Abstract
The sarcoplasmic reticulum of skeletal muscle cells is a highly ordered structure consisting of an intricate network of tubules and cisternae specialized for regulating Ca2+ homeostasis in the context of muscle contraction. The sarcoplasmic reticulum contains several proteins, some of which support Ca2+ storage and release, while others regulate the formation and maintenance of this highly convoluted organelle and mediate the interaction with other components of the muscle fiber. In this review, some of the main issues concerning the biology of the sarcoplasmic reticulum will be described and discussed; particular attention will be addressed to the structure and function of the two domains of the sarcoplasmic reticulum supporting the excitation–contraction coupling and Ca2+-uptake mechanisms.
Keywords: muscle, Ca2+, myopathy, intracellular membrane
1. Introduction
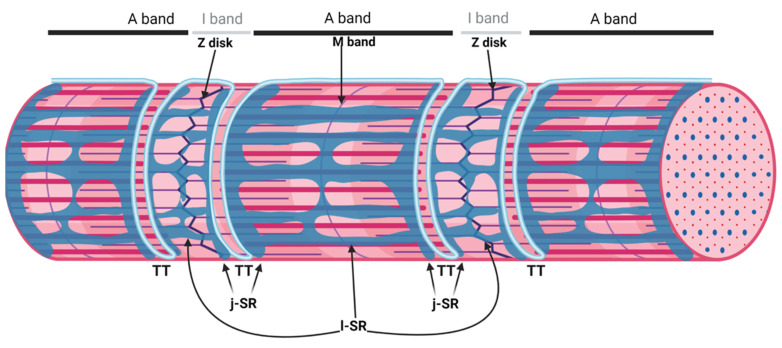
The sarcoplasmic reticulum (SR) is a specialized form of the endoplasmic reticulum of muscle cells, dedicated to calcium ion (Ca2+) handling, necessary for muscle contraction and relaxation. Studies with electron microscopes (EMs) have revealed that, in striated muscle cells, the SR is organized into numerous interconnected tubules forming a network of longitudinal elements surrounding each myofibril [1]. These elongated tubules, known as longitudinal SR (l-SR), are dedicated to the removal of Ca2+ from the cytosol, an activity operated by the sarcoplasmic/endoplasmic reticulum Ca2+ ATPases (SERCA) that actively pump Ca2+ from the cytoplasm to the lumen of the SR. In skeletal muscle fibers, the l-SR is localized around the A and I bands of each sarcomere. At regular intervals, which correspond to the borders between A and I bands of sarcomeres, the tubules of the l-SR merge into enlarged sac-shaped structures, known as terminal cisternae. Here, two terminal cisternae are positioned at the opposite sides of tubular infoldings in direct continuity with the plasma membrane, named transverse tubules (TT) (Figure 1). The structure formed by two terminal cisternae and one TT, called a "triad", represents the membrane platform where several dedicated proteins operate in transducing the depolarization of the plasma membrane into Ca2+ release from the SR, a mechanism known as excitation–contraction coupling (ECC). The region of the terminal cisternae that opposes the TT membrane is called junctional SR (j-SR), and, in this region, the Ryanodine Receptors type 1 (RyR1) Ca2+ release channels are localized.
Figure 1.
Organization of the sarcoplasmic reticulum in skeletal muscle cells. The regular alternation of anisotropic (A) and isotropic (I) bands along a single myofibril is depicted in the upper part of the image. A and I bands are bisected by the M band and the Z disk, respectively. The portion of the myofibril between two Z disks is the sarcomere. The SR is composed of tubules and cisternae surrounding each myofibril. The elongated tubules are known as longitudinal SR (l-SR); they are dedicated to the removal of Ca2+ from the cytosol and are localized around the A and I band of each sarcomere. At the borders between the A and I band, the l-SR merges to form the terminal cisternae. These are positioned at the opposite sides of a transverse tubule (TT); the structure formed by two terminal cisternae and one TT is called a "triad". The region of the terminal cisternae that opposes the TT membrane is called junctional SR (j-SR). Adapted from “Myofibril Structure” by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates, accessed on 14 March 2022.
sAnk1.5 and Obscurin Stabilize the SR around the Myofibrils
The proper relationship between the elongated tubules of the l-SR and myofibrils is a pivotal structural prerequisite to organize the different domains of the SR and allow an efficient muscle contraction [1,2]. As of today, the only two known proteins responsible for the correct SR juxtaposition and stabilization around myofibrils are the small muscle-specific isoform of ankyrin 1 (sAnk1.5) localized on the SR membrane, and obscurin, a giant sarcomeric protein [3,4,5,6]. sAnk1.5 is a striated muscle-specific isoform encoded by the ANK1 gene, containing a transmembrane domain that allows its insertion in the SR membrane, in correspondence with the M bands and, to a lesser extent, the Z disks. The C-terminal cytosolic tail of sAnk1.5 contains an ankyrin-like repeat that mediates binding to the non-modular region at the COOH-terminal of obscurin [3,4,5,6]. Similar to sAnk1.5, obscurin shows localization at the M band and Z disk of the sarcomere [6,7,8,9,10,11]. Several pieces of evidence obtained in mice knockout for either obscurin or sAnk1.5 show a significant reduction in the volume of l-SR tubules around the myofibrils even when only one of the two proteins is absent, confirming that the interaction between sAnk1.5 and obscurin is crucial to stabilize the SR around the myofibrils [7,8].
2. The Triad, a Unique Membrane System of the Skeletal Muscle
The triad is the membrane structure that supports ECC in skeletal muscle fibers by allowing the interaction between the dihydropyridine receptor (DHPR) and RyR1 [12]. The DHPR is a voltage-dependent L-type Ca2+ channel located on the TT that, following sarcolemma depolarization induced by motor-neuron stimulation, undergoes a conformational change that allows the opening of RyR1, the Ca2+ channel located on the terminal cisternae of the j-SR, resulting in a massive Ca2+ efflux from the SR into the myoplasm and activation of muscle contraction [13].
Triad Biogenesis, Repair, and Maintenance
The events that characterize TT maturation and triads formation have been morphologically defined by electron and confocal microscopy studies. These revealed that the juxtaposition of j-SR and TT begins in the embryonic (E) period of development and evolves through sequential steps in the postnatal life [14,15]. In mammals, the first step in triads biogenesis is represented by the formation of a tubular SR adjacent to myofibrils, which will subsequently develop into reticular structures surrounding the myofibrils [16]. In mouse embryos, a detectable SR is observed as early as day E14; by day E15, immunohistochemical analysis reveals the presence of RyR1 clusters positioned at the periphery of developing myotubes, and, by day E16, RyR1 clusters start aligning with the A-I borders of the sarcomeres. On day E17 and continuing after birth, the j-SR progressively acquires a transverse orientation, with RyR1 showing its typical double-row distribution [14,15].
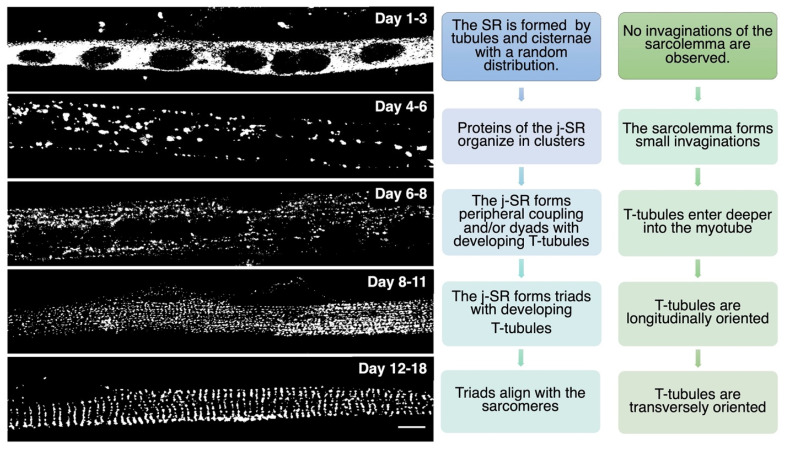
TT development appears to start immediately after that of the SR. At day E15, small tubular invaginations can be observed at the periphery of developing muscle fibers. By day E19, these invaginations expand deeper into the fiber, although remaining almost longitudinally oriented. After birth, the TT system starts acquiring its final transverse position over the borders between the A and I bands, achieving its definitive localization at around three weeks of age in mice [2,14]. A recent study performed in zebrafish that combined live imaging with three-dimensional electron microscopy proposes that membrane-derived tubules start forming at transient sarcolemma endocytic nucleation sites. These structures further grow by membrane traffic-based mechanisms and interact with the sarcomere, likely via the SR, finally resulting in an ordered transverse organization [17]. The association between SR and sarcolemma membranes is already evident as early as E14–E15, mainly at the periphery of the fiber. Starting on E16, internal junction sites appear as diads, formed by one developing TT flanked by a single terminal cisterna. On days E17–E18, most SR/TT junctions are structured as triads [18]. During in vitro differentiation of skeletal muscle, myoblasts recapitulate the main steps of in vivo SR and TT development, as reported in Figure 2.
Figure 2.
In vitro differentiation of primary rat myocytes.Primary rat myocytes induced to differentiate for 1 to 18 days were decorated with antibodies against RyR1 to label the j-SR. At the beginning of differentiation, RyRs show a diffuse distribution in the SR. Starting from day 4 of differentiation, RyRs form clusters, and the SR progressively forms peripheral couplings and/or diads with the TT. Triads form during the following days, and at the end of differentiation, they acquire their transverse orientation and localize at the borders between the A and I bands of the sarcomeres.
While several details of the morphological events that characterize TT and SR maturation have been defined, the molecular mechanisms that drive this process remain more elusive. RyR1 and DHPR, although essential for ECC, have been shown to be dispensable for the formation of triads; indeed, triad formation was observed in knockout mice for RyR1, also called “dyspedic” mice since RyRs were defined as “junctional feet”. Triad formation was also observed in mice carrying muscular dysgenesis in DHPR, a natural mutation that causes lethal paralysis in mice. Finally, mice lacking both RyR1 and DHPR also show triad formation, further supporting the idea that these two proteins are not directly involved in triad formation or maintenance [19,20,21]. Several lipid-binding proteins and enzymes regulating phospholipid metabolism have been proposed to participate in TT and j-SR biogenesis and maintenance, such as amphiphysin 2, Caveolin-3, Dysferlin, Mitsugumins, Myotubularin, and Junctophilins [22] (Figure 3).
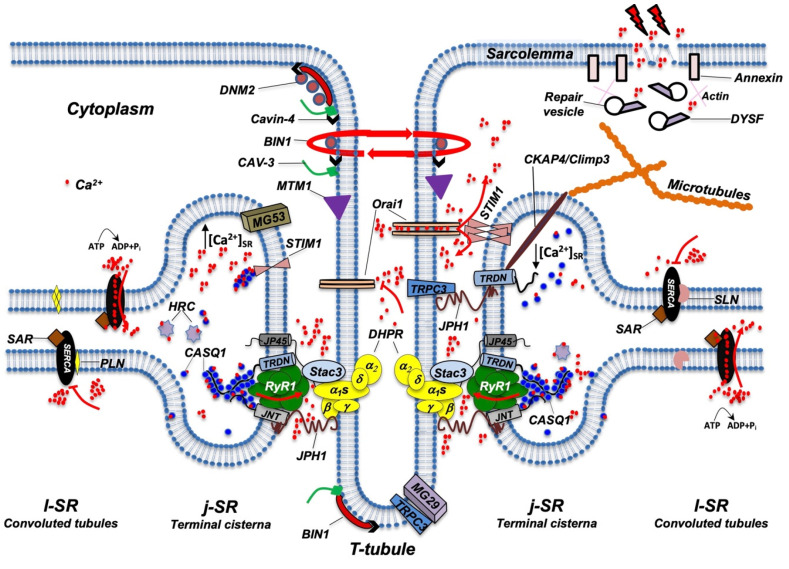
Figure 3.
Schematic representation of the main proteins accommodated in TT, j-SR, and l-SR. Protein localization and reciprocal interactions are schematized as detailed in the text. Red arrows indicate Ca2+ fluxes (red dots) through RyR1, Orai1, and SERCA pumps. RyR1 opens following interaction with DHPR; Orai1 opens following interaction with STIM1 aggregates, which in turn are induced by a reduction in Ca2+ levels in the SR; SERCA pumps actively transport Ca2+ from the cytosol to l-SR; PLN or SLN act as SERCA inhibitors. DNM2, Cavin-4, BIN1, CAV-3, and MTM1 are involved in the maintenance of TT architecture and stability. They also participate in TT formation (not shown) and, together with DYSF, contribute to vesicle trafficking during the repair of the damaged plasma membrane (see text for additional details). For simplicity, not all proteins and/or protein complexes depicted, including cytoskeleton components, are positioned on both sides of the triad, as it occurs physiologically. The following is a list of acronyms depicted in Figure 3: BIN1 (Bridging integrator-1/Amphiphysin 2); CASQ1 (Calsequestrin 1); CAV-3 (Caveolin-3); CKAP4 (Cytoskeleton-associated protein 4/Climp63); DHPR (dihydropyridine receptor); DNM2 (Dynamin 2); DYSF (Dysferlin); HRC (Histidine-Rich Calcium binding protein); JNT (Junctin); JP45 (J-SR protein 1); JPH1 (Junctophilin 1); j-SR (junctional sarcoplasmic reticulum); l-SR (longitudinal sarcoplasmic reticulum); MG29 (Mitsugumin-29); MG53 (Mitsugumin-53); MTM1 (Myotubularin); PLN (Phospholamban); RyR1 (Type 1 Ryanodine Receptor); SAR (Sarcalumenin); SERCA (Sarco/Endoplasmic Reticulum Calcium ATPase); SLN (Sarcolipin); STIM1 (Stromal Interaction Molecule 1); TRDN (Triadin); TRPC3 (Transient Receptor Potential Cation Channel 3); T-tubule (transverse tubule). Adapted from [23].
Amphiphysin 2, also known as Bridging integrator-1 (Bin1), is a member of a family of ubiquitous proteins able to shape lipid membranes; thanks to their BAR (Bin/Amphiphysin/Rvs) domain, they can interact with membrane lipids and detect and/or generate membrane curvatures [24]. Bin1 can drive membrane infoldings, thus supporting the generation of TT in muscle cells [25]. In skeletal muscle cell differentiation, Bin1 shuttles between the nucleus and cytoplasm [26] and can recruit and partially inhibit the embryonic isoform of Dynamin2 (DNM2), a GTPase involved in membrane fission and endocytosis [27,28]. Interestingly, Bin1 is not able to affect the activity of the adult isoform of DNM2, suggesting that it supports the first steps of membrane tubulation while, in the adult, it contributes to stabilizing the TT system [27]. The concurrent role of Bin1 and DNM2 in TT biogenesis has been confirmed by studies performed in Bin1 knockout mice [29] and by the identification of mutations in both genes that are causative of human centronuclear myopathy [30,31]. The SH3 domain of Bin1 also forms a complex with N-WASP that regulates the assembly of the actin cytoskeleton required for structural stability and organization of triads. This complex appears to include gamma-actin and tropomyosin Tm5NM1 in the proximity of the triads [32,33].
Caveolin-3 (Cav-3) is the muscle-specific isoform of caveolins, which mediates the formation of peripheral invaginations in cholesterol-rich regions of the plasma membrane [27,34,35]. Cav-3 knockout mice develop morphological abnormalities in the TT system consisting of tubule dilation and loss of transversal orientation [36]. In the adult, caveolae concentrate at sites of membrane injury where the resorption of damaged material is mediated through caveolae-dependent endocytosis [37]. The process of caveolae-mediated repair is inhibited or impaired in caveolinopathies, including limb–girdle muscular dystrophy (LGMD1C) [38], rippling muscle disease [39], and distal myopathy [40], all characterized by TT structural alterations. In this context, a key role is also played by another protein family, the cavins, which have been described as essential structural components of caveolae [41]. Cavin-4 is expressed predominantly in muscle, and its distribution is perturbed in human muscle diseases associated with Cav-3 dysfunction [42]. Cavin-1 (also known as Polymerase I and transcript release factor) is also expressed in skeletal muscle, where it works as a docking protein, together with Mitsugumin 53, during acute cell damage and membrane repair [43]. In addition to cavins, Cav-3 also interacts with Dysferlin and Mitsugumin 53 during membrane repair [44,45,46].
Dystrophy-associated fer-1-like protein (DYSF) is an integral protein belonging to the ferlin family, characterized by multiple C2 domains able to bind membrane phospholipids in the presence of Ca2+ [47]. It is mainly involved in membrane repair. Indeed, DYSF-deficient myofibers show a slower sarcolemma resealing and contain more dilated, irregularly shaped, and longitudinally oriented TT, with abnormal accumulation of vesicles at damaged sarcolemma sites [48,49]. Mutations in the human DYSF gene cause myopathies, including limb–girdle muscular dystrophy type 2B (LGMD2B), Miyoshi myopathy (MM), and distal anterior compartment myopathy [50,51,52]. DYSF, together with the membrane-associated actin-binding proteins annexin A6, annexin A2a, and annexin A1a, is concentrated in the vicinity of membrane injury, where it regulates Ca2+-dependent vesicle fusion with the plasma membrane [53]. Annexins are also recruited at the sarcolemma by Anoctamin 5 (ANO5/TMEM16E), a Ca2+-activated chloride channel, which was also suggested to display phospholipid scramblase activity [54]. Mutations in ANO5/TMEM16E are associated with autosomal recessive limb–girdle muscular dystrophy-12 [55,56]. More recently, Chandra and co-workers proposed that ANO5/TMEM16E may play a dual role in membrane repair since, in addition to recruiting annexins to the injury sites, it also mediates counter anion influx into the SR, supporting the activity of SERCA pumps in clearing the cytoplasm from Ca2+ overload due to membrane damage [57].
Mitsugumin-53 (MG53), also known as tripartite motif 72 (TRIM72), is a skeletal and cardiac muscle-specific protein that contributes to vesicle trafficking involved in the membrane repair machinery [46,58]. MG53 expression increases during myogenesis, and, during membrane damage repair, it is recruited at damaged membrane sites by cavin-1 where, by binding to phosphatidylserine, it acts as a scaffold to recruit additional proteins, such as Cav-3, DYSF, and annexin V, to start vesicle trafficking [49,59,60]. Indeed, MG53 knockout mice develop progressive muscle pathologies, likely correlated with an impediment in the membrane repair mechanism [49]. Surprisingly, no MG53 mutation associated with the onset of skeletal muscle pathologies has been described so far.
Mitsugumin-29 (MG29), which belongs to the synaptophysin family and is characterized by early expression during myogenesis, associates with newly formed SR vesicles [61,62]. During triad maturation, MG29 can be detected on both the SR and TT, while, in adult skeletal muscle, it localizes exclusively at triads [63]. MG29 knockout mice are characterized by decreased muscle mass and overall muscle weakness, likely due to the presence of swollen TT and less organized triads [64,65]. In vitro studies on mouse primary skeletal myotubes also showed that MG29 interacts with the canonical-type transient receptor potential cation channel 3 (TRPC3), localized at the TT, suggesting the existence of an additional mechanism able to regulate Ca2+ transients during skeletal muscle contraction [66].
Myotubularin (MTM1) is a lipid phosphatase localized at triads. Mutations in human MTM1 are associated with X-linked myotubular myopathy, a skeletal muscle disorder that results in severe muscle weakness with abnormal nuclei positioning [67]. Mice knockout for MTM1 present with progressive centronuclear myopathy characterized by SR and TT disorganization, centrally positioned nuclei, and disorganized mitochondria distribution. Interestingly, these structural alterations are more evident with aging, suggesting that MTM1 might play a role in later stages of TT formation and/or in maintaining the stability of TT, rather than in the early biogenesis of triads [68]. MTM1 dephosphorylates phosphatidyl-inositol-3-phosphate (PtdIns3P) and phosphatidyl-inositol-3,5-bisphosphate (PtdIns3,5P2), two second messengers that regulate docking and fusion of vesicles with the plasma membrane [69]. This suggests that muscle damage observed in MTM1 knockout mice might be due to abnormal endosomal vesicle trafficking and autophagic fluxes [70]. On the other hand, overexpression of MTM1 also led to the formation of abnormal membrane structures that stained positive for Cav-3, dystrophin, and DHPR [71], indicating that balanced expression levels of MTM1 are required for the proper assembly of SR and TT into triads. Interestingly, MTM1 co-localizes and interacts with BIN1 and can enhance BIN1-mediated membrane tubulation, indicating that MTM1 may also be involved in regulating membrane curvature by acting on PtdIns3P-rich membrane subdomains [72,73]. Finally, MTM1 was found to interact with the striated muscle enriched protein kinase SPEG [74]; mutations in SPEG have been linked to centronuclear myopathies with disruptions of triadic structures and impairment of ECC [75].
Junctophilins mediate the apposition between ER/SR and plasma membrane in different cell types. In mammals, Junctophilin 1 (JPH1) and Junctophilin 2 (JPH2) are expressed in striated muscles [76,77,78]. All isoforms share a similar protein structure consisting of two groups of membrane occupation and recognition nexus (MORN) motifs, separated by a joining region followed by an α-helix, a divergent region, and a transmembrane domain at their C-terminus [79]. MORN motifs can recognize and interact with membrane phospholipids [76,80]. The α-helical region is highly conserved and provides mechanical elasticity to the protein, while the divergent region, which displays the lowest similarity among the four isoforms, still lacks a clear functional role [80]. The short transmembrane domain anchors JPHs to the ER/SR membrane and, together with MORN motifs—which interact with plasma membrane phospholipids—allows JPHs to establish a stable contact site keeping the two cell membranes 12–15 nm apart [77,80].
In skeletal muscle, both JPH1 and JPH2 are present at triads, where they are essential for the formation and maintenance of these contact sites. In addition to their structural role, JPHs can also bind and regulate various proteins at triads, acting as a scaffold for assembly of the Ca2+ release complex [81,82,83]. JPH1 and JPH2 form homo- and heterodimers that can interact with several proteins on the SR and the sarcolemma, including RyRs, DHPR, Cav-3, and the TRPC3 channel [84,85,86,87,88,89,90,91]. JPHs are also required to support the organization of the Store Operated Calcium Entry (SOCE) pathway [92]. Downregulation of JPHs results in the defective formation of triads and diads and altered Ca2+ homeostasis due to a partial loss in RyR1 and DHPR co-localization [84,85,86]. JPH1 knockout mice die shortly after birth, presenting triad alterations and impaired ECC [93,94], while JPH2 knockout mice show embryonic mortality due to significant alterations in diads and cardiac ECC [77]. In the heart, remodeling and loss of TT in chronic heart failure, in aging or in cardiomyopathies correlate with downregulation of JPH2 protein expression and/or loss of JPH2 localization at TT; in addition, mutations in JPH2 are linked to the development of cardiomyopathies [95,96,97,98,99,100]. Two mechanisms have been suggested to explain JPH2 downregulation in cardiac diseases. The first is associated with the up-regulation of microRNA-24 (miR-24) observed in failing hearts; this microRNA can bind the 3′ untranslated region of JPH2 mRNA, resulting in the downregulation of JPH2 translation [95]. The second mechanism is due to calpain-mediated protein cleavage [95]; several studies showed that both JPH1 and JPH2 are targets of the calcium-depended protease μ-calpain (or calpain-1) and m-calpain (or calpain-2), whose activity increases in response to stress and/or unbalanced Ca2+ homeostasis [101,102,103,104,105]. In cardiac muscle, calpain-dependent proteolysis may explain the downregulation of JPH2 observed in heart failure [103,104]. Nevertheless, while JPH degradation results in the modification of the geometry of triads and diads, leading to ECC alterations, the N-terminal fragment of JPH2 generated by calpain digestion in stressed cardiac muscle was found to translocate to the cell nucleus, where it acts as a transcriptional regulator mitigating the hypertrophic response to cardiac disease [101,102,103,104,105].
3. The Protein Complex of the ECC
At triads, several proteins participate in the ECC process, including integral membrane proteins such as RyR1, DHPR, triadin, junctin, j-SR protein 1 (jp45), and mitsugumin-56, the STAC3 adaptor protein, or luminal proteins such as the Ca2+ binding proteins calsequestrin (CASQ) and histidine-rich calcium (HRC) binding protein [14,23,106,107] (Figure 3).
3.1. RyR1, DHPR, and STAC3 Are Essential for ECC
RyRs are a family of Ca2+ release channels that represent the largest ion channels known to date. In skeletal muscle, two RyR isoforms, RyR1 and RyR3, are expressed, although only RyR1 is essential for ECC activation. In skeletal muscle, RyR1 is primarily activated by a conformational coupling with the DHPR, a mechanism defined as depolarization-induced Ca2+ release (DICR) [13]. At variance with RyR1 channels, RyR3 channels are mainly expressed in neonatal versus adult muscles and are activated by a Ca2+-induced Ca2+-release (CICR) mechanism, whereby an increase in cytosolic Ca2+ causes RyR3 opening and, thus, Ca2+ release from intracellular stores [13,108,109]. RyR2 channels are mainly expressed in cardiac muscle, where they are involved in cardiac ECC through a CICR mechanism [106]. RyRs have a tetrameric structure, made by the assembly of four identical monomers, each consisting of a large N-terminal region of about 4300 amino acids extending in the sarcoplasm, while the remaining region contains the six transmembrane helices that anchor each monomer to the SR membrane and contribute to forming the channel pore region, followed by a short cytoplasmic C-terminal tail [110,111]. EM reconstruction studies have shown that RyRs display a mushroom-like form, with the cap in the cytoplasm representing 80% of the volume, and the stalk anchored in the j-SR membrane. In the cytoplasmic region, many domains have been described, referred to as subregions, that represent binding sites for several auxiliary proteins and molecules that contribute to regulating the opening and closing of the channel [110,111,112]. Calmodulin (CaM) was proposed to bind at different regions of RyR1 channels, acting either as a weak activator, at nanomolar Ca2+ concentration, or, in the micromolar range, as a channel inhibitor [113]. One of the proposed binding sites for CaM also binds the EF-hand protein S100, a protein also able to activate the channel [114]. RyR1 is also regulated by a member of the FK506-binding protein family, namely, FKBP12, which stabilizes the channel in a closed state [110,111,115,116]. Additional physiological or pharmacological RyR1 regulators include Mg2+, Homer 1c, the scorpion venom imperatoxin, halothane, dantrolene, PCB95, ryanodine, ruthenium red, methylxanthines, and 4-chloro-m-cresol [111,117,118]. Among these, dantrolene deserves special attention since it is now the only available agent for the treatment of malignant hyperthermia (MH), a pharmacogenetic disorder that results in a hypermetabolic state following exposure to volatile anesthetics or succinylcholine. MH is mainly associated with dominant mutations in human RYR1 that result in RyR1 hyperactivation, leading to massive and uncontrolled Ca2+ efflux from the SR. Dantrolene was historically used as a skeletal muscle relaxant, and later, it was demonstrated to be able to block calcium release from the SR. Although the molecular mechanism responsible for dantrolene action on RYR1 is still not completely defined, its introduction for the clinical treatment of MH led to a significant decrease in mortality, from more than 90% to less than 5% [119]. Finally, RyRs can also be regulated by post-translational modification, such as oxidation, nitrosylation, and phosphorylation/dephosphorylation cycles that occur via several kinases, such as PKA, PKG, Ca2+/CaM-dependent protein kinase II (CaMKII), and phosphatases (PP1, PP2A, and PDE4D3) [111,120].
DHPRs are L-type voltage-gated Ca2+ channels located on the TT. The skeletal muscle DHPR is composed of a heteromultimeric complex that includes the α1s, α2, and δ, β1α, and γ1 subunits [121]. The α1s subunit (also referred to as CaV1.1) is an integral membrane protein containing four transmembrane domains, each composed of six alpha helices, acting as the pore-forming and the voltage-sensing unit. The voltage-sensing domain of α1s interacts with the γ1 subunit that regulates channel inactivation [122]. The β1a subunit is also located in proximity to the voltage-sensing domain of the α1s subunit and supports the assembly of DHPRs in tetrads, where one DHPR tetrad faces one of the four subunits of the tetrameric RyR1 channel [121,123,124]. Mice knockout for either α1s or β1a subunits die perinatally, indicating that these subunits are essential for ECC [125,126]. The α2-δ subunits associate through four disulfide bonds and interact with the extracellular loops of the α1s subunit [121]. Despite its function as a voltage-gated calcium channel, the role of DHPR in skeletal muscle ECC is to induce Ca2+ release from the SR by physically interacting with RyR1, according to “orthograde” signaling [127]. RyR1 is also able to regulate DHPR activity by the so-called "retrograde" coupling effect [17,128]. In particular, it was demonstrated that, in the absence of RyR1, the Ca2+ current mediated by DHPR is smaller than that measured in the presence of RyR1, suggesting that RyR1 can promote the Ca2+ channel activity of DHPR and accelerates DHPR activation. Interestingly, this activity is unique to RyR1 since RyR2 was found not to be able to restore either orthograde or retrograde signaling. Different regions in DHPR and RyR1 have been proposed to be important for their reciprocal association and regulation; the II-III loop of the α1s subunit is generally accepted to be involved in both orthograde and retrograde coupling, while the site of interaction on RyR1 has not yet been completely defined [14,127,129].
STAC3 is a recently identified component of the ECC machinery. It is an adaptor protein that supports the trafficking of the α1s subunit of DHPR and regulates the coupling of DHPR with RyR1 [129,130,131,132]. STAC3 knockout mice present severe defects in muscle development, mass, and morphology; they are completely paralyzed and die shortly after birth [132]. Mutations in RYR1, DHPR, and STAC3 were identified in several human congenital myopathies, as summarized in Table 1 [119,133,134,135,136].
Table 1.
Major subtypes of RyR1-related congenital myopathies.
| Causative Gene (S) * | Inheritance | Histological Features |
Clinical Features |
|
|---|---|---|---|---|
| Central Core Disease (CCD) |
RyR1 > 90% CACNA1S |
AD or AR |
|
|
| Multiminicore Disease (MMD) |
RyR1
CACNA1S |
AR |
|
|
| Centronuclear Myopathy (CNM) | RyR1~12% | AR |
|
|
| Congenital Fibre Type Disproportion (CFTD) | RyR1~20% | AR |
|
|
| Dusty Core Disease (DUCD) | RyR1 | AR |
|
|
| Core Rod Myopathy (CRM) | RyR1 | AD or AR |
|
|
| Malignant Hyperthermia (MH) |
RyR1
CACNA1S STAC3 |
AD |
|
|
3.2. Triadin and Junctin (JNT), Two Integral Membrane Proteins That Regulate ECC
Triadin is encoded by the TRDN gene that generates, via alternative splicing, four different isoforms named according to their molecular weight: the skeletal muscle isoforms are Trisk 95 and Trisk 51, both displaying a triadic localization; the main cardiac isoform Trisk 32 (also known as CT1) and Trisk 49, which does not localize at the j-SR in skeletal muscle fibers [137,138]. Triadin isoforms share a common cytoplasmic N-terminal and transmembrane domain but differ in the length and composition of their luminal C-terminal segment [139]. Trisk 95 contains two cysteine residues in its luminal domain, C270 and C649, that enable self-multimerization and is targeted at triads by specific domains localized both in the cytoplasmic and the luminal regions [140,141]. Triadin acts as a functional regulator of ECC by interacting with RyRs [142,143], junctin [144,145], calsequestrin [141,146,147,148], and the histidine-rich Ca2+-binding protein (HRC) [149]. Triadin may also play a structural role in supporting triad architecture by interacting with the microtubule-binding protein Climp-63, also known as Cytoskeleton-associated protein 4 (CKAP4) [150]. Triadin knockout mice present both ECC alterations and abnormal triads [151,152]. In contrast to heart muscle, where mutations in triadin are associated with heart disease, no skeletal muscle disease has been associated with triadin so far.
Junctin is structurally similar to triadin and, like triadin, can bind calsequestrin and the ryanodine receptor [144,145,153,154,155]. Junctin knockout mice exhibited increased contractility and Ca2+-cycling parameters in cardiac muscle [156], but no significant changes were observed in skeletal muscle [157]. Similarly, acute junctin downregulation in cardiomyocytes resulted in a significant increase in contraction [158], while in skeletal myotubes, it resulted in a decrease in Ca2+ release, suggesting that junctin may display a different role in cardiac and skeletal muscles [159].
3.3. Calsequestrin- and Histidine-Rich Calcium-Binding Protein Store Ca2+ in the SR Lumen
In skeletal muscle, two isoforms of calsequestrin (CASQ) have been identified: CASQ1 and CASQ2. CASQ1 is expressed in fast- and slow-twitch skeletal muscle fibers, whereas CASQ2 is expressed in slow-twitch skeletal muscle fibers and in cardiac muscle [160]. The two isoforms present a high sequence homology and basically only differ in their acidic C-terminus [161]. CASQs are intra-luminal SR soluble proteins with high capacity and low-affinity Ca2+-binding properties. [162,163]. CASQ1 exists as either a monomer or polymers, whose assembly depends on the SR Ca2+ concentration [164,165]. CASQ1 is formed by three negative thioredoxin-like domains surrounding a hydrophilic core [166]. Park and collaborators proposed a model where, when the ionic strength in the lumen of the SR increases, these domains fold together, and CASQ monomers start to polymerize. Following a further increase in Ca2+ concentration, front-to-front and back-to-back interactions take place. This finally results in the assembly of large ribbon-like structures, where negatively charged cavities may accommodate additional Ca2+ [167]. CASQ1 polymerization is a reversible process depending on the luminal Ca2+ concentration and the presence of CASQ-binding proteins or post-translational modification [168,169,170,171]. During sustained SR Ca2+ depletion, it was shown that almost all CASQ is present in its depolymerized status. In this condition, progressive closure of the RyRs was reported, with a consequent reduction in Ca2+ release aimed to prevent dangerous levels of SR Ca2+ depletion. Accordingly, it has been proposed that CASQ1 depolymerization may represent the intracellular switch that induces RyR1 closing [172].
In skeletal muscle, CASQs interact with RyR1, triadin, and junctin, forming a quaternary Ca2+ release complex [144,145,148,173,174]. However, how these interactions regulate ECC still remains to be defined, since different studies report that CASQs may either inhibit or activate or even have no effect on the opening of RyRs [169,174,175,176,177,178,179,180,181]. CASQ1 knockout mice show mild atrophy, narrower terminal cisternae, and proliferation of multilayered junctions, as well as mitochondria alterations. They exhibit significantly reduced SR Ca2+ content and marked SR Ca2+ depletion during high-frequency stimulation [182,183]. Recently, it has been demonstrated that CASQ1 acts as a regulator of SOCE by binding to STIM1. In particular, CASQ1 overexpression was found to inhibit SOCE by reducing STIM1 clustering and STIM1/OraiI interaction [184,185]. Indeed, in CASQ1 knockout mice, SOCE is constitutively active, resulting in enhanced Ca2+ entry that may compensate for the reduced total SR Ca2+ content [186,187]. Mutations in the CASQ1 gene have been identified in patients affected by a rare vacuolar myopathy [188] and in patients with tubular Aggregate Myopathy, TAM [189,190].
HRC is far less abundant than CASQs, suggesting that it works as a secondary calcium-binding protein [191,192]. It is composed of a conserved N-terminal domain, a C-terminal cysteine-rich region, and a central histidine-rich region, which allow HRC to bind Ca2+ and to interact with triadin [193,194]. Under physiological Ca2+ levels, HRC has a pentameric structure, but when Ca2+-binding sites are saturated due to increased SR Ca2+ concentration, the protein shifts to a monomeric form [195]. Although considered as a secondary calcium-binding protein, HRC appears to play a non-secondary role in regulating Ca2+ homeostasis. Indeed, HRC may function as a negative regulator of RyRs and, in cardiac muscle, it was suggested to interact with SERCA pumps, acting as a negative regulator of Ca2+ re-uptake [149,192,196,197].
4. SERCA Pumps at the l-SR Are Responsible for Ca2+ Re-Uptake from the Sarcoplasm
The l-SR is mainly involved in Ca2+ re-uptake, although some ligand-gated Ca2+ channels, such as inositol 1,4,5-trisphosphate receptors (InsP3R), which release Ca2+ to regulate signaling pathways apart from myofibrils contraction, are also present here [198]. SERCA pumps play the most important role in SR Ca2+ replenishing. SERCAs belong to the P-type ATPases family, a large superfamily of integral membrane proteins that pump ions and lipids across cellular membranes using the energy derived from ATP hydrolysis. The P-type ATPases family is composed of five subfamilies, defined as P1–P5. SERCAs belong to the P2A-ATPases subfamily and are specific for Ca2+ transport, while P4-ATPases act as phospholipid flippases to maintain lipid asymmetry in a variety of membranes [199,200]. SERCA pumps can transport two Ca2+ ions for each hydrolyzed ATP. In vertebrates, SERCA pumps are encoded by three different genes, named ATP2A1-3. More than 10 protein variants are generated through alternative splicing occurring in the 3′-end of the main transcripts [201,202].
ATP2A1 encodes two major skeletal muscle proteins: SERCA1a and SERCA1b, highly expressed in fast-twitch skeletal muscle fibers. SERCA1b is expressed during neonatal stages and in regenerating muscles, while it is replaced by SERCA1a in adult skeletal muscles [203,204,205]. Four isoforms of SERCA2 (a–d) are generated by alternative splicing of the ATP2A2 gene. SERCA 2a is expressed in cardiac muscle, slow-twitch skeletal muscle fibers, and smooth muscle cells. SERCA2b is reported to be ubiquitously expressed, and it represents the main SERCA isoform in the brain [206,207,208]. mRNA of SERCA2c was found in epithelial, mesenchymal, and hematopoietic cell lines, as well as in primary human monocytes [206] and cardiac muscle, while SERCA 2d has been identified in skeletal muscle, although its role has not yet been fully elucidated [209]. ATP2A3 codes for six isoforms (SERCA3a–f), mostly identified in non-muscle cells, although SERCA3a, 3d, and 3f were also detected in cardiac muscle cells [209,210]. The primary structure of the different SERCA isoforms is highly conserved. Nevertheless, the SERCA variants differ in their enzymatic properties. For instance, SERCA1a displays a maximal activity that is two-fold higher than that of SERCA2a [211,212], and differences in Ca2+ affinities have also been reported between SERCA2b and SERCA3 [212]. SERCA activity is finely regulated by numerous small proteins. The first identified regulator of SERCA is Phospholamban (PLN), which exerts an inhibitory effect on SERCA in cardiac muscle [213,214,215] and, to a lesser extent, in smooth muscle and slow-twitch oxidative skeletal muscle fibers where, however, its functional role appears to be marginal [216,217,218]. PLN exists in two forms, monomeric and pentameric. The first is considered the “active” inhibitory form, while the pentameric state is considered the “inactive” storage form of PLN. PLN regulation occurs through one-to-one interaction with SERCA [218]. At high cytosolic Ca2+ concentrations, the inhibitory effect of PLN is mitigated due to Ca2+/calmodulin kinase (CaMKII)- and/or Protein Kinase A (PKA)-dependent phosphorylation [219,220,221]. A second inhibitor of SERCA in skeletal muscle and atria is represented by sarcolipin (SLN). In contrast to PLN, which inhibits SERCA activity by reducing its affinity for Ca2+, SLN uncouples ATP hydrolysis from Ca2+ transport [222], thus promoting futile cycling, resulting in ATP consumption and increased thermogenesis. Indeed, SLN knockout mice do not show any visible phenotype but show compromised thermogenic capacity when exposed to cold [222,223]. This suggests that the role of SLN may be related to muscle functions linked to body metabolism and heat production rather than contraction [223]. Similar to the j-SR, the l-SR also contains a Ca2+ binding protein, namely, Sarcalumenin (SAR) [224], which colocalizes and interacts with SERCA2 [225]. Furthermore, SAR has been shown to play an essential role in maintaining cardiac function under biomechanical stresses and endurance exercise training [226,227]. Mutations in SERCA1 are associated with Brody disease, an extremely rare autosomal recessive myopathy (with a prevalence estimated at around 1 in 10 million births) characterized by muscle stiffness, myalgia and muscle cramps, aggravation of symptoms upon exposure to cold temperatures, and, in a smaller percentage of patients, muscle weakness. It is basically characterized by exercise-induced impairment of fast-twitch skeletal muscle relaxation due to diminished SERCA1 activity. This increased levels of cytosolic Ca2+ due to SERCA1 impairment directly contribute to muscle stiffness [228,229,230].
Although SERCAs represent the key enzymes that contribute to sarcoplasmic Ca2+ clearance following muscle contraction, a trans-sarcolemmal flux toward the extracellular environment also occurs in striated muscles. This is mediated by Plasma Membrane Calcium ATPases (PMCAs) and Na+/Ca2+ exchangers localized in the sarcolemma. Similar to SERCAs, PMCAs also belong to the P-type ATPase superfamily. They can transport one Ca2+ for each hydrolyzed ATP and are directly activated by interaction with Ca2+-calmodulin, which enhances PMCAs affinity for Ca2+ [231]. PMCAs are encoded by four different genes (ATP2B1-4) that, by alternative splicing, generate multiple isoforms [232]. Among these, PMCA3 and PMCA1 were found to be expressed in skeletal muscle, and PMCA1 was specifically localized on the T-tubule membrane [233,234]. NCX catalyzes the exchange of three Na+ ions and one Ca2+ ion across the sarcolemma in a high capacity and low Ca2+ affinity manner. Depending on the electrochemical gradient across the plasma membrane, NCX can either extrude intracellular Ca2+ (according to the so-called forward mechanism) or take up extracellular Ca2+ (a reverse mechanism) [235]. In mammals, three genes, NCX1, NCX2, and NCX3, are present; NCX3 is expressed in brain and adult skeletal muscle [236,237], while NCX1 is expressed in developing skeletal muscles and in cardiac muscle [237]. The role of NCX has been mainly defined in cardiac muscle cells, where it represents the major mechanism of Ca2+ extrusion from the cytoplasm during diastole [238,239].
5. Ca2+ Entry Units (CEU): Novel SR/Plasma Membrane Contact Sites to Refill Intracellular Ca2+ Stores
Entry of Ca2+ from the extracellular environment in response to a decrease in the endoplasmic reticulum Ca2+ content is a ubiquitous mechanism present in all cell types. The key elements of this pathway, named Store Operated Calcium Entry (SOCE), are STIM1 and Orai1 proteins [240]. STIM1 is a transmembrane protein localized in the ER membrane that acts as a sensor of luminal Ca2+ content; when intracellular Ca2+ stores are full, it is in a Ca2+-bound monomeric conformation. Depletion of the intracellular Ca2+ store results in dissociation of Ca2+ from STIM1, which forms aggregates that relocate at ER–plasma membrane contact sites, where they interact with and activate the Orai1 channels on the plasma membrane, allowing Ca2+ entry from the extracellular environment and refill of intracellular stores. Ca2+ entry from the extracellular environment through the SOCE mechanism is also present in skeletal muscle [241]. More recently, the requirement of extracellular Ca2+ to refill the SR Ca2+ stores, after intense, prolonged activity, has been shown to result in the assembly of additional intracellular junctions between the SR and the plasma membrane, providing additional space for STIM1 and Orai1 interaction. These newly identified sites, named Ca2+ Entry Units (CEU), support increased Ca2+ entry via Orai1, and their activity appears to contribute to improving fatigue resistance under continuous muscle activity [242,243,244] (Figure 4). Mutations in Orai1 and STIM1 have been identified in patients with tubular aggregate myopathy (TAM), a rare genetic disease characterized by accumulation, in type-2 fibers, of highly ordered membrane tubules containing SR proteins, such as CASQ1, SERCA, triadin, RyR1, and STIM1 [245,246,247,248,249]. More recently, mutations in CASQ1 have also been identified in TAM patients [189,190].
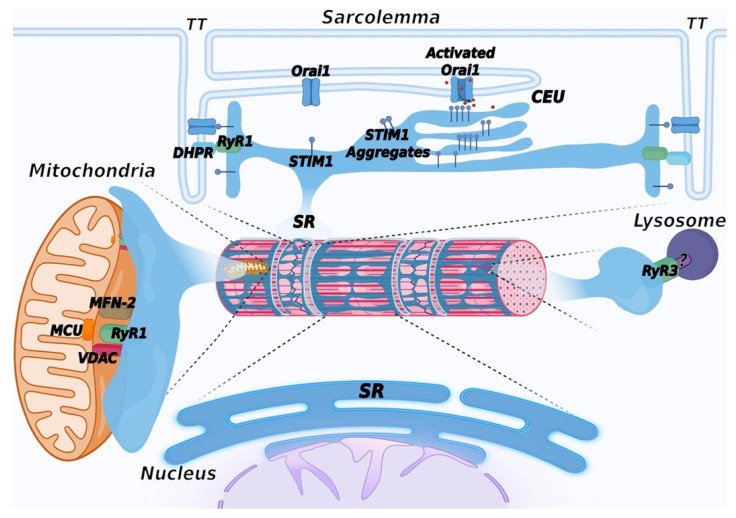
Figure 4.
Schematic representation of membrane contact sites contributed by the SR. In addition to triads, the SR contributes to the formation of additional membrane contact sites in muscle cells. Depletion of intracellular Ca2+ stores in the SR induces activation of SOCE, mediated by a physical interaction between STIM1, a Ca2+ sensor of the SR and Orai1, a Ca2+ channel located in TT, allowing entry of Ca2+ from the extracellular space. Repetitive stimulation of muscle contraction was found to promote SR and TT remodeling to form additional sites of interaction between STIM1 and Orai1, called calcium entry units (CEU). These are formed by stacks of flat cisternae of the SR that make contact with elongated tubules extending from TT. In striated muscles, mitochondria are mostly positioned adjacent to triads. Association with the SR is mediated by the voltage-dependent anion channel 1 (VDAC) and RyR. Mitofusin-2 (MFN-2) also contributes to tethering SR and mitochondria. These contact sites are aligned with the inner mitochondrial membrane, where the mitochondrial Ca2+ uniporter (MCU) is located. Lysosome–SR nanojunctions mediated by RyR3 channels have been first described in pulmonary arterial myocytes; the RyR3 interactor present on the lysosomal membrane is not known and is therefore indicated with a question mark (?). More recently, these junctions have also been observed in cardiac muscle, while no data are currently available concerning skeletal muscle. The outer nuclear membrane is continuous with the membrane of the SR, allowing the arrangement of a continuous Ca2+ storage system between the SR and the nuclear envelope. The inner nuclear membrane forms invaginations that enter the nuclear matrix to support intra-nuclear Ca2+ signaling. Created with BioRender.com, accessed on 14 March 2022.
6. Mitochondria-Associated Membranes (MAM)
Interaction between the ER/SR and mitochondria occurs at specific membrane contact sites, generally defined as Mitochondria Associated Membranes (MAMs), populated by several proteins and protein complexes that act as tethers between the SR and the outer mitochondrial membrane [250]. In striated muscles, mitochondria are mostly positioned in the inter-myofibrillar spaces adjacent to triads, although contacts between mitochondria and the longitudinal SR have also been observed [251,252,253,254]. Association with the SR is mediated by the voltage-dependent anion channel 1 (VDAC) [255]. In cardiac muscle, it has been proposed that VDAC interacts with RyR2 [256]. Mitofusin-2 (MFN-2) also contributes to tethering SR and mitochondria in skeletal muscles [257,258]. Interestingly, during mouse skeletal muscle development, mitochondria redistribute from a longitudinal arrangement towards a triad-associated distribution, with an increase in the frequency of contacts until the first four months after birth [258]. These contacts are aligned with domains of the inner mitochondrial membrane enriched in the mitochondrial Ca2+ uniporter (MCU), a low-affinity Ca2+ transport protein (Figure 4). It has been shown that RyR1 opening results in the formation of local micro-domains, where Ca2+ concentration may be up to 10 folds higher than in the rest of the cytoplasm [259]. This high Ca2+ concentration allows the transfer of Ca2+ to the mitochondrial matrix through the uniporter [260]. From a functional point of view, increases in mitochondrial Ca2+ concentration regulate ATP production by enhancing the synthesis of NADH and FADH2 [261,262], thus linking ATP production to muscle demand. Accordingly, slow-twitch fibers in skeletal muscle that are highly oxidative have the highest mitochondrial content compared to fast-twitch fibers; as a result, MCU deletion results in impairment in muscle force and in oxidative metabolism [263]. In cardiac muscle, the MCU-mediated Ca2+ influx was shown to play an important role during the fight-or-flight response following catecholaminergic stimulation [264].
Alterations of SR/mitochondria interaction or of mitochondrial morphology and function in striated muscles are often observed in human muscle diseases or mouse models of muscle diseases characterized by alterations in Ca2+ signaling [251,265,266,267]. However, the exact mechanisms linking alterations of MAM, mitochondrial function, and muscle diseases are still to be defined.
7. The SR and the ER: Two Faces of a Single Organelle
This review started with the statement that the SR is a specialized form of ER of muscle cells dedicated to Ca2+ handling. One may therefore ask what happened to the ER in striated muscle cells. In eukaryotic cells, the ER is dedicated to Ca2+ storage, protein synthesis and folding, and lipid and sterol synthesis. Although ER-related functions are clearly present in striated muscles, the distribution of the ER within the SR membranes is less evident. The recognized idea is that the SR and the ER represent a continuous membrane system made of different specialized subdomains [268]. Compartmentalization of ER and SR markers was demonstrated in skeletal muscle fibers, where ER-specific proteins were detected at the perinuclear region and in two distinct rough ER sub-compartments: the first, in correspondence of the I band that does not apparently contain ER exit sites, and the second, near the Z disk, which sustains export activity towards the Golgi [106,269]. Similarly, in cardiac muscle cells, protein synthesis was found to occur not only around the nuclei but also within ER/SR membranes surrounding the sarcomeres; interestingly, translation of some SR membrane proteins has been proposed to occur starting from pools of mRNAs that are transported from the perinuclear region towards ER/SR protein-synthesis sites through a microtubule-dependent system [270]. A less-clear distinction can be observed in smooth muscle, where ER and SR markers are basically overlapping [271].
Additional Contact Sites Contributed by the ER/SR
In eukaryotic cells, the ER is engaged in a complex network of interactions with many intracellular components, such as the nuclear envelope (NE), the plasma membrane, mitochondria, the Golgi complex, peroxisomes, endosomes, lysosomes, and lipid droplets [272]. All these interactions have been less characterized in striated muscle cells. Lysosome–SR nanojunctions were described in pulmonary arterial myocytes (Figure 4). These junctions were identified between clusters of lysosomes and perinuclear regions of the SR rich in RyR3 and are believed to provide an intracellular structure involved in the regulation of specific Ca2+-signaling events. These junctions were identified based on the observation that nicotinic acid adenine dinucleotide phosphate (NAADP), a Ca2+-mobilizing second messenger, induced the release of Ca2+ from a lysosome-related Ca2+ store, and this was amplified by the generation of intracellular Ca2+ waves supported by RyR3 opening, following a CICR mechanism [273]. According to these studies, lysosomes act as a trigger zone through which Ca2+ is released with a quantal pattern; upon reaching an activation threshold, RyR3 channels open, and calcium oscillations are generated. More recently, nanojunctions between lysosomes, SR, and mitochondria have been described at the ultrastructural level in rabbit ventricular myocytes. At difference with junctions observed in pulmonary arterial myocytes, in cardiac myocytes, lysosomes are distributed with a periodicity consistent with the length of a sarcomere, allowing the association with specific areas of the SR [274]. The role of NAADP-mediated signaling in cardiac muscle cells is suggested to be involved in intracellular Ca2+ regulation, mainly following beta-adrenergic stimulation [275].
In striated muscle cells, stacks of ER/SR cisternae distribute in the perinuclear regions and connect with the nuclear envelope (NE) with functions correlated with protein synthesis and the regulation of intra-nuclear Ca2+ signaling (Figure 4). The NE is formed by two phospholipid bilayers that form the inner nuclear membrane and the outer nuclear membrane, outlining an internal space called the perinuclear cisternae. The outer nuclear membrane is continuous with the membrane of the rough ER; this connection results in the formation of common luminal space with the ER, which, in muscle cells, allows the arrangement of a continuous Ca2+ storage system between the SR and the NE [276]. EM analysis showed the existence of membrane invaginations of the inner nuclear membrane that enter the nuclear matrix to support different functions, including intranuclear Ca2+ signaling [277]. InsP3R, RyRs, and SERCA have been described at these sites; in striated muscle cells, Ca2+ release is apparently mainly regulated by RyRs [278,279], while both InsP3R and RyRs are present in smooth muscle cells [280]. In cardiac myocytes, STIM1 and Orai1 have also been described in the inner nuclear membrane, suggesting that the intranuclear Ca2+ signaling may be more complex than previously expected [281].
An additional issue is represented by the SR/nuclei juxtaposition within the overall skeletal muscle architecture. In skeletal muscle, nuclei are positioned at the fiber periphery between myofibrils and the sarcolemma, in proximity to T-tubules. The positioning of nuclei is regulated by different mechanisms, including the formation of a specific microtubule network, and is critical for muscle function; indeed, improper myonuclear localization is observed in muscle diseases, such as centronuclear myopathy and muscular dystrophies [282]. The molecular link supporting nuclear juxtaposition with the SR is currently not known. A recent work performed in skeletal muscles of Drosophila melanogaster suggests that this interaction may be mediated by a three-partner connection where amphiphysin 2 on TT binds, at the same time, Ma2/d, a protein present on both TT and SR membranes, and, at the nuclear membrane, Msp30, a protein previously described to mediate connections between the NE and the Z disks [283,284,285].
8. Concluding Remarks
This review provides an updated view of the structural, molecular, and functional organization of the SR in skeletal muscle, highlighting some of the specialized domains whereby this organelle supports activation of muscle contraction. The historical knowledge that the terminal cisternae of the SR engage with the T-tubules in forming the triad, which represents the first identified membrane contact site, is now expanded by evidence that in skeletal muscle, the SR participates in the assembly of at least two more types of membrane-contact sites.
In one case, the identification of CEUs, dynamic membrane contact sites consisting of stacks of SR cisternae and the extension of T tubules, structurally and functionally different from triads, provides evidence of how skeletal muscle fibers can sustain refilling of intracellular Ca2+ stores under conditions of prolonged activity. The second contact site, MAM, between the SR and mitochondria, has the functional role of synchronizing the mitochondrial energy production rate with the metabolic demand of muscle fibers. Altogether, these new findings are changing the traditional view of the SR as a static organelle by depicting an unexpected dynamic ability to extend its complex structure to support muscle function.
At the same time, there are several points that remain to be clarified. A major open question relates to the precise positioning of triads at the boundary of the I and A bands of the sarcomere. This highly precise pattern, evident in EM micrographs for about a half-century, is waiting for the identification of the proteins that tether triads to the sarcomere with such a regularly repetitive precision. Concerning CEU, we need to identify the molecules that support the dynamic assembly and disassembly of CEU and how Orai1 and STIM1 are recruited at these sites. Mitochondria, like many other organelles, in skeletal muscle, are localized with a well-defined pattern with respect to triads and sarcolemma, a pattern that differs in slow- and fast-twitch fibers. Additionally, in this case, the molecules that anchor mitochondria to these domains of muscle fibers remain to be identified.
In conclusion, we can anticipate that the above list is, certainly, still missing additional SR domains that are yet to be identified; these might probably involve activities related to the regulation of Ca2+ homeostasis, but more likely, additional functions necessary to keep skeletal muscle fibers healthy and fully functional.
Author Contributions
D.O.A., S.B. and E.M.R. wrote the manuscript draft, D.R., E.P. and V.S. supervised, revised, and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Work reported in this article was funded by Fondazione Telethon (grant number GGP19291).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Franzini-Armstrong C. Miology. 3rd ed. Mc Graw-Hill; New York, NY, USA: 2004. The membrane systems of muscle cells; pp. 232–256. [Google Scholar]
- 2.Franzini-Armstrong C. The relationship between form and function throughout the history of excitation-contraction coupling. J. Gen. Physiol. 2018;150:189–210. doi: 10.1085/jgp.201711889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnato P., Barone V., Giacomello E., Rossi D., Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J. Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kontrogianni-Konstantopoulos A., Jones E.M., van Rossum D.B., Bloch R.J. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell. 2003;14:1138–1148. doi: 10.1091/mbc.e02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kontrogianni-Konstantopoulos A., Catino D.H., Strong J.C., Sutter S., Borisov A.B., Pumplin D.W., Russell M.K., Bloch R.J. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- 6.Giacomello E., Sorrentino V. Localization of ank1.5 in the sarcoplasmic reticulum precedes that of SERCA and RyR: Relationship with the organization of obscurin in developing sarcomeres. Histochem. Cell Biol. 2009;131:371–382. doi: 10.1007/s00418-008-0534-4. [DOI] [PubMed] [Google Scholar]
- 7.Lange S., Ouyang K., Meyer G., Cui L., Cheng H., Lieber R.L., Chen J. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J. Cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giacomello E., Quarta M., Paolini C., Squecco R., Fusco P., Toniolo L., Blaauw B., Formoso L., Rossi D., Birkenmeier C., et al. Deletion of small ankyrin 1 (sAnk1) isoforms results in structural and functional alterations in aging skeletal muscle fibers. Am. J. Physiol.-Cell Physiol. 2015;308:C123–C138. doi: 10.1152/ajpcell.00090.2014. [DOI] [PubMed] [Google Scholar]
- 9.Armani A., Galli S., Giacomello E., Bagnato P., Barone V., Rossi D., Sorrentino V. Molecular interactions with obscurin are involved in the localization of muscle-specific small ankyrin1 isoforms to subcompartments of the sarcoplasmic reticulum. Exp. Cell Res. 2006;312:3546–3558. doi: 10.1016/j.yexcr.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Randazzo D., Giacomello E., Lorenzini S., Rossi D., Pierantozzi E., Blaauw B., Reggiani C., Lange S., Peter A.K., Chen J., et al. Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J. Cell Biol. 2013;200:523–536. doi: 10.1083/jcb.201205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randazzo D., Blaauw B., Paolini C., Pierantozzi E., Spinozzi S., Lange S., Chen J., Protasi F., Reggiani C., Sorrentino V. Exercise-induced alterations and loss of sarcomeric M-line organization in the diaphragm muscle of obscurin knockout mice. Am. J. Physiol.-Cell Physiol. 2017;312:C16–C28. doi: 10.1152/ajpcell.00098.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dulhunty A.F. Excitation-contraction coupling from the 1950s into the new millennium. Clin. Exp. Pharmacol. Physiol. 2006;33:763–772. doi: 10.1111/j.1440-1681.2006.04441.x. [DOI] [PubMed] [Google Scholar]
- 13.Rios E. Calcium-induced release of calcium in muscle: 50 years of work and the emerging consensus. J. Gen. Physiol. 2018;150:521–537. doi: 10.1085/jgp.201711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takekura H., Flucher B.E., Franzini-Armstrong C. Sequential docking, molecular differentiation, and positioning of T-Tubule/SR junctions in developing mouse skeletal muscle. Dev. Biol. 2001;239:204–214. doi: 10.1006/dbio.2001.0437. [DOI] [PubMed] [Google Scholar]
- 15.Flucher B.E., Takekura H., Franzini-Armstrong C. Development of the excitation-contraction coupling apparatus in skeletal muscle: Association of sarcoplasmic reticulum and transverse tubules with myofibrils. Dev. Biol. 1993;160:135–147. doi: 10.1006/dbio.1993.1292. [DOI] [PubMed] [Google Scholar]
- 16.Luff A.R., Atwood H.L. Changes in the sarcoplasmic reticulum and transverse tubular system of fast and slow skeletal muscles of the mouse during postnatal development. J. Cell Biol. 1971;51:369–383. doi: 10.1083/jcb.51.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall T.E., Martel N., Ariotti N., Xiong Z., Lo H.P., Ferguson C., Rae J., Lim Y.W., Parton R.G. In Vivo cell biological screening identifies an endocytic capture mechanism for T-tubule formation. Nat. Commun. 2020;11:3711. doi: 10.1038/s41467-020-17486-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan S.H., Arnold W., Jorgensen A.O. Biogenesis of transverse tubules and triads: Immunolocalization of the 1,4-dihydropyridine receptor, TS28, and the ryanodine receptor in rabbit skeletal muscle developing in situ. J. Cell Biol. 1991;112:289–301. doi: 10.1083/jcb.112.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franzini-Armstrong C., Pincon-Raymond M., Rieger F. Muscle fibers from dysgenic mouse In Vivo lack a surface component of peripheral couplings. Dev. Biol. 1991;146:364–376. doi: 10.1016/0012-1606(91)90238-X. [DOI] [PubMed] [Google Scholar]
- 20.Powell J.A., Petherbridge L., Flucher B.E. Formation of triads without the dihydropyridine receptor alpha subunits in cell lines from dysgenic skeletal muscle. J. Cell Biol. 1996;134:375–387. doi: 10.1083/jcb.134.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felder E., Protasi F., Hirsch R., Franzini-Armstrong C., Allen P.D. Morphology and molecular composition of sarcoplasmic reticulum surface junctions in the absence of DHPR and RyR in mouse skeletal muscle. Biophys. J. 2002;82:3144–3149. doi: 10.1016/S0006-3495(02)75656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Qusairi L., Laporte J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet. Muscle. 2011;1:26. doi: 10.1186/2044-5040-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barone V., Randazzo D., Del Re V., Sorrentino V., Rossi D. Organization of junctional sarcoplasmic reticulum proteins in skeletal muscle fibers. J. Muscle Res. Cell Motil. 2015;36:501–515. doi: 10.1007/s10974-015-9421-5. [DOI] [PubMed] [Google Scholar]
- 24.Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J., Evans P.R., McMahon H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 25.Lee E., Marcucci M., Daniell L., Pypaert M., Weisz O.A., Ochoa G.C., Farsad K., Wenk M.R., De Camilli P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 26.Wechsler-Reya R.J., Elliott K.J., Prendergast G.C. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell Biol. 1998;18:566–575. doi: 10.1128/MCB.18.1.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowling B.S., Prokic I., Tasfaout H., Rabai A., Humbert F., Rinaldi B., Nicot A.S., Kretz C., Friant S., Roux A., et al. Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J. Clin. Investig. 2017;127:4477–4487. doi: 10.1172/JCI90542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujise K., Okubo M., Abe T., Yamada H., Nishino I., Noguchi S., Takei K., Takeda T. Mutant BIN1-Dynamin 2 complexes dysregulate membrane remodeling in the pathogenesis of centronuclear myopathy. J. Biol. Chem. 2020;296:100077. doi: 10.1074/jbc.RA120.015184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva-Rojas R., Nattarayan V., Jaque-Fernandez F., Gomez-Oca R., Menuet A., Reiss D., Goret M., Messaddeq N., Lionello V.M., Kretz C., et al. Mice with muscle-specific deletion of Bin1 recapitulate centronuclear myopathy and acute downregulation of dynamin 2 improves their phenotypes. Mol. Ther. 2022;30:868–880. doi: 10.1016/j.ymthe.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toussaint A., Cowling B.S., Hnia K., Mohr M., Oldfors A., Schwab Y., Yis U., Maisonobe T., Stojkovic T., Wallgren-Pettersson C., et al. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta Neuropathol. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 31.Jungbluth H., Gautel M. Pathogenic Mechanisms in Centronuclear Myopathies. Front. Aging Neurosci. 2014;6:339. doi: 10.3389/fnagi.2014.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlahovich N., Kee A.J., Van der Poel C., Kettle E., Hernandez-Deviez D., Lucas C., Lynch G.S., Parton R.G., Gunning P.W., Hardeman E.C. Cytoskeletal Tropomyosin Tm5NM1 Is Required for Normal Excitation–Contraction Coupling in Skeletal Muscle. Mol. Biol. Cell. 2009;20:400–409. doi: 10.1091/mbc.e08-06-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falcone S., Roman W., Hnia K., Gache V., Didier N., Lainé J., Auradé F., Marty I., Nishino I., Charlet-Berguerand N., et al. N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol. Med. 2014;6:1455–1475. doi: 10.15252/emmm.201404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z., Scherer P.E., Okamoto T., Song K., Chu C., Kohtz D.S., Nishimoto I., Lodish H.F., Lisanti M.P. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J. Biol. Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 35.Parton R.G., Way M., Zorzi N., Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J. Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galbiati F., Engelman J.A., Volonte D., Zhang X.L., Minetti C., Li M., Hou H., Jr., Kneitz B., Edelmann W., Lisanti M.P. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophinglycoprotein complex, and t-tubule abnormalities. J. Biol. Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- 37.Corrotte M., Almeida P.E., Tam C., Castro-Gomes T., Fernandes M.C., Millis B.A., Cortez M., Miller H., Song W., Maugel T.K., et al. Caveolae internalization repairs wounded cells and muscle fibers. eLife. 2013;2:e00926. doi: 10.7554/eLife.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minetti C., Sotgia F., Bruno C., Scartezzini P., Broda P., Bado M., Masetti E., Mazzocco M., Egeo A., Donati M.A., et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat. Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 39.Betz R.C., Schoser B.G., Kasper D., Ricker K., Ramirez A., Stein V., Torbergsen T., Lee Y.A., Nothen M.M., Wienker T.F., et al. Mutations in CAV3 cause mechanical hyperirritability of skeletal muscle in rippling muscle disease. Nat. Genet. 2001;28:218–219. doi: 10.1038/90050. [DOI] [PubMed] [Google Scholar]
- 40.Woodman S.E., Sotgia F., Galbiati F., Minetti C., Lisanti M.P. Mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases. Neurology. 2004;62:538–543. doi: 10.1212/WNL.62.4.538. [DOI] [PubMed] [Google Scholar]
- 41.Kovtun O., Tillu V.A., Ariotti N., Parton R.G., Collins B.M. Cavin family proteins and the assembly of caveolae. J. Cell Sci. 2015;128:1269–1278. doi: 10.1242/jcs.167866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastiani M., Liu L., Hill M.M., Jedrychowski M.P., Nixon S.J., Lo H.P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M.R., et al. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 2009;185:1259–1273. doi: 10.1083/jcb.200903053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H., Lin P., De G., Choi K.H., Takeshima H., Weisleder N., Ma J. Polymerase transcriptase release factor (PTRF) anchors MG53 protein to cell injury site for initiation of membrane repair. J. Biol. Chem. 2011;286:12820–12824. doi: 10.1074/jbc.C111.221440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández-Deviez D.J., Martin S., Laval S.H., Lo H.P., Cooper S.T., North K.N., Bushby K., Parton R.G. Aberrant dysferlin trafficking in cells lacking caveolin or expressing dystrophy mutants of caveolin-3. Hum. Mol. Genet. 2006;15:129–142. doi: 10.1093/hmg/ddi434. [DOI] [PubMed] [Google Scholar]
- 45.Matsuda C., Hayashi Y.K., Ogawa M., Aoki M., Murayama K., Nishino I., Nonaka I., Arahata K., Brown R.H., Jr. The sarcolemmal proteins dysferlin and caveolin-3 interact in skeletal muscle. Hum. Mol. Genet. 2001;10:1761–1766. doi: 10.1093/hmg/10.17.1761. [DOI] [PubMed] [Google Scholar]
- 46.Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J.K., Lin P., Thornton A., Zhao X., et al. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 2009;11:56–64. doi: 10.1038/ncb1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson L.V., Davison K., Moss J.A., Young C., Cullen M.J., Walsh J., Johnson M.A., Bashir R., Britton S., Keers S., et al. Dysferlin is a plasma membrane protein and is expressed early in human development. Hum. Mol. Genet. 1999;8:855–861. doi: 10.1093/hmg/8.5.855. [DOI] [PubMed] [Google Scholar]
- 48.Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., Campbell K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 49.Klinge L., Harris J., Sewry C., Charlton R., Anderson L., Laval S., Chiu Y.H., Hornsey M., Straub V., Barresi R., et al. Dysferlin associates with the developing T-tubule system in rodent and human skeletal muscle. Muscle Nerve. 2010;41:166–173. doi: 10.1002/mus.21166. [DOI] [PubMed] [Google Scholar]
- 50.Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 51.Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B., et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 52.Illa I., Serrano-Munuera C., Gallardo E., Lasa A., Rojas-García R., Palmer J., Gallano P., Baiget M., Matsuda C., Brown R.H. Distal anterior compartment myopathy: A dysferlin mutation causing a new muscular dystrophy phenotype. Ann. Neurol. 2001;49:130–134. doi: 10.1002/1531-8249(200101)49:1<130::AID-ANA22>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 53.Roostalu U., Strähle U. In Vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell. 2012;22:515–529. doi: 10.1016/j.devcel.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Di Zanni E., Gradogna A., Scholz-Starke J., Boccaccio A. Gain of function of TMEM16E/ANO5 scrambling activity caused by a mutation associated with gnathodiaphyseal dysplasia. Cell Mol. Life Sci. 2018;75:1657–1670. doi: 10.1007/s00018-017-2704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolduc V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., et al. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 2010;86:213–221. doi: 10.1016/j.ajhg.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foltz S.J., Cui Y.Y., Choo H.J., Hartzell H.C. ANO5 ensures trafficking of annexins in wounded myofibers. J. Cell Biol. 2021;220:e202007059. doi: 10.1083/jcb.202007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chandra G., Sreetama S.C., Mázala D.A.G., Charton K., VanderMeulen J.H., Richard I., Jaiswal J.K. Endoplasmic reticulum maintains ion homeostasis required for plasma membrane repair. J. Cell Biol. 2021;220:e202006035. doi: 10.1083/jcb.202006035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Wang L., Yue H., Whitson B.A., Haggard E., Xu X., Ma J. MG53, A Tissue Repair Protein with Broad Applications in Regenerative Medicine. Cells. 2021;10:122. doi: 10.3390/cells10010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McNeil P.L., Kirchhausen T. An emergency response team for membrane repair. Nat. Rev. Mol. Cell Biol. 2005;6:499–505. doi: 10.1038/nrm1665. [DOI] [PubMed] [Google Scholar]
- 60.Cai C., Weisleder N., Ko J.K., Komazaki S., Sunada Y., Nishi M., Takeshima H., Ma J. Membrane repair defects in muscular dystrophy are linked to altered interaction between MG53, caveolin-3, and dysferlin. J. Biol. Chem. 2009;284:15894–15902. doi: 10.1074/jbc.M109.009589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshima H., Shimuta M., Komazaki S., Ohmi K., Nishi M., Iino M., Miyata A., Kangawa K. Mitsugumin29, a novel synaptophysin family member from the triad junction in skeletal muscle. Biochem. J. 1998;331:317–322. doi: 10.1042/bj3310317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimuta M., Komazaki S., Nishi M., Iino M., Nakagawara K.-I., Takeshima H. Structure and expression of mitsugumin29 gene. FEBS Lett. 1998;431:263–267. doi: 10.1016/S0014-5793(98)00770-4. [DOI] [PubMed] [Google Scholar]
- 63.Komazaki S., Nishi M., Kangawa K., Takeshima H. Immunolocalization of mitsugumin29 in developing skeletal muscle and effects of the protein expressed in amphibian embryonic cells. Dev. Dyn. 1999;215:87–95. doi: 10.1002/(SICI)1097-0177(199906)215:2<87::AID-DVDY1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 64.Nishi M., Komazaki S., Kurebayashi N., Ogawa Y., Noda T., Iino M., Takeshima H. Abnormal features in skeletal muscle from mice lacking mitsugumin29. J. Cell Biol. 1999;147:1473–1480. doi: 10.1083/jcb.147.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komazaki S., Nishi M., Takeshima H., Nakamura H. Abnormal formation of sarcoplasmic reticulum networks and triads during early development of skeletal muscle cells in mitsugumin 29-deficient mice. Dev. Growth Differ. 2001;43:717–723. doi: 10.1046/j.1440-169X.2001.00609.x. [DOI] [PubMed] [Google Scholar]
- 66.Woo J.S., Hwang J.H., Huang M., Ahn M.K., Cho C.H., Ma J., Lee E.H. Interaction between mitsugumin 29 and TRPC3 participates in regulating Ca2+ transients in skeletal muscle. Biochem. Biophys. Res. Commun. 2015;464:133–139. doi: 10.1016/j.bbrc.2015.06.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raess M.A., Friant S., Cowling B.S., Laporte J. WANTED-Dead or alive: Myotubularins, a large disease-associated protein family. Adv. Biol. Regul. 2017;63:49–58. doi: 10.1016/j.jbior.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A.H., Allard B., Mandel J.L., et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl. Acad. Sci. USA. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hnia K., Vaccari I., Bolino A., Laporte J. Myotubularin phosphoinositide phosphatases: Cellular functions and disease pathophysiology. Trends Mol. Med. 2012;18:317–327. doi: 10.1016/j.molmed.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Amoasii L., Bertazzi D.L., Tronchère H., Hnia K., Chicanne G., Rinaldi B., Cowling B.S., Ferry A., Klaholz B., Payrastre B., et al. Phosphatase-dead myotubularin ameliorates X-linked centronuclear myopathy phenotypes in mice. PLoS Genet. 2012;8:e1002965. doi: 10.1371/journal.pgen.1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C.R., Durand M., Kretz C., Danos O., Douar A.M., et al. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in p.plasma membrane homeostasis. Hum. Mol. Genet. 2008;17:2132–2143. doi: 10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amoasii L., Hnia K., Chicanne G., Brech A., Cowling B.S., Muller M.M., Schwab Y., Koebel P., Ferry A., Payrastre B., et al. Myotubularin and PtdIns3P remodel the sarcoplasmic reticulum in muscle In Vivo. J. Cell Sci. 2013;126:1806–1819. doi: 10.1242/jcs.118505. [DOI] [PubMed] [Google Scholar]
- 73.Royer B., Hnia K., Gavriilidis C., Tronchère H., Tosch V., Laporte J. The myotubularin-amphiphysin 2 complex in membrane tubulation and centronuclear myopathies. EMBO Rep. 2013;14:907–915. doi: 10.1038/embor.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agrawal P.B., Pierson C.R., Joshi M., Liu X., Ravenscroft G., Moghadaszadeh B., Talabere T., Viola M., Swanson L.C., Haliloğlu G., et al. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am. J. Hum. Genet. 2014;95:218–226. doi: 10.1016/j.ajhg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo S., Rosen S.M., Li Q., Agrawal P.B. Striated Preferentially Expressed Protein Kinase (SPEG) in Muscle Development, Function, and Disease. Int. J. Mol. Sci. 2021;22:5732. doi: 10.3390/ijms22115732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garbino A., van Oort R.J., Dixit S.S., Landstrom A.P., Ackerman M.J., Wehrens X.H.T. Molecular evolution of the junctophilin gene family. Physiol. Genomics. 2009;37:175–186. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 78.Nishi H., Sakagami S., Komazaki H., Kondo H., Takeshima H. Coexpression of junctophilin type 3 and type 4 in brain. Mol. Brain Res. 2003;118:102–110. doi: 10.1016/S0169-328X(03)00341-3. [DOI] [PubMed] [Google Scholar]
- 79.Landstrom A.P., Beavers D.L., Wehrens X.H.T. The junctophilin family of proteins: From bench to bedside. Trends Mol. Med. 2014;20:353–362. doi: 10.1016/j.molmed.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takeshima H., Hoshijima M., Song L.S. Ca²⁺ microdomains organized by junctophilins. Cell Calcium. 2015;58:349–356. doi: 10.1016/j.ceca.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rossi D., Scarcella A.M., Liguori E., Lorenzini S., Pierantozzi E., Kutchukian C., Jacquemond V., Messa M., De Camilli P., Sorrentino V. Molecular determinants of homo- and heteromeric interactions of Junctophilin-1 at triads in adult skeletal muscle fibers. Proc. Natl. Acad. Sci. USA. 2019;116:15716–15724. doi: 10.1073/pnas.1820980116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perni S., Lavorato M., Beam K.G. De novo reconstitution reveals the proteins required for skeletal muscle voltage-induced Ca2+ release. Proc. Natl. Acad. Sci. USA. 2017;114:13822–13827. doi: 10.1073/pnas.1716461115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perni S., Beam K.G. Voltage-induced Ca2+ release is supported by junctophilins 1, 2 and 3, and not by junctophilin 4. J. Gen. Physiol. 2022;154:e2021ecc22. doi: 10.1085/jgp.2021ecc22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirata Y., Brotto M., Weisleder N., Chu Y., Lin P., Zhao X., Thornton A., Komazaki S., Takeshima H., Ma J., et al. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys. J. 2006;90:4418–4427. doi: 10.1529/biophysj.105.076570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Golini L., Chouabe C., Berthier C., Cusimano V., Fornaro M., Bonvallet R., Formoso L., Giacomello E., Jacquemond V., Sorrentino V. Junctophilin 1 and 2 proteins interact with the L-type Ca2+ channel dihydropyridine receptors (DHPRs) in skeletal muscle. J. Biol. Chem. 2011;286:43717–43725. doi: 10.1074/jbc.M111.292755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Oort R.J., Garbino A., Wang W., Dixit S.S., Landstrom A.P., Gaur N., De Almeida A.C., Skapura D.G., Rudy Y., Burns A.R., et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phimister A.J., Lango J., Lee E.H., Ernst-Russell M.A., Takeshima H., Ma J., Allen P.D., Pessah I.N. Conformation-dependent stability of junctophilin 1 (JP1) and ryanodine receptor type 1 (RyR1) channel complex is mediated by their hyper-reactive thiols. J. Biol. Chem. 2007;282:8667–8677. doi: 10.1074/jbc.M609936200. [DOI] [PubMed] [Google Scholar]
- 88.Nakada T., Kashihara T., Komatsu M., Kojima K., Takeshita T., Yamada M. Physical interaction of junctophilin and the CaV1.1 C terminus is crucial for skeletal muscle contraction. Proc. Natl. Acad. Sci. USA. 2018;115:4507–4512. doi: 10.1073/pnas.1716649115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gross P., Johnson J., Romero C.M., Eaton D.M., Poulet C., Sanchez-Alonso J., Lucarelli C., Ross J., Gibb A.A., Garbincius J.F., et al. Interaction of the Joining Region in Junctophilin-2 With the L-Type Ca2+ Channel Is Pivotal for Cardiac Dyad Assembly and Intracellular Ca2+ Dynamics. Circ. Res. 2021;128:92–114. doi: 10.1161/CIRCRESAHA.119.315715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poulet C., Sanchez-Alonso J., Swiatlowska P., Mouy F., Lucarelli C., Alvarez-Laviada A., Gross P., Terracciano C., Houser S., Gorelik J. Junctophilin-2 tethers T-tubules and recruits functional L-type calcium channels to lipid rafts in adult cardiomyocytes. Cardiovasc. Res. 2021;117:149–161. doi: 10.1093/cvr/cvaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woo J.S., Hwang J.H., Ko J.K., Kim D.H., Ma J., Lee E.H. Glutamate at position 227 of junctophilin-2 is involved in binding to TRPC3. Mol. Cell Biochem. 2009;328:25–32. doi: 10.1007/s11010-009-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H., Ding X., Lopez J.R., Takeshima H., Ma J., Allen P.D., Eltit J.M. Impaired Orai1-mediated resting Ca2+ entry reduces the cytosolic [Ca2+] and sarcoplasmic reticulum Ca2+ loading in quiescent junctophilin 1 knock-out myotubes. J. Biol. Chem. 2010;285:39171–39179. doi: 10.1074/jbc.M110.149690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ito K., Komazaki S., Sasamoto K., Yoshida M., Nishi M., Kitamura K., Takeshima H. Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J. Cell Biol. 2001;154:1059–1067. doi: 10.1083/jcb.200105040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Komazaki S., Ito K., Takeshima H., Nakamura H. Deficiency of triad formation in developing skeletal muscle cells lacking junctophilin type 1. FEBS Lett. 2002;524:225–229. doi: 10.1016/S0014-5793(02)03042-9. [DOI] [PubMed] [Google Scholar]
- 95.Beavers D.L., Landstrom A.P., Chiang D.Y., Wehrens X.H.T. Emerging roles of junctophilin-2 in the heart and implications for cardiac diseases. Cardiovasc. Res. 2014;103:198–205. doi: 10.1093/cvr/cvu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bennett H.J., Davenport J.B., Collins R.F., Trafford A.W., Pinali C., Kitmitto A. Human junctophilin-2 undergoes a structural rearrangement upon binding PtdIns(3,4,5)P3 and the S101R mutation identified in hypertrophic cardiomyopathy obviates this response. Biochem. J. 2013;456:205–217. doi: 10.1042/BJ20130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quick A.P., Landstrom A.P., Wang Q., Beavers D.L., Reynolds J.O., Barreto-Torres G., Tran V., Showell J., Philippen L.E., Morris S.A., et al. Novel junctophilin-2 mutation A405S is associated with basal septal hypertrophy and diastolic dysfunction. JACC Basic Transl. Sci. 2017;2:56–67. doi: 10.1016/j.jacbts.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vanninen S.U.M., Leivo K., Seppälä E.H., Aalto-Setälä K., Pitkänen O., Suursalmi P., Annala A.P., Anttila I., Alastalo T.P., Myllykangas S., et al. Heterozygous junctophilin-2 (JPH2) p.(Thr161Lys) is a monogenic cause for HCM with heart failure. PLoS ONE. 2018;13:e0203422. doi: 10.1371/journal.pone.0203422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lyu Y., Verma V.K., Lee Y., Taleb I., Badolia R., Shankar T.S., Kyriakopoulos C.P., Selzman C.H., Caine W., Alharethi R., et al. Remodeling of t-system and proteins underlying excitation-contraction coupling in aging versus failing human heart. NPJ Aging Mech. Dis. 2021;7:16. doi: 10.1038/s41514-021-00066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Minamisawa S., Oshikawa J., Takeshima H., Hoshijima M., Wang Y., Chien K.R., Ishikawa Y., Matsuoka R. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem. Biophys. Res. Commun. 2004;325:852–856. doi: 10.1016/j.bbrc.2004.10.107. [DOI] [PubMed] [Google Scholar]
- 101.Murphy R.M., Dutka T.L., Horvath D., Bell J.R., Delbridge L.M., Lamb G.D. Ca2+-dependent proteolysis of junctophilin-1 and junctophilin-2 in skeletal and cardiac muscle. J. Physiol. 2013;591:719–729. doi: 10.1113/jphysiol.2012.243279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo A., Wang Y., Chen B., Wang Y., Yuan J., Zhang L., Hall D., Wu J., Shi Y., Zhu Q., et al. E-C coupling structural protein junctophilin-2 encodes a stress-adaptive transcription regulator. Science. 2018;362:eaan3303. doi: 10.1126/science.aan3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo A., Hall D., Zhang C., Peng T., Miller J.D., Kutschke W., Grueter C.E., Johnson F.L., Lin R.Z., Song L.S. Molecular Determinants of Calpain-dependent Cleavage of Junctophilin-2 Protein in Cardiomyocytes. J. Biol. Chem. 2015;290:17946–17955. doi: 10.1074/jbc.M115.652396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lahiri S.K., Quick A.P., Samson-Couterie B., Hulsurkar M., Elzenaar I., van Oort R.J., Wehrens X.H.T. Nuclear localization of a novel calpain-2 mediated junctophilin-2 C-terminal cleavage peptide promotes cardiomyocyte remodeling. Basic Res. Cardiol. 2020;115:49. doi: 10.1007/s00395-020-0807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo A., Fang W., Gibson S. Sequence determinants of human junctophilin-2 protein nuclear localization and phase separation. Biochem. Biophys. Res. Commun. 2021;563:79–84. doi: 10.1016/j.bbrc.2021.05.078. [DOI] [PubMed] [Google Scholar]
- 106.Rossi D., Barone V., Giacomello E., Cusimano V., Sorrentino V. The sarcoplasmic reticulum: An organized patchwork of specialized domains. Traffic. 2008;9:1044–1049. doi: 10.1111/j.1600-0854.2008.00717.x. [DOI] [PubMed] [Google Scholar]
- 107.Rossi D., Simeoni I., Micheli M., Bootman M., Lipp P., Allen P.D., Sorrentino V. RyR1 and RyR3 isoforms provide distinct intracellular Ca2+ signals in HEK 293 cells. J. Cell Sci. 2002;115:2497–2504. doi: 10.1242/jcs.115.12.2497. [DOI] [PubMed] [Google Scholar]
- 108.Rossi D., Sorrentino V. Molecular genetics of ryanodine receptors Ca2+-release channels. Cell Calcium. 2002;32:307–319. doi: 10.1016/S0143416002001987. [DOI] [PubMed] [Google Scholar]
- 109.Van Petegem F. Ryanodine Receptors: Structure and Function. J. Biol. Chem. 2012;287:31624–31632. doi: 10.1074/jbc.R112.349068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Des Georges A., Clarke O.B., Zalk R., Yuan Q., Condon K.J., Grassucci R.A., Frank J. Structural basis for gating and activation of RyR1. Cell. 2016;167:145–157. doi: 10.1016/j.cell.2016.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Caprara G.A., Morabito C., Perni S., Navarra R., Guarnieri S., Mariggiò M.A. Evidence for Altered Ca2+ Handling in Growth Associated Protein 43-Knockout Skeletal Muscle. Front. Physiol. 2016;7:493. doi: 10.3389/fphys.2016.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gong D., Chi X., Wei J., Zhou G., Huang G., Zhang L., Wang R., Lei J., Chen S.R.W., Yan N. Modulation of cardiac ryanodine receptor 2 by calmodulin. Nature. 2019;572:347–351. doi: 10.1038/s41586-019-1377-y. [DOI] [PubMed] [Google Scholar]
- 113.Prosser B.L., Hernández-Ochoa E.O., and Schneider M.F. S100A1 and calmodulin regulation of ryanodine receptor in striated muscle. Cell Calcium. 2011;50:323–331. doi: 10.1016/j.ceca.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brillantes A.B., Ondrias K., Scott A., Kobrinsky E., Ondriašová E., Moschella M.C., Jayaraman T., Landers M., Ehrlich B.E., Marks A.R. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 115.Bultynck G., De Smet P., Rossi D., Callewaert G., Missiaen L., Sorrentino V., De Smedt H., Parys J.B. Characterization and mapping of the 12 kDa FK506-binding protein (FKBP12)-binding site on different isoforms of the ryanodine receptor and of the inositol 1,4,5-trisphosphate receptor. Biochem. J. 2001;354:413–422. doi: 10.1042/bj3540413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pessah I.N., Cherednichenko G., Lein P.J. Minding the calcium store: Ryanodine receptor activation as a convergent mechanism of PCB toxicity. Pharmacol. Ther. 2010;125:260–285. doi: 10.1016/j.pharmthera.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gurrola G.B., Capes E.M., Zamudio F.Z., Possani L.D., Valdivia H.H. Imperatoxin A, a Cell-Penetrating Peptide from Scorpion Venom, as a Probe of Ca-Release Channels/Ryanodine Receptors. Pharmaceuticals. 2010;3:1093–1107. doi: 10.3390/ph3041093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santulli G., Lewis D., des Georges A., Marks A.R., Frank J. Ryanodine Receptor Structure and Function in Health and Disease. Subcell Biochem. 2018;87:329–352. doi: 10.1007/978-981-10-7757-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hopkins P.M. Malignant hyperthermia: Pharmacology of triggering. Br. J. Anaesth. 2011;107:48–56. doi: 10.1093/bja/aer132. [DOI] [PubMed] [Google Scholar]
- 120.Hu H., Wang Z., Wei R., Fan G., Wang Q., Zhang K., Yin C.C. The molecular architecture of dihydropyrindine receptor/L-type Ca2+ channel complex. Sci. Rep. 2015;5:8370. doi: 10.1038/srep08370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Flucher B.E. Skeletal muscle CaV1.1 channelopathies. Pflügers Arch.-Eur. J. Physiol. 2020;472:739–754. doi: 10.1007/s00424-020-02368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Petegem F., Clark K.A., Chatelain F.C., Minor D.L. Structure of a complex between a voltage-gated calcium channel b-subunit and an a-subunit domain. Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Buraei Z., Yang J. The β Subunit of Voltage-Gated Ca2+ Channels. Physiol. Rev. 2010;90:1461–1506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gregg R.G., Messing A., Strube C., Beurg M., Moss R., Behan M., Sukhareva M., Haynes S., Powell J.A., Coronado R., et al. Absence of the beta subunit (cchb1) of the skeletal muscle dihydropyridine receptor alters expression of the alpha 1 subunit and eliminates excitation-contraction coupling. Proc. Natl. Acad. Sci. USA. 1996;93:13961–13966. doi: 10.1073/pnas.93.24.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tanabe T., Beam K.G., Powell J.A., Numa S. Restoration of excitation-contraction coupling and slow calcium current in dysgenic muscle by dihydropyridine receptor complementary DNA. Nature. 1988;336:134–139. doi: 10.1038/336134a0. [DOI] [PubMed] [Google Scholar]
- 126.Bannister R.A. Bridging the myoplasmic gap II: More recent advances in skeletal muscle excitation-contraction coupling. J. Exp. Biol. 2016;219:175–182. doi: 10.1242/jeb.124123. [DOI] [PubMed] [Google Scholar]
- 127.Nakai J., Dirksen R.T., Nguyen H.T., Pessah I.N., Beam K.G., Allen P.D. Enhanced dihydropyridine receptor channel activity in the presence of ryanodine receptor. Nature. 1996;380:72–75. doi: 10.1038/380072a0. [DOI] [PubMed] [Google Scholar]
- 128.Horstick E.J., Linsley J.W., Dowling J.J., Hauser M.A., McDonald K.K., Ashley-Koch A., Saint-Amant L., Satish A., Cui W.W., Zhou W., et al. Stac3 is a component of the excitation–contraction coupling machinery and mutated in Native American myopathy. Nat. Commun. 2013;4:1952. doi: 10.1038/ncomms2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Polster A., Nelson B.R., Olson E.N., Beam K.G. Stac3 has a direct role in skeletal muscle-type excitation-contraction coupling that is disrupted by a myopathy-causing mutation. Proc. Natl. Acad. Sci. USA. 2016;113:10986–10991. doi: 10.1073/pnas.1612441113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Flucher B.E., Campiglio M. STAC proteins: The missing link in skeletal muscle EC coupling and new regulators of calcium channel function. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:1101–1110. doi: 10.1016/j.bbamcr.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 131.Rufenach B., Van Petegem F. Structure and function of STAC proteins: Calcium channel modulators and critical components of muscle excitation-contraction coupling. J. Biol. Chem. 2021;297:100874. doi: 10.1016/j.jbc.2021.100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nelson B.R., Wu F., Liu Y., Anderson D.M., McAnally J., Lin W., Cannon S.C., Bassel-Duby R., Olson E.N. Skeletal muscle-specific T-tubule protein STAC3 mediates voltage-induced Ca2+ release and contractility. Proc. Natl. Acad. Sci. USA. 2013;110:11881–11886. doi: 10.1073/pnas.1310571110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ogasawara M., Nishino I. A review of core myopathy: Central core disease, multiminicore disease, dusty core disease, and core-rod myopathy. Neuromuscul. Disord. 2021;31:968–977. doi: 10.1016/j.nmd.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 134.Abath Neto O., Moreno C.A.M., Malfatti E., Donkervoort S., Böhm J., Guimarães J.B., Foley A.R., Mohassel P., Dastgir J., Bharucha-Goebel D.X., et al. Common and variable clinical, histological, and imaging findings of recessive RYR1-related centronuclear myopathy patients. Neuromuscul. Disord. 2017;27:975–985. doi: 10.1016/j.nmd.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 135.Lawal T.A., Todd J.J., Witherspoon J.W., Bönnemann C.G., Dowling J.J., Hamilton S.L., Meilleur K.G., Dirksen R.T. Ryanodine receptor 1-related disorders: An historical perspective and proposal for a unified nomenclature. Skelet. Muscle. 2020;10:32. doi: 10.1186/s13395-020-00243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.North K.N., Wang C.H., Clarke N., Jungbluth H., Vainzof M., Dowling J.J., Amburgey K., Quijano-Roy S., Beggs A.H., Sewry C., et al. International Standard of Care Committee for Congenital Myopathies. Approach to the diagnosis of congenital myopathies. Neuromuscul. Disord. 2014;24:97–116. doi: 10.1016/j.nmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kobayashi Y.M., Jones L.R. Identification of triadin 1 as the predominant triadin isoform expressed in mammalian myocardium. J. Biol. Chem. 1999;274:28660–28668. doi: 10.1074/jbc.274.40.28660. [DOI] [PubMed] [Google Scholar]
- 138.Vassilopoulos S., Thevenon D., Rezgui S.S., Brocard J., Chapel A., Lacampagne A., Lunardi J., Dewaard M., Marty I. Triadins are not triad-specific proteins: Two new skeletal muscle triadins possibly involved in the architecture of sarcoplasmic reticulum. J. Biol. Chem. 2005;280:28601–28609. doi: 10.1074/jbc.M501484200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marty I. Triadin regulation of the ryanodine receptor complex. J. Physiol. 2015;593:3261–3266. doi: 10.1113/jphysiol.2014.281147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fourest-Lieuvin A., Rendu J., Osseni A., Pernet-Gallay K., Rossi D., Oddoux S., Brocard J., Sorrentino V., Marty I., Fauré J. Role of triadin in the organization of reticulum membrane at the muscle triad. J. Cell Sci. 2012;125:3443–3453. doi: 10.1242/jcs.100958. [DOI] [PubMed] [Google Scholar]
- 141.Rossi D., Bencini C., Maritati M., Benini F., Lorenzini S., Pierantozzi E., Scarcella A.M., Paolini C., Protasi F., Sorrentino V. Distinct regions of triadin are required for targeting and retention at the junctional domain of the sarcoplasmic reticulum. Biochem. J. 2014;458:407–417. doi: 10.1042/BJ20130719. [DOI] [PubMed] [Google Scholar]
- 142.Caswell H., Motoike H.K., Fan H., Brandt N.R. Location of ryanodine receptor binding site on skeletal muscle triadin. Biochemistry. 1999;38:90–97. doi: 10.1021/bi981306+. [DOI] [PubMed] [Google Scholar]
- 143.Groh S., Marty I., Ottolia M., Prestipino G., Chapel A., Villaz M., Ronjat M. Functional interaction of the cytoplasmic domain of triadin with the skeletal ryanodine receptor. J. Biol. Chem. 1999;274:12278–12283. doi: 10.1074/jbc.274.18.12278. [DOI] [PubMed] [Google Scholar]
- 144.Zhang L., Kelley J., Schmeisser G., Kobayashi Y.M., Jones L.R. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 1997;272:23389–23397. doi: 10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 145.Rossi D., Lorenzini S., Pierantozzi E., Van Petegem F., Amadsun D.O., Sorrentino V. Multiple regions within junctin drive its interaction with calsequestrin-1 and its localization to triads in skeletal muscle. J. Cell Sci. 2022;135:jcs259185. doi: 10.1242/jcs.259185. [DOI] [PubMed] [Google Scholar]
- 146.Guo W., Campbell K.P. Association of triadin with the ryanodine receptor and calsequestrin in the lumen of the sarcoplasmic reticulum. J. Biol. Chem. 1995;270:9027–9030. doi: 10.1074/jbc.270.16.9027. [DOI] [PubMed] [Google Scholar]
- 147.Kobayashi Y.M., Alseikhan B.A., Jones L.R. Localization and characterization of the calsequestrin-binding domain of triadin 1. Evidence for a charged beta-strand in mediating the protein-protein interaction. J. Biol. Chem. 2000;275:17639–17646. doi: 10.1074/jbc.M002091200. [DOI] [PubMed] [Google Scholar]
- 148.Shin D.W., Ma J., Kim D.H. The asp-rich region at the carboxyl-terminus of calsequestrin binds to Ca2+ and interacts with triadin. FEBS Lett. 2000;486:178–182. doi: 10.1016/S0014-5793(00)02246-8. [DOI] [PubMed] [Google Scholar]
- 149.Lee H.G., Kang H., Kim D.H., Park W.J. Interaction of HRC (histidine-rich Ca2+-binding protein) and triadin in the lumen of sarcoplasmic reticulum. J. Biol. Chem. 2001;276:39533–39538. doi: 10.1074/jbc.M010664200. [DOI] [PubMed] [Google Scholar]
- 150.Osseni A., Sébastien M., Sarrault O., Baudet M., Couté Y., Fauré J., Fourest-Lieuvin A., Marty I. Triadin and CLIMP-63 form a link between triads and microtubules in muscle cells. J. Cell Sci. 2016;129:3744–3755. doi: 10.1242/jcs.188862. [DOI] [PubMed] [Google Scholar]
- 151.Shen X., Franzini-Armstrong C., Lopez J.R., Jones L.R., Kobayashi Y.M., Wang Y., Kerrick W.G.L., Caswell A.H., Potter J.D., Miller T., et al. Triadins modulate intracellular Ca2+ homeostasis but are not essential for excitation-contraction coupling in skeletal muscle. J. Biol. Chem. 2007;282:37864–37874. doi: 10.1074/jbc.M705702200. [DOI] [PubMed] [Google Scholar]
- 152.Oddoux S., Brocard J., Schweitzer A., Szentesi P., Giannesini B., Brocard J., Fauré J., Pernet-Gallay K., Bendahan D., Lunardi J., et al. Triadin deletion induces impaired skeletal muscle function. J. Biol. Chem. 2009;284:34918–34929. doi: 10.1074/jbc.M109.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dulhunty A., Wei L., Beard N. Junctin—The quiet achiever. J. Physiol. 2009;587:3135–3137. doi: 10.1113/jphysiol.2009.171959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dulhunty A.F., Wei-LaPierre L., Casarotto M.G., Beard N.A. Core skeletal muscle ryanodine receptor calcium release complex. Clin. Exp. Pharmacol. Physiol. 2017;44:3–12. doi: 10.1111/1440-1681.12676. [DOI] [PubMed] [Google Scholar]
- 155.Li L., Mirza S., Richardson S.J., Gallant E.M., Thekkedam C., Pace S.M., Zorzato F., Liu D., Beard N.A., Dulhunty A.F. A new cytoplasmic interaction between junctin and ryanodine receptor Ca2+ release channels. J. Cell Sci. 2015;128:951–963. doi: 10.1242/jcs.160689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yuan Q., Fan G.C., Dong M., Altschafl B., Diwan A., Ren X., Hahn H.H., Zhao W., Waggoner J.R., Jones L.R., et al. Sarcoplasmic reticulum calcium overloading in junctin deficiency enhances cardiac contractility but increases ventricular automaticity. Circulation. 2007;115:300–309. doi: 10.1161/CIRCULATIONAHA.106.654699. [DOI] [PubMed] [Google Scholar]
- 157.Boncompagni S., Thomas M., Lopez J.R., Allen P.D., Yuan Q., Kranias E.G., Franzini-Armstrong C., Perez C.F. Triadin/junctin double null mouse reveals a differential role for triadin and junctin in anchoring CASQ to the jSR and regulating Ca2+ homeostasis. PLoS ONE. 2012;7:e3962. doi: 10.1371/journal.pone.0039962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fan G.C., Yuan Q., Kranias E.G. Regulatory roles of junctin in sarcoplasmic reticulum calcium cycling and myocardial function. Trends Cardiovasc. Med. 2008;18:1–5. doi: 10.1016/j.tcm.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Y., Li X., Duan H., Fulton T.R., Eu J.P., Meissner G. Altered stored calcium release in skeletal myotubes deficient of triadin and junctin. Cell Calcium. 2009;45:29–37. doi: 10.1016/j.ceca.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Biral D., Volpe P., Damiani E., Margreth A. Coexistence of two calsequestrin isoforms in rabbit slow-twitch skeletal muscle fibers. FEBS Lett. 1992;299:175–178. doi: 10.1016/0014-5793(92)80241-8. [DOI] [PubMed] [Google Scholar]
- 161.Beard N.A., Laver D.R., Dulhunty A.F. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog. Biophys. Mol. Biol. 2004;85:33–69. doi: 10.1016/j.pbiomolbio.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 162.MacLennan D.H., Wong P.T. Isolation of a calcium-sequestering protein from sarcoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1971;68:1231–1235. doi: 10.1073/pnas.68.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Murphy R.M., Larkins N.T., Mollica J.P., Beard N.A., Lamb G.D. Calsequestrin content and SERCA determine normal and maximal Ca2+ storage levels in sarcoplasmic reticulum of fast- and slow-twitch fibres of rat. J. Physiol. 2009;587:443–460. doi: 10.1113/jphysiol.2008.163162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maguire P.B., Briggs F.N., Lennon N.J., Ohlendieck K. Oligomerization is an intrinsic property of calsequestrin in normal and transformed skeletal muscle. Biochem. Biophys. Res. Commun. 1997;240:721–727. doi: 10.1006/bbrc.1997.7729. [DOI] [PubMed] [Google Scholar]
- 165.Kim E., Youn B., Kemper L., Campbell C., Milting H., Varsanyi M., Kang C. Characterization of human cardiac calsequestrin and its deleterious mutants. J. Mol. Biol. 2007;373:1047–1057. doi: 10.1016/j.jmb.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 166.Wang S., Trumble W.R., Liao H., Wesson C.R., Dunker A.K., Kang C.H. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat. Struct. Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
- 167.Park H., Wu S., Dunker A.K., Kang C. Polymerization of calsequestrin. Implications for Ca2+ regulation. J. Biol. Chem. 2003;278:16176–16182. doi: 10.1074/jbc.M300120200. [DOI] [PubMed] [Google Scholar]
- 168.Cala S.E., Jones L.R. Phosphorylation of cardiac and skeletal muscle calsequestrin isoforms by casein kinase II. Demonstration of a cluster of unique rapidly phosphorylated sites in cardiac calsequestrin. J. Biol. Chem. 1991;266:391–398. doi: 10.1016/S0021-9258(18)52447-9. [DOI] [PubMed] [Google Scholar]
- 169.Beard N.A., Wei L., Cheung S.N., Kimura T., Varsányi M., Dulhunty A.F. Phosphorylation of skeletal muscle calsequestrin enhances its Ca2+ binding capacity and promotes its association with junctin. Cell Calcium. 2008;44:363–373. doi: 10.1016/j.ceca.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 170.Kumar A., Chakravarty H., Bal N.C., Balaraju T., Jena N., Misra G., Bal C., Pieroni E., Periasamy M., Sharon A. Identification of calcium binding sites on calsequestrin 1 and their implications for polymerization. Mol. Biosyst. 2013;9:1949–1957. doi: 10.1039/c3mb25588c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mitchell R.D., Simmerman H.K., Jones L.R. Ca2+ binding effects on protein conformation and protein interactions of canine cardiac calsequestrin. J. Biol. Chem. 1988;263:1376–1381. doi: 10.1016/S0021-9258(19)57313-6. [DOI] [PubMed] [Google Scholar]
- 172.Manno C., Figueroa L.C., Gillespie D., Fitts R., Kang C., Franzini-Armstrong C., Rios E. Calsequestrin depolymerizes when calcium is depleted in the sarcoplasmic reticulum of working muscle. Proc. Natl. Acad. Sci. USA. 2017;114:E638–E647. doi: 10.1073/pnas.1620265114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jones L.R., Zhang L., Sanborn K., Jorgensen A.O., Kelley J. Purification, primary structure, and immunological characterization of the 26-kDa calsequestrin binding protein (junctin) from cardiac junctional sarcoplasmic reticulum. J. Biol. Chem. 1995;270:30787–30796. doi: 10.1074/jbc.270.51.30787. [DOI] [PubMed] [Google Scholar]
- 174.Herzog A., Szegedi C., Jona I., Herberg F.W., Varsanyi M. Surface plasmon resonance studies prove the interaction of skeletal muscle sarcoplasmic reticular Ca2+ release channel/ryanodine receptor with calsequestrin. FEBS Lett. 2000;472:73–77. doi: 10.1016/S0014-5793(00)01431-9. [DOI] [PubMed] [Google Scholar]
- 175.Wei L., Varsányi M., Dulhunty A.F., Beard N.A. The conformation of calsequestrin determines its ability to regulate skeletal ryanodine receptors. Biophys. J. 2006;91:1288–1301. doi: 10.1529/biophysj.106.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kawasaki T., Kasai M. Regulation of calcium channel in sarcoplasmic reticulum by calsequestrin. Biochem. Biophys. Res. Commun. 1994;199:1120–1127. doi: 10.1006/bbrc.1994.1347. [DOI] [PubMed] [Google Scholar]
- 177.Beard N.A., Sakowska M.M., Dulhunty A.F., Laver D.R. Calsequestrin is an inhibitor of skeletal muscle ryanodine receptor calcium release channels. Biophys. J. 2002;82:310–320. doi: 10.1016/S0006-3495(02)75396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Beard N.A., Casarotto M.G., Wei L., Varsányi M., Laver D.R., Dulhunty A.F. Regulation of ryanodine receptors by calsequestrin: Effect of high luminal Ca2+ and phosphorylation. Biophys. J. 2005;88:3444–3454. doi: 10.1529/biophysj.104.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Qin J., Valle G., Nani A., Chen H., Ramos-Franco J., Nori A., Volpe P., Fill M. Ryanodine receptor luminal Ca2+ regulation: Swapping calsequestrin and channel isoforms. Biophys. J. 2009;97:1961–1970. doi: 10.1016/j.bpj.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Beard N.A., Dulhunty A.F. C-terminal residues of skeletal muscle calsequestrin are essential for calcium binding and for skeletal ryanodine receptor inhibition. Skelet. Muscle. 2015;22:5–6. doi: 10.1186/s13395-015-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen H., Valle G., Furlan S., Nani A., Gyorke S., Fill M., Volpe P. Mechanism of calsequestrin regulation of single cardiac ryanodine receptor in normal and pathological conditions. J. Gen. Physiol. 2013;142:127–136. doi: 10.1085/jgp.201311022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Canato M., Scorzeto M., Giacomello M., Protasi F., Reggiani C., Stienen G.J.M. Massive alterations of sarcoplasmic reticulum free calcium in skeletal muscle fibers lacking calsequestrin revealed by a genetically encoded probe. Proc. Natl. Acad. Sci. USA. 2010;107:22326–22331. doi: 10.1073/pnas.1009168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Protasi F., Paolini C., Canato M., Reggiani C., Quarta M. Lessons from calsequestrin-1 ablation In Vivo: Much more than a Ca2+ buffer after all. J. Muscle Res. Cell Motil. 2011;32:257–270. doi: 10.1007/s10974-011-9277-2. [DOI] [PubMed] [Google Scholar]
- 184.Shin D.W., Pan Z., Kim E.K., Lee J.M., Bhat M.B., Parness J., Kim D.H., Ma J. A retrograde signal from calsequestrin for the regulation of store-operated Ca2+ entry in skeletal muscle. J. Biol. Chem. 2003;278:3286–3292. doi: 10.1074/jbc.M209045200. [DOI] [PubMed] [Google Scholar]
- 185.Zhang L., Wang L., Li S., Xue J., Luo D. Calsequestrin-1 Regulates Store-Operated Ca2+ Entry by Inhibiting STIM1 Aggregation. Cell Physiol. Biochem. 2016;38:2183–2193. doi: 10.1159/000445574. [DOI] [PubMed] [Google Scholar]
- 186.Zhao X., Min C.K., Ko J., Parness J., Kim D.H., Weisleder N., Ma J. Increased store-operated Ca2+ entry in skeletal muscle with reduced calsequestrin-1 expression. Biophys. J. 2010;99:1556–1564. doi: 10.1016/j.bpj.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Michelucci A., Boncompagni S., Pietrangelo L., Takano T., Protasi F., Dirksen R.T. Pre-assembled Ca2+ entry units and constitutively active Ca2+ entry in skeletal muscle of calsequestrin-1 knockout mice. J. Gen. Physiol. 2020;152:e202012617. doi: 10.1085/jgp.202012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rossi D., Vezzani B., Galli L., Paolini C., Toniolo L., Pierantozzi E., Spinozzi S., Barone V., Pegoraro E., Bello L., et al. A mutation in the CASQ1 gene causes a vacuolar myopathy with accumulation of sarcoplasmic reticulum protein aggregates. Hum. Mutat. 2014;35:1163–1170. doi: 10.1002/humu.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.D’Adamo M.C., Sforna L., Visentin S., Grottesi A., Servettini L., Guglielmi L., Macchioni L., Saredi S., Curcio M., De Nuccio C., et al. A Calsequestrin-1 Mutation Associated with a Skeletal Muscle Disease Alters Sarcoplasmic Ca2+ Release. PLoS ONE. 2016;11:e0155516. doi: 10.1371/journal.pone.0155516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Barone V., Del Re V., Gamberucci A., Polverino V., Galli L., Rossi D., Costanzi E., Toniolo L., Berti G., Malandrini A., et al. Identification and characterization of three novel mutations in the CASQ1 gene in four patients with tubular aggregate myopathy. Hum. Mutat. 2017;38:1761–1773. doi: 10.1002/humu.23338. [DOI] [PubMed] [Google Scholar]
- 191.Böhm J., Lornage X., Chevessier F., Birck C., Zanotti S., Cudia P., Bulla M., Granger F., Bui M.T., Sartori M., et al. CASQ1 mutations impair calsequestrin polymerization and cause tubular aggregate myopathy. Acta Neuropathol. 2018;135:149–151. doi: 10.1007/s00401-017-1775-x. [DOI] [PubMed] [Google Scholar]
- 192.Arvanitis D.A., Vafiadaki E., Sanoudou D., Kranias E.G. Histidine-rich calcium binding protein: The new regulator of sarcoplasmic reticulum calcium cycling. J. Mol. Cell Cardiol. 2011;50:43–49. doi: 10.1016/j.yjmcc.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hofmann S.L., Goldstein J.L., Orth K., Moomaw C.R., Slaughter C.A., Brown M.S. Molecular cloning of a histidine-rich Ca2+-binding protein of sarcoplasmic reticulum that contains highly conserved repeated elements. J. Biol. Chem. 1989;264:18083–18090. doi: 10.1016/S0021-9258(19)84681-1. [DOI] [PubMed] [Google Scholar]
- 194.Sacchetto R., Damiani E., Turcato F., Nori A., Margreth A. Ca2+-dependent interaction of triadin with histidine-rich Ca2+-binding protein carboxyl-terminal region. Biochem. Biophys. Res. Commun. 2001;289:1125–1134. doi: 10.1006/bbrc.2001.6126. [DOI] [PubMed] [Google Scholar]
- 195.Suk J.Y., Kim Y.S., Park W.J. HRC (histidine-rich Ca2+ binding protein) resides in the lumen of sarcoplasmic reticulum as a multimer. Biochem. Biophys. Res. Commun. 1999;263:667–671. doi: 10.1006/bbrc.1999.1432. [DOI] [PubMed] [Google Scholar]
- 196.Orr I., Shoshan-Barmatz V. Modulation of the skeletal muscle ryanodine receptor by endogenous phosphorylation of 160/150-kDa proteins of the sarcoplasmic reticulum. Biochim. Biophys. Acta-Biomembr. 1996;1283:80–88. doi: 10.1016/0005-2736(96)00078-8. [DOI] [PubMed] [Google Scholar]
- 197.Park C.S., Chen S., Lee H., Cha H., Oh J.G., Hong S., Han P., Ginsburg K.S., Jin S., Park I., et al. Targeted ablation of the histidine-rich Ca2+-binding protein (HRC) gene is associated with abnormal SR Ca2+-cycling and severe pathology under pressure-overload stress. Basic Res. Cardiol. 2013;108:344. doi: 10.1007/s00395-013-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Salanova M., Priori G., Barone V., Intravaia E., Flucher B., Ciruela F., McIlhinney R.A.J., Parys J.B., Mikoshiba K., Sorrentino V. Homer proteins and InsP(3) receptors co-localise in the longitudinal sarcoplasmic reticulum of skeletal muscle fibres. Cell Calcium. 2002;32:193–200. doi: 10.1016/S0143416002001549. [DOI] [PubMed] [Google Scholar]
- 199.López-Marqués R.L., Gourdon P., Günther Pomorski T., Palmgren M. The transport mechanism of P4 ATPase lipid flippases. Biochem. J. 2020;477:3769–3790. doi: 10.1042/BCJ20200249. [DOI] [PubMed] [Google Scholar]
- 200.Thever M.D., Saier M.H., Jr. Bioinformatic characterization of p-type ATPases encoded within the fully sequenced genomes of 26 eukaryotes. J. Membr. Biol. 2009;229:115–130. doi: 10.1007/s00232-009-9176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Periasamy M., Kalyanasundaram A. SERCA pump isoforms: Their role in calcium transport and disease. Muscle Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- 202.Brini M., Calì T., Ottolini D., Carafoli E. Calcium pumps: Why so many? Compr. Physiol. 2012;2:1045–1060. doi: 10.1002/cphy.c110034. [DOI] [PubMed] [Google Scholar]
- 203.Brandl C.J., de Leon S., Martin D.R., MacLennan D.H. Adult forms of the Ca2+-ATPase of sarcoplasmic reticulum. Expression in developing skeletal muscle. J. Biol. Chem. 1987;262:3768–3774. doi: 10.1016/S0021-9258(18)61421-8. [DOI] [PubMed] [Google Scholar]
- 204.Zádor E., Vangheluwe P., Wuytack F. The expression of the neonatal sarcoplasmic reticulum Ca2+ pump (SERCA1b) hints to a role in muscle growth and development. Cell Calcium. 2007;41:379–388. doi: 10.1016/j.ceca.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 205.Kósa M., Brinyiczki K., van Damme P., Goemans N., Hancsák K., Mendler L., Zádor E. The neonatal sarcoplasmic reticulum Ca2+-ATPase gives a clue to development and pathology in human muscles. J. Muscle Res. Cell Motil. 2015;36:195–203. doi: 10.1007/s10974-014-9403-z. [DOI] [PubMed] [Google Scholar]
- 206.Gélébart P., Martin V., Enouf J., Papp B. Identification of a new SERCA2 splice variant regulated during monocytic differentiation. Biochem. Biophys. Res. Commun. 2003;303:676–684. doi: 10.1016/S0006-291X(03)00405-4. [DOI] [PubMed] [Google Scholar]
- 207.Vandecaetsbeek I., Vangheluwe P., Raeymaekers L., Wuytack F., Vanoevelen J. The Ca2+ pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011;1:a004184. doi: 10.1101/cshperspect.a004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wu K.D., Lee W.S., Wey J., Bungard D., Lytton J. Localization and quantification of endoplasmic reticulum Ca2+-ATPase isoform transcripts. Am. J. Physiol. 1995;269:C775–C784. doi: 10.1152/ajpcell.1995.269.3.C775. [DOI] [PubMed] [Google Scholar]
- 209.Dally S., Corvazier E., Bredoux R., Bobe R., Enouf J. Multiple and diverse coexpression, location, and regulation of additional SERCA2 and SERCA3 isoforms in nonfailing and failing human heart. J. Mol. Cell Cardiol. 2010;48:633–644. doi: 10.1016/j.yjmcc.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 210.Wuytack F., Dode L., Baba-Aissa F., Raeymaekers L. The SERCA3-type of organellar Ca2+ pumps. Biosci. Rep. 1995;15:299–306. doi: 10.1007/BF01788362. [DOI] [PubMed] [Google Scholar]
- 211.Sumbilla C., Cavagna M., Zhong L., Ma H., Lewis D., Farrance I., Inesi G. Comparison of SERCA1 and SERCA2a expressed in COS-1 cells and cardiac myocytes. Am. J. Physiol. 1999;277:H2381–H2391. doi: 10.1152/ajpheart.1999.277.6.H2381. [DOI] [PubMed] [Google Scholar]
- 212.Lytton J., Westlin M., Burk S.E., Shull G.E., MacLennan D.H. Functional comparisons between isoforms of the sarcoplasmic or endoplasmic reticulum family of calcium pumps. J. Biol. Chem. 1992;267:14483–14489. doi: 10.1016/S0021-9258(19)49738-X. [DOI] [PubMed] [Google Scholar]
- 213.Kirchberber M.A., Tada M., Katz A.M. Phospholamban: A regulatory protein of the cardiac sarcoplasmic reticulum. Recent Adv. Stud. Card. Struct. Metab. 1975;5:103–115. [PubMed] [Google Scholar]
- 214.Kranias E.G., Hajjar R.J. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ. Res. 2012;110:1646–1660. doi: 10.1161/CIRCRESAHA.111.259754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Slack J.P., Grupp I.L., Luo W., Kranias E.G. Phospholamban ablation enhances relaxation in the murine soleus. Am. J. Physiol. 1997;273:C1–C6. doi: 10.1152/ajpcell.1997.273.1.C1. [DOI] [PubMed] [Google Scholar]
- 216.Gamu D., Juracic E.S., Fajardo V.A., Rietze B.A., Tran K., Bombardier E., Tupling A.R. Phospholamban deficiency does not alter skeletal muscle SERCA pumping efficiency or predispose mice to diet-induced obesity. Am. J. Physiol.-Endocrinol. Metab. 2019;316:E432–E442. doi: 10.1152/ajpendo.00288.2018. [DOI] [PubMed] [Google Scholar]
- 217.Periasamy M., Bhupathy P., Babu G.J. Regulation of sarcoplasmic reticulum Ca2+-ATPase pump expression and its relevance to cardiac muscle physiology and pathology. Cardiovasc. Res. 2008;77:265–273. doi: 10.1093/cvr/cvm056. [DOI] [PubMed] [Google Scholar]
- 218.Bhupathy P., Babu G.J., Periasamy M. Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+-ATPase. J. Mol. Cell Cardiol. 2007;42:903–911. doi: 10.1016/j.yjmcc.2007.03.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.MacLennan D.H., Kranias E.G. Phospholamban: A crucial regulator of cardiac contractility. Na.t Rev. Mol. Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 220.Wittmann T., Lohse M.J., Schmitt J.P. Phospholamban pentamers attenuate PKA-dependent phosphorylation of monomers. J. Mol. Cell Cardiol. 2015;80:90–97. doi: 10.1016/j.yjmcc.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 221.Shaikh S.A., Sahoo S.K., Periasamy M. Phospholamban and sarcolipin: Are they functionally redundant or distinct regulators of the Sarco(Endo)Plasmic Reticulum Calcium ATPase? J. Mol. Cell Cardiol. 2016;91:81–91. doi: 10.1016/j.yjmcc.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bal N.C., Maurya S.K., Sopariwala D.H., Sahoo S.K., Gupta S.C., Shaikh S.A., Pant M., Rowland L.A., Bombardier E., Goonasekera S.A., et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat. Med. 2012;18:1575–1579. doi: 10.1038/nm.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Maurya S.K., Bal N.C., Sopariwala D.H., Pant M., Rowland L.A., Shaikh S.A., Periasamy M. Sarcolipin Is a Key Determinant of the Basal Metabolic Rate, and Its Overexpression Enhances Energy Expenditure and Resistance against Diet-induced Obesity. J. Biol. Chem. 2015;290:10840–10849. doi: 10.1074/jbc.M115.636878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Leberer E., Timms B.G., Campbell K.P., MacLennan D.H. Purification, calcium binding properties, and ultrastructural localization of the 53,000- and 160,000 (sarcalumenin)-dalton glycoproteins of the sarcoplasmic reticulum. J. Biol. Chem. 1990;265:10118–10124. doi: 10.1016/S0021-9258(19)38787-3. [DOI] [PubMed] [Google Scholar]
- 225.Yoshida M., Minamisawa S., Shimura M., Komazaki S., Kume H., Zhang M., Matsumura K., Nishi M., Saito M., Saeki Y., et al. Impaired Ca2+ store functions in skeletal and cardiac muscle cells from sarcalumenin-deficient mice. J. Biol. Chem. 2005;280:3500–3506. doi: 10.1074/jbc.M406618200. [DOI] [PubMed] [Google Scholar]
- 226.Shimura M., Minamisawa S., Takeshima H., Jiao Q., Bai Y., Umemura S., Ishikawa Y. Sarcalumenin alleviates stress-induced cardiac dysfunction by improving Ca2+ handling of the sarcoplasmic reticulum. Cardiovasc. Res. 2008;77:362–370. doi: 10.1093/cvr/cvm019. [DOI] [PubMed] [Google Scholar]
- 227.Jiao Q., Bai Y., Akaike T., Takeshima H., Ishikawa Y., Minamisawa S. Sarcalumenin is essential for maintaining cardiac function during endurance exercise training. Am. J. Physiol.-Heart Circ. Physiol. 2009;297:H576–H582. doi: 10.1152/ajpheart.00946.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.MacLennan D.H. Ca2+ signalling and muscle disease. Eur. J. Biochem. 2000;267:5291–5297. doi: 10.1046/j.1432-1327.2000.01566.x. [DOI] [PubMed] [Google Scholar]
- 229.Molenaar J.P., Verhoeven J.I., Rodenburg R.J., Kamsteeg E.J., Erasmus C.E., Vicart S., Behin A., Bassez G., Magot A., Péréon Y., et al. Clinical, morphological and genetic characterization of Brody disease: An international study of 40 patients. Brain. 2020;143:452–466. doi: 10.1093/brain/awz410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Close M., Perni S., Franzini-Armstrong C., Cundall D. Highly extensible skeletal muscle in snakes. J. Exp. Biol. 2014;217:2445–2448. doi: 10.1242/jeb.097634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tibbits G.F., Thomas M.J. Ca2+ transport across the plasma membrane of striated muscle. Med. Sci. Sports Exerc. 1989;1:399–405. doi: 10.1249/00005768-198908000-00010. [DOI] [PubMed] [Google Scholar]
- 232.Krebs J. The plethora of PMCA isoforms: Alternative splicing and differential expression. Biochim. Biophys. Acta-Mol. Cell Res. 2015;1853:2018–2024. doi: 10.1016/j.bbamcr.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 233.Greeb J., Shull G.E. Molecular cloning of a third isoform of the calmodulin-sensitive plasma membrane Ca2+-transporting ATPase that is expressed predominantly in brain and skeletal muscle. J. Biol. Chem. 1989;264:18569–18576. doi: 10.1016/S0021-9258(18)51505-2. [DOI] [PubMed] [Google Scholar]
- 234.Sacchetto R., Margreth A., Pelosi M., Carafoli E. Colocalization of the dihydropyridine receptor, the plasma-membrane calcium ATPase isoform 1 and the sodium/calcium exchanger to the junctional-membrane domain of transverse tubules of rabbit skeletal muscle. Eur. J. Biochem. 1996;237:483–488. doi: 10.1111/j.1432-1033.1996.0483k.x. [DOI] [PubMed] [Google Scholar]
- 235.Kimura J., Noma A., Irisawa H. Na-Ca exchange current in mammalian heart cells. Nature. 1986;319:596–597. doi: 10.1038/319596a0. [DOI] [PubMed] [Google Scholar]
- 236.Michel L.Y.M., Verkaart S., Koopman W.J.H., Willems P.H.G.M., Hoenderop J.G.J., Bindels R.J.M. Function and regulation of the Na+-Ca2+ exchanger NCX3 splice variants in brain and skeletal muscle. J. Biol. Chem. 2014;289:11293–11303. doi: 10.1074/jbc.M113.529388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fraysse B., Rouaud T., Millour M., Fontaine-Pérus J., Gardahaut M.F., Levitsky D.O. Expression of the Na+/Ca2+ exchanger in skeletal muscle. Am. J. Physiol.-Cell Physiol. 2001;280:C146–C154. doi: 10.1152/ajpcell.2001.280.1.C146. [DOI] [PubMed] [Google Scholar]
- 238.Reeves J.P., Condrescu M., Chernaya G., Gardner J.P. Na+/Ca2+ antiport in the mammalian heart. J. Exp. Biol. 1994;196:375–388. doi: 10.1242/jeb.196.1.375. [DOI] [PubMed] [Google Scholar]
- 239.Shigekawa M., Iwamoto T. Cardiac Na+-Ca2+ exchange: Molecular and pharmacological aspects. Circ. Res. 2001;88:864–876. doi: 10.1161/hh0901.090298. [DOI] [PubMed] [Google Scholar]
- 240.Shen Y., Thillaiappan N.B., Taylor C.W. The store-operated Ca2+ entry complex comprises a small cluster of STIM1 associated with one Orai1 channel. Proc. Natl. Acad. Sci. USA. 2021;118:e2010789118. doi: 10.1073/pnas.2010789118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kurebayashi N., Ogawa Y. Depletion of Ca2+ in the sarcoplasmic reticulum stimulates Ca2+ entry into mouse skeletal muscle fibres. J. Physiol. 2001;533:185–199. doi: 10.1111/j.1469-7793.2001.0185b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Boncompagni S. Discovery of new intracellular junctions: The calcium entry units (CEUs) J. Gen. Physiol. 2022;154:e2021ecc39. doi: 10.1085/jgp.2021ecc39. [DOI] [Google Scholar]
- 243.Boncompagni S., Michelucci A., Pietrangelo P., Dirksen R.T., Protasi F. Exercise-dependent formation of new junctions that promote STIM1-Orai1 assembly in skeletal muscle. Sci. Rep. 2017;7:14286. doi: 10.1038/s41598-017-14134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Protasi F., Pietrangelo L., Boncompagni S. Calcium entry units (CEUs): Perspectives in skeletal muscle function and disease. J. Muscle Res. Cell Motil. 2021;42:233–249. doi: 10.1007/s10974-020-09586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Chevessier F., Bauché-Godard S., Leroy J.P., Koenig J., Paturneau-Jouas M., Eymard B., Hantaï D., Verdière-Sahuqué M. The origin of tubular aggregates in human myopathies. J. Pathol. 2015;207:313–323. doi: 10.1002/path.1832. [DOI] [PubMed] [Google Scholar]
- 246.Silva-Rojas R., Laporte J., Böhm J. STIM1/ORAI1 Loss-of-Function and Gain-of-Function Mutations Inversely Impact on SOCE and Calcium Homeostasis and Cause Multi-Systemic Mirror Diseases. Front. Physiol. 2020;11:604941. doi: 10.3389/fphys.2020.604941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Böhm J., Bulla M., Urquhart J.E., Malfatti E., Williams S.G., O’Sullivan J., Szlauer A., Koch C., Baranello G., Mora M., et al. ORAI1 Mutations with Distinct Channel Gating Defects in Tubular Aggregate Myopathy. Hum. Mutat. 2017;38:426–438. doi: 10.1002/humu.23172. [DOI] [PubMed] [Google Scholar]
- 248.Böhm J., Laporte J. Gain-of-function mutations in STIM1 and ORAI1 causing tubular aggregate myopathy and Stormorken syndrome. Cell Calcium. 2018;76:1–9. doi: 10.1016/j.ceca.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 249.Lacruz R.S., Feske S. Diseases caused by mutations in ORAI1 and STIM1. Ann. N. Y. Acad. Sci. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Eisner V., Csordás G., Hajnóczky G. Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle—pivotal roles in Ca²⁺ and reactive oxygen species signaling. J. Cell Sci. 2013;126:2965–2978. doi: 10.1242/jcs.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Boncompagni S., Rossi A.E., Micaroni M., Hamilton S.L., Dirksen R.T., Franzini-Armstrong C., Protasi F. Characterization and temporal development of cores in a mouse model of malignant hyperthermia. Proc. Natl. Acad. Sci. USA. 2009;106:21996–22001. doi: 10.1073/pnas.0911496106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Boncompagni S., Pozzer D., Viscomi C., Ferreiro A., Zito E. Physical and Functional Cross Talk Between Endo-Sarcoplasmic Reticulum and Mitochondria in Skeletal Muscle. Antioxid. Redox Signal. 2020;32:873–883. doi: 10.1089/ars.2019.7934. [DOI] [PubMed] [Google Scholar]
- 253.Sharma V.K., Ramesh V., Franzini-Armstrong C., Sheu S.S. Transport of Ca2+ from sarcoplasmic reticulum to mitochondria in rat ventricular myocytes. J. Bioenerg. Biomembr. 2000;32:97–104. doi: 10.1023/A:1005520714221. [DOI] [PubMed] [Google Scholar]
- 254.García-Pérez C., Schneider T.G., Hajnóczky G., Csordás G. Alignment of sarcoplasmic reticulum-mitochondrial junctions with mitochondrial contact points. Am. J. Physiol.-Heart Circ. Physiol. 2011;301:H1907–H1915. doi: 10.1152/ajpheart.00397.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Min C.K., Yeom D.R., Lee K., Kwon H., Kang M., Kim Y., Park Z.Y., Jeon H., Kim D.O. Coupling of ryanodine receptor 2 and voltage-dependent anion channel 2 is essential for Ca2+ transfer from the sarcoplasmic reticulum to the mitochondria in the heart. Biochem. J. 2012;447:371–379. doi: 10.1042/BJ20120705. [DOI] [PubMed] [Google Scholar]
- 256.Chen Y., Csordás G., Jowdy C., Schneider T.G., Csordás N., Wang W., Liu Y., Kohlhaas M., Meiser M., Bergem S., et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca2+ crosstalk. Circ. Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ainbinder A., Boncompagni S., Protasi F., Dirksen R.T. Role of Mitofusin-2 in mitochondrial localization and calcium uptake in skeletal muscle. Cell Calcium. 2015;57:14–24. doi: 10.1016/j.ceca.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Giacomello M., Drago I., Bortolozzi M., Scorzeto M., Gianelle A., Pizzo P., Pozzan T. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell. 2010;38:280–290. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 259.Penna E., Espino J., De Stefani D., Rizzuto R. The MCU complex in cell death. Cell Calcium. 2018;69:73–80. doi: 10.1016/j.ceca.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 260.Mammucari C., Raffaello A., Vecellio Reane D., Gherardi G., De Mario A., Rizzuto R. Mitochondrial calcium uptake in organ physiology: From molecular mechanism to animal models. Pflugers Arch. 2018;470:1165–1179. doi: 10.1007/s00424-018-2123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rossi A., Pizzo P., Filadi R. Calcium, mitochondria and cell metabolism: A functional triangle in bioenergetics. Biochim. Biophys. Acta.-Mol. Cell Res. 2019;1866:1068–1078. doi: 10.1016/j.bbamcr.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 262.Wu Y., Rasmussen T.P., Koval O.M., Joiner M.L., Hall D.D., Chen B., Luczak E.D., Wang Q., Rokita A.G., Wehrens X.H., et al. The mitochondrial uniporter controls fight or flight heart rate increases. Nat. Commun. 2015;6:6081. doi: 10.1038/ncomms7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gherardi G., Nogara L., Ciciliot S., Fadini G.P., Blaauw B., Braghetta P., Bonaldo P., De Stefani D., Rizzuto R., Mammucari C. Loss of mitochondrial calcium uniporter rewires skeletal muscle metabolism and substrate preference. Cell Death Differ. 2019;26:362–381. doi: 10.1038/s41418-018-0191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Robert V., Massimino M.L., Tosello V., Marsault R., Cantini M., Sorrentino V., Pozzan T. Alteration in calcium handling at the subcellular level in mdx myotubes. J. Biol. Chem. 2001;276:4647–4651. doi: 10.1074/jbc.M006337200. [DOI] [PubMed] [Google Scholar]
- 265.Durham W.J., Aracena-Parks P., Long C., Rossi A.E., Goonasekera S.A., Boncompagni S., Galvan D.L., Gilman C.P., Baker M.R., Shirokova N., et al. RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell. 2008;133:53–65. doi: 10.1016/j.cell.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hamilton S., Terentyeva R., Clements R.T., Belevych A.E., Terentyev D. Sarcoplasmic reticulum-mitochondria communication; implications for cardiac arrhythmia. J. Mol. Cell Cardiol. 2021;156:105–113. doi: 10.1016/j.yjmcc.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lee C.S., Hanna A.D., Wang H., Dagnino-Acosta A., Joshi A.D., Knoblauch M., Xia Y., Georgiou D.K., Xu J., Long C., et al. A chemical chaperone improves muscle function in mice with a RyR1 mutation. Nat. Commun. 2017;8:14659. doi: 10.1038/ncomms14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Volpe P., Villa A., Podini P., Martini A., Nori A., Panzeri M.C., Meldolesi J. The endoplasmic reticulum-sarcoplasmic reticulum connection: Distribution of endoplasmic reticulum markers in the sarcoplasmic reticulum of skeletal muscle fibers. Proc. Natl. Acad. Sci. USA. 1992;89:6142–6146. doi: 10.1073/pnas.89.13.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kaisto T., Metsikkö K. Distribution of the endoplasmic reticulum and its relationship with the sarcoplasmic reticulum in skeletal myofibers. Exp. Cell Res. 2003;289:47–57. doi: 10.1016/S0014-4827(03)00231-3. [DOI] [PubMed] [Google Scholar]
- 270.Bogdanov V., Soltisz A.M., Moise N., Sakuta G., Orengo B.H., Janssen P.M.-L., Weinberg S.H., Davis J.P., Veeraraghavan R., Györke S. Distributed synthesis of sarcolemmal and sarcoplasmic reticulum membrane proteins in cardiac myocytes. Basic Res. Cardiol. 2021;116:1–16. doi: 10.1007/s00395-021-00895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Villa A., Podini P., Panzeri M.C., Söling H.D., Volpe P., Meldolesi J. The endoplasmic-sarcoplasmic reticulum of smooth muscle: Immunocytochemistry of vas deferens fibers reveals specialized subcompartments differently equipped for the control of Ca2+ homeostasis. J. Cell Biol. 1993;121:1041–1051. doi: 10.1083/jcb.121.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Prinz W.A., Toulmay A., Balla T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020;21:7–24. doi: 10.1038/s41580-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kinnear N.P., Wyatt C.N., Clark J.H., Calcraft P.J., Fleischer S., Jeyakumar L.H., Nixon G.F., Evans A.M. Lysosomes co-localize with ryanodine receptor subtype 3 to form a trigger zone for calcium signalling by NAADP in rat pulmonary arterial smooth muscle. Cell Calcium. 2008;44:190–201. doi: 10.1016/j.ceca.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Aston D., Capel R.A., Ford K.L., Christian H.C., Mirams G.R., Rog-Zielinska E.A., Kohl P., Galione A., Burton R.A., Terrar D.A. High resolution structural evidence suggests the Sarcoplasmic Reticulum forms microdomains with Acidic Stores (lysosomes) in the heart. Sci. Rep. 2017;7:1–15. doi: 10.1038/srep40620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Capel R.A., Bolton E.L., Lin W.K., Aston D., Wang Y., Liu W., Wang X., Burton R.A., Bloor-Young D., Shade K.T., et al. Two-pore Channels (TPC2s) and Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) at Lysosomal-Sarcoplasmic Reticular Junctions Contribute to Acute and Chronic β-Adrenoceptor Signaling in the Heart. J. Biol. Chem. 2015;290:30087–30098. doi: 10.1074/jbc.M115.684076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wu X., Bers D.M. Sarcoplasmic reticulum and nuclear envelope are one highly interconnected Ca2+ store throughout cardiac myocyte. Circ. Res. 2006;99:283–291. doi: 10.1161/01.RES.0000233386.02708.72. [DOI] [PubMed] [Google Scholar]
- 277.Malhas A., Goulbourne C., Vaux D.J. The nucleoplasmic reticulum: Form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 278.Marius P., Guerra M.T., Nathanson M.H., Ehrlich B.E., Leite M.F. Calcium release from ryanodine receptors in the nucleoplasmic reticulum. Cell Calcium. 2006;39:65–73. doi: 10.1016/j.ceca.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 279.Irvine R.F. Nuclear inositide signalling-expansion, structures and clarification. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids. 2006;1761:505–508. doi: 10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Avedanian L., Jacques D., Bkaily G. Presence of tubular and reticular structures in the nucleus of human vascular smooth muscle cells. J. Mol. Cell Cardiol. 2011;50:175–186. doi: 10.1016/j.yjmcc.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 281.Lee S.H., Hadipour-Lakmehsari S., Miyake T., Gramolini A.O. Three-dimensional imaging reveals endo(sarco)plasmic reticulum-containing invaginations within the nucleoplasm of muscle. Am. J. Physiol.-Cell Physiol. 2018;314:C257–C267. doi: 10.1152/ajpcell.00141.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Roman W., Gomes E.R. Nuclear positioning in skeletal muscle. Semin. Cell Dev. Biol. 2018;82:51–56. doi: 10.1016/j.semcdb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 283.Reuveny A., Shnayder M., Lorber D., Wang S., Volk T. Ma2/d promotes myonuclear positioning and association with the sarcoplasmic reticulum. Development. 2018;145:dev159558. doi: 10.1242/dev.159558. [DOI] [PubMed] [Google Scholar]
- 284.Azevedo M., Baylies M.K. Getting into Position: Nuclear Movement in Muscle Cells. Trends Cell Biol. 2020;30:303–316. doi: 10.1016/j.tcb.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wang S., Reuveny A., Volk T. Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. J. Cell Biol. 2015;209:529–538. doi: 10.1083/jcb.201408098. [DOI] [PMC free article] [PubMed] [Google Scholar]