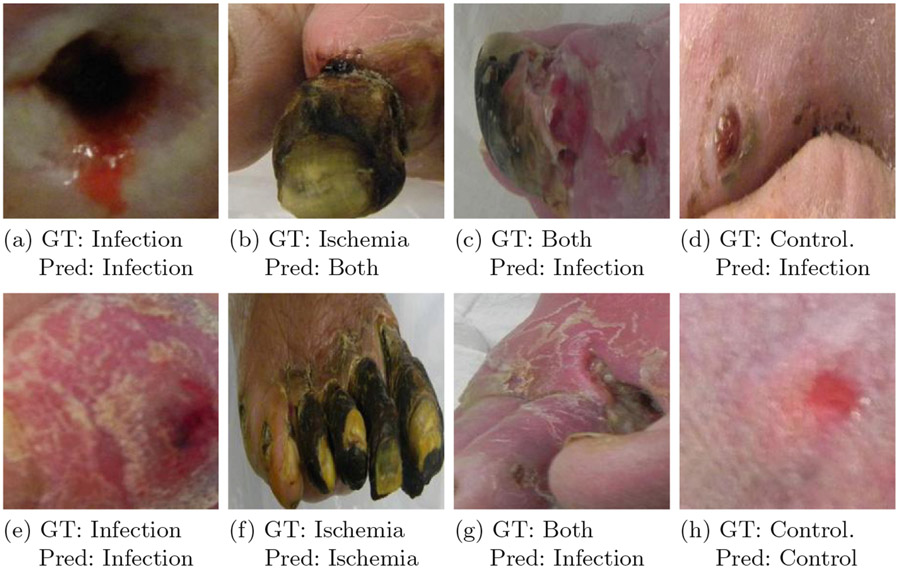
This study aims to solve a multi-class (4-class) classification problem for Diabetic Foot Ulceration (DFU), by leveraging the GenerAlly Nuanced Deep Learning Framework (GaNDLF)1 [39]. The specific 4 classes considered are: i) infection, ii) ischemia, iii) both infection & ischemia, and iv) controls (i.e., neither infection, nor ischemia) (Fig. 1), as defined by the DFU Challenge (DFUC) 2021 [40], conducted in conjunction with the Annual Scientific Meeting of Medical Image Computing and Computer Assisted Interventions (MICCAI) 2021. GaNDLF facilitated our work by providing simple application programming interfaces to rapidly and robustly incorporate techniques such as cross-validation [41], data pre-processing, data augmentation, and weighted loss calculation into our experimental design. Our best model, with which we participated at the DFUC 2021, was compared to the baseline models provided by Yap et al. [40], and was ranked in the 2nd, 5th, and 7th place in the DFUC 2021, based on the macro-averaged AUC (area under the curve), macro-averaged F1 score, and macro-averaged recall metrics, respectively.
Fig. 1.
Examples from each class considered in the DFUC2021 challenge. GT: ground truth, pred: prediction of our best model VGG11 5-fold
2. Materials and Methods
In this section, we describe the provided dataset in detail, illustrate the examples of infection, ischemia, both infection & ischemia, and control cases from DFU patients with images. Additionally, we explain the methods used in the work, their configuration, the overall VGG architecture, and the various training strategies we followed.
2.1. DFU Dataset
The DFUC2021 dataset describes a multi-institutional collection for analysis of specific pathologies, focusing on infection and ischemia [40]. Specifically, Manchester Metropolitan University and Lancashire Teaching Hospitals established a repository that contains DFU images of infection and ischemia cases for the purpose of supporting research on more advanced methods of pathology detection and recognition of DFU. These DFU images are collected from Lancashire Teaching Hospitals, where photographs were taken from patients during their clinical visits. The three cameras used for capturing the foot images are Kodak DX4530, Nikon D3300 and Nikon COOLPIX P100. The images taken were close-ups of the whole foot from a distance of approximately 30–40 cm with parallel inclination to the ulcer plane. Thereafter, the DFU regions are cropped from the original images and natural data augmentation is performed by preserving the case ids and splitting them to train and test sets.
The complete DFUC2021 dataset comprises of a total of n = 15, 683 DFU images. The provided ground truth labels, defining the four classes considered by DFUC2021 are i) infection, ii) ischemia, iii) both infection & ischemia, and iv) controls (i.e., neither infection, nor ischemia). Representative example cases for each class are shown in Fig. 1. To quantitatively evaluate the performance of algorithms developed for this task, the complete set of n = 15, 863 images are partitioned into three distinct independent subsets. The training set includes n = 5, 955 images, provided with their ground truth labels dividing the training set in n = 2, 555 cases with only infection, n = 227 cases with only ischemia, n = 621 cases with both infection & ischemia, and n = 2, 552 control cases. For algorithmic evaluation on unseen data, the DFUC2021 dataset further provides n = 5, 734 cases for testing, and n = 3, 994 unlabeled cases. The utilization of unlabelled data for the ischemia and infection classification is left for future work.
2.2. DL Framework
We leveraged the GenerAlly Nuanced Deep Learning Framework (GaNDLF)2 [39] to conduct all experimentation and training for this study. GaNDLF has been developed in Python using the well-known DL library PyTorch [42]. It enables researchers to target various machine learning (ML) and artificial intelligent (AI) workloads (such as segmentation, regression, classification, and synthesis) using different types of imaging modalities (such as RGB, radiographic, and histopathologic imaging), by providing a complete end-to-end solution for training and deploying robust DL models [39]. GaNDLF makes DL accessible for researchers who do not have extensive experience in designing and implementing DL pipelines, while making it straightforward for computational researchers to make their algorithms available for a wider array of applications. It also aims to deploy DL workflows in clinical environments with relative ease. With properties such as an end-to-end application programming interface, ease of training robust and generalizable models with different configurations, robust data pre-processing & augmentation techniques, and nested k-fold cross validation, GaNDLF aims to fill gaps of other popular DL libraries. This allows a user to easily design different experiments by simply editing text parameter files without requiring any additional coding, and when combined with the rich metrics library for validating trained models, GaNDLF provides deeper insights into model robustness.
2.3. Architecture
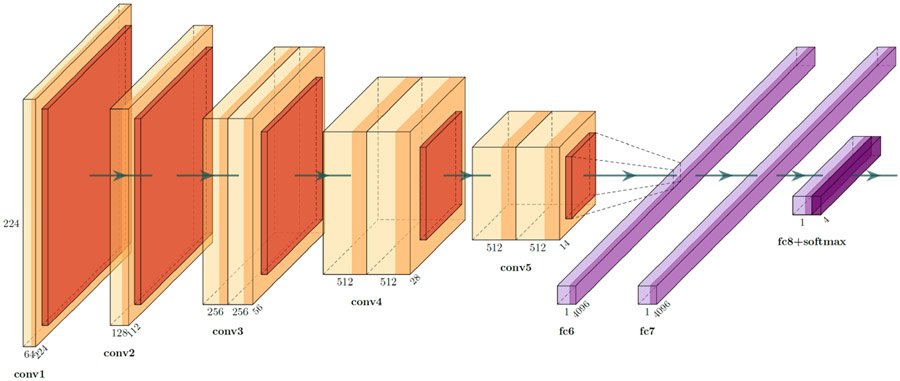
We trained three versions of the VGG architecture [43] for the DFUC2021, namely the VGG11, VGG16, and VGG19 [43]. VGG architectures use very small convolutional filters, and apply spatial padding with the intention of preserving the original resolution of the input image. A total of 5 max-pooling operations are performed over a 2 × 2 window size, with a stride of 2, to ensure that each max-pooling operation reduces both the image height and width in half. The classifier component of our VGG variants use the ReLU activation function [44], along with a global average pooling [45], two drop-out layers, and a penultimate linear layer. To ensure that the network performs classification, a softmax layer is added as the final layer, which enables us to extract the likelihood for each class. Schematic representations for the overall architecture of VGG11, VGG16, and VGG19, can be found in Figs. 2, 3, and 4, respectively.
Fig. 2.
VGG11 architecture (Figure constructed using PlotNeuralNet [46])
2.4. Training
We used two distinct approaches to train the model. First, we consider a more clinically oriented paradigm, and we split the training data into two equal halves, using one as a retrospective/discovery cohort (i.e., training) and the other as an unseen prospective/replication cohort (i.e., blinded validation). For this partitioning, all subjects of each label/class were proportionally and randomly divided across the 2 halves (retrospective/prospective). This approach was used to yield our baseline results and enabled us to tune the hyperparameters of the model for the task at hand.
Once we obtained these baseline results, we considered a more computationally oriented paradigm, to ensure generalizability of the trained model and prevent overfitting. We specifically employed a k-fold cross validation schema [41], which is a technique widely-used in ML to ensure reporting unbiased performance estimates, and help capture information from an entire dataset by training k different models on k corresponding non-overlapping folds/subsets of the complete training data. Using k-fold cross-validation one can test the model’s ability to make accurate predictions on unseen data, detect problems like overfitting or selection bias [47], and provide an understanding on how well the model will generalize to the real distribution of data. For all experiments that used cross-validation, we set the number of folds as k = 5. We have performed an equal non-intersecting 5-way split of the training data, ensuring that the model trains on the full training data without overfitting.
Based on the knowledge of the model hyperparameters obtained from our initial experiment using the labelled training data split in equal halves, we proceeded to split the training data into a set of n = 5 randomized splits. Each split was used to train the VGG11, VGG16, and VGG19 variants of the network architectures. During the training step, a single patch of 128 × 128 is extracted from a random location from each image and processed by the model for loss back-propagation. For the forward pass of the model (i.e., the validation/inference phases), enough patches of the specified size are generated from each image to ensure that every pixel is processed at least once by the model, and the final prediction is generated by averaging all the predictions from each patch. To generate the final predictions during the testing phase, we have averaged predictions from every fold. The batch size was chosen as b = 256 for VGG11 and VGG16 architectures, and b = 128 for VGG19 to ensure maximal utilization of the available hardware resources. For each model architecture, configurations with a standard set of data augmentations and data pre-processing techniques were also evaluated. For data augmentation, bias, blur, noise, and swapping techniques are used with a maximum probability p = 0.5. For data pre-processing, a z-scoring normalization mechanism was used. The choice of the loss function stayed the same across these experiments, as the original VGG architecture, which is softmax followed by categorical cross entropy loss, and has shown to work better for multi-class classification problems [48].
To increase the variability of our experiments, we further used weighted cross-entropy loss [49], which ensures that misclassifications of the class with the smallest number of labels generates the largest loss, and could thus be better suited for datasets with imbalanced classes. We used ADAM [50] as the optimizer with an initial learning rate of 0.001. For the half split configurations, the set the total number of epochs to n = 200 and set the patience value to n = 50. For the 5-fold cross validation configurations, we set the number of epochs to n = 300 and patience value to n = 50. All of the experimentation was performed on a high performance computing cluster with NVIDIA P100 GPU (which has 11 GB of dedicated video memory), using 32 GB RAM, and 10 CPU threads.
3. Results
In this section, we first analyze the validation performance of the models trained with the different training strategies. We perform our final model selection based on the performance of our partitioned validation set, and submit the inference results from the best model for the challenge evaluation and ranking. Thereafter, we analyze our best models’ performance in terms of macro-averaged F1 score, and Macro-AUC, macro-averaged recall and accuracy.
3.1. Training/Validation Dataset Performance
We conducted n = 12 different experiments with various training strategies and VGG architectures (Table 1). The models trained without any data augmentation and pre-processing and without using the weighted cross entropy loss outperformed the rest. The overall best model, in terms of validation loss and validation accuracy, was the standard VGG11 architecture trained using 5-fold cross-validation. This model, trained without any data augmentation and pre-processing, was able to reach an average cross entropy loss of 0.24 after averaging over the outputs of all the folds. Table 1 includes a detailed overview of the training results.
Table 1.
Validation performance of VGG variations. DA: Data augmentation, DP: Data pre-processing, WCE: Weighted Cross-Entropy. Bold values imply the best performance and the underlined values imply the models that are selected for experimenting in real test set
| Model architecture | Training strategy | Validation loss |
|---|---|---|
| VGG11 | Half-Split | 0.49 |
| VGG16 | Half-Split | 0.51 |
| VGG19 | Half-Split | 0.59 |
| VGG11 | 5-fold (DA, DP) | 0.52 |
| VGG16 | 5-fold (DA, DP) | 0.63 |
| VGG19 | 5-fold (DA, DP) | 0.72 |
| VGG11 | 5-fold | 0.24 |
| VGG16 | 5-fold | 0.30 |
| VGG19 | 5-fold | 0.31 |
| VGG11 | 5-fold (WCE) | 0.27 |
| VGG16 | 5-fold (WCE) | 0.39 |
| VGG19 | 5-fold (WCE) | 0.52 |
3.2. Testing Dataset Performance
Once we obtained results for all our cross-validated experiments (Table 1), we measured the performance of our best 6 models with the DFUC2021 testing data, as provided by the challenge organizers. The best configuration, in terms of accuracy, macro-averaged F1 score, and Macro-AUC, was the standard VGG11 architecture with 5-fold cross validation. Our best configuration placed in the 5th place in the DFU Challenge 2021, where the participants were ranked according to macro-averaged F1 score. In addition, we analyzed our results according to macro-averaged AUC and macro-averaged recall metrics. Our model ranked in the 2nd and 7th place, respectively (for details, please see the official DFU leaderboard3). Considering that the DFUC2021 dataset is an imbalanced set (i.e., the number of samples for each of the classes are not similar), it was surprising to us that standard VGG11 model trained without any pre-processing, data augmentation, and weighted cross-entropy outperformed the other configurations we tested during both validation and testing phases of the challenge. This may require some future meta-analysis to gain a deeper understanding on the exact driving factors. We conducted a visual demonstration of our model’s performance on the test set with example samples given by the providers. Samples with ground truth values and predictions can be found in Fig. 1. A detailed illustration of these results can be found at Table 2.
Table 2.
Test set performance of the 6 best models, selected according to the average performance of during cross validation training.
| Model | Accuracy | Macro-AUC | Macro-Recall | Macro F1-Score |
|---|---|---|---|---|
| VGG11 5-fold | 0.640 | 0.870 | 0.576 | 0.561 |
| VGG16 5-fold | 0.617 | 0.869 | 0.575 | 0.543 |
| VGG19 5-fold | 0.615 | 0.870 | 0.572 | 0.551 |
| VGG11 5-fold WCE | 0.624 | 0.845 | 0.561 | 0.541 |
| VGG16 5-fold WCE | 0.601 | 0.858 | 0.583 | 0.534 |
| VGG19 5-fold WCE | 0.595 | 0.859 | 0.562 | 0.521 |
We also conducted an analysis based on the class-individual F1 scores. The standard VGG11 model performed well based on the ischemia F1 score and control F1 score, achieving ranks of 3rd and 5th, respectively. On the other hand, the performance based on infection F1 score and both F1 score was relatively poor, with rank of 8th for both metrics. Additionally, we compared the VGG19 5-fold model’s performance with the other models in the challenge leaderboard and observe that it had the highest 2nd and 5th score based on the results on ischemia F1 score and both F1 score, respectively. Additional details can be found in Table 3.
Table 3.
Class-individual F1 scores of the selected models.
| Model | Infection F1-Score |
Ischemia F1-Score |
Both F1-Score |
None F1-Score |
|---|---|---|---|---|
| VGG11 5-fold | 0.547 | 0.522 | 0.440 | 0.736 |
| VGG16 5-fold | 0.493 | 0.531 | 0.424 | 0.723 |
| VGG19 5-fold | 0.475 | 0.548 | 0.461 | 0.719 |
| VGG11 5-fold WCE | 0.521 | 0.488 | 0.428 | 0.726 |
| VGG16 5-fold WCE | 0.448 | 0.515 | 0.462 | 0.712 |
| VGG19 5-fold WCE | 0.417 | 0.502 | 0.450 | 0.716 |
4. Discussion
In this study, we modified a well-known DL neural network architecture, namely VGG, to classify images containing diabetic foot ulcerations (DFUs), into infection, ischemia, both infection & ischemia, and control. Classification of ischemia and infection of DFU patients is an important task which would help early diagnosis and prevent serious illness in the future. Although 4 classes can be considered as small, the training dataset is not balanced, which makes it harder to learn for the classes with small number of samples. Our results indicate that the best approach, in the set of experiments we performed, was the one that did not rely upon weighted loss calculation or any pre-processing methods.
All architectures evaluated in this study were implemented in GaNDLF, which is a high level framework for training robust DL models. For training, we used n = 12 different configurations of the general VGG architecture, with different number of weight layers and different training strategies. The number of weight layers were 11, 16 and 19. The two major training strategies we leveraged was 1) splitting the training data into retrospective/training and prospective/-validation datasets, as halves, and 2) following a 5-fold cross validation schema. Effect of data pre-processing and augmentation was explored and quantified in these experiments. It is observed that the 5-fold configuration of the VGG11 without any data augmentation or pre-processing performed the best out all experiments, with an average loss of 0.24 across 5 folds. We hypothesise that VGG16 and VGG19 architectures performed worse than VGG11 because they are simply too large for the given task, and addition of more data is required to properly optimize their weights. The best model in our experimentation was ranked as the 5th place in DFU Challenge 2021, in which the participants were ranked according to macro-averaged F1 score. In addition, our best model was ranked as 2nd and 7th place based on macro-averaged AUC and macro-averaged recall metrics, respectively. Additionally, we further analyzed the class-individual F1 scores and observe that even though our model has performed better on ischemia F1 score and both F1 score, it has performed poorly on infection F1 score and both F1 score metrics.
We believe that there is plenty of room for improvement and further analysis in related future work. Especially, by taking RGB-specific augmentations [51] and pre-processing methods [52] into account, we would expect to significantly improve the model performance. Considering DFU 2021 dataset is imbalanced, exploration of the reasons why weighted cross-entropy loss did not increase the performance would be insightful. Additionally, exploration of custom loss functions, which are specifically designed for optimizing the macro-averaged F1 score could improve the performance [53]. We would also like to see how other popular architectures would perform on this task. Experimentation with architectures like residual networks [54], Efficient-Net [55], Xception-Net [56], InceptionRes-Net [57] and vision transformers [58] would be insightful. Finally, the use of the unlabelled datasets (n = 3, 994) to augment the training using weakly supervised training [59] could also help the model performance.
5. Conclusions
DFU can cause critical health problems when it is combined with ischemia and infection. Thus, early diagnosis of potential severe can save lives, and contribute to improvement in the quality of life for all stakeholders in the healthcare system. Automated computational approaches targeting on providing classification suggestions could contribute in early disease detection and the management of DFU patients. Our proposed VGG11 model, trained using a 5-fold cross validation configuration, without any data augmentation or pre-processing, demonstrated superior performance when compared to the other evaluated models, and placed in the 5th place on DFU Challenge 2021, where the rankings were determined by the macro-averaged F1 score. Future work, towards further improving our obtained results, should explore custom loss functions, RGB-specific augmentations (for example, using contrast, brightness, and scale augmentations) along with RGB-specific pre-processing.
Acknowledgments
Research reported in this publication was partly supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH), under award number NCI:U01CA242871. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
A. Illustrations of Various VGG Variants
A.1. VGG16
Fig. 3.
VGG16 architecture (Figure constructed using PlotNeuralNet [46])
A.2. VGG19
Fig. 4.
VGG19 architecture (Figure constructed using PlotNeuralNet [46])
Footnotes
References
- 1.Yazdanpanah L, Nasiri M, Adarvishi S: Literature review on the management of diabetic foot ulcer. World J. Diabetes 6(1), 37 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shahbazian H, Yazdanpanah L, Latifi SM: Risk assessment of patients with diabetes for foot ulcers according to risk classification consensus of international working group on diabetic foot (IWGDF). Pak. J. Med. Sci 29(3), 730 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snyder RJ, Hanft JR: Diabetic foot ulcers-effects on QOL, costs, and mortality and the role of standard wound care and advanced-care therapies. Ostomy Wound Manage. 55, 28–38 (2009) [PubMed] [Google Scholar]
- 4.Vileikyte L: Diabetic foot ulcers: a quality of life issue. Diabetes Metab. Res. Rev 17(4), 246–249 (2001) [DOI] [PubMed] [Google Scholar]
- 5.Brown R, Ploderer B, Da Seng LS, Lazzarini P, Van Netten J: Myfootcare: a mobile self-tracking tool to promote self-care amongst people with diabetic foot ulcers. In: Proceedings of the 29th Australian Conference on Computer-Human Interaction, pp. 462–466 (2017) [Google Scholar]
- 6.Ploderer B, Brown R, Da Seng LS, Lazzarini PA, van Netten JJ: Promoting self-care of diabetic foot ulcers through a mobile phone app: user-centered design and evaluation. JMIR Diabetes 3(4), e10105 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap MH, et al. : A new mobile application for standardizing diabetic foot images. J. Diabetes Sci. Technol 12(1), 169–173 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogrin R, Viswanathan R, Aylen T, Wallace F, Scott J, Kumar D: Co-design of an evidence-based health education diabetes foot app to prevent serious foot complications: a feasibility study. Pract. Diabetes 35(6), 203–209d (2018) [Google Scholar]
- 9.Lundervold AS, Lundervold A: An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys 29(2), 102–127 (2019) [DOI] [PubMed] [Google Scholar]
- 10.Cheng J-Z, et al. : Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in CT scans. Sci. Rep 6(1), 1–13 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, et al. : Deep learning in medical ultrasound analysis: a review. Engineering 5(2), 261–275 (2019) [Google Scholar]
- 12.Bakas S, et al. : Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge, arXiv preprint arXiv:1811.02629 (2018) [Google Scholar]
- 13.Akbari H, et al. : Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudoprogression in glioblastoma. Cancer 126, 2625–2636 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akbari H, et al. : Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology 273(2), 502–510 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder ZA, et al. : Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell 34(1), 163–177 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakas S, et al. : In vivo detection of egfrviii in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index. Clin. Cancer Res 23(16), 4724–4734 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurc T, et al. : Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front. Neurosci 14, 27 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mang A, Bakas S, Subramanian S, Davatzikos C, Biros G: Integrated biophysical modeling and image analysis: application to neuro-oncology. Annu. Rev. Biomed. Eng 22, 309–341, (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakas S, et al. : Overall survival prediction in glioblastoma patients using structural magnetic resonance imaging (MRI): advanced radiomic features may compensate for lack of advanced mri modalities. J. Med. Imaging 7(3), 031505 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akbari H et al. : Survival prediction in glioblastoma patients using multi-parametric MRI biomarkers and machine learning methods. ASNR, Chicago, IL: (2015) [Google Scholar]
- 21.Akbari H: et al. : Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 78(4), 572–580 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari H, Bakas S, Martinez-Lage M, et al. : Quantitative radiomics and machine learning to distinguish true progression from pseudoprogression in patients with GBM. In: 56th Annual Meeting of the American Society for Neuroradiology, Vancouver, BC, Canada (2018) [Google Scholar]
- 23.Rathore S, et al. : Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J. Med. Imaging 5(2), 021219 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathore S, Bakas S, Akbari H, Shukla G, Rozycki M, Davatzikos C: Deriving stable multi-parametric MRI radiomic signatures in the presence of interscanner variations: survival prediction of glioblastoma via imaging pattern analysis and machine learning techniques. In: Medical Imaging 2018: Computer-Aided Diagnosis, vol. 10575, p. 1057509, International Society for Optics and Photonics; (2018) [Google Scholar]
- 25.Li H, Galperin-Aizenberg M, Pryma D, Simone CB II, Fan Y: Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother. Oncol 129(2), 218–226 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur S, et al. : Brain extraction on MRI scans in presence of diffuse glioma: multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training. Neuroimage 220, 117081 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz-Vega I, Hernandez-Contreras D, Peregrina-Barreto H, Rangel-Magdaleno J.d.J., Ramirez-Cortes JM: Deep learning classification for diabetic foot thermograms. Sensors 20(6), 1762 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng K, et al. : Segmentation of gliomas in pre-operative and post-operative multimodal magnetic resonance imaging volumes based on a hybrid generative-discriminative framework. In: Crimi A, Menze B, Maier O, Reyes M, Winzeck S, Handels H (eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 184–194. Springer, Cham: (2016). 10.1007/978-3-319-55524-9_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S: Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds.) BrainLes 2018. LNCS, vol. 11383, pp. 92–104. Springer, Cham: (2019). 10.1007/978-3-030-11723-8_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashyam VM, et al. : MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain 143(7), 2312–2324 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheller MJ, et al. : Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep 10(1), 1–12 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal M, Reeves ND, Rajbhandari S, Yap MH: Robust methods for real-time diabetic foot ulcer detection and localization on mobile devices. IEEE J. Biomed. Health Inform 23, 1730–1741 (2019) [DOI] [PubMed] [Google Scholar]
- 33.Goyal M, Reeves ND, Rajbhandari S, Ahmad N, Wang C, Yap MH: Recognition of ischaemia and infection in diabetic foot ulcers: dataset and techniques. Comput. Biol. Med 117, 103616 (2020) [DOI] [PubMed] [Google Scholar]
- 34.Goyal M, Yap MH, Reeves ND, Rajbhandari S, Spragg J: Fully convolutional networks for diabetic foot ulcer segmentation. In: 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), pp. 618–623. IEEE; (2017) [Google Scholar]
- 35.Goyal M, Reeves ND, Davison AK, Rajbhandari S, Spragg J, Yap MH: DFUNet: convolutional neural networks for diabetic foot ulcer classification. IEEE Trans. Emerg. Top. Comput. Intell 4(5), 728–739 (2018) [Google Scholar]
- 36.Yap MH, et al. : Deep learning in diabetic foot ulcers detection: a comprehensive evaluation. Comput. Biol. Med 135, 104596 (2021) [DOI] [PubMed] [Google Scholar]
- 37.Goyal M, Hassanpour S: A refined deep learning architecture for diabetic foot ulcers detection, arXiv preprint arXiv:2007.07922 (2020) [Google Scholar]
- 38.Alzubaidi L, Fadhel MA, Oleiwi SR, Al-Shamma O, Zhang J: DFU QUT-Net: diabetic foot ulcer classification using novel deep convolutional neural network. Multimedia Tools and Appl. 79, 15655–15677 (2019) [Google Scholar]
- 39.Pati S, et al. : GANDLF: a generally nuanced deep learning framework for scalable end-to-end clinical workflows in medical imaging (2021) [Google Scholar]
- 40.Yap MH, Cassidy B, Pappachan JM, O’Shea C, Gillespie D, Reeves N: Analysis towards classification of infection and ischaemia of diabetic foot ulcers arXiv preprint arXiv:2104.03068 (2021) [Google Scholar]
- 41.Allen DM: The relationship between variable selection and data agumentation and a method for prediction. Technometrics 16(1), 125–127 (1974) [Google Scholar]
- 42.Paszke A, et al. : PyTorch: an imperative style, high-performance deep learning library. Adv. Neural. Inf. Process. Syst 32, 8026–8037 (2019) [Google Scholar]
- 43.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition (2014) [Google Scholar]
- 44.Agarap AF: Deep learning using rectified linear units (ReLu), arXiv preprint arXiv:1803.08375 (2018) [Google Scholar]
- 45.Lin M, Chen Q, Yan S: Network in network, arXiv preprint arXiv:1312.4400 (2013) [Google Scholar]
- 46.Iqbal H: Harisiqbal88/plotneuralnet v1.0.0, December 2018 [Google Scholar]
- 47.Cawley GC, Talbot NL: On over-fitting in model selection and subsequent selection bias in performance evaluation. J. Mach. Learn. Res 11, 2079–2107 (2010) [Google Scholar]
- 48.Mahajan D, et al. : Exploring the limits of weakly supervised pretraining. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y (eds.) ECCV 2018. LNCS, vol. 11206, pp. 185–201. Springer, Cham: (2018). 10.1007/978-3-030-01216-8.12 [DOI] [Google Scholar]
- 49.Fernando KRM, Tsokos CP: Dynamically weighted balanced loss: class imbalanced learning and confidence calibration of deep neural networks. IEEE Trans. Neural Netw. Learn. Syst, 1–12 (2021) [DOI] [PubMed] [Google Scholar]
- 50.Kingma DP, Ba J: Adam: a method for stochastic optimization, arXiv preprint arXiv:1412.6980 (2014) [Google Scholar]
- 51.Buslaev A, Iglovikov VI, Khvedchenya E, Parinov A, Druzhinin M, Kalinin AA: Albumentations: fast and flexible image augmentations. Information 11(2), 125 (2020) [Google Scholar]
- 52.Finlayson GD, Schiele B, Crowley JL: Comprehensive colour image normalization. In: Burkhardt H, Neumann B (eds.) ECCV 1998. LNCS, vol. 1406, pp. 475–490. Springer, Heidelberg: (1998). 10.1007/BFb0055685 [DOI] [Google Scholar]
- 53.Li F, Yang Y: A loss function analysis for classification methods in text categorization. In: Proceedings of the 20th International Conference on Machine Learning (ICML 2003), pp. 472–479 (2003) [Google Scholar]
- 54.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016) [Google Scholar]
- 55.Tan M, Le Q: EfficientNet: rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114. PMLR; (2019) [Google Scholar]
- 56.Chollet F: Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258 (2017) [Google Scholar]
- 57.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA: Inception-v4, inception-ResNet and the impact of residual connections on learning. In: Thirty-First AAAI Conference on Artificial Intelligence (2017) [Google Scholar]
- 58.Dosovitskiy A, et al. : An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020) [Google Scholar]
- 59.Wang D, et al. : Deep-segmentation of plantar pressure images incorporating fully convolutional neural networks. Biocybern. Biomed. Eng 40(1), 546–558 (2020) [Google Scholar]