Abstract
Echinoderms are important experimental models for analyzing embryonic development, but a lack of spatial and temporal control over gene perturbations has hindered developmental studies using these animals. Morpholino antisense oligonucleotides (MOs) have been used successfully by the echinoderm research community for almost two decades, and MOs remain the most widely used tool for acute gene knockdowns in these organisms. Echinoderm embryos develop externally and are optically transparent, making them ideally-suited to many light-based approaches for analyzing and manipulating development. Studies using zebrafish embryos have demonstrated the effectiveness of photoactivatable (caged) MOs for conditional gene knockdowns. Here we show that caged MOs, synthesized using nucleobase-caged monomers, provide light-regulated control over gene expression in sea urchin embryos. Our work provides the first robust approach for conditional gene silencing in this prominent model system.
Keywords: Echinoderm, Sea urchin, Embryogenesis, Morpholino, Caged morpholino, Conditional gene knockdown, Regulated gene knockdown, Photoactivation
1. Introduction
Sea urchins and other echinoderms are prominent experimental models for analyzing developmental mechanisms (Angerer and Angerer, 2003; Wilt, 2005; McClay, 2011, 2016; Wessel, 2016; Cary et al., 2019). The rapid, external development of sea urchin embryos, their relatively simple structure and optical transparency, and the ease with which large numbers of synchronously developing embryos can be obtained, have historically lent value to these organisms as developmental models. More recently, echinoderms have been pre-eminent models for systems-level analysis of gene regulation during embryogenesis, including the elucidation of developmental gene regulatory networks (GRNs) (Davidson, 2009; Martik et al., 2016; Cary and Hinman, 2017; Lowe et al., 2017; Peter, 2017; Shashikant et al., 2018).
Several different experimental approaches, each with its own advantages and disadvantages, are currently used to perturb gene function in sea urchin embryos. Chemical inhibitors are easy to use and the timing of their application can be controlled (e.g., Fernandez-Serra et al., 2004; Duboc et al., 2005; Luo and Su, 2012; Warner et al., 2016; Mellot et al., 2017; Foster et al., 2019, and many other studies), but they often lack specificity, exist for only a fraction of the proteome, and cannot be targeted to specific embryonic tissues. Microinjection of mRNAs encoding dominant negative constructs can also be highly effective (Molina et al., 2019 and references therein), but this approach is limited to proteins with well-characterized dominant negative forms and leads to the ubiquitous expression of the dominant negative protein in tissues. Moreover, the limited stability of injected mRNAs limits this strategy to early developmental stages. More recently, CRISPR/Cas9-mediated gene editing has been successfully applied to sea urchins (Lin and Su, 2016; Mellot et al., 2017; Ouhlen et al., 2017; Lin et al., 2019; Liu et al., 2019; Yaguchi et al., 2020). This approach targets mutations to genes of interest with a high degree of specificity, but F0 embryos are genetic mosaics (Mehravar et al., 2019) and CRISPR/Cas9-mediated gene editing can result in compensatory gene expression changes (Rossi et al., 2015; Zhu et al., 2017; El-Brolosy et al., 2019; Ma et al., 2019; Peng, 2019). Although strategies for inducible gene editing are currently being developed (Zhang et al., 2019; Liu et al., 2020; Zhou et al., 2020) these have not been tested in sea urchins.
The most widely used reagents for perturbing gene function in echinoderms are morpholino antisense oligonucleotides (MOs) (reviewed by Heasman, 2002; Angerer and Angerer, 2004). When used with appropriate controls, both translation- and splice-blocking MOs have been shown to reliably produce highly specific and effective gene knockdowns. Notably, MOs have been critically important reagents for the construction of detailed GRN models in sea urchins and other echinoderms (Materna et al., 2017; Peter, 2019). MO-based knockdowns of regulatory (i.e., transcription factor-encoding) genes are typically combined with gene expression profiling to elucidate the gene regulatory circuits that comprise GRNs.
A major limitation of sea urchins and other echinoderms as experimental models for developmental studies is that gene perturbations cannot be regulated temporally or spatially. MOs, mRNAs that encode dominant negative proteins, and CRISPR components are typically microinjected into fertilized eggs, resulting in perturbation of the targeted gene throughout embryogenesis and in all tissues. Microinjection into individual cells at later stages becomes progressively more difficult as the size of the cells decreases during cleavage. Although MOs with enhanced cell-permeability (vivo-MOs) have been developed, these have not generally been useful in sea urchins because of their toxicity and low solubility (Cui et al., 2017). The lack of conditional gene perturbations makes it difficult to explore the late developmental functions of genes that have essential, early functions or to tease apart the tissue-specific roles of genes that are expressed in multiple embryonic territories. With respect to GRN biology, the lack of conditional approaches has made it extremely difficult to analyze developmental (temporal) changes in GRN circuitry. Moreover, most regulatory genes are expressed in multiple territories, confounding the interpretation of embryo-wide knockdowns and hampering the construction of accurate, cell-type specific GRN models. The ability to regulate gene knockdowns in sea urchins would make possible the direct interrogation of GRN circuitry at late developmental stages and in specific tissues.
Light-based methodologies provide unparalleled temporal and spatial control of molecular processes. The transparency of echinoderm embryos makes them ideally suited to such approaches. Recently, considerable progress has been made in developing light-activated, caged MOs (cMOs), and several studies have demonstrated the effectiveness of cMOs in zebrafish embryos, another optically transparent animal model. Various design strategies have been explored with the goal of simplifying the synthesis of these reagents, minimizing their leakiness and toxicity, and optimizing their activity after decaging. The production of cMOs has involved a) the tethering of a complementary, inhibitory oligonucleotide to the MO through a photocleavable linker, creating a hairpin structure (Shestoplaov et al., 2007; Ouyang et al., 2009), b) duplexing the MO with an inhibitory strand that contains an internal photolabile linker (Tomasini et al., 2009; Tallafuss et al., 2012), or c) circularizing MOs with photocleavable linkers that can be activated with light of different wavelengths (Wang et al., 2012; Yamazoe et al., 2012, 2014; Pattanayak et al., 2019). The first two approaches require careful design of the inhibitory strand and its binding properties in addition to releasing “waste” MO strands upon photolysis. Circular MOs have good synthetic yield and rely on the use of a single caging group, but can display background activity. A more direct approach involves the generation of MOs that contain multiple, evenly spaced, nucleobase-caged monomers, a strategy that has been used successfully for conditional gene knockdowns in zebrafish (Deiters et al., 2010). The inclusion of caged nucleobases fully blocks MO:mRNA hybridization and ensures that no background gene silencing is observed before photoactivation (Liu and Deiters, 2014). Here, we show that this direct MO-caging strategy can be used to provide effective, light-controlled gene perturbations in echinoderms, with important implications for future research using these model organisms.
2. Materials and methods
2.1. General chemical methods
All reagents were purchased from commercial suppliers and used without further purification. Flash chromatography was performed using an ISCO CombiFlash Rf (Lincoln, NE) with normal phase silica gel cartridges. NMR spectra were recorded on Bruker 400 MHz NMR spectrometer, Bruker (Billerica, MA). Analytical LCMS data were collected on a Shimadzu LCMS-2020, Shimadzu Corporation (Santa Clara, CA) and a Q-Exactive Orbitrap, ThermoFisher Scientific (Waltham, MA).
2.2. Synthesis of cMOs
The NPOM-caged thymidine-MO chlorophosphoramidate 7 (see Supporting Information) was site-specifically installed on Lv-vegf3 and Lv-kirrelL MO sequences by Gene Tools (Philomath, OR) to generate the caged morpholinos. Successful syntheses were confirmed by MALDI-MS. The MO sequences used in this study are shown below (bold and underlined Ts indicate caged nucleobases in the cMO sequences):
Lv-vegf3: 5′-TCGACTGAAGGTCCCATCGTGCTTT-3′
Lv-kirrelL: 5′-GGTTCATTCTTGCTGGACTGAATTT-3′
2.3. Gel-shift assays
Gel-shift assays of caged (T4) and non-caged morpholinos (T0) were adopted from previous protocols (Deiters et al., 2010; Ouyang et al., 2009) with slight modifications. Complementary DNA (cDNA) was used in the assays instead of RNA for stability reasons. MOs were annealed to their corresponding cDNA in 1× TE/Mg2+ buffer (0.01 M Tris-HCl, 100 mM EDTA and 12.5 mM MgCl2, pH = 7.8) at 37 °C. Sample sets that required UV illumination were irradiated with 365 nm LED light (P = 25 mW). Samples were analyzed by 16% native PAGE using 1× TBE (0.13 M Tris, 45 mM boric acid and 2.5 mM EDTA, pH = 7.6) as the running buffer and imaged with ChemiDoc™ MP Imaging System, Bio-Rad Laboratories (Hercules, CA) after staining with SYBR™ Gold Nucleic Acid Gel Stain, Invitrogen (Carlsbad, CA). Gel-shift experiments were performed in triplicates.
2.4. Background binding assays
For background binding analysis, 100 nM of target cDNA was incubated with increasing concentrations of the corresponding T0 and T4-MO (0, 25, 50, 100, 150 and 200 nM) in 1× TE/Mg2+ buffer (pH = 7.8) at 37 °C for 2 h. An equimolar mixture of cDNA with the standard control oligo (Gene Tools) was used as a negative control. Unbound cDNA was then resolved from the MO:cDNA duplex by 16% native PAGE.
2.5. Irradiation time course assays
Target cDNA complementary to the corresponding MO sequence was used at 100 nM while the concentration of the T4-MO was 200 nM for a 2:1 T4-MO:cDNA ratio. Duplex of cDNA with non-caged ssDNA (T0-DNA), the latter resembling the corresponding MO sequence, was used as a positive control. The T4-MO:cDNA mixtures were incubated in TE/Mg2+ buffer at 37 °C for 2 h. Sample sets were then either kept in the dark or subjected to 365 nm irradiation (1 s - 10 min) followed by incubation at 37 °C for an additional 1 h. Unbound cDNA was then separated from the MO:cDNA duplex by 16% native PAGE.
2.6. Adult animals and embryo culture
Gravid, adult Lytechinus variegatus were obtained from the Duke University Marine Laboratory (Beaufort, NC, USA) and Reeftopia, Inc. (Key West, FL, USA). Spawning by intracoelomic injection of 0.5 M KCl and fertilizations were carried out as described by Foltz et al. (2004). Embryos were cultured in artificial seawater (ASW) at 23 °C in temperature-controlled incubators.
2.7. Microinjection of MOs
Stock solutions of MOs (2–5 mM) were prepared in sterile, double-deionized water and stored at −20 °C. Injection solutions were prepared by warming frozen stock solutions for 1.5 h at 37 °C and adding an appropriate volume of sterile, double-deionized water, glycerol (to a final concentration of 20%, vol/vol) and 10 kDa MW fluorescein dextran (to a final concentration of 0.5%, wt/vol). Final concentrations of MOs in the injection solutions were 2 mM (Lv-vegf3, non-caged and caged forms) and 500 μM (Lv-kirrelL, caged and non-caged forms) cMO solutions were continuously protected from light by wrapping tubes in foil or by working under red light. MOs were injected into freshly fertilized eggs as previously described (Cheers and Ettensohn, 2004).
2.8. In vivo photoactivation
Control (uninjected) and MO-injected embryos were irradiated by placing them ~2 cm away from a 4.6 W, 365 nm LED light source (UV LED Gen 2 Emitter, LED Engin, Osram Sylvania, powered at 1 A) for 15 min. When photoactivation was carried out prior to hatching, 60 mm protamine sulfate-coated injection dishes containing 10 ml of seawater and with embryos attached were placed beneath the LED source. For such experiments, unfertilized eggs were positioned in three short, parallel rows near the middle of the injection dish to ensure that all embryos were illuminated evenly. When photoactivation was carried out after hatching, swimming embryos were collected by centrifugation and transferred to ~1 ml of seawater in a glass depression slide, which was placed beneath the LED source. After photoactivation, injection dishes and depression slides were placed in humid chambers inside temperature-controlled incubators.
2.9. Analysis of morphant embryos
Embryos were examined 24–28 hpf (hours post-fertilization) to assess skeletal morphology. Living embryos were collected by centrifugation and immobilized on poly-l-lysine-coated coverslips. Skeletal phenotypes were assessed using differential interference contrast (DIC) and polarization optics. Larval skeletal elements are composed of calcite and appear bright when viewed under crossed polarizers due to the natural birefringence of the biomineral. In some experiments, embryos were fixed at the late gastrula stage (16 hpf) and immunostained with monoclonal antibody 6a9 as described previously (Ettensohn and Dey, 2017). This antibody was generated in-house and specifically labels skeletogenic primary mesenchyme cells (PMCs) (Ettensohn and McClay, 1988).
3. Results
3.1. Synthesis of cMOs and EMSA analysis
To explore the utility of cMOs for gene knockdowns in sea urchins, we focused on two well-characterized proteins: Vegf3 and KirrelL. These proteins play essential but distinct roles in skeletogenesis: Vegf3 is an ectoderm-derived growth factor that regulates the differentiation of skeleton-forming cells (primary mesenchyme cells, or PMCs) and KirrelL is a PMC-specific adhesion protein required for cell-cell fusion. Conventional translation-blocking MOs have been used to individually block the expression of both proteins and each knockdown produces a well-characterized, highly penetrant phenotype (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013; Ettensohn and Dey, 2017; Ettensohn and Adomako-Ankomah, 2019).
While numerous caging groups are available for optical control of biological systems (Bardhan and Deiters, 2019), for our caged morpholinos, we utilized the nitrobenzyl-based NPOM group due to its robustness, ease of photolysis, and stability under MO synthesis and biological conditions, as demonstrated in a wide range of biological studies (Govan et al., 2013; Hemphill et al., 2014, 2015; Naro et al., 2020; Zhou et al., 2020). Based on the even distribution of thymidine bases in the selected MO sequences, we decided to synthesize a caged thymidine subunit. The synthesis of the NPOM-caged thymidine-MO monomer was adopted from a previous report (Deiters et al., 2010) with slight modifications to improve yields and to increase the scale (see Supporting Information and Supp. Fig. 1). The activated thymidine monomer 7 was generated and then site-specifically incorporated into the desired sequences by Gene Tools, LLC (Philomath, OR) via solid-phase synthesis.
For background binding analysis, the caged MOs (T4) along with their non-caged analogs (T0) were tested for hybridization to their complementary target DNAs (cDNA) (Fig. 1). The non-caged MOs exhibited almost complete binding to target cDNA at equimolar concentrations, as expected. More importantly, we observed almost no binding of cMOs to cDNA at equimolar concentrations or when the cMOs were present in excess (200 nM) (Fig. 1a and b). This highlights the importance of even distribution of caging groups in the sequence for complete abolishment of target hybridization and similar trends have been previously observed in the context of caged antisense oligonucleotides (Young et al., 2008a, 2008b) and caged MOs (Deiters et al., 2010).
Fig. 1.
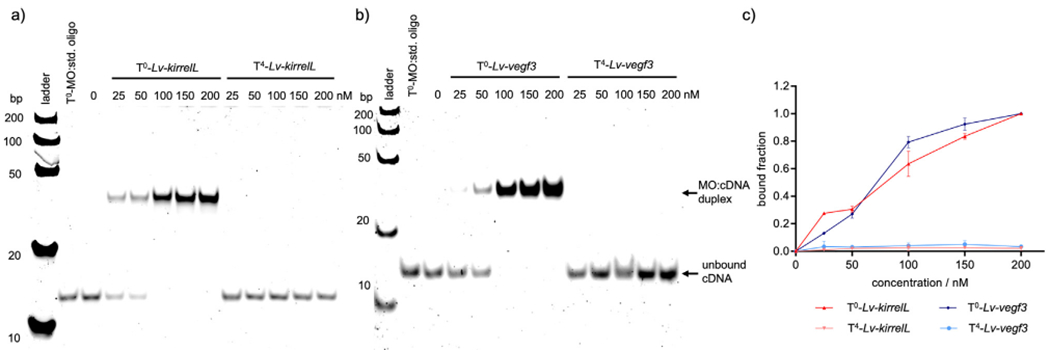
Representative gel shift assays of non-caged (T0) and caged (T4) MOs interacting with their corresponding complementary target DNA (100 nM) sequences. a) T0-Lv-kirrelL and T4-Lv-kirrelL, b) T0- Lv-vegf3 and T4- Lv-vegf3 and c) quantification of the gel shift assay. A Fast Ruler Ultra Low Range ds-DNA Ladder (SM1233, ThermoFisher Scientific) was used. A standard control oligo from Gene Tools (denoted as “std. oligo”) was incubated with T0-MOs in equimolar amounts as a negative control. Gel images were quantified with ImageLab, and the bound MO:cDNA fraction was plotted against MO concentration. Error bars represent the standard deviation of three independent trials.
Additional gel-shift assays were carried out to determine the effect of 365 nm LED irradiation on caging group cleavage and subsequent MO:cDNA duplex formation (Fig. 2). These assays were carried out using a 2:1 ratio of T4-MO:cDNA, mimicking the presence of an excess probe as compared to the endogenous mRNA target in vivo (Ekker, 2004) upon MO/cMO injection. We either kept the samples in dark or subjected them to 5 or 10 min of irradiation to regulate duplex formation. While we observed almost no background binding in the absence of irradiation, complete binding of the decaged MO to cDNA was observed after only 5 min of irradiation. A shorter time-course experiment showed that, under these in vitro conditions, both cMOs were robustly activated with only a 1 s exposure to 4.6 W, 365 nm LED illumination (Supp. Fig. 2).
Fig. 2.
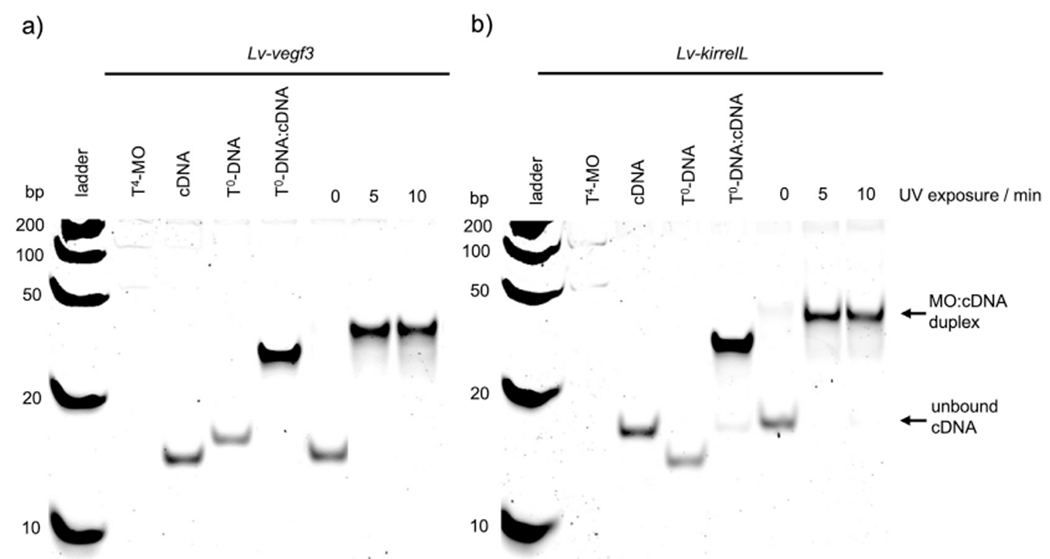
Irradiation time course gel-shift assay of a) T4-Lv-vegf3 and b) T4-Lv-kirrelL MOs. The concentration of target cDNA was 100 nM while cMOs (T4-MOs) were used at 200 nM to achieve 2:1 T4-MO:cDNA ratio. Non-caged DNA with the same sequence as the corresponding MO (T0-DNA) was used as positive control in a 1:1 T0-DNA:cDNA ratio. A Fast Ruler Ultra Low Range ds-DNA Ladder (SM1233, ThermoFisher Scientific) was used.
3.2. In vivo validation of cMOs
To test the efficacy of cMOs in vivo, we microinjected Lv-vegf3 and Lv-kirrelL cMOs individually into fertilized L. variegatus eggs and photo-activated the cMOs immediately after injection or at later developmental stages. Previous studies have shown that constitutive knockdown of Lv-vegf3 using the non-caged form of the same MO results in larvae that lack skeletons, while constitutive knockdown of Lv-kirrelL using the non-caged MO blocks PMC fusion and leads to the formation of small, disconnected skeletal elements (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013; Ettensohn and Dey, 2017; Ettensohn and Adomako-Ankomah, 2019). These well-characterized morphant phenotypes provided a basis for assessing the potential leakiness of the cMOs in vivo and their efficacy following photoactivation.
We injected Lv-vegf3 cMO at a concentration of 2 mM based on previous studies, which used the non-caged form at the same concentration (Adomako-Ankomah and Ettensohn, 2013) and at 4 mM (Ettensohn and Adomalo-Ankomah, 2019). To decage the cMO, we placed embryos ~2 cm away from a 365 nm, 4.6 W LED light source for 15 min (see Methods). In multiple control experiments, we found that this dosage of 365 nm light by itself had no detectable effect on embryonic development. Uninjected embryos irradiated for 15 min at the 1-cell stage developed in synchrony with sibling, dark-reared embryos and in both cases 97% of the resulting swimming larvae appeared normal (137/140 UV-treated larvae and 100/103 sibling, dark-reared larvae). In a dose-response experiment, we found that irradiation for as long as 30 min had no discernible effect on embryogenesis (Supp. Fig. 2). We also found that Lv-vegf3 cMO had little or no effect on skeletogenesis when injected embryos were reared in the dark, as ~96% of these embryos developed into normal plutei (50/52 larvae). These findings indicated that, in the absence of decaging, the cMO did not inhibit the translation of Lv-vegf3 mRNA, consistent with our EMSA data (Fig. 1), which also pointed to minimal background activity of the cMO.
When embryos that had been injected with 2.0 mM Lv-vegf3 cMO were irradiated at the 1-cell stage, they exhibited a striking, specific inhibition of skeletal development characteristic of Vegf3 knockdown. We assessed skeletal morphology in living larvae (24 hpf) using polarization optics to directly visualize birefringent calcite and classified the phenotypes of morphant embryos into four categories based on the extent to which skeletogenesis was perturbed (Fig. 3). The four classes were: 1) Strong phenotype (no detectable skeletal elements or only 1–2 tiny, birefringent granules; 2) Moderate phenotype (small, linear skeletal elements); 3) Weak phenotype (highly reduced skeleton but with branched skeletal rods); and 4) No effect (normal, branched skeleton). In three separate trials, we found that the great majority of embryos (70–80%) exhibited the most extreme phenotype (Fig. 4, Trials 1–3). A minority of embryos showed a variety of weaker phenotypes, consistent with the effects of conventional Vegf3 knockdown using the non-caged form of the MO (Fig. 4, Trial 5).
Fig. 3.
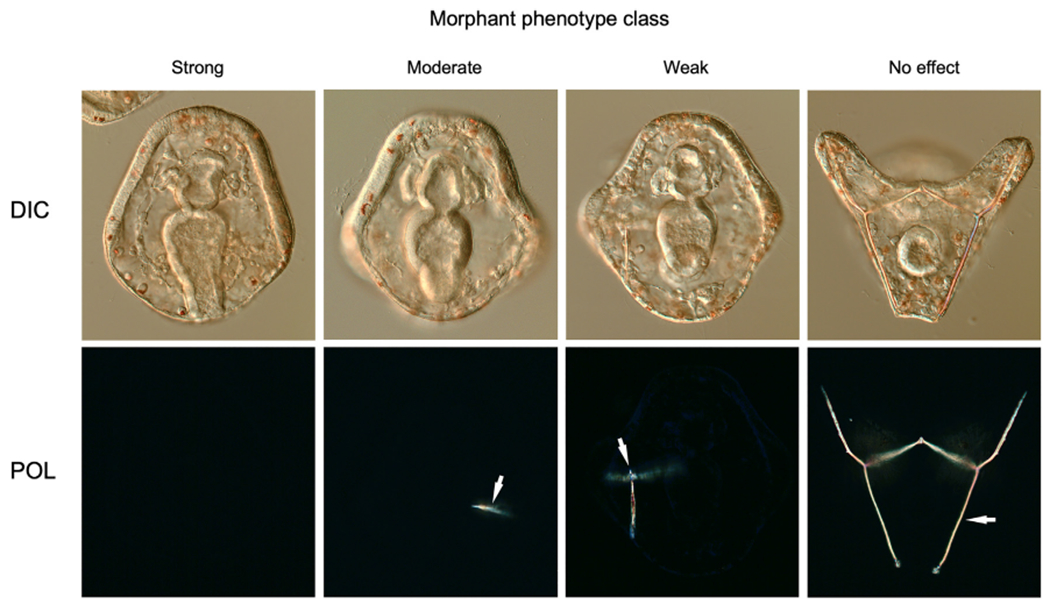
Classification of morphant phenotypes. We examined skeletal morphology at 24 hpf using differential interference contrast (DIC) and polarization (POL) optics and classified morphant phenotypes into four categories based on the extent to which skeletogenesis was perturbed: 1) Strong phenotype (no detectable skeletal elements or only 1–2 tiny, birefringent granules; 2) Moderate phenotype (small, linear skeletal elements); 3) Weak phenotype (highly reduced skeleton but with branched skeletal rods); and 4) No effect (normal, branched larval skeleton). Arrows indicate birefringent skeletal elements.
Fig. 4.
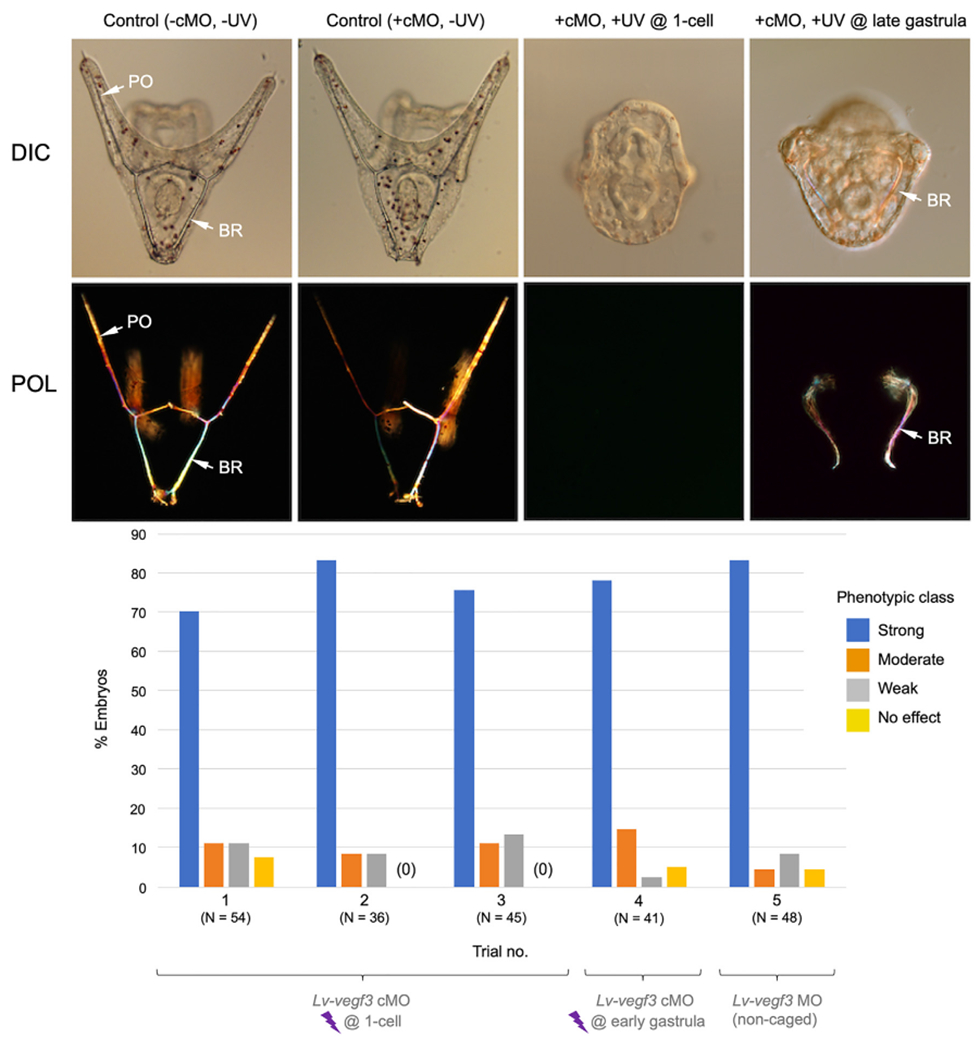
In vivo analysis of Lv-vegf3 cMO. The top two rows of panels show living embryos (24 hpf) viewed with differential interference contrast (DIC) and polarization (POL) optics. Control embryos (−cMO, −UV and +cMO, −UV) developed extensive, branched skeletons that contained elongated, paired body rods (BR) and postoral rods (PO). Decaging of the Lv-vegf3 cMO at the 1-cell stage (+cMO, +UV @ 1-cell; aboral view) completely blocked skeleton formation in most embryos, phenocopying the effects of the non-caged form of the MO. Decaging at the late gastrula stage (16 hpf) (+cMO, +UV @ late gastrula; blastoporal view) blocked the growth of postoral rods but not that of body rods, mimicking the effect of treating late gastrula stage embryos with axitinib, a highly specific inhibitor of Vegf signaling (Adomako-Ankomah and Ettensohn, 2013). Bottom panel- Quantification of phenotypes of embryos injected with Lv-vegf3 cMO and irradiated at the 1-cell (Trials 1–3) or early gastrula (12 hpf; Trial 4) stages, or injected at the 1-cell stage with the non-caged form of the same MO sequence (Trial 5). Morphant phenotypes were scored as shown in Fig. 3.
Temporal control over Vegf3 signaling has been achieved with a highly specific Vegfr inhibitor, axitinib (Adomako-Ankomah and Ettensohn, 2013). Continuous treatment of L. variegatus embryos with axitinib after fertilization phenocopies constitutive Vegf3 knockdown. In addition, a time-course (“wash in”) analysis of axitinib sensitivity has shown that addition of the drug at or before the early gastrula stage results in an almost complete inhibition of skeletogenesis, while addition at later developmental stages has a selective effect on the growth of specific skeletal rods; viz., treatment at the late gastrula stage selectively inhibits the growth of the postoral rods but not that of the body rods (Adomako-Ankomah and Ettensohn, 2013).
We used this information to explore the utility of the Lv-vegf3 cMO for temporal control of gene knockdowns. We reared Lv-vegf3 cMO-injected embryos in the dark until the early gastrula stage (12 hpf) and then collected and irradiated the embryos. The resulting larvae exhibited severe skeletal defects that closely mimicked those produced by irradiation at the 1-cell stage (Fig. 4, Trial 4). When we dark-reared Lv-vegf3 cMO-injected embryos to the late gastrula stage (16 hpf) before irradiation, however, we observed a selective inhibition of postoral rod growth without any detectable effect on the growth of the body rods (Fig. 4, +cMO, +UV @ late gastrula). Of 28 larvae scored, 16 (57%) exhibited complete, bilateral absence of the two postoral rods and another 10 (36%) had severely shortened postoral rods, often with either the right or left rod entirely absent. Thus, the effects of late photoactivation of Lv-vegf3n cMO closely mimicked those observed following the inhibition of Vegf signaling by axitinib at the same developmental stage.
To confirm the efficacy and broad applicability of cMOs in sea urchins we tested a second, distinct MO sequence, one complementary to Lv-kirrelL mRNA. For these studies, we chose to use a relatively low working concentration of the cMO (500 μM), as previous dose-response studies had shown that at this concentration the non-caged form of the MO produces a characteristic spectrum of skeletal phenotypes (Ettensohn and Dey, 2017). This facilitated a comparison of the distribution of the skeletal phenotypes produced by the caged and non-caged forms of the Lv-kirrelL MO. The non-caged form of the MO was re-tested in the present study to provide a direct comparison.
As with the Lv-vegf3 cMO, we observed little or no leakiness in vivo with Lv-kirrelL cMO; ~97% of injected, dark-reared embryos (33/34 embryos) gave rise to pluteus larvae with normal skeletons. Decaging of the cMO at the 1-cell stage, however, produced a distribution of skeletal phenotypes very similar to that produced by constitutive knockdown using the non-caged form (Fig. 5). The extreme class of phenotype produced by decaging at the 1-cell stage was indistinguishable from that of the non-caged form of the MO (Ettensohn and Dey, 2017). PMCs ingressed and arranged themselves in a loose, ring-like pattern but did not align in chains and secreted only small, linear skeletal rods. One distinctive feature of KirrelL morphant embryos is the absence of the filopodial cable that links the PMCs in a syncytial network. We used a monoclonal antibody (mAb) 6a9, a PMC-specific marker (Ettensohn and McClay, 1988), to immunostain late gastrula stage embryos after decaging the Lv-kirrelL cMO at the 1-cell stage and confirmed that PMCs did not fuse, as shown by the absence of the filopodial cable and the failure of PMCs to align in chains (Fig. 5).
Fig. 5.
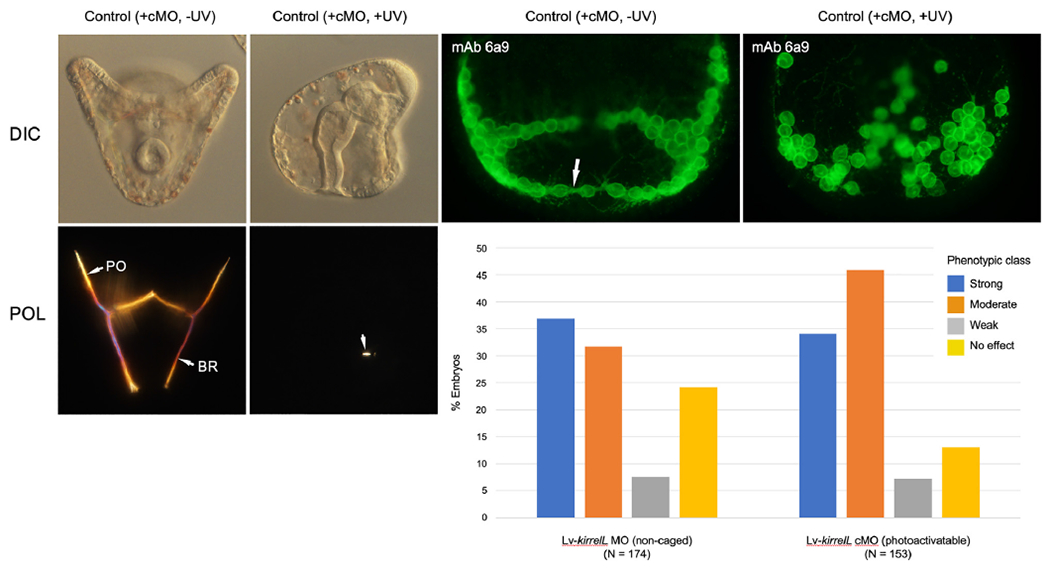
In vivo analysis of Lv-kirrelL cMO. Left panels- Living embryos (24 hpf) viewed with differential interference contrast (DIC) and polarization (POL) optics. Control embryos (+cMO, −UV) developed complete skeletons that contained elongated, paired body rods (BR) and postoral rods (PO). Decaging of the Lv-kirrelL cMO at the 1-cell stage (+cMO, +UV; lateral view) resulted in highly reduced skeletons, phenocoping the effects of the non-caged form of the MO (Ettensohn and Dey, 2017). The arrowhead indicates one small, birefringent spicule rod that formed in this embryo. Top right panels- Late gastrula stage embryos (16 hpf) immunostained with monoclonal antibody (mAb) 6a9, which specifically labels PMCs (Ettensohn and McClay, 1988). In control embryos (+cMO, −UV), PMCs became aligned in strands and their cell bodies were joined by a prominent filopodal cable (arrow). Decaging of the Lv-kirrelL cMO at the 1-cell stage (+cMO, +UV) blocked PMC fusion, as indicated by the scattered arrangement of the cells and absence of a filopodial cable. Bottom right- Quantification of phenotypes of embryos injected with the non-caged or caged form of Lv-kirrelL MO. Embryos injected with cMO were irradiated at the 1-cell stage. Morphant phenotypes were scored at 24 hpf as shown in Fig. 3.
In one trial, cMO-injected embryos were allowed to develop in the dark to the early blastula stage (6.5 hpf) before decaging. The resultant larvae exhibited a similar distribution of phenotypes (i.e., ~60% exhibited strong or moderate phenotypes and 23% had no detectable phenotype). These findings were consistent with evidence that the zygotic activation of kirrelL occurs later in development (at the late blastula stage) and that the protein is required for PMC fusion at the early gastrula stage (Hodor and Ettensohn, 1998; Tu et al., 2014; Ettensohn and Dey, 2017).
4. Discussion
Although sea urchins and other echinoderms have been prominent experimental models for many decades, a lack of spatial or temporal control over gene perturbations currently hinders developmental studies using these animals. It is well known from work on many model organisms that embryonically expressed genes often have both early and late developmental functions and that silencing of genes at early embryonic stages can produce severe developmental defects that make late gene functions difficult to study. It is also well established that most genes are expressed in multiple embryonic territories or cell types, making it challenging to determine assign the effects of gene perturbations to specific tissues or to distinguish direct effects on a particular tissue from indirect effects.
These limitations of non-conditional gene perturbations, while they apply to all developmental studies, are especially significant with respect to GRN analysis, a prominent area of current biological research using echinoderms. For example, a fundamental aspect of GRN analysis is to identify epistatic gene interactions by silencing the expression of regulatory (transcription factor-encoding) genes and assessing changes in the expression of putative target genes. Most regulatory genes are expressed in multiple embryonic territories, however, making it difficult to assign gene expression changes to a specific tissue. In addition, those changes in gene expression that can be unambiguously assigned to a particular tissue can be indirect consequences of the silencing of the regulatory gene in a different, interacting tissue. These issues can produce glaring errors in models of network circuitry; e.g., they can lead to the artificial amalgamation of networks that are actually deployed in separate embryonic territories. A second major issue is that current approaches are not sufficient to dissect temporal changes in GRN circuitry. For example, it cannot be determined whether a particular transcription factor provide continuous inputs into its repertoire of target genes or instead “hands off” its regulatory functions to other transcription factors later in development. Conditional perturbations of regulatory genes would therefore advance GRN analysis in several important ways.
For nearly two decades, MOs have been the reagent of choice for gene silencing in sea urchins and other echinoderms, and more than a hundred studies have been published using conventional MOs to effect gene knockdowns in these animals. Although some reports have raised concerns about possible off-target effects of MOs (Robu et al., 2007; Kok et al., 2015; Gentsch et al., 2018), a large and compelling body of evidence in multiple model systems has confirmed their value when combined with appropriate controls for efficacy and specificity (Blum et al., 2015; Rossi et al., 2015; Stanier et al., 2015; Paraiso et al., 2019). Theoretically, temporal control of MO-based gene knockdowns could be achieved using cell-permeant morpholinos (vivo-MOs), which have been commercially available for several years. The experience of our own lab and others (Cui et al., 2017), however, has been that the utility of vivo-MOs is hampered by their toxicity and insolubility, and there have been only two published reports using vivo-MOs in echinoderms (Luo and Su, 2012; Heyland et al., 2014).
Here we have demonstrated the effectiveness of light-activated MOs for manipulating gene expression in sea urchins. We targeted two different genes, Lv-vegf3 and Lv-kirrelL, which were chosen for proof-of-concept studies because their developmental functions are well documented. The results of in vitro studies (gel-shift assays) and in vivo studies (microinjection into fertilized eggs) were consistent and showed that decaged Lv-vegf3 and Lv-kirrelL cMOs were as effective as their non-caged counterparts in silencing target gene function. Temporal control was demonstrated by irradiating embryos at different developmental stages (1-cell, early gastrula, and late gastrula stages) which recapitulated previously published results obtained with use of a chemical inhibitor. The cMOs used in this study did not exhibit any toxicity at the concentrations used. More importantly, we found that cMOs exhibited little or no background activity in the absence of light. We also found 365 nm irradiation alone had no detectable effect on embryogenesis at dosages sufficient to produce efficient photoactivation of NPOM-caged MOs. These findings are consistent with studies in zebrafish and Xenopus (Deiters et al., 2010) and suggest that cMOs will be effective reagents for facilitating conditional gene knockdowns in many externally-developing organisms.
The effective use of cMOs requires careful attention to several factors-some of these are important considerations when using non-caged MOs, while others are unique to cMOs and conditional gene silencing. Regardless of whether non-caged or caged MOs are used, proper controls for specificity must be included, including mRNA rescue experiments and/or the confirmation of phenotypes using multiple MOs directed at the same molecular target. The effectiveness of MOs (caged or non-caged forms) in blocking expression of the targeted gene can be assessed through the use of antibodies or, in the case of splice-blocking MOs, RT-PCR. This assessment is especially important if a morphant phenotype is not evident and an argument is being made that a targeted gene product is dispensable for a particular developmental process. In this study, we did not directly assess protein levels due to the lack of antibodies against either Lv-Vegf3 or Lv-KirrelL, but we observed morphant phenotypes that were documented in previous studies (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013; Ettensohn and Dey, 2017; Ettensohn and Adomako-Ankomah, 2019). An additional challenge is presented when cMOs or other methods are used to silence genes after the onset of expression. In such cases, perdurance of protein that was present at the time of silencing may mask the effect of a knockdown and it may be necessary to directly monitor perdurance (which will be different for each protein) through the use of antibodies. Here, we found that late knockdown of Lv-Vegf3 mimicked acute, inhibitor (axitinib)-based inactivation of Vegf3 function at the same stage. Thus, at least in this case, perdurance of the protein after knockdown did not prevent us from detecting the developmental consequences of inhibiting the function of the protein. A final issue to be considered is that some degree of phenotypic variability is typically seen in populations of morphant embryos. In this study, we found that the distributions of morphant phenotypes were the same when caged and non-caged forms were used at equivalent concentrations. Therefore, cMOs are as effective as non-caged forms and no more prone to produce phenotypic variability.
The photocaging of MO oligomers is a very direct strategy for producing light-activated MOs. In theory, this approach could be applied to any MO sequence that has been shown to effectively mediate gene knockdown. In practice, however, our use of caged T monomers currently limits this approach to MO sequences that contain at least three or four T residues (Young et al., 2008b; Deiters et al., 2010). Since MOs are usually GC-rich, we are currently applying similar synthetic chemistry to develop caged G-bases, which will provide even greater flexibility in the design of cMOs. Another useful extension of our work will be to cage MOs with other photo-cleavable groups (Bardhan and Deiters, 2019), especially red-shifted caging groups, which will provide better tissue penetration and allow multiplexed spatiotemporal gene perturbations on single specimens when used in combination with NPOM-caged MOs.
Supplementary Material
Acknowledgements
This work was supported by NIH Grant R24 OD023046.
Footnotes
Appendix A.: Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2021.02.014.
References
- Adomako-Ankomah A, Ettensohn CA, 2013. Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Angerer RC, 2003. Patterning the sea urchin embryo: gene regulatory networks, signaling pathways, and cellular interactions. Curr. Top. Dev. Biol 53, 159–198. [DOI] [PubMed] [Google Scholar]
- Angerer LM, Angerer RC, 2004. Disruption of gene function using antisense morpholinos. Methods Cell Biol. 74, 699–711. 10.1016/s0091-679x(04)74028-5. [DOI] [PubMed] [Google Scholar]
- Bardhan A, Deiters A, 2019. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol 57, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, De Robertis EM, Wallingford JB, Niehrs C, 2015. Morpholinos: antisense and sensibility. Dev. Cell 35, 145–149. [DOI] [PubMed] [Google Scholar]
- Cary GA, Cameron RA, Hinman VF, 2019. Genomic resources for the study of echinoderm development and evolution. Methods Cell Biol. 151, 65–88. [DOI] [PubMed] [Google Scholar]
- Cary GA, Hinman VF, 2017. Echinoderm development and evolution in the postgenomic era. Dev. Biol 427, 203–211. [DOI] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA, 2004. Rapid microinjection of fertilized eggs. Methods Cell Biol. 74, 287–310. 10.1016/s0091-679x(04)74013-3. [DOI] [PubMed] [Google Scholar]
- Cui M, Lin CY, Su YH, 2017. Recent advances in functional perturbation and genome editing techniques in studying sea urchin development. Brief. Funct. Genomics 16, 309–318. [DOI] [PubMed] [Google Scholar]
- Davidson EH, 2009. Network design principles from the sea urchin embryo. Curr. Opin. Genet. Dev 19, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A, Garner RA, Lusic H, Govan JM, Dush M, Nascone-Yoder NM, Yoder JA, 2010. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc 132, 15644–15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Röttinger E, Lapraz F, Besnardeau L, Lepage T, 2005. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev. Cell 9, 147–158. [DOI] [PubMed] [Google Scholar]
- Duloquin L, Lhomond G, Gache C, 2007. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134, 2293–2302. [DOI] [PubMed] [Google Scholar]
- Ekker SC, 2004. Nonconventional antisense in zebrafish for functional genomics applications. Methods Cell Biol. 77, 121–136. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR, 2019. Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettensohn CA, Adomako-Ankomah A, 2019. The evolution of a new cell type was associated with competition for a signaling ligand. PLoS Biol. 17, e3000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettensohn CA, Dey D, 2017. KirrelL, a member of the Ig-domain superfamily of adhesion proteins, is essential for fusion of primary mesenchyme cells in the sea urchin embryo. Dev. Biol 421, 258–270. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, McClay DR, 1988. Cell lineage conversion in the sea urchin embryo. Dev. Biol 125, 396–409. [DOI] [PubMed] [Google Scholar]
- Fernandez-Serra M, Consales C, Livigni A, Arnone MI, 2004. Role of the ERK-mediated signaling pathway in mesenchyme formation and differentiation in the sea urchin embryo. Dev. Biol 268, 384–402. [DOI] [PubMed] [Google Scholar]
- Foltz KR, Adams NL, Runft LL, 2004. Echinoderm eggs and embryos: procurement and culture. Methods Cell Biol. 74, 39–74. [DOI] [PubMed] [Google Scholar]
- Foster S, Teo YV, Neretti N, Oulhen N, Wessel GM, 2019. Single cell RNA-seq in the sea urchin embryo show marked cell-type specificity in the Delta/Notch pathway. Mol. Reprod. Dev 86, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Spruce T, Monteiro RS, Owens NDL, Martin SR, Smith JC, 2018. Innate immune response and off-target mis-splicing are common morpholino-induced side effects in Xenopus. Dev. Cell 44, 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JM, Young DD, Lusic H, Liu Q, Lively MO, Deiters A, 2013. Optochemical control of RNA interference in mammalian cells. Nucleic Acids Res. 41, 10518–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, 2002. Morpholino oligos: making sense of antisense? Dev. Biol 243, 209–214. [DOI] [PubMed] [Google Scholar]
- Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A, 2015. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc 137, 5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill J, Govan J, Uprety R, Tsang M, Deiters A, 2014. Site-specific promoter caging enables optochemical gene activation in cells and animals. J. Am. Chem. Soc 136, 7152–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyland A, Hodin J, Bishop C, 2014. Manipulation of developing juvenile structures in purple sea urchins (Strongylocentrotus purpuratus) by morpholino injection into late stage larvae. PloS One 9, e113866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodor PG, Ettensohn CA, 1998. The dynamics and regulation of mesenchymal cell fusion in the sea urchin embryo. Dev Biol 199, 111–124. 10.1006/dbio.1998.8924. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND, 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Oulhen N, Wessel G, Su YH, 2019. CRISPR/Cas9-mediated genome editing in sea urchins. Methods Cell Biol. 151, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Su YH, 2016. Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Dev. Biol 409, 420–428. [DOI] [PubMed] [Google Scholar]
- Liu D, Awazu A, Sakuma T, Yamamoto T, Sakamoto N, 2019. Establishment of knockout adult sea urchins by using a CRISPR-Cas9 system. Dev. Growth Differ 61, 378–388. [DOI] [PubMed] [Google Scholar]
- Liu Q, Deiters A, 2014. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc. Chem. Res 47, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zou RS, He S, Nihongaki Y, Li X, Razavi S, Wu B, Ha T, 2020. Very fast CRISPR on demand. Science 368, 1265–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe EK, Cuomo C, Arnone MI, 2017. Omics approaches to study gene regulatory networks for development in echinoderms. Brief. Funct. Genomics 16, 299–308. [DOI] [PubMed] [Google Scholar]
- Luo YJ, Su YH, 2012. Opposing nodal and BMP signals regulate left-right asymmetry in the sea urchin larva. PLoS Biol. 10, e1001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Zhu P, Shi H, Guo L, Zhang Q, Chen Y, Chen S, Zhang Z, Peng J, Chen J, 2019. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. [DOI] [PubMed] [Google Scholar]
- Martik ML, Lyons DC, McClay DR, 2016. Developmental Gene Regulatory Networks in Sea Urchins and what We Can Learn from Them. F1000Res. 5: F1000 Faculty Rev-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, 2017. Using morpholinos to probe gene networks in sea urchin. Methods Mol. Biol 1565, 87–104. [DOI] [PubMed] [Google Scholar]
- McClay DR, 2011. Evolutionary crossroads in developmental biology: sea urchins. Development 138, 2639–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR, 2016. Sea urchin morphogenesis. Curr. Top. Dev. Biol 117, 15–29. [DOI] [PubMed] [Google Scholar]
- Mehravar M, Shirazi A, Nazari M, Banan M, 2019. Mosaicism in CRISPR/Cas9-mediated genome editing. Dev. Biol 445, 156–162. [DOI] [PubMed] [Google Scholar]
- Mellott DO, Thisdelle J, Burke RD, 2017. Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Development 144, 3602–3611. [DOI] [PubMed] [Google Scholar]
- Molina MD, Gache C, Lepage T, 2019. Expression of exogenous mRNAs to study gene function in echinoderm embryos. Methods Cell Biol. 151, 239–282. [DOI] [PubMed] [Google Scholar]
- Naro Y, Darrah K, Deiters A, 2020. Optical control of small molecule-induced protein degradation. J. Am. Chem. Soc 142, 2193–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N, Swartz SZ, Laird J, Mascaro A, Wessel GM, 2017. Transient translational quiescence in primordial germ cells. Development 144, 1201–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK, 2009. Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc 131, 13255–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KD, Blitz IL, Zhou JJ, Cho KWY, 2019. Morpholinos do not elicit an innate immune response during early Xenopus embryogenesis. Dev. Cell 49, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak S, Vázquez-Maldonado LA, Deiters A, Chen JK, 2019. Combinatorial control of gene function with wavelength-selective caged morpholinos. Methods Enzymol. 624, 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, 2019. Gene redundancy and gene compensation: an updated view. J. Genet. Genom 46, 329–333. [DOI] [PubMed] [Google Scholar]
- Peter IS, 2017. Regulatory states in the developmental control of gene expression. Brief. Funct. Genomics 16, 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, 2019. Methods for the experimental and computational analysis of gene regulatory networks in sea urchins. Methods Cell Biol. 151, 89–113. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC, 2007. p53 activation by knockdown technologies. PLoS Genet. 3, e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY, 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233. [DOI] [PubMed] [Google Scholar]
- Shashikant T, Khor JM, Ettensohn CA, 2018. From genome to anatomy: the architecture and evolution of the skeletogenic gene regulatory network of sea urchins and other echinoderms. Genesis 56, e23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestopalov IA, Sinha S, Chen JK, 2007. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol 3, 650–651. [DOI] [PubMed] [Google Scholar]
- Stainier DY, Kontarakis Z, Rossi A, 2015. Making sense of anti-sense data. Dev. Cell 32, 7–8. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Gibson D, Morcos P, Li Y, Seredick S, Eisen J, Washbourne P, 2012. Turning gene function ON and OFF using sense and antisense photo- morpholinos in zebrafish. Development 139, 1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini AJ, Schuler AD, Zebala JA, Mayer AN, 2009. PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis 47, 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Davidson EH, 2014. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev. Biol 385, 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu L, Wang P, Lv C, Yang Z, Tang X, 2012. Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. (2012). Nucleic Acids Res. 40, 11155–11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JF, Miranda EL, McClay DR, 2016. Contribution of hedgehog signaling to the establishment of left-right asymmetry in the sea urchin. Dev. Biol 411, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, 2016. Germ line mechanics—and unfinished business. Curr. Top. Dev. Biol 117, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt FH, 2005. Developmental biology meets materials science: morphogenesis of biomineralized structures. Dev. Biol 280, 15–25. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Suzuki H, Kinjo S, Kiyomoto M, Ikeo K, Yamamoto T, 2020. Establishment of homozygous knock-out sea urchins. Curr. Biol 30, R427–R429. [DOI] [PubMed] [Google Scholar]
- Yamazoe S, Liu Q, McQuade LE, Deiters A, Chen JK, 2014. Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem., Int. Ed. Engl 53, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe S, Shestopalov IA, Provost E, Leach SD, Chen JK, 2012. Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem., Int. Ed. Engl 51, 6908–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DD, Edwards WF, Lusic H, Lively MO, Deiters A, 2008a. Light-triggered polymerase chain reaction. Chem. Commun. (Camb) 28 (4), 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DD, Lusic H, Lively MO, Yoder JA, Deiters A, 2008b. Gene silencing in mammalian cells with light-activated antisense agents. Chembiochem 9, 2937–2940. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen L, Zhang J, Wang Y, 2019. Drug inducible CRISPR/Cas systems. Comput. Struct. Biotechnol. J 17, 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Brown W, Bardhan A, Delaney M, Ilk AS, Rauen RR, Kahn SI, Tsang M, Deiters A, 2020. Spatiotemporal control of CRISPR/Cas9 function in cells and zebrafish using light-activated guide RNA. Angew. Chem., Int. Ed. Engl 59, 8998–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Ma Z, Guo L, Zhang W, Zhang Q, Zhao T, Jiang K, Peng J, Chen J, 2017. Short body length phenotype is compensated by the upregulation of nidogen family members in a deleterious nid1a mutation of zebrafish. J. Genet. Genom 44, 553–556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


