Abstract
Background: Despite investigation, 95% of thyroid nodules are ultimately benign. Radiomics is a field that uses radiological features to inform individualized patient care. We aimed to evaluate the diagnostic utility of radiomics in classifying undetermined thyroid nodules into benign and malignant using ultrasonography (US). Methods: A diagnostic test accuracy systematic review and meta-analysis was performed in accordance with PRISMA guidelines. Sensitivity, specificity, and area under curve (AUC) delineating benign and malignant lesions were recorded. Results: Seventy-five studies including 26,373 patients and 46,175 thyroid nodules met inclusion criteria. Males accounted for 24.6% of patients, while 75.4% of patients were female. Radiomics provided a pooled sensitivity of 0.87 (95% CI: 0.86–0.87) and a pooled specificity of 0.84 (95% CI: 0.84–0.85) for characterizing benign and malignant lesions. Using convolutional neural network (CNN) methods, pooled sensitivity was 0.85 (95% CI: 0.84–0.86) and pooled specificity was 0.82 (95% CI: 0.82–0.83); significantly lower than studies using non-CNN: sensitivity 0.90 (95% CI: 0.89–0.90) and specificity 0.88 (95% CI: 0.87–0.89) (p < 0.05). The diagnostic ability of radiologists and radiomics were comparable for both sensitivity (OR 0.98) and specificity (OR 0.95). Conclusions: Radiomic analysis using US provides a reproducible, reliable evaluation of undetermined thyroid nodules when compared to current best practice.
Keywords: thyroid nodules, radiomics, radiogenomics, ultrasound, personalized medicine
1. Introduction
Thyroid nodules occur commonly within the general population, with studies suggesting a prevalence of 20–67%, with an increased propensity in females and the elderly [1,2]. Increased access to healthcare and availability of modern imaging techniques such as ultrasonography (US) have led to the markedly increased detection of thyroid nodules [3]. The American Thyroid Association (ATA), British Thyroid Association (BTA), and European Society for Medical Oncology (ESMO) guidelines recommend US as the primary imaging modality for the assessment of thyroid nodules [4,5,6]. Several classification systems (e.g., ATA, BTA, and Thyroid Imaging Reporting and Data System (TIRADS)) are utilized by radiologists to stratify the risk of malignancy for each thyroid nodule based on US features [4,5,7]. These systems classify lesions on a scale ranging from benign to malignant based on sonographic parameters such as size, echogenicity, degree of margin irregularity, nodule height to width ratio, extra nodular extension, and calcification [4,5,7]. Suspicious nodules then proceed to fine-needle aspiration cytology (FNAC), where they are reassessed and graded as non-diagnostic, benign, atypical, suspicious for malignancy, or definitively malignant, using cytological reporting systems such as the Bethesda or Thy classification systems [8,9]. At present, patients with undetermined nodules following FNAC undergo surgery in order to obtain a definitive histological diagnosis, with 95% of nodules subsequently being stratified as “benign” [10], leading to the conceptualization that the present paradigm is guilty of overexposing patients to unnecessary over-investigation and overtreatment. Thus, it is vital for translational research efforts to focus on means of accurately screening and sub stratifying detected thyroid nodules into benign and malignant categories [11].
Precision medicine builds on the mantra that every patient, cancer, and disease process possesses its own characteristics with individualized diagnoses, prognoses, and management strategies. The clinical application of artificial intelligence (AI) has advanced the field of precision medicine through the exploration of hypotheses in large data sets [12]. Radiomics (or radiogenomics) is a rapidly evolving field that uses AI to extract vast quantities of data from medical imaging [13]. It represents a quantitative approach to medical imaging through mathematical extraction of the spatial distribution of signal intensities and pixel interrelationships, quantifying textural information by using AI analysis methods. Various radiomic methods exist at present, including radiomic AI, machine learning (ML), convolutional neural networks (CNN), and other deep-learning techniques. The radiomic process involves numerous steps incorporating image acquisition, image segmentation, quantitative feature extraction, computational analysis, and finally, computational modeling [14]. Through this use of vast amounts of data, radiomics provides a quick, reproducible, and objective analysis that can inform individualized diagnostics, sub stratification, prognostication, and future management of disease [13,14].
Due to the ever-increasing number of thyroid nodules detected, there is significant interest within the literature to develop novel strategies to inform diagnostics within the clinical workup of thyroid nodules [15]. Current data suggests radiomic imaging analysis may be capable of accurately stratifying thyroid lesions into benign and malignant based on data captured using sonographic imaging. Accordingly, the aim of the present study was to determine whether radiomic imaging analysis can provide an accurate evaluation of thyroid nodules undergoing diagnostic US evaluation.
2. Materials and Methods
A systematic review was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [16] and in accordance with the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy [17]. Local institutional ethical approval was not required.
2.1. Population, Intervention, Comparison, Outcomes (PICO)
Population: Patients who have undergone preoperative US and definitive thyroid nodule diagnosis as benign or malignant.
Intervention: Radiomic analyses applied to preoperative US used to inform whether thyroid nodules are benign or malignant.
Comparison: The discriminative ability of radiomics compared to confirmation of benign and malignant nodules. Nodules were determined as benign by either cytological or histological means, while malignancy was confirmed by histological analysis only.
Outcomes: Primary outcomes included the evaluation of the clinical utility of preoperative US imaging to stratify thyroid nodules as either benign or malignant. Generated pooled sensitivity, specificity, and receiver operating characteristic (ROC) curve analyses will be representative of our primary outcomes. Secondary outcomes include comparing the ability of different radiomic methods to differentiate such nodules and to compare radiologists and radiomics in correctly discriminating benign versus malignant thyroid nodules.
2.2. Search Strategy
An electronic search of the PubMed Medline, EMBASE, and Scopus databases was performed on 16 January 2021 for relevant studies. This search was performed for the following headings: (Thyroid Cancer) and (Radiomics) linked using the boolean operator “AND”. Included studies were limited to those published in the English language and were not restricted based on the year of publication. All duplicate studies were manually removed before titles were screened, and studies deemed appropriate had their abstracts reviewed. Studies remaining had their full texts reviewed for eligibility.
2.3. Inclusion and Exclusion Criteria
Studies meeting the following inclusion criteria were included: (1) Studies with thyroid nodules confirmed as benign or malignant following US imaging; (2) imaging of tumors had to have been performed pre-diagnosis; (3) either stated study numbers of true positive, true negative, false positive, false negative, sensitivity, specificity, or accuracy data in relation radiomic tests or the ability to calculate these figures based on study data. In some cases, sensitivity and specificity were calculated from ROC curve analyses. Studies comparing the diagnostic ability of radiologists with radiomics were also included. Studies meeting any of the following exclusion criteria were excluded from this study: (1) studies not providing radiomic validation or “test” data, (2) studies outlining the diagnostic ability of radiomics differentiating benign and malignant lesions in other cancers (e.g., breast carcinoma, skin cancers, etc.), (3) studies with no full English text, (4) review articles, (5) studies including less than five patients in their series or case reports, and (6) editorial articles.
2.4. Data Extraction and Quality Assessment
This literature search was performed by two independent reviewers (E.F.C. and S.O.) using the aforementioned search strategy. Where discrepancies in opinion occurred between the reviewers, a third reviewer was asked to arbitrate (M.G.D.). As described, duplicate studies were removed. Both reviewers reviewed all retrieved manuscripts to ensure all inclusion criteria were met before extracting the following data: (1) first author name, (2) year of publication, (3) study design, (4) country, (5) level of evidence, (6) study title, (7) number of patients, (8) number of benign and malignant nodules confirmed though cytologic or histopathologic analysis, (9) sensitivity, specificity, and area under curve (AUC) scores from the ROC curve analyses obtained from radiomic “test” data and (10) sensitivity, specificity, and AUC scores from the ROC analyses from radiologists within studies where available. Sensitivity and specificity were directly extracted from tables and study text. When not provided as discrete data in tables or the text, specific estimates of sensitivity and specificity were calculated from ROC curves with the most accurate and appropriate sensitivity prioritized. Where studies tested the diagnostic ability of multiple radiomic methods (i.e., CNN, ML, etc.), only data for the best performing radiomic method within that study was extracted. Similarly, where studies detailed data on multiple radiologists’ ability to discriminate benign versus malignant nodules, data from the best performing radiologist from that particular study was included. Appraisal of the quality of each study was performed using the radiomics quality score (RQS), as outlined previously by Lambin et al. [18].
2.5. Statistical Analysis
Statistical analysis was performed according to the Cochrane guidelines. Pooled sensitivity and specificity and summary ROC analysis were calculated for included studies to demonstrate to convey the diagnostic test performance of radiomics in differentiating malignant thyroid nodules from benign thyroid nodules. We then performed a comparison between studies using CNNs (incorporating both CNNs and other deep learning methods) versus those using either ML or Radiomic AI analyses (together termed non-CNNs). For comparing radiologist and radiomic diagnostic test accuracy, sensitivity and specificity data were expressed as dichotomous data and reported as odds ratios (ORs) with 95% confidence intervals (CIs) following estimation using the Mantel–Haenszel method using random effects. The symmetry of funnel plots was used to assess publication bias. Statistical heterogeneity was determined using I2 statistics. Statistical significance was determined to be p < 0.05. Statistical analysis was performed using Review Manager (RevMan), Version 5.4 (Nordic Cochrane Centre, Copenhagen, Denmark).
3. Results
3.1. Literature Search
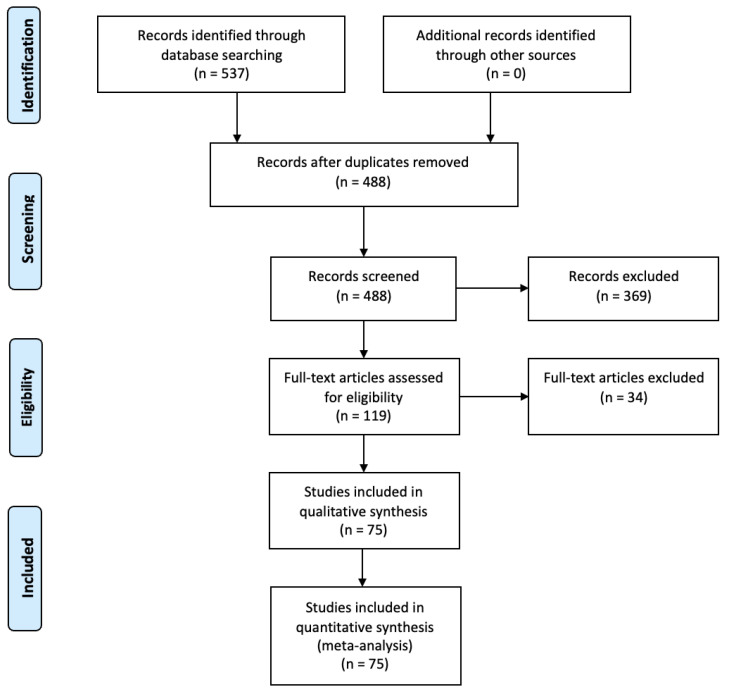
The initial search of PUBMED, SCOPUS, and EMBASE resulted in a total of 537 studies identified. Following the removal of duplicates, 488 studies remained. These studies were then screened by title and abstract for relevance, after which 119 studies remained—all had their full text analyzed for eligibility. Finally, 75 studies remained for inclusion in the analysis as depicted by Figure 1 [19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93].
Figure 1.
PRISMA flow diagram detailing the systematic search process.
3.2. Study Characteristics
Overall, 75 studies arising from 15 different countries met inclusion and exclusion criteria (19–93), 8 studies were prospective in nature (25, 31, 38, 55, 57, 81, 83, 84) while the remaining 67 studies were retrospective. Of the included studies, 46 used convolutional neural networking (CNN) to analyse thyroid nodule US images (19, 25, 26, 29, 30, 33, 36–38, 40–45, 47, 48, 50–55, 57, 59, 63–65, 67–71, 73, 74, 77, 79, 81–84, 89–93), 29 studies used non CNN methods (20–24, 27, 28, 31, 32, 34, 35, 39, 46, 49, 56, 58, 60–62, 66, 72, 75, 76, 78, 80, 85–88) (Table 1).
Table 1.
Study characteristics and demographics.
| Author | Year | Study Type (LOE) | Radiomics | Country | US Device Brand | N Patients | Male | Female | Mean Age |
|---|---|---|---|---|---|---|---|---|---|
| Zhou | 2020 | RC (III) | CNN | China | Esaote/Phillips | 105 | 25 | 80 | 47.9 |
| Nguyen | 2019 | RC (III) | CNN | Korea | NS | 61 | NS | NS | NS |
| Wei | 2020 | RC (III) | CNN | China | NS | 2489 | 614 | 1875 | 45.3 |
| Park | 2019 | PC (II) | CNN | Korea | Samsung | 265 | 52 | 213 | 47.1 |
| Thomas | 2020 | RC (III) | CNN | USA | 4 brands | 103 | NS | NS | NS |
| Wei (2) | 2020 | RC (III) | Non-CNN | China | 5 brands | NS | NS | NS | 47 |
| Liu | 2019 | RC (III) | CNN | China | Vinno | 131 | 54 | 77 | 46.7 |
| Stib | 2020 | RC (III) | CNN | USA | Siemens/GE/Phillips | 571 | 234 | 337 | 52.9 |
| Ye | 2020 | RC (III) | CNN | China | 5 brands | 166 | 46 | 100 | 44.6 |
| Ma | 2020 | RC (III) | CNN | China | NS | 211 | 34 | 177 | NS |
| Koh | 2020 | RC (III) | CNN | Korea | 11 brands | 200 | 49 | 151 | 49.6 |
| Kwon | 2020 | RC (III) | CNN | Korea | Phillips/Hitachi | 762 | NS | NS | NS |
| Kim | 2019 | RC (III) | Non-CNN | Korea | Samsung | 106 | 29 | 77 | 48 |
| Zhao | 2020 | RC (III) | Non-CNN | China | Phillips/Hitachi | 174 | 44 | 130 | 45 |
| Qin | 2019 | RC (III) | CNN | China | NS | 233 | NS | NS | NS |
| Zhu | 2021 | RC (III) | CNN | China | 4 brands | 102 | 0 | 102 | 54.8 |
| Liu (2) | 2019 | RC (III) | CNN | China | GE | 376 | NS | NS | NS |
| Xia | 2019 | PC (II) | CNN | China | Samsung | 171 | 32 | 139 | 47.2 |
| Zhao | 2021 | RC (III) | Non-CNN | China | SuperSonic | 102 | 25 | 77 | 50.6 |
| Lee | 2019 | RC (III) | CNN | Korea | Phillips/Hitachi | 519 | 93 | 426 | 47.5 |
| Ataide | 2020 | RC (III) | Non-CNN | Germany | NS | 99 | NS | NS | NS |
| Chen | 2020 | RC (III) | Non-CNN | China | GE/Hitachi | 1480 | 302 | 1178 | 45.6 |
| Zhu (2) | 2021 | RC (III) | CNN | China | Phillips/GE/Toshiba | 261 | 64 | 197 | 52 |
| Shi | 2020 | RC (III) | CNN | China | Esaote/Hitachi/Toshiba | NS | NS | NS | NS |
| Barczyński | 2020 | PC (II) | CNN | Poland | Samsung | 50 | 9 | 41 | 47.5 |
| Zhang | 2020 | RC (III) | Non-CNN | Korea | Siemens | 303 | 59 | 244 | 46.4 |
| Wei (3) | 2020 | RC (III) | Non-CNN | China | Samsung | 181 | 35 | 146 | 46 |
| Colakoglu | 2019 | RC (III) | Non-CNN | Turkey | GE | 198 | 48 | 150 | 44.5 |
| Park | 2021 | RC (III) | Non-CNN | Korea | Phillips | 325 | 61 | 264 | 50.1 |
| Nguyen | 2020 | RC (III) | CNN | Korea | NS | 61 | NS | NS | NS |
| Sun | 2020 | RC (III) | CNN | China | GE | 338 | 134 | 416 | 43.8 |
| Peng | 2021 | PC (II) | CNN | China | 13 brands | 2775 | 726 | 2049 | 42.2 |
| Liu | 2021 | RC (III) | CNN | China | Siemens | 163 | 48 | 115 | 44.3 |
| Han | 2021 | RC (III) | CNN | Korea | Samsung | 372 | NS | NS | NS |
| Shin | 2020 | RC (III) | CNN | Korea | Samsung | 340 | 79 | 261 | 47.2 |
| Wang | 2019 | RC (III) | CNN | China | GE/Phillips | 276 | 53 | 223 | 46.3 |
| Zhu | 2019 | RC (III) | CNN | China | 4 brands | 467 | 97 | 370 | 45.3 |
| Zhang (2) | 2019 | RC (III) | Non-CNN | China | Hitachi | 2032 | 695 | 1337 | 42.3 |
| Ko | 2019 | RC (III) | CNN | Korea | Phillips/Hitachi | 150 | 23 | 127 | 49.7 |
| Song | 2019 | RC (III) | CNN | Korea | Toshiba | 100 | NS | NS | NS |
| Li | 2019 | RC (III) | CNN | China | Phillips/Toshiba/GE | 154 | 34 | 120 | 51 |
| Wildman-Tobriner | 2019 | RC (III) | Non-CNN | UK | Siemens/GE/Phillips | 94 | 21 | 73 | 52.6 |
| Yu | 2017 | PC (II) | CNN | China | Phillips/Siemens | 50 | 9 | 41 | 48.4 |
| Buda | 2019 | RC (III) | CNN | USA | Siemens/GE/Phillips | 91 | NS | NS | 52.3 |
| Raghavendra | 2018 | RC (III) | Non-CNN | India | 4 brands | 344 | NS | NS | 44.1 |
| Li | 2018 | RC (III) | CNN | China | NS | 300 | 53 | 247 | NS |
| Ma | 2017 | RC (III) | CNN | china | 7 brands | 4782 | NS | NS | 52 |
| Raghavendra | 2017 | RC (III) | Non-CNN | India | GE | 242 | 63 | 179 | 44.1 |
| Zhu | 2013 | RC (III) | CNN | China | Siemens | 618 | 161 | 528 | 47.7 |
| Choi | 2017 | PC (II) | Non-CNN | Korea | Samsung | 89 | 18 | 71 | 43.5 |
| Gao | 2017 | RC (III) | CNN | China | Phillips/GE | 342 | 70 | 272 | 44.8 |
| Choi | 2015 | RC (III) | CNN | Korea | Phillips | 85 | 24 | 61 | 52 |
| Chi | 2017 | RC (III) | CNN | Canada | Toshiba | 61 | NS | NS | NS |
| Jeong | 2019 | PC (II) | CNN | Korea | Samsung | 76 | NS | NS | NS |
| Acharya | 2012 | RC (III) | Non-CNN | Singapore | NS | 20 | 10 | 10 | NS |
| Liang | 2018 | RC (III) | Non-CNN | China | Phillips | 95 | 20 | 75 | 43.2 |
| Prochazka (2) | 2019 | RC (III) | Non-CNN | Czechia | Phillips/GE | 60 | 11 | 49 | 55.7 |
| Song | 2015 | RC (III) | Non-CNN | China | GE | 147 | 32 | 115 | NS |
| Ardakani | 2015 | RC (III) | Non-CNN | Iran | Medison | 60 | NS | NS | NS |
| Guan | 2019 | RC (III) | CNN | China | NS | 399 | NS | NS | NS |
| Xia | 2017 | RC (III) | Non-CNN | China | Siemens | 187 | 36 | 151 | 50.8 |
| Yoo | 2018 | PC (II) | CNN | Korea | Samsung | 50 | 10 | 40 | 43.2 |
| Tsantis | 2009 | RC (III) | Non-CNN | Greece | Phillips | 85 | NS | NS | NS |
| Liu | 2008 | RC (III) | Non-CNN | USA | NS | 37 | NS | NS | NS |
| Acharya | 2013 | RC (III) | Non-CNN | Italy | Esaote | 20 | 10 | 10 | 52.8 |
| Acharya (2) | 2012 | RC (III) | Non-CNN | Italy | Esaote | 20 | 10 | 10 | 52.8 |
| Acharya | 2011 | RC (III) | Non-CNN | Italy | Esaote | 20 | 10 | 10 | 52.8 |
| Kweon Seo | 2017 | RC (III) | CNN | Korea | NS | 230 | 51 | 179 | 48.7 |
| Ardakani (2) | 2015 | RC (III) | CNN | Iran | Medison | 60 | NS | NS | NS |
| Wu | 2016 | RC (III) | CNN | China | Phillips | 970 | 214 | 756 | 46.7 |
| Cao | 2019 | RC (III) | Non-CNN | China | NS | 120 | NS | NS | NS |
| Wang (2) | 2020 | RC (III) | CNN | China | NS | 1040 | NS | NS | NS |
| Sun (2) | 2020 | RC (III) | CNN | China | NS | 245 | NS | NS | NS |
| Reverter | 2019 | RC (III) | Non-CNN | Spain | GE | 300 | 45 | 255 | 55.5 |
| Gitto | 2019 | RC (III) | Non-CNN | Italy | Samsung | 62 | 12 | 50 | 60 |
NS: not specified, LOE: level of evidence, RC: retrospective cohort, PC: prospective cohort, CNN: convolutional neural network, non-CNN: analysis performed using a method other than a convolutional neural network, GE: General Electric.
3.3. Clinicopathological Characteristics
Overall, there were 28,373 patients with 46,175 thyroid nodules included from the 75 studies. Males accounted for 24.6% of patients, while 75.4% of patients were female. There were 51 studies reporting mean patient age; within these studies, mean patient age was 48.3 years (range: 42.2–69.0 years) (Table 1).
Overall, 22,814 (49.4%) nodules were benign while 23,361 (50.6%) of nodules were malignant. Within included studies, 35 reported mean nodule size; mean nodule size in these studies was 19.7 mm (range 8.3–31.7 mm). We found 34 studies provided a breakdown of malignant nodules by subtype. Papillary thyroid carcinoma (PTC) was the most prevalent subtype of malignant thyroid nodule within these studies, representing 94.7% of malignant thyroid nodules (Table 2).
Table 2.
Study characteristics and demographics.
| Author | Year | N Nodules | Mean Nodule Size (mm) | N Benign Nodules | N Malignant Nodules | Papillary Ca | Follicular Ca | Medullary Ca | Other Thyroid Ca |
|---|---|---|---|---|---|---|---|---|---|
| Zhou | 2020 | 105 | NS | 75 | 30 | NS | NS | NS | NS |
| Nguyen | 2019 | 61 | NS | 11 | 50 | NS | NS | NS | NS |
| Wei | 2020 | 2489 | NS | 1021 | 1468 | 1442 | 11 | 15 | 0 |
| Park | 2019 | 286 | 16.2 | 130 | 156 | 149 | 6 | 1 | 0 |
| Thomas | 2020 | 103 | NS | 70 | 33 | 24 | 3 | 2 | 4 |
| Wei (2) | 2020 | 7560 | NS | 3063 | 4587 | NS | NS | NS | NS |
| Liu | 2019 | 131 | 16.1 | 59 | 72 | 72 | 0 | 0 | 0 |
| Stib | 2020 | 651 | NS | 500 | 151 | NS | NS | NS | NS |
| Ye | 2020 | 209 | NS | 109 | 100 | NS | NS | NS | NS |
| Ma | 2020 | 846 | NS | 360 | 486 | NS | NS | NS | NS |
| Koh | 2020 | 200 | 22.4 | 102 | 98 | 97 | 0 | 0 | 1 |
| Kwon | 2020 | 762 | NS | 325 | 437 | 437 | 0 | 0 | 0 |
| Kim | 2019 | 218 | 12 | 132 | 86 | 86 | 0 | 0 | 0 |
| Zhao | 2020 | 177 | 21.8 | 96 | 81 | 81 | 0 | 0 | 0 |
| Qin | 2019 | 248 | NS | 115 | 133 | NS | NS | NS | NS |
| Zhu | 2021 | NS | NS | 57 | 45 | NS | NS | NS | NS |
| Liu (2) | 2019 | 450 | NS | 128 | 322 | NS | NS | NS | NS |
| Xia | 2019 | 180 | 10.3 | 85 | 95 | 91 | 4 | 0 | 0 |
| Zhao | 2021 | 106 | 17.3 | 73 | 33 | NS | NS | NS | NS |
| Lee | 2019 | 589 | 12.9 | 193 | 396 | 395 | 1 | 0 | 0 |
| Ataide | 2020 | 99 | NS | 17 | 82 | NS | NS | NS | NS |
| Chen | 2020 | 1558 | NS | 347 | 1211 | NS | NS | NS | NS |
| Zhu (2) | 2021 | 1032 | NS | 502 | 530 | NS | NS | NS | NS |
| Shi | 2020 | 1937 | NS | 1032 | 905 | NS | NS | NS | NS |
| Barczyński | 2020 | NS | 30.5 | 40 | 10 | 10 | 0 | 0 | 0 |
| Zhang | 2020 | 365 | 18.3 | 179 | 186 | 168 | 11 | 7 | 0 |
| Wei (3) | 2020 | 204 | 15 | 112 | 92 | 90 | 1 | 0 | 1 |
| Colakoglu | 2019 | 235 | NS | 133 | 102 | 102 | 0 | 0 | 0 |
| Park | 2021 | 325 | 21 | 257 | 68 | NS | NS | NS | NS |
| Nguyen | 2020 | NS | NS | 11 | 50 | NS | NS | NS | NS |
| Sun | 2020 | 550 | 14 | 128 | 422 | NS | NS | NS | NS |
| Peng | 2021 | 2775 | NS | 2472 | 303 | 299 | 4 | 0 | 0 |
| Liu | 2021 | 175 | 11.9 | 67 | 108 | 103 | 5 | 0 | 0 |
| Han | 2021 | 454 | 17.8 | 287 | 167 | 161 | 4 | 2 | 0 |
| Shin | 2020 | 348 | 31 | 252 | 96 | 0 | 96 | 0 | 0 |
| Wang | 2019 | NS | 18.5 | 95 | 181 | NS | NS | NS | NS |
| Zhu | 2019 | 467 | 8.3 | 128 | 339 | NS | NS | NS | NS |
| Zhang (2) | 2019 | 2064 | NS | 1314 | 750 | NS | NS | NS | NS |
| Ko | 2019 | 150 | 12.9 | 50 | 100 | NS | NS | NS | NS |
| Song | 2019 | 100 | NS | 50 | 50 | NS | NS | NS | NS |
| Li | 2019 | 154 | NS | 70 | 84 | NS | NS | NS | NS |
| Wildman-Tobriner | 2019 | 100 | 27.1 | 85 | 15 | NS | NS | NS | NS |
| Yu | 2017 | 50 | NS | 33 | 17 | 16 | 0 | 1 | 0 |
| Buda | 2019 | 99 | 27 | 84 | 15 | NS | NS | NS | NS |
| Raghavendra | 2018 | 344 | NS | 288 | 56 | NS | NS | NS | NS |
| Li | 2018 | NS | NS | 50 | 250 | 250 | 0 | 0 | 0 |
| Ma | 2017 | 8148 | 25 | 4126 | 4022 | NS | NS | NS | NS |
| Raghavendra | 2017 | 242 | NS | 211 | 31 | NS | NS | NS | NS |
| Zhu | 2013 | 689 | 13.3 | 265 | 465 | NS | NS | NS | NS |
| Choi | 2017 | 102 | 12 | 59 | 43 | 43 | 0 | 0 | 0 |
| Gao | 2017 | 342 | 12.1 | 103 | 239 | NS | NS | NS | NS |
| Choi | 2015 | 99 | NS | 21 | 78 | 77 | 1 | 0 | 9 |
| Chi | 2017 | NS | NS | 11 | 50 | NS | NS | NS | NS |
| Jeong | 2019 | 100 | 17 | 56 | 44 | 43 | 1 | 0 | 0 |
| Acharya | 2012 | 20 | NS | 10 | 10 | 7 | 1 | 0 | 2 |
| Liang | 2018 | 95 | 16 | 43 | 52 | 51 | 1 | 0 | 0 |
| Prochazka (2) | 2019 | 60 | NS | 40 | 20 | NS | NS | NS | NS |
| Song | 2015 | 155 | NS | 76 | 79 | NS | NS | NS | NS |
| Ardakani | 2015 | 60 | NS | 26 | 34 | NS | NS | NS | NS |
| Guan | 2019 | 399 | NS | 190 | 209 | 209 | 0 | 0 | 0 |
| Xia | 2017 | 203 | 24.8 | 114 | 89 | NS | NS | NS | NS |
| Yoo | 2018 | 117 | 15 | 67 | 50 | 50 | 0 | 0 | 0 |
| Tsantis | 2009 | 85 | NS | 54 | 31 | NS | NS | NS | NS |
| Liu | 2008 | 41 | NS | 21 | 20 | 18 | 0 | 0 | 2 |
| Acharya | 2013 | 20 | 31.7 | 10 | 10 | 7 | 1 | 0 | 2 |
| Acharya (2) | 2012 | 20 | 31.7 | 10 | 10 | 7 | 1 | 0 | 2 |
| Acharya | 2011 | 20 | 31.7 | 10 | 10 | 7 | 1 | 0 | 2 |
| Kweon Seo | 2017 | 230 | 29.4 | 191 | 39 | 0 | 39 | 0 | 0 |
| Ardakani (2) | 2015 | 60 | NS | 26 | 34 | NS | NS | NS | NS |
| Wu | 2016 | 970 | NS | 463 | 507 | 487 | 12 | 4 | 4 |
| Cao | 2019 | 120 | NS | 73 | 47 | NS | NS | NS | NS |
| Wang (2) | 2020 | 3120 | NS | 1393 | 1841 | NS | NS | NS | NS |
| Sun (2) | 2020 | 245 | NS | 145 | 100 | NS | NS | NS | NS |
| Reverter | 2019 | 300 | 29.8 | 165 | 135 | 112 | 15 | 3 | 5 |
| Gitto | 2019 | 62 | 18 | 48 | 14 | NS | NS | NS | NS |
NS: not specified, Ca: cancer.
3.4. Diagnostic Ability of Radiomics
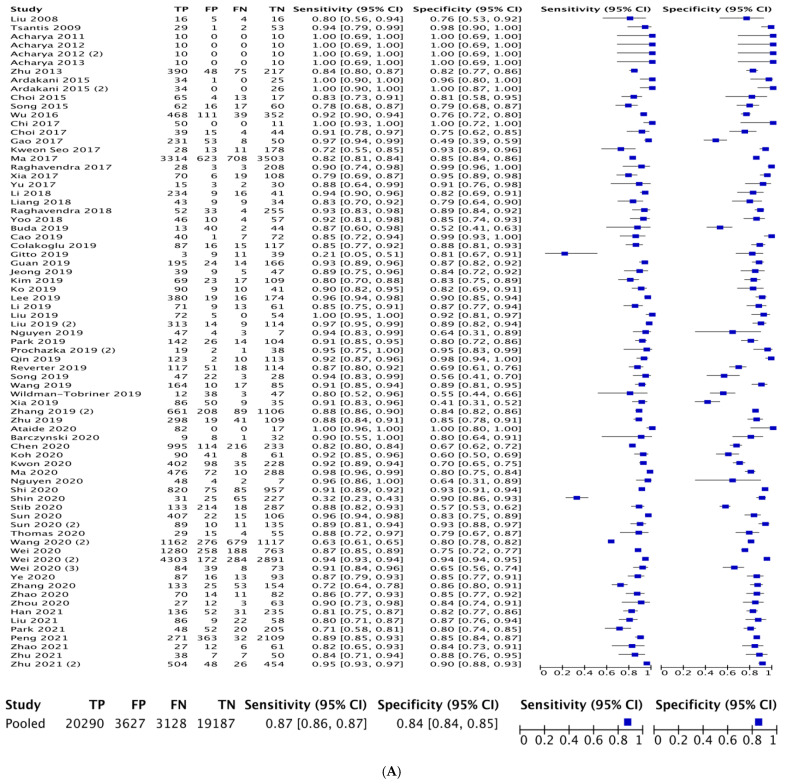
The mean AUC calculated from independent ROC curve analyses within included studies was 0.88 (range: 0.61–1.00). Individual study sensitivity and specificity for determining malignant versus benign thyroid nodules is demonstrated in Figure 2A. Pooled sensitivity for radiomics in distinguishing thyroid nodules was 0.87 (95% CI: 0.86–0.87). Pooled specificity for radiomics in distinguishing thyroid nodules was 0.84 (95% CI: 0.84–0.85). A combined ROC curve for radiomics of thyroid nodules by ultrasound sonography is demonstrated in Figure 2B.
Figure 2.
(A) Overall sensitivity and specificity of radiomics. (B) Receiver operating characteristic (ROC) curve of malignant versus benign thyroid nodules based on radiomic analyses.
3.5. Comparison of CNN versus Non-CNN Radiomics
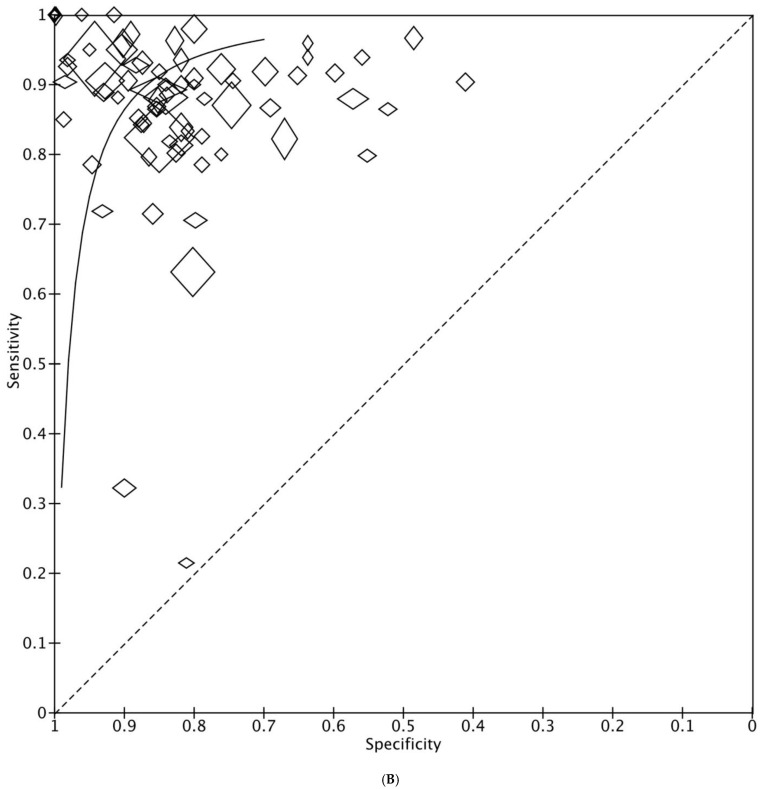
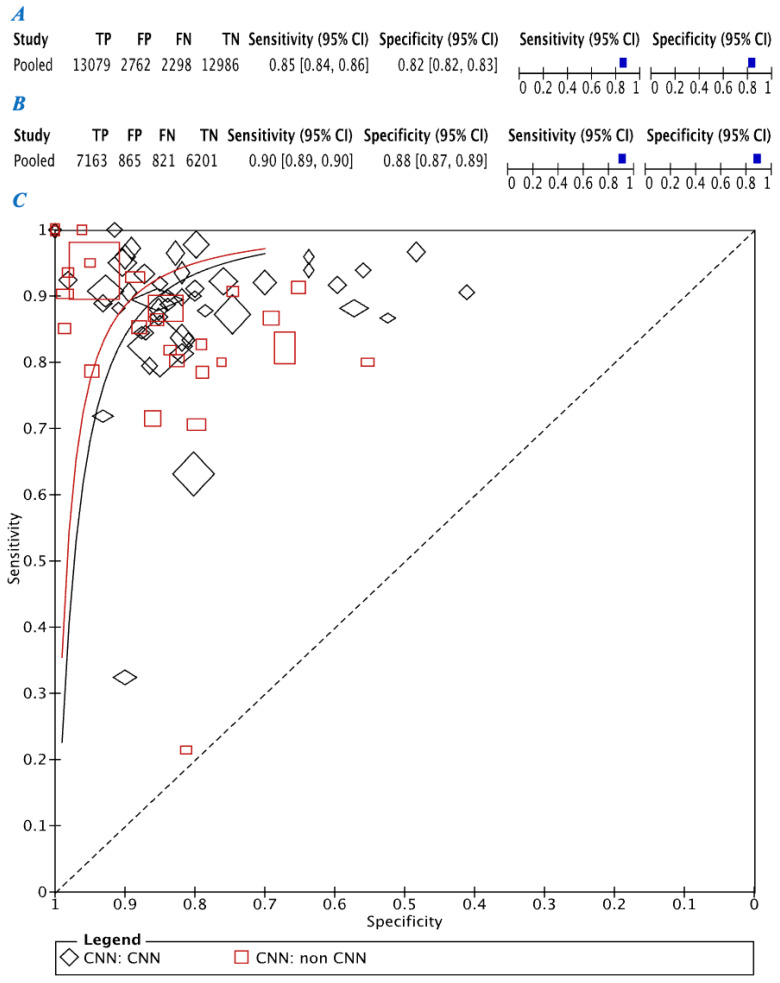
For studies using CNN pooled sensitivity was 0.85 (95% CI: 0.84–0.86) and pooled specificity was 0.82 (95% CI: 0.82–0.83). Pooled sensitivity 0.90 (95% CI: 0.89–0.90) and specificity 0.88 (95% CI: 0.87–0.89) was significantly higher in studies using non-CNN radiomics (p < 0.05) (Figure 3A,B). ROC curve comparison between CNN and non-CNN methods is outlined in Figure 3C.
Figure 3.
(A) Pooled sensitivity and specificity of convolutional neural network (CNN) analyses and (B) represents pooled sensitivity and specificity of non-CNN analyses. (C) Depicts the receiver operating characteristic (ROC) curve for CNN analyses (black) versus non-CNN analyses (red).
3.6. Comparison of Radiomic Analysis of Thyroid Nodule US versus Radiologists Analysis of Thyroid Nodule US
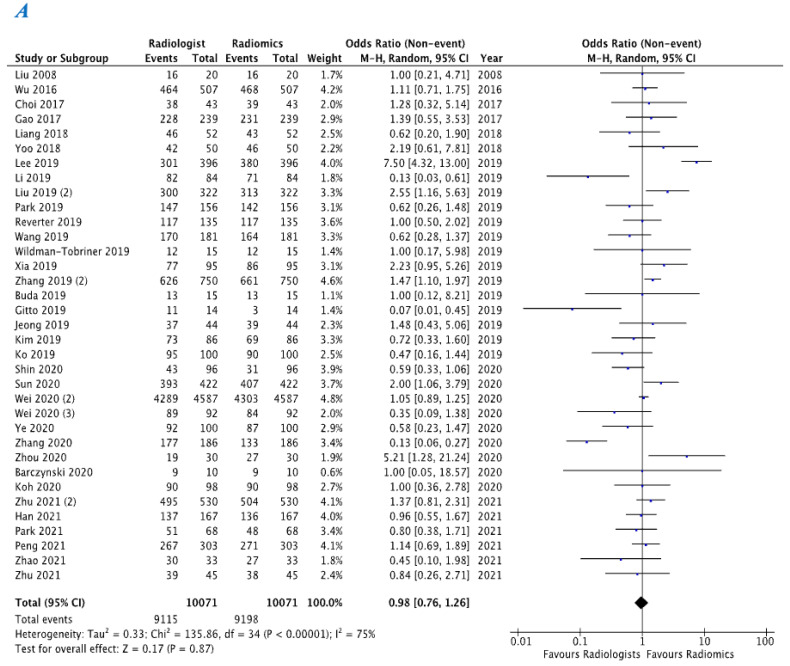
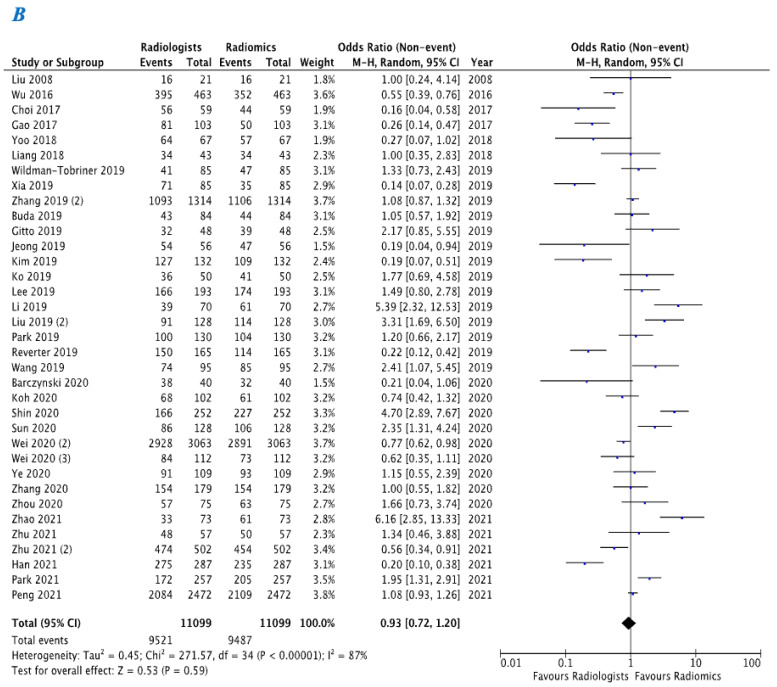
Within the studies included in the meta-analysis, 35 studies provided a comparison between radiologists and radiomics in differentiating malignant versus benign thyroid nodules using thyroid US. Radiomics demonstrated similar sensitivity for detection of malignancy within a given thyroid nodule (OR 0.98, 95% CI 0.76–1.26) when compared with radiologists (Figure 4A). Radiomics also demonstrated similar specificity (OR 0.93, 95% CI 0.72–1.20) when compared with radiologists for this purpose (Figure 4B).
Figure 4.
(A) Represents sensitivity comparison between radiologists and radiomics. (B) Represents specificity comparison between radiologists and radiomics.
4. Discussion
To the best of our knowledge, the current systematic review and meta-analysis is the first to evaluate the diagnostic test accuracy of radiomic imaging analysis in differentiating malignant from benign thyroid nodules using US. Due to the increasing prevalence of thyroid nodules now detected within the general population and the rising incidence of thyroid malignancy (which has tripled since 1975), accurate risk stratification is paramount to the enhancement of clinical outcomes [3]. The most important finding in this analysis of over 28,000 patients possessing over 46,000 thyroid nodules is the data supporting the utility of radiomic analysis in correctly stratifying undetermined thyroid nodules correctly into benign and malignant lesions (sensitivity: 0.87, specificity: 0.84). This is promising as we look to enhance diagnostics in this field of oncology, all the while promoting minimally invasive techniques in order to reduce morbidity and mortality for prospective patients. These results come at the timely promotion of precision oncology as a rapidly evolving field, which manipulates individual patient, cancer, or disease process characteristics in order to develop a personalized diagnosis, prognosis, and treatment strategies [12]. Data from this analysis support radiomic imaging analysis using US as a means of quantification of malignancy in thyroid nodules, without exposing patients to the risks associated with invasive FNAC sampling or surgical specimen assessment. For some patients, the use of radiomics could possibly circumvent the need for FNAC and surgical resection, providing a potentially more cost and time-efficient assessment of thyroid nodules than what is currently practiced [20,94].
Results of this analysis indicate that radiomics is a novel avenue worth exploring in the differentiation of benign and malignant thyroid lesions. CNN provided a pooled sensitivity of 85% and specificity of 82% compared to a pooled sensitivity of 90% and pooled specificity of 88% in non-CNN. CNN is designed as an automated means to adaptively learn spatial hierarchies of features through backpropagation by using multiple building blocks: convolution layers, pooling layers, and fully connected layers of data processing [95]. CNN has powerful pattern recognition capabilities due to the fact that they can approximate any continuous function, given an appropriate network structure [96]. In neural networking, high variance gives networks the ability to learn complex patterns, although it also runs the risk of overfitting since models will learn peculiarities, or noise, from a data set [96]. The noise phenomenon incorporates features into the model which are not generalizable outside of the training set [95]. This makes the model appear to perform well in training but fail to perform in a true clinical environment. Such overfitting in the setting of CNN has been noted in studies evaluating papillary thyroid nodules on US [44]. The margin between benign thyroidal tissue and malignant tissue may be unclear or blurry on US imaging, with significant overlap between cancerous and normal or benign regions. Thus, it is then challenging for the CNN model to perform accurate textural feature extraction of the malignant tissue, possibly contributing to poor model performance [44]. Ideally, a CNN should have a large training set to mitigate the risk of overfitting, but this is not always feasible due to cost, time, and other factors limiting available data [95]. Non-CNN incorporates a number of methods such as support vector machines (SVM), random forest (RF), k-nearest neighbor (k-nn), and Bayesian classifiers [97]. Each method has its own strengths and weakness. For example, SVM classifiers are based on decision planes that define decision boundaries. SVM is often used for the principle of structural risk minimization, which allows robust analysis of test data without the need for a large training set through margin maximization [98]. Another popular ML method is RF, which consists of a large network of individual decision trees that allows for ensemble learning, providing the benefit of human-readable data and the ability to adjust the classifiers’ decision trees where appropriate [97]. Ultimately, the randomness of this model makes it robust, generalizable, and less prone to overfitting, although large numbers of decision trees make this approach more time-consuming. Within our analysis, small-data and overfitting within individual studies may have contributed to the overall worse performance of CNN versus non-CNN. Based on the results of this meta-analysis non-CNN radiomics should be the preferred methods for evaluating the risk of malignancy in an undetermined thyroid nodule using US.
For detecting malignancy within a given thyroid nodule radiomic methods had similar sensitivity (0.98, 95% CI 0.76–1.26) and specificity (0.93, 95% CI 0.72–1.20) when compared with radiologists. However, acknowledgment for the strengths maintained by radiologists compared to radiomics: At present, radiomic models are dependent on high-quality image acquisition and segmentation by radiologists. Without good imaging data to analyze, the radiomic model is unable to correctly stratify nodules [99]. Radiologists also maintain the innate ability to incorporate the global context of patients and the ability to maintain subjective associations based on experience, which current radiomic models are unable to perform. Radiomics can face issues with model fitting, poor input data, and subsequent suboptimal performance [100]. However, human assessment of medical imaging and, in particular, US suffers from significant inter-observer variability [101,102]. Radiomics, on the other hand, provides the benefit of an objective, quick, and reproducible analysis with the ability to analyze features of the nodule that are both visible to the radiologist and textural features occult to human perception [13,14]. Studies have attempted to blend the strengths of both radiologists and radiomic models to form computer-assisted diagnosis (CAD) tools. While CAD was not evaluated within the confines of the current meta-analysis, CAD has shown to be of benefit in the evaluation of thyroid nodules within the literature [39].
The present analysis is subject to a number of limitations. Primarily, radiomics involves a broad spectrum of analysis methods, ranging from the radiomic AI methods to deep-learning techniques; we have included all of these under the umbrella term “radiomics” despite variance in their reproducibility of data [103]. Secondly, the authors wish to highlight the inter-user variability of US due to this imaging acquisition being operator dependent. Radiomic analyses are dependent on high-quality images of thyroid nodules being obtained and nodules being correctly selected by ultra-sonographers. Thirdly, when extracting data, we selected the highest performing radiomic method within any given study. This may have led to over-estimation of overall sensitivity and specificity for radiomic evaluation of thyroid nodules on US as a whole. To combat this potential bias when comparing radiomics to radiologists, we selected data for the highest performing radiologist. Finally, prospective validation evaluating the utility of AI in the field of radiological diagnostics typically necessitates buying from large, international corporations in order to finance developing the evidence base in this field.
5. Conclusions
In conclusion, this meta-analysis of current evidence demonstrates an almost 90% reliability of radiomic imaging analyses to US in detecting malignancy within undetermined thyroid nodules. At present, radiomic analyses demonstrate equal diagnostic sensitivity and specificity of identifying malignant lesions when compared to radiologists. Within the field of radiomics, at present, non-CNN methods may be considered the preferred radiomic means of classifying thyroid nodules. Based on this meta-analysis, AI offers promising results as an avenue to be explored as we look to enhance the diagnostic accuracy and risk stratification of thyroid nodules in the era of personalized medical and oncological patient care. We advocate for rigorous experimentation in this field, given the potential for this technology to bolster diagnostic workflows, enhance clinical outcomes, and minimize patient morbidity; all while mitigating associated healthcare costs.
Author Contributions
Conceptualization, E.F.C., M.G.D. and M.J.K.; methodology, E.F.C., M.G.D. and M.J.K.; formal analysis, E.F.C., M.G.D. and S.H.; investigation, E.F.C., J.P.O. and M.G.D.; data curation, E.F.C., S.O. and M.C.; writing—original draft preparation, E.F.C. and M.G.D.; writing—review and editing all authors; supervision, I.J.K., A.J.L., and M.J.K. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was supported by National University of Ireland Galway.
Institutional Review Board Statement
Not applicable for systematic review and meta-analysis.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data used within this manuscript is available online via Pubmed, EMBASE, and Scopus.
Conflicts of Interest
S.H. is a cofounder and shareholder of Cloud Path Diagnostics LLC, New York.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tan G.H., Gharib H. Thyroid incidentalomas: Management approaches to nonpalpable nodules discovered incidentally on thyroid imaging. Ann. Intern. Med. 1997;126:226–231. doi: 10.7326/0003-4819-126-3-199702010-00009. [DOI] [PubMed] [Google Scholar]
- 2.Guth S., Theune U., Aberle J., Galach A., Bamberger C.M. Very high prevalence of thyroid nodules detected by high frequency (13 MHz) ultrasound examination. Eur. J. Clin. Investig. 2009;39:699–706. doi: 10.1111/j.1365-2362.2009.02162.x. [DOI] [PubMed] [Google Scholar]
- 3.Davies L., Welch H.G. Current thyroid cancer trends in the United States. JAMA Otolaryngol. Head Neck Surg. 2014;140:317–322. doi: 10.1001/jamaoto.2014.1. [DOI] [PubMed] [Google Scholar]
- 4.Haugen B.R., Alexander E.K., Bible K.C., Doherty G.M., Mandel S.J., Nikiforov Y.E., Pacini F., Randolph G.W., Sawka A.M., Schlumberger M., et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perros P., Boelaert K., Colley S., Evans C., Evans R.M., Gerrard B.G., Gilbert J., Harrison B., Johnson S.J., Giles T.E., et al. Guidelines for the management of thyroid cancer. Clin. Endocrinol. 2014;81((Suppl. S1)):1–122. doi: 10.1111/cen.12515. [DOI] [PubMed] [Google Scholar]
- 6.Filetti S., Durante C., Hartl D., Leboulleux S., Locati L.D., Newbold K., Papotti M., Berruti A. Thyroid cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. 2019;30:1856–1883. doi: 10.1093/annonc/mdz400. [DOI] [PubMed] [Google Scholar]
- 7.Tessler F.N., Middleton W.D., Grant E.G., Hoang J.K., Berland L.L., Teefey S.A., Cronan J.J., Beland M.D., Desser T.S., Frates M.C., et al. ACR Thyroid Imaging, Reporting and Data System (TI-RADS): White Paper of the ACR TI-RADS Committee. J. Am. Coll. Radiol. 2017;14:587–595. doi: 10.1016/j.jacr.2017.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Cibas E.S., Ali S.Z. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid. 2017;27:1341–1346. doi: 10.1089/thy.2017.0500. [DOI] [PubMed] [Google Scholar]
- 9.Cowling P., Chandra A., Giles T. Guidance on the Reporting of Thyroid Cytology Specimens. Royal College of Pathologists; London, UK: 2016. [Google Scholar]
- 10.Hegedus L. Clinical practice. The thyroid nodule. N. Engl. J. Med. 2004;351:1764–1771. doi: 10.1056/NEJMcp031436. [DOI] [PubMed] [Google Scholar]
- 11.Poller D.N., Baloch Z.W., Fadda G., Johnson S.J., Bongiovanni M., Pontecorvi A., Cochand-Priollet B. Thyroid FNA: New classifications and new interpretations. Cancer Cytopathol. 2016;124:457–466. doi: 10.1002/cncy.21703. [DOI] [PubMed] [Google Scholar]
- 12.Konig I.R., Fuchs O., Hansen G., Mutius E.v., Kopp M.V. What is precision medicine? Eur. Respir. J. 2017;50:1700391. doi: 10.1183/13993003.00391-2017. [DOI] [PubMed] [Google Scholar]
- 13.Santos M.K., Ferreira J.R., Wada D.T., Tenorio A.P.M., Barbosa M.H.N., Marques P.M.A. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: Advances in imaging towards to precision medicine. Radiol. Bras. 2019;52:387–396. doi: 10.1590/0100-3984.2019.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzo S., Botta F., Raimondi S., Origgi D., Fanciullo C., Morganti A.G., Bellomi M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grani G., Sponziello M., Pecce V., Ramundo V., Durante C. Contemporary Thyroid Nodule Evaluation and Management. J. Clin. Endocrinol. Metab. 2020;105:2869–2883. doi: 10.1210/clinem/dgaa322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leeflang M.M., Deeks J.J., Takwoingi Y., Macaskill P. Cochrane diagnostic test accuracy reviews. Syst. Rev. 2013;2:82. doi: 10.1186/2046-4053-2-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R.T.H.M., Even A.J.G., Jochems A., et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 19.Ardakani A.A., Gharbali A., Mohammadi A. Application of texture analysis method for classification of benign and malignant thyroid nodules in ultrasound images. Iran J. Cancer Prev. 2015;8:116–124. [PMC free article] [PubMed] [Google Scholar]
- 20.Acharya U.R., Faust O., Sree S.V., Molinari F., Garberoglio R., Suri J.S. Cost-effective and non-invasive automated benign and malignant thyroid lesion classification in 3D contrast-enhanced ultrasound using combination of wavelets and textures: A class of ThyroScan algorithms. Technol. Cancer Res. Treat. 2011;10:371–380. doi: 10.7785/tcrt.2012.500214. [DOI] [PubMed] [Google Scholar]
- 21.Acharya U.R., Molinari F., Garberoglio R., Witkowska A., Suri J.S. Automated benign & malignant thyroid lesion characterization and classification in 3D contrast-enhanced ultrasound. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2012;2012:452–455. doi: 10.1109/EMBC.2012.6345965. [DOI] [PubMed] [Google Scholar]
- 22.Acharya U.R., Sree S.V., Swapna G., Gupta S., Molinari F., Garberoglio R., Witkowska A., Suri J.S. Effect of complex wavelet transform filter on thyroid tumor classification in three-dimensional ultrasound. Proc. Inst. Mech. Eng. H. 2013;227:284–292. doi: 10.1177/0954411912472422. [DOI] [PubMed] [Google Scholar]
- 23.Acharya U.R., Sree S.V., Krishnan M.M., Molinari F., Garberoglio R., Suri J.S. Non-invasive automated 3D thyroid lesion classification in ultrasound: A class of ThyroScan systems. Ultrasonics. 2012;52:508–520. doi: 10.1016/j.ultras.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Ardakani A.A., Gharbali A., Mohammadi A. Classification of Benign and Malignant Thyroid Nodules Using Wavelet Texture Analysis of Sonograms. J. Ultrasound Med. 2015;34:1983–1989. doi: 10.7863/ultra.14.09057. [DOI] [PubMed] [Google Scholar]
- 25.Barczynski M., Stopa-Barczynska M., Wojtczak B., Czarniecka A., Konturek A. Clinical validation of S-Detect(TM) mode in semi-automated ultrasound classification of thyroid lesions in surgical office. Gland. Surg. 2020;9((Suppl. S2)):S77–S85. doi: 10.21037/gs.2019.12.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buda M., Wildman-Tobriner B., Hoang J.K., Thayer D., Tessler F.N., Middleton W.D., Mazurowski M.A. Management of Thyroid Nodules Seen on US Images: Deep Learning May Match Performance of Radiologists. Radiology. 2019;292:695–701. doi: 10.1148/radiol.2019181343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y., Fu Y., Yang G. Sparse Representation-Based Radiomics in the Diagnosis of Thyroid Nodules; Proceedings of the Artificial Intelligence and Security; Budapest, Hungary. 14–19 July 2019; pp. 391–401. [Google Scholar]
- 28.Chen D., Hu J., Zhu M., Tang N., Yang Y., Feng Y. Diagnosis of thyroid nodules for ultrasonographic characteristics indicative of malignancy using random forest. Bio. Data Min. 2020;13:14. doi: 10.1186/s13040-020-00223-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi J., Walia E., Babyn P., Wang J., Groot G., Eramian M. Thyroid Nodule Classification in Ultrasound Images by Fine-Tuning Deep Convolutional Neural Network. J. Digit. Imaging. 2017;30:477–486. doi: 10.1007/s10278-017-9997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi W.J., Park J.S., Kim K.G., Kim S.Y., Koo H.R., Lee Y.J. Computerized analysis of calcification of thyroid nodules as visualized by ultrasonography. Eur. J. Radiol. 2015;84:1949–1953. doi: 10.1016/j.ejrad.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Choi Y.J., Baek J.H., Park H.S., Shim W.H., Kim T.Y., Shong Y.K., Lee J.H. A Computer-Aided Diagnosis System Using Artificial Intelligence for the Diagnosis and Characterization of Thyroid Nodules on Ultrasound: Initial Clinical Assessment. Thyroid. 2017;27:546–552. doi: 10.1089/thy.2016.0372. [DOI] [PubMed] [Google Scholar]
- 32.Colakoglu B., Alis D., Yergin M. Diagnostic Value of Machine Learning-Based Quantitative Texture Analysis in Differentiating Benign and Malignant Thyroid Nodules. J. Oncol. 2019;2019:6328329. doi: 10.1155/2019/6328329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao L., Liu R., Jiang Y., Song W., Wang Y., Liu J., Wang J., Wu D., Li S., Hao A., et al. Computer-aided system for diagnosing thyroid nodules on ultrasound: A comparison with radiologist-based clinical assessments. Head Neck. 2018;40:778–783. doi: 10.1002/hed.25049. [DOI] [PubMed] [Google Scholar]
- 34.Gitto S., Grassi G., Angelis C.D., Monaco C.G., Sdao S., Sardanelli F., Sconfienza L.M., Mauri G. A computer-aided diagnosis system for the assessment and characterization of low-to-high suspicion thyroid nodules on ultrasound. Radiol. Med. 2019;124:118–125. doi: 10.1007/s11547-018-0942-z. [DOI] [PubMed] [Google Scholar]
- 35.Ataide E.J.G., Ponugoti N., Illanes A., Schenke S., Kreissl M., Friebe M. Thyroid Nodule Classification for Physician Decision Support Using Machine Learning-Evaluated Geometric and Morphological Features. Sensors. 2020;20:6110. doi: 10.3390/s20216110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan Q., Wang Y., Du J., Qin Y., Lu H., Xiang J., Wang F. Deep learning based classification of ultrasound images for thyroid nodules: A large scale of pilot study. Ann. Transl. Med. 2019;7:137. doi: 10.21037/atm.2019.04.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han M., Ha E.J., Park J.H. Computer-Aided Diagnostic System for Thyroid Nodules on Ultrasonography: Diagnostic Performance Based on the Thyroid Imaging Reporting and Data System Classification and Dichotomous Outcomes. AJNR Am. J. Neuroradiol. 2021;42:559–565. doi: 10.3174/ajnr.A6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong E.Y., Kim H.L., Ha E.J., Park S.Y., Cho Y.J., Han M. Computer-aided diagnosis system for thyroid nodules on ultrasonography: Diagnostic performance and reproducibility based on the experience level of operators. Eur. Radiol. 2019;29:1978–1985. doi: 10.1007/s00330-018-5772-9. [DOI] [PubMed] [Google Scholar]
- 39.Kim H.L., Ha E.J., Han M. Real-World Performance of Computer-Aided Diagnosis System for Thyroid Nodules Using Ultrasonography. Ultrasound Med. Biol. 2019;45:2672–2678. doi: 10.1016/j.ultrasmedbio.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 40.Ko S.Y., Lee J.H., Yoon J.H., Na H., Hong E., Han K., Jung I., Kim E., Moon H.J., Park V.Y., et al. Deep convolutional neural network for the diagnosis of thyroid nodules on ultrasound. Head Neck. 2019;41:885–891. doi: 10.1002/hed.25415. [DOI] [PubMed] [Google Scholar]
- 41.Koh J., Lee E., Han K., Kim E.K., Son E.J., Sohn Y.M., Seo M., Kwon M.-R., Yoon J.H., Lee J.H., et al. Diagnosis of thyroid nodules on ultrasonography by a deep convolutional neural network. Sci. Rep. 2020;10:15245. doi: 10.1038/s41598-020-72270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon S.W., Choi I.J., Kang J.Y., Jang W.I., Lee G.H., Lee M.C. Ultrasonographic Thyroid Nodule Classification Using a Deep Convolutional Neural Network with Surgical Pathology. J. Digit. Imaging. 2020;33:1202–1208. doi: 10.1007/s10278-020-00362-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee E., Ha H., Kim H.J., Moon H.J., Byon J.H., Huh S., Son J., Yoon J., Han K., Kwak J.Y., et al. Differentiation of thyroid nodules on US using features learned and extracted from various convolutional neural networks. Sci. Rep. 2019;9:19854. doi: 10.1038/s41598-019-56395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Weng J., Shi Y., Gu W., Mao Y., Wang Y., Liu W., Zhang J. An improved deep learning approach for detection of thyroid papillary cancer in ultrasound images. Sci. Rep. 2018;8:6600. doi: 10.1038/s41598-018-25005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Zhang S., Zhang Q., Wei X., Pan Y., Zhao J., Xin X., Qin C., Wang X., Li J., et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: A retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20:193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang J., Huang X., Hu H., Liu Y., Zhou Q., Cao Q., Wang W., Liu B., Zheng Y., Li X., et al. Predicting Malignancy in Thyroid Nodules: Radiomics Score Versus 2017 American College of Radiology Thyroid Imaging, Reporting and Data System. Thyroid. 2018;28:1024–1033. doi: 10.1089/thy.2017.0525. [DOI] [PubMed] [Google Scholar]
- 47.Liu C., Xie L., Kong W., Lu X., Zhang D., Wu M., Zhang L., Yang B. Prediction of suspicious thyroid nodule using artificial neural network based on radiofrequency ultrasound and conventional ultrasound: A preliminary study. Ultrasonics. 2019;99:105951. doi: 10.1016/j.ultras.2019.105951. [DOI] [PubMed] [Google Scholar]
- 48.Liu T., Guo Q., Lian C., Ren X., Liang S., Yu J., Niu L., Sun W., Shen D. Automated detection and classification of thyroid nodules in ultrasound images using clinical-knowledge-guided convolutional neural networks. Med. Image Anal. 2019;58:101555. doi: 10.1016/j.media.2019.101555. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y.I., Kamaya A., Desser T.S., Rubin D.L. A Bayesian classifier for differentiating benign versus malignant thyroid nodules using sonographic features. AMIA Annu. Symp. Proc. 2008;2008:419–423. [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z., Zhong S., Liu Q., Xie C., Dai Y., Peng C., Chen X., Zou R. Thyroid nodule recognition using a joint convolutional neural network with information fusion of ultrasound images and radiofrequency data. Eur. Radiol. 2021;31:5001–5011. doi: 10.1007/s00330-020-07585-z. [DOI] [PubMed] [Google Scholar]
- 51.Ma J., Duan S., Zhang Y., Wang J., Wang Z., Li R., Li Y., Zhang L., Ma H. Efficient Deep Learning Architecture for Detection and Recognition of Thyroid Nodules. Comput. Intell. Neurosci. 2020;2020:1242781. doi: 10.1155/2020/1242781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma J., Wu F., Zhu J., Xu D., Kong D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. 2017;73:221–230. doi: 10.1016/j.ultras.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen D.T., Kang J.K., Pham T.D., Batchuluun G., Park K.R. Ultrasound Image-Based Diagnosis of Malignant Thyroid Nodule Using Artificial Intelligence. Sensors. 2020;20:1822. doi: 10.3390/s20071822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen D.T., Pham T.D., Batchuluun G., Yoon H.S., Park K.R. Artificial Intelligence-Based Thyroid Nodule Classification Using Information from Spatial and Frequency Domains. J. Clin. Med. 2019;8:1976. doi: 10.3390/jcm8111976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park V.Y., Han K., Seong Y.K., Park M.H., Kim E.K., Moon H.J., Yoon J.H., Kwak J.Y. Diagnosis of Thyroid Nodules: Performance of a Deep Learning Convolutional Neural Network Model vs. Radiologists. Sci. Rep. 2019;9:17843. doi: 10.1038/s41598-019-54434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park V.Y., Lee E., Lee H.S., Kim H.J., Yoon J., Son J., Song K., Moon H.J., Yoon J.H., Kim G.R., et al. Combining radiomics with ultrasound-based risk stratification systems for thyroid nodules: An approach for improving performance. Eur. Radiol. 2021;31:2405–2413. doi: 10.1007/s00330-020-07365-9. [DOI] [PubMed] [Google Scholar]
- 57.Peng S., Liu Y., Lv W., Liu L., Zhou Q., Yang H., Ren J., Liu G., Wang X., Zhang X., et al. Deep learning-based artificial intelligence model to assist thyroid nodule diagnosis and management: A multicentre diagnostic study. Lancet Digit. Health. 2021;3:e250–e259. doi: 10.1016/S2589-7500(21)00041-8. [DOI] [PubMed] [Google Scholar]
- 58.Prochazka A., Gulati S., Holinka S., Smutek D. Patch-based classification of thyroid nodules in ultrasound images using direction independent features extracted by two-threshold binary decomposition. Comput. Med. Imaging Graph. 2019;71:9–18. doi: 10.1016/j.compmedimag.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Qin P., Wu K., Hu Y., Zeng J., Chai X. Diagnosis of Benign and Malignant Thyroid Nodules Using Combined Conventional Ultrasound and Ultrasound Elasticity Imaging. IEEE J. Biomed. Health Inform. 2020;24:1028–1036. doi: 10.1109/JBHI.2019.2950994. [DOI] [PubMed] [Google Scholar]
- 60.Raghavendra U., Gudigar A., Maithri M., Gertych A., Meiburger K.M., Yeong C.H., Madla C., Kongmebhol P., Molinari F., Ng K.H., et al. Optimized multi-level elongated quinary patterns for the assessment of thyroid nodules in ultrasound images. Comput. Biol. Med. 2018;95:55–62. doi: 10.1016/j.compbiomed.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Raghavendra U., Acharya U.R., Gudigar A., Tan J.H., Fujita H., Hagiwara Y., Molinari F., Kongmebhol P., Ng K.H. Fusion of spatial gray level dependency and fractal texture features for the characterization of thyroid lesions. Ultrasonics. 2017;77:110–120. doi: 10.1016/j.ultras.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 62.Reverter J.L., Vazquez F., Puig-Domingo M. Diagnostic Performance Evaluation of a Computer-Assisted Imaging Analysis System for Ultrasound Risk Stratification of Thyroid Nodules. AJR Am. J. Roentgenol. 2019;213:169–174. doi: 10.2214/AJR.18.20740. [DOI] [PubMed] [Google Scholar]
- 63.Seo J.K., Kim Y.J., Kim K.G., Shin I., Shin J.H., Kwak J.Y. Differentiation of the Follicular Neoplasm on the Gray-Scale US by Image Selection Subsampling along with the Marginal Outline Using Convolutional Neural Network. Biomed. Res. Int. 2017;2017:3098293. doi: 10.1155/2017/3098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi G., Wang J., Qiang Y., Yang X., Zhao J., Hao R., Yang W., Du Q., Kazihise N.G.-F. Knowledge-guided synthetic medical image adversarial augmentation for ultrasonography thyroid nodule classification. Comput. Methods Programs Biomed. 2020;196:105611. doi: 10.1016/j.cmpb.2020.105611. [DOI] [PubMed] [Google Scholar]
- 65.Shin I., Kim Y.J., Han K., Lee E., Kim H.J., Shin J.H., Moon H.J., Youk J.H., Kim K.G., Kwak J.Y. Application of machine learning to ultrasound images to differentiate follicular neoplasms of the thyroid gland. Ultrasonography. 2020;39:257–265. doi: 10.14366/usg.19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song G., Xue F., Zhang C. A Model Using Texture Features to Differentiate the Nature of Thyroid Nodules on Sonography. J. Ultrasound Med. 2015;34:1753–1760. doi: 10.7863/ultra.15.14.10045. [DOI] [PubMed] [Google Scholar]
- 67.Song J., Chai Y.J., Masuoka H., Park S.W., Kim S.J., Choi J.Y., Kong H.-J., Lee K.E., Lee J., Kwak N., et al. Ultrasound image analysis using deep learning algorithm for the diagnosis of thyroid nodules. Medicine. 2019;98:e15133. doi: 10.1097/MD.0000000000015133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stib M.T., Pan I., Merck D., Middleton W.D., Beland M.D. Thyroid Nodule Malignancy Risk Stratification Using a Convolutional Neural Network. Ultrasound Q. 2020;36:164–172. doi: 10.1097/RUQ.0000000000000501. [DOI] [PubMed] [Google Scholar]
- 69.Sun C., Zhang Y., Chang Q., Liu T., Zhang S., Wang X., Guo Q., Yao J., Sun W., Niu L. Evaluation of a deep learning-based computer-aided diagnosis system for distinguishing benign from malignant thyroid nodules in ultrasound images. Med. Phys. 2020;47:3952–3960. doi: 10.1002/mp.14301. [DOI] [PubMed] [Google Scholar]
- 70.Sun H.Y.F., Xu H. Discriminating the Nature of Thyroid Nodules Using the Hybrid Method. Math. Probl. Engineering. 2020;2020:6147037. doi: 10.1155/2020/6147037. [DOI] [Google Scholar]
- 71.Thomas J., Haertling T. AIBx, Artificial Intelligence Model to Risk Stratify Thyroid Nodules. Thyroid. 2020;30:878–884. doi: 10.1089/thy.2019.0752. [DOI] [PubMed] [Google Scholar]
- 72.Tsantis S., Dimitropoulos N., Cavouras D., Nikiforidis G. Morphological and wavelet features towards sonographic thyroid nodules evaluation. Comput. Med. Imaging Graph. 2009;33:91–99. doi: 10.1016/j.compmedimag.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 73.Wang L., Yang S., Yang S., Zhao C., Tian G., Gao Y., Chen Y., Lu Y. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J. Surg. Oncol. 2019;17:12. doi: 10.1186/s12957-019-1558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y.Y.W., Li X., Liu S., Guo L., Xu H., Zhang H., Yang G. Comparison Study of Radiomics and Deep Learning-Based Methods for Thyroid Nodules Classification Using Ultrasound Images. IEEE Access. 2020;8:52010–52017. doi: 10.1109/ACCESS.2020.2980290. [DOI] [Google Scholar]
- 75.Wei Q., Zeng S.E., Wang L.P., Yan Y.J., Wang T., Xu J.W., Zhang M.-Y., Lv W.-Z., Cui X.-W., Dietrich C.F. The value of S-Detect in improving the diagnostic performance of radiologists for the differential diagnosis of thyroid nodules. Med. Ultrason. 2020;22:415–423. doi: 10.11152/mu-2501. [DOI] [PubMed] [Google Scholar]
- 76.Wei X., Gao M., Yu R., Liu Z., Gu Q., Liu X., Zheng Z., Zheng X., Zhu J., Zhang S. Ensemble Deep Learning Model for Multicenter Classification of Thyroid Nodules on Ultrasound Images. Med. Sci. Monit. 2020;26:e926096. doi: 10.12659/MSM.926096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei X., Zhu J., Zhang H., Gao H., Yu R., Liu Z., Zheng X., Gao M., Zhang S. Visual Interpretability in Computer-Assisted Diagnosis of Thyroid Nodules Using Ultrasound Images. Med. Sci. Monit. 2020;26:e927007. doi: 10.12659/MSM.927007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wildman-Tobriner B., Buda M., Hoang J.K., Middleton W.D., Thayer D., Short R.G., Tessler F.N., Mazurowski M.A. Using Artificial Intelligence to Revise ACR TI-RADS Risk Stratification of Thyroid Nodules: Diagnostic Accuracy and Utility. Radiology. 2019;292:112–119. doi: 10.1148/radiol.2019182128. [DOI] [PubMed] [Google Scholar]
- 79.Wu H., Deng Z., Zhang B., Liu Q., Chen J. Classifier Model Based on Machine Learning Algorithms: Application to Differential Diagnosis of Suspicious Thyroid Nodules via Sonography. AJR Am. J. Roentgenol. 2016;207:859–864. doi: 10.2214/AJR.15.15813. [DOI] [PubMed] [Google Scholar]
- 80.Xia J., Chen H., Li Q., Zhou M., Chen L., Cai Z., Fang Y., Zhou H. Ultrasound-based differentiation of malignant and benign thyroid Nodules: An extreme learning machine approach. Comput. Methods Programs Biomed. 2017;147:37–49. doi: 10.1016/j.cmpb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 81.Xia S., Yao J., Zhou W., Dong Y., Xu S., Zhou J., Zhan W. A computer-aided diagnosing system in the evaluation of thyroid nodules-experience in a specialized thyroid center. World J. Surg. Oncol. 2019;17:210. doi: 10.1186/s12957-019-1752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye H., Hang J., Chen X., Di X., Chen J., Ye X., Zhang D. An intelligent platform for ultrasound diagnosis of thyroid nodules. Sci. Rep. 2020;10:13223. doi: 10.1038/s41598-020-70159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoo Y.J., Ha E.J., Cho Y.J., Kim H.L., Han M., Kang S.Y. Computer-Aided Diagnosis of Thyroid Nodules via Ultrasonography: Initial Clinical Experience. Korean J. Radiol. 2018;19:665–672. doi: 10.3348/kjr.2018.19.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Q., Jiang T., Zhou A., Zhang L., Zhang C., Xu P. Computer-aided diagnosis of malignant or benign thyroid nodes based on ultrasound images. Eur. Arch. Otorhinolaryngol. 2017;274:2891–2897. doi: 10.1007/s00405-017-4562-3. [DOI] [PubMed] [Google Scholar]
- 85.Zhang B., Tian J., Pei S., Chen Y., He X., Dong Y., Zhang L., Mo X., Huang W., Cong S., et al. Machine Learning-Assisted System for Thyroid Nodule Diagnosis. Thyroid. 2019;29:858–867. doi: 10.1089/thy.2018.0380. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y., Wu Q., Chen Y., Wang Y. A Clinical Assessment of an Ultrasound Computer-Aided Diagnosis System in Differentiating Thyroid Nodules With Radiologists of Different Diagnostic Experience. Front. Oncol. 2020;10:557169. doi: 10.3389/fonc.2020.557169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao C.K., Ren T.T., Yin Y.F., Shi H., Wang H.X., Zhou B.Y., Wang X.-R., Li X., Zhang Y.-F., Liu C., et al. A Comparative Analysis of Two Machine Learning-Based Diagnostic Patterns with Thyroid Imaging Reporting and Data System for Thyroid Nodules: Diagnostic Performance and Unnecessary Biopsy Rate. Thyroid. 2021;31:470–481. doi: 10.1089/thy.2020.0305. [DOI] [PubMed] [Google Scholar]
- 88.Zhao H.N., Liu J.Y., Lin Q.Z., He Y.S., Luo H.H., Peng Y.L., Ma B.-Y. Partially cystic thyroid cancer on conventional and elastographic ultrasound: A retrospective study and a machine learning-assisted system. Ann. Transl. Med. 2020;8:495. doi: 10.21037/atm.2020.03.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou H., Jin Y., Dai L., Zhang M., Qiu Y., Wang K., Tian J., Zheng J. Differential Diagnosis of Benign and Malignant Thyroid Nodules Using Deep Learning Radiomics of Thyroid Ultrasound Images. Eur. J. Radiol. 2020;127:108992. doi: 10.1016/j.ejrad.2020.108992. [DOI] [PubMed] [Google Scholar]
- 90.Zhu J., Zhang S., Yu R., Liu Z., Gao H., Yue B., Liu X., Zheng X., Gao M., Wei X. An efficient deep convolutional neural network model for visual localization and automatic diagnosis of thyroid nodules on ultrasound images. Quant. Imaging Med. Surg. 2021;11:1368–1380. doi: 10.21037/qims-20-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu L.C., Ye Y.L., Luo W.H., Su M., Wei H.P., Zhang X.B., Wei J., Zou C.-L. A model to discriminate malignant from benign thyroid nodules using artificial neural network. PLoS ONE. 2013;8:e82211. doi: 10.1371/journal.pone.0082211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu Y., Sang Q., Jia S., Wang Y., Deyer T. Deep neural networks could differentiate Bethesda class III versus class IV/V/VI. Ann. Transl, Med. 2019;7:231. doi: 10.21037/atm.2018.07.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhu Y.C., Al-Zoubi A., Jassim S., Jiang Q., Zhang Y., Wang Y.B., Ye X.-D., Du H. A generic deep learning framework to classify thyroid and breast lesions in ultrasound images. Ultrasonics. 2021;110:106300. doi: 10.1016/j.ultras.2020.106300. [DOI] [PubMed] [Google Scholar]
- 94.Yoon J., Lee E., Kang S.W., Han K., Park V.Y., Kwak J.Y. Implications of US radiomics signature for predicting malignancy in thyroid nodules with indeterminate cytology. Eur. Radiol. 2021;31:5059–5067. doi: 10.1007/s00330-020-07670-3. [DOI] [PubMed] [Google Scholar]
- 95.Yamashita R., Nishio M., Do R.K.G., Togashi K. Convolutional neural networks: An overview and application in radiology. Insights Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berardi V.L., Zhang G.P. An empirical investigation of bias and variance in time series forecasting: Modeling considerations and error evaluation. IEEE Trans. Neural Netw. 2003;14:668–679. doi: 10.1109/TNN.2003.810601. [DOI] [PubMed] [Google Scholar]
- 97.Erickson B.J., Korfiatis P., Akkus Z., Kline T.L. Machine Learning for Medical Imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.El-Naqa I., Yang Y., Wernick M.N., Galatsanos N.P., Nishikawa R.M. A support vector machine approach for detection of microcalcifications. IEEE Trans. Med. Imaging. 2002;21:1552–1563. doi: 10.1109/TMI.2002.806569. [DOI] [PubMed] [Google Scholar]
- 99.Yip S.S., Aerts H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016;61:R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rogers W., Seetha S.T., Refaee T.A.G., Lieverse R.I.Y., Granzier R.W.Y., Ibrahim A., Keek S.A., Sanduleanu S., Primakov S.P., Beuque M.P.L., et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020;93:20190948. doi: 10.1259/bjr.20190948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim H.G., Kwak J.Y., Kim E.K., Choi S.H., Moon H.J. Man to man training: Can it help improve the diagnostic performances and interobserver variabilities of thyroid ultrasonography in residents? Eur. J. Radiol. 2012;81:e352–e356. doi: 10.1016/j.ejrad.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 102.Choi S.H., Kim E.K., Kwak J.Y., Kim M.J., Son E.J. Interobserver and intraobserver variations in ultrasound assessment of thyroid nodules. Thyroid. 2010;20:167–172. doi: 10.1089/thy.2008.0354. [DOI] [PubMed] [Google Scholar]
- 103.Kocak B., Durmaz E.S., Ates E., Kilickesmez O. Radiomics with artificial intelligence: A practical guide for beginners. Diagn. Interv. Radiol. 2019;25:485–495. doi: 10.5152/dir.2019.19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used within this manuscript is available online via Pubmed, EMBASE, and Scopus.