Abstract
The 3C-like protease (Mpro, 3CLpro) plays a key role in the replication process in coronaviruses (CoVs). The Mpro is an essential enzyme mediates CoVs replication and is a promising target for development of antiviral drugs. Until now, baicalein has been shown the specific activity for SARS-CoV Mpro in vitro experiments. In this study, we resolved the SARS-CoV Mpro with baicalein by X-ray diffraction at 2.25 Å (PDB code 7XAX), which provided a structural basis for the research and development of baicalein as an anti-CoVs drug.
Keywords: SARS-CoV, Baicalein, 3C-like protease
1. Introduction
CoVs are the largest RNA viruses, which have a positive-sense, single-stranded RNA genome [1]. The severe acute respiratory syndrome (SARS) is caused by a novel species of CoVs (SARS-CoV) [2]. The symptoms of SARS-CoV infection are the lower respiratory tract disease including fever, lymphopenia, malaise and mildly elevated serum hepatic enzymes etc. [3,4]. At present, no anti-viral drugs have been found to be beneficial for SARS. The world is facing with a pandemic caused by SARS-CoV-2 now, which is a strain of CoVs spreading rapidly across the globe. But SARS-CoV-2 resulted in less widespread morbidity and mortality compared to SARS-CoV [5]. Although vaccination campaigns are underway globally, the efficacy is reduced because of the variants of concern (VOCs) [6]. Potential risk exists for SARS-CoV-2 VOCs to develop and gain some mutations similar to life threaten SARS happened in 2003. There is a need to fully understand the SARS-CoV and even the whole sub-type of coronavirus.
Mpro is an attractive drug target among CoVs due to its essential role in processing the polyproteins which were translated from the viral RNA [7]. Studies show Mpro is an essential target for inhibition by interaction with Cys145 of its catalytic site [[8], [9], [10]]. The substrate-binding site and active site of the SARS-CoV-2 Mpro crystal structure in the apo state was more flexible than the ligand-binding mode [11,12]. Various complexes of the Mpro structure of SARS-CoV-2 with natural products and novel inhibitors have emerged. The elucidation of the mechanism of shikonin against CoVs laid the foundation for more natural products and traditional Chinese medicines as a source for antivirus drug candidates [[13], [14], [15]].
Flavonoids, found in various plants, are a class of polyphenolic compounds which have a structural unit of 2-phenylchromone [16]. Some flavonoid compounds have antiviral activity against CoVs by inhibiting the activity of Mpro. Studies showed herbacetin, gallocatechin gallate and rhoifolin can block the enzymatic activity of SARS-CoV Mpro due to S1, S2 and S3 sites [17,18]. Baicalein is an ingredient of Shuanghuanglian, mainly derived from the root of Scutellaria baicalensis. Baicalein shown superior binding effect to Mpro. Previous data showed baicalein was identified as potential noncovalent inhibitors for SARS-CoV-2 Mpro of IC50 values at 0.94 μM [19,20]. In this paper, we resolved the crystal structure of SARS-CoV Mpro-baicalein at 2.25 Å, analyzed and compared with the structure of SARS-CoV-2 Mpro-baicalein. It provides a structural basis and theoretical basis for the drug research and development of treating CoVs in the near future.
2. Materials and methods
2.1. Expression and purification of human SARS-CoV
The codon-optimized cDNAs for the SARS-CoV was synthesized fused with 6_His at the N terminus. Synthesized gene was subcloned into the pET-28a vector. The expression and purification of protease was performed by a standard method previously described [21].
2.2. X-ray crystallography
Details of the crystallization, data collection, structure solution, and refinement are provided in Table 1 . Briefly, all crystallization trials were conducted using a sitting-drop vapor diffusion method at 20 °C. Baicalein was soaked with the crystal of SARS-CoV-apo within 12 h, and the X-ray diffraction data were collected at beamline02U1 (BL02U1) at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). The structure solution was conducted by molecular replacement using SARS-CoV-apo (PDB code 7DQZ) as an initial model. Refinement and model building were carried out using Phenix [22] and Coot [23], respectively.
Table 1.
Statistics for data processing and model refinement of SARS-CoV Mpro with baicalein.
| PDB code | 7XAX |
|---|---|
| Synchrotron | SSRF |
| Beam line | BL02U1 |
| Wavelength (Å) | 0.97919 |
| Space group | P1 |
| a, b, c (Å) | 55.51, 60.55, 68.28 |
| α, β, γ (°) | 90.95, 120.71, 108.65 |
| Total reflections | 129,291 |
| Unique reflections | 37,931 |
| Resolution (Å) | 2.25 (2.31–2.25) |
| R-merge (%) | 7.1 (66.2) |
| Mean I/σ (I) | 11.0/1.8 |
| Completeness (%) | 97.7 (96.2) |
| Redundancy | 3.4 (3.5) |
| Resolution (Å) | 66.31–2.25 |
| Rwork/Rfree (%) | 22.92/27.74 |
| Atoms | 4442 |
| Mean temperature factor (Å2) | 46.1 |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.973 |
| Ramachandran plot (%) | |
| Preferred | 97.11 |
| Allowed | 2.89 |
| outliers | 0 |
2.3. Data availability
Coordinates for SARS-CoV-Mpro-baicalein complexe has been deposited in the Protein Data Bank (PDB) under accession numbers 7XAX.
3. Results
3.1. Structures of SARS-CoV Mpro-baicalein
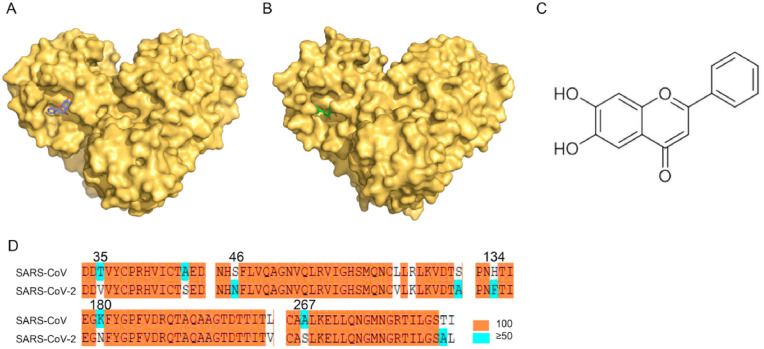
The binding modes of SARS-CoV Mpro-baicalein were compared with structure of SARS-CoV-2 Mpro-baicalein. In order to identify the key residues binding to baicalein, we obtained the crystal structure of SARS-CoV Mpro with baicalein at 2.25 Å (PDB code 7XAX) (Fig. 1 A). SARS-CoV-2 Mpro with baicalein at 2.20 Å (PDB code 6M2N) (Fig. 1B). The structure of baicalein has been shown in (Fig. 1C). The Mpro of SARS-CoV and SARS-CoV-2 had 96% similarity and 95% amino acid homology [24,25]. A comparison of the sequences shows that twelve residues are different between the Mpro of SARS-CoV and SARS-CoV-2 (Fig. 1D).
Fig. 1.
Comparison of the binding pocket of Mprofrom different CoVs. (A) Surface of SARS-CoV Mpro. Baicalein is shown in purple. Oxygen atoms are shown in red. (B) Surface of SARS-CoV-2 Mpro. Baicalein is shown in green. Oxygen atoms are shown in red. (C) The structure of baicalein. (D) Alignment of the Mpro of SARS-CoV and SARS-CoV-2. The conserved precent has been shown orange (100),blue (≥50). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Crystal structure of SARS-CoV Mpro with baicalein
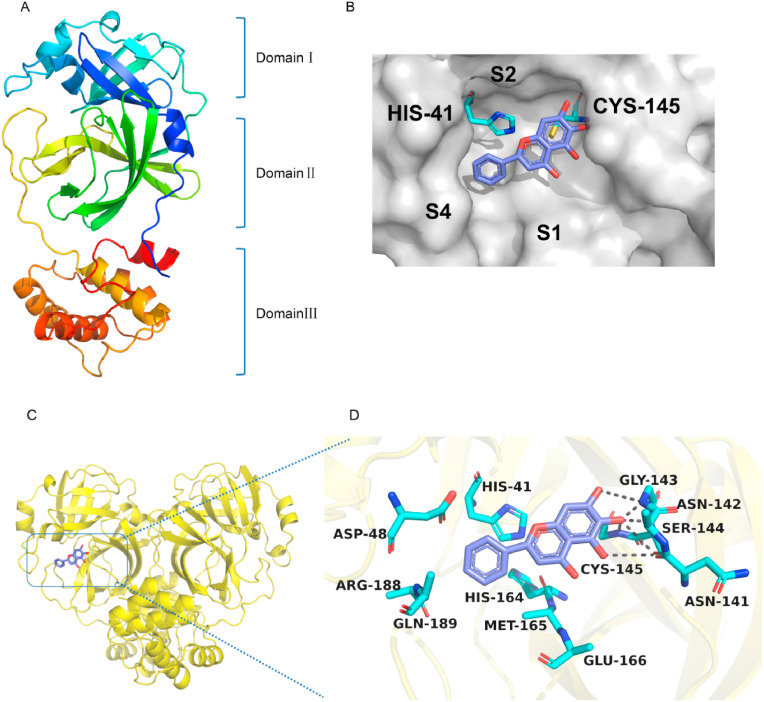
The protomer is composed of three domains. Domain I and domain II have an antiparallelβ-barrel structure. Domain III contains five α-helices arranged into a largely antiparallel globular cluster, and it is connected to domain II by a long loop region [13](Fig. 2 A). From the structure of SARS-CoV Mpro, the S2 and S2′subsites are critical for substrate binding to the SARS-CoV Mpro [[26], [27], [28]]. SARS-CoV Mpro with baicalein had a Cys145-His41 catalytic dyad in the S2 subsite, which located in a cleft between domain I and domain II [19,[29], [30], [31]](Fig. 2B). SARS-CoV Mpro with baicalein is shown in (Fig. 2C). Three phenolic hydroxyl groups of baicalein form hydrogen bonds with the main chains of Cys145/Ser144/Gly143 as well as the side chains of Asn142/Asn141. The free benzene ring inserted into S2 subsite by making hydrophobic interactions with multiple residues Gln189/Arg188/Glu166/Met165/His164/Asp48/His41. With the aid of an array of direct hydrogen bonds with Cys145/Ser144/Gly143, baicalein served to stabilize the tetrahedral transition state of the proteolytic reaction (Fig. 2D).
Fig. 2.
Crystal structure of SARS-CoV Mprowith baicalein (PDB code7XAX). (A) Overview of homodimers shown as cartoon. (B) The binding pocket of SARS-CoV Mpro with baicalein. Baicalein is shown in purple. (C) The structure of SARS-CoV Mpro with baicalein. SARS-CoV is shown in yellow and baicalein is shown in purple. Oxygen atoms are shown in red. (D) Interactions of SARS-CoV with baicalein (purple). Residues as well as the baicalein are shown as sticks and hydrogen bonds are represented by dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Comparison of structure of SARS-CoV Mpro-baicalein and SARS-CoV-2 Mpro-baicalein
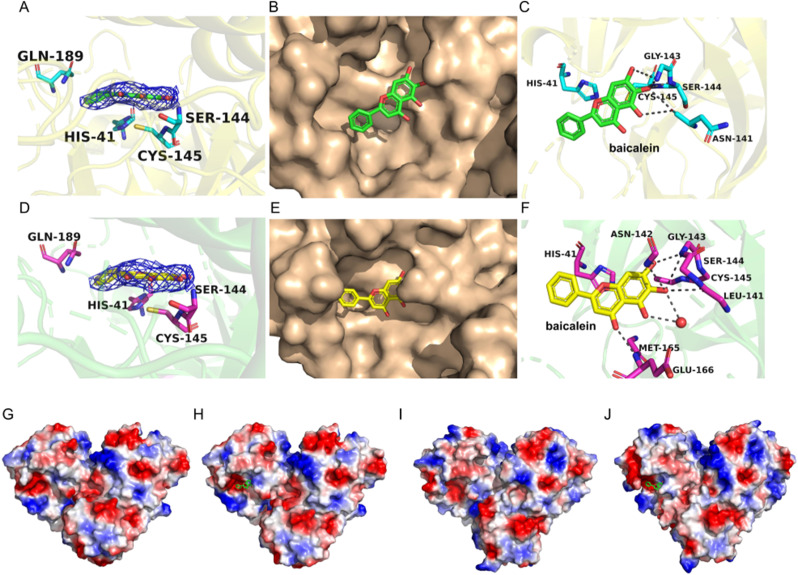
SARS-CoV Mpro and SARS-CoV-2 Mpro have the same binding mode with baicalein, both have the binding sites include the Cys145-His41 catalytic dyad (Fig. 3 A and D). Baicalein in the active site of SARS-CoV Mpro and SARS-CoV-2 Mpro (Fig. 3B and E). The phenolic hydroxyl group of baicalein in SARS-CoV Mpro forms hydrogen bonds with mains chains and side chains has been shown in (Fig. 3D). The SARS-CoV-2 Mpro with baicalein complexes are hydrogen-bonded to the Ser144/Gly143/Leu141 and side chains via the water molecule, where the only carbonyl group established a hydrogen bond with the main chain of Glu166 [[32], [33], [34]]. The free benzene ring also inserted into the S2 subsite by hydrophobic interactions with His41 residue (Fig. 3F). The electrostatic potential surface surrounding the active pocket in SARS-CoVs with baicalein are also shown in Fig. 3. It was revealed that SARS-CoV-apo (Fig. 3G) is different from that of SARS-CoV Mpro-baicalein(Fig. 3H). SARS-CoV-2-apo (Fig. 3I) is different from SARS-CoV-2 Mpro-baicalein (Fig. 3J).
Fig. 3.
Crystal structures of SARS-CoVs with baicalein. (A) Electron density maps (2Fo-Fc) of baicalein at 1.0 σ (7XAX). (B) Baicalein (green) in the active site of SARS-CoV Mpro. (C) Hydrogen bonding (dashed lines) interactions between SARS-CoV Mpro and baicalein. (D) Electron density maps (2Fo-Fc) of baicalein at 1.0 σ (6M2N). (E) Baicalein (yellow) in the active site of SARS-CoV-2 Mpro. (F) Hydrogen bonding (dashed lines) interactions between SARS-CoV-2 Mpro and baicalein. (G) Electrostatic potential surface distribution of SARS-CoV-apo. (H) Electrostatic potential surface distribution of SARS-CoV Mpro-baicalein. (I) Electrostatic potential surface distribution of SARS-CoV-2-apo. (J) Electrostatic potential surface distribution of SARS-CoV-2 Mpro-baicalein. The color of the surface denotes the electrostatic potential, while red signififies negative charge and blue signififies positive charge. Baicalein is shown in sticks (green). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Recently, as the cases of SASR-CoV-2 infections, the effective drugs and vaccines has already been found [35]. But there are currently no antivirus drugs approved for the prevention or treatment of highly virulent SARS-CoV infection. Mpro plays a key role involved in the replication and transcription of CoVs among the few available targets for anti-CoVs drugs, which has become an essential and relatively mature drug target in anti-CoVs drug research. Mpro inhibitors mainly exhibit reversible binding with the amino acid residues in S1, S2, and S4 pockets. The inhibitors contain unsymmetrical aromatic disulphides showing inhibitory activity including the flavonoids compounds [36].
Flavonoid compounds displayed good inhibition toward Mpro [37]. Correspondingly, baicalein which belongs to flavonoid compounds has been shown the specific activity for SARS-CoV Mpro in vitro experiments. SARS-CoV Mpro has a Cys145-His41 catalytic dyad in the cleft between domains I and II, can recognize the eleven cleavage sites of nsp4-16 specifically and exhibit self-hydrolytic cleavage activity [38]. Here, we resolved the crystal structure of SARS-CoV Mpro with baicalein that can bind to the substrate pocket between domain I and domain II. Three phenolic hydroxyl groups of baicalein make hydrogen bonds with the main chains of Cys145/Ser144/Gly143 as well as the side chains of Asn142/Asn141, providing a structural basis and theoretical basis for baicalein to inhibit the replication of SARS-CoV.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgements
This work was supported by Jiangxi natural science Foundation for distinguished young scholar (20212ACB216001), Gannan Medical University (QD201910), Jiangxi key research and development program (20203BBG73063) and Jiangxi "Double Thousand Plan (jxsq2019101064)", Central government funds for guiding local scientific and Technological Development(2021JH6/10500225), Construction of Liaoning technological innovation center (1590826279052).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2022.04.086.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Stanley Perlman, Dandekar Ajai A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7097326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris Joseph S.M., Yuen Kwok Y., Osterhaus Albert D.M. E., et al. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. https://pubmed.ncbi.nlm.nih.gov/14681510 [DOI] [PubMed] [Google Scholar]
- 3.Namita Satija, Lal Sunil K. The molecular biology of SARS coronavirus. Ann. N. Y. Acad. Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. https://pubmed.ncbi.nlm.nih.gov/17470909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7096017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand K.B., Karade S., Sen S., et al. SARS-CoV-2SARS-CoV-2: camazotz's curse. Med. J. Armed Forces India. 2020;76:136–141. doi: 10.1016/j.mjafi.2020.04.008. https://pubmed.ncbi.nlm.nih.gov/32341622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tirosh Shapira, Abrrey Monreal I., Dion Sébastien P., et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature. 2022 doi: 10.1038/s41586-022-04661-w. https://pubmed.ncbi.nlm.nih.gov/35344983 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Linlin, Lin Daizong, Sun Xinyuanyuan, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020:368. doi: 10.1126/science.abb3405. https://pubmed.ncbi.nlm.nih.gov/32198291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akinori Kidera, Kei Moritsugu, Toru Ekimoto, et al. Allosteric regulation of 3CL protease of SARS-CoV-2 and SARS-CoV observed in the crystal structure ensemble. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167324. https://pubmed.ncbi.nlm.nih.gov/34717972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niesor Eric J., Guy Boivin, Eric Rhéaume, et al. Inhibition of the 3CL protease and SARS-CoV-2 replication by dalcetrapib. ACS Omega. 2021;6:16584–16591. doi: 10.1021/acsomega.1c01797. https://pubmed.ncbi.nlm.nih.gov/34235330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhenming Jin, Yao Zhao, Yuan Sun et al Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur, Nat. Struct. Mol. Biol. 27, 529–532. https://pubmed.ncbi.nlm.nih.gov/32382072. [DOI] [PubMed]
- 11.Jin Zhenming, Du Xiaoyu, Xu Yechun, et al. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. https://pubmed.ncbi.nlm.nih.gov/32272481 [DOI] [PubMed] [Google Scholar]
- 12.Zhou Xuelan, Zhong Fanglin, Cheng Lin, et al. Structure of SARS-CoV-2 main protease in the apo state. Sci. China Life Sci. 2021;64:656–659. doi: 10.1007/s11427-020-1791-3. https://pubmed.ncbi.nlm.nih.gov/32880863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee Riddhidev, Perera Lalith, Viranga Tillekeratne L.M. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today. 2021 Mar doi: 10.1016/j.drudis.2020.12.005. https://pubmed.ncbi.nlm.nih.gov/33309533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jian Li, Zhou Xuelan, Zhang Yan, et al. Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci. Bull. 2021;66:661–663. doi: 10.1016/j.scib.2020.10.018. https://pubmed.ncbi.nlm.nih.gov/33163253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Yuting, Gao Hongxia, Hu Xiaohui, et al. Structure-Based discovery and structural basis of a novel broad-spectrum natural product against the main protease of coronavirus. J. Virol. 2022;96 doi: 10.1128/JVI.01253-21. https://pubmed.ncbi.nlm.nih.gov/34586857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen Kangmei, Fang Xiaochuan, Yang Junli, et al. Recent research on flavonoids and their biomedical applications. Curr. Med. Chem. 2021;28:1042–1066. doi: 10.2174/0929867327666200713184138. https://pubmed.ncbi.nlm.nih.gov/32660393 [DOI] [PubMed] [Google Scholar]
- 17.Jo Seri, Kim Suwon, Shin Dong Hae, Kim Mi-Sun. Inhibition of SARS-CoV 3CL protease by flavonoids. Enzyme Inhib. Med. Chem. 2020 Dec doi: 10.1080/14756366.2019.1690480. https://pubmed.ncbi.nlm.nih.gov/31724441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen Thi Thanh Hanh, Woo Hye-Jin, Kang Hee-Kyoung, et al. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34:831–838. doi: 10.1007/s10529-011-0845-8. https://pubmed.ncbi.nlm.nih.gov/22350287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su Hai-Xia, Yao Sheng, Zhao Wen-Feng, et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. https://pubmed.ncbi.nlm.nih.gov/32737471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Dafu, Su Haixia, Ke Changqiang, et al. Efficient discovery of potential inhibitors for SARS-CoV-2 3C-like protease from herbal extracts using a native MS-based affinity-selection method. Pharm. Biomed. Anal. 2022 Feb 5;209 doi: 10.1016/j.jpba.2021.114538. https://pubmed.ncbi.nlm.nih.gov/34929567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H., Zhang Y., Jiang H., et al. Crystal structures of human coronavirus NL63 main protease at different pH values. Acta Crystallogr. F Struct. Biol. Commun. 2021 Oct doi: 10.1107/S2053230X21009523. https://pubmed.ncbi.nlm.nih.gov/34605439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams Paul D., Grosse-Kunstleve Ralf W., Hung Li Wei, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. https://pubmed.ncbi.nlm.nih.gov/12393927 [DOI] [PubMed] [Google Scholar]
- 23.Paul Emsley, Kevin Cowtan. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. https://pubmed.ncbi.nlm.nih.gov/15572765 [DOI] [PubMed] [Google Scholar]
- 24.Anand Kanchan, John Ziebuhr, Parvesh Wadhwani, et al. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. https://pubmed.ncbi.nlm.nih.gov/12746549 Science.2003. [DOI] [PubMed] [Google Scholar]
- 25.Prakash Archisha, Subhomoi Borkotoky, Kumar Dubey Vikash. Targeting two potential sites of SARS-CoV-2 main protease through computational drug repurposing. J. Biomol. Struct. Dyn. 2022:1–11. doi: 10.1080/07391102.2022.2044907. https://pubmed.ncbi.nlm.nih.gov/35266856 undefined. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Yao, Zhu Yan, Liu Xiang, et al. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2117142119. https://pubmed.ncbi.nlm.nih.gov/35380892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue Xiaoyu, Yang Haitao, Shen Wei, et al. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. https://pubmed.ncbi.nlm.nih.gov/17189639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Achutha A.S., Pushpa V.L., Surendran Suchitra. Theoretical insights into the anti-SARS-CoV-2 activity of chloroquine and its analogs and in silico screening of main protease inhibitors. J. Proteome Res. 2020;19:4706–4717. doi: 10.1021/acs.jproteome.0c00683. https://pubmed.ncbi.nlm.nih.gov/32960061 [DOI] [PubMed] [Google Scholar]
- 29.Dai W., Zhang B., Jiang X.M., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. https://pubmed.ncbi.nlm.nih.gov/32321856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan M., Wu N.C., Zhu X. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 May 8 doi: 10.1126/science.abb7269. https://pubmed.ncbi.nlm.nih.gov/32245784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alimuddin Zumla, Chan Jasper F.W., Azhar Esam I., et al. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. https://pubmed.ncbi.nlm.nih.gov/26868298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anna Pavlova, Lynch Diane L., Isabella Daidone, et al. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021;12:1513–1527. doi: 10.1039/d0sc04942e. https://pubmed.ncbi.nlm.nih.gov/35356437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. https://pubmed.ncbi.nlm.nih.gov/32198291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Io Antonopoulou, Eleftheria Sapountzaki, Ulrika Rova, et al. Inhibition of the main protease of SARS-CoV-2 (M) by repurposing/designing drug-like substances and utilizing nature's toolbox of bioactive compounds. Comput. Struct. Biotechnol. J. 2022;20:1306–1344. doi: 10.1016/j.csbj.2022.03.009. https://pubmed.ncbi.nlm.nih.gov/35308802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinesh Mohanraj, Alison Whitelegg. Trilogy of COVID-19: infection, vaccination, and immunosuppression. Int. Arch. Allergy Immunol. 2022:1–19. doi: 10.1159/000524056. https://pubmed.ncbi.nlm.nih.gov/35390803 undefined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Yuzhi, Liang Chengyuan, Liang Xin, et al. The development of Coronavirus 3C-Like protease (3CL) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7409838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo Seri, Kim Suwon, Kim Dae Yong, et al. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. Enzyme Inhib. Med. Chem. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. https://pubmed.ncbi.nlm.nih.gov/32746637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan Keqiang, Liang Ma, Han Xiaofeng, et al. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2005;329:934–940. doi: 10.1016/j.bbrc.2005.02.061. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7092912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for SARS-CoV-Mpro-baicalein complexe has been deposited in the Protein Data Bank (PDB) under accession numbers 7XAX.