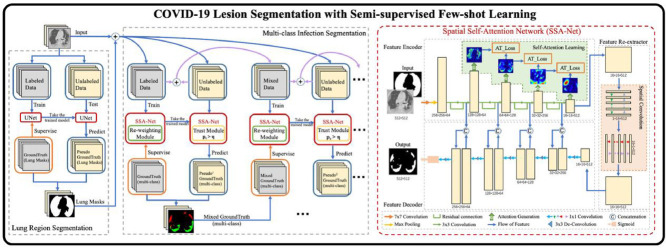
Graphical abstract
Keywords: COVID-19, Lesion segmentation, Semi-supervised, Few-shot learning
Abstract
Coronavirus disease (COVID-19) broke out at the end of 2019, and has resulted in an ongoing global pandemic. Segmentation of pneumonia infections from chest computed tomography (CT) scans of COVID-19 patients is significant for accurate diagnosis and quantitative analysis. Deep learning-based methods can be developed for automatic segmentation and offer a great potential to strengthen timely quarantine and medical treatment. Unfortunately, due to the urgent nature of the COVID-19 pandemic, a systematic collection of CT data sets for deep neural network training is quite difficult, especially high-quality annotations of multi-category infections are limited. In addition, it is still a challenge to segment the infected areas from CT slices because of the irregular shapes and fuzzy boundaries. To solve these issues, we propose a novel COVID-19 pneumonia lesion segmentation network, called Spatial Self-Attention network (SSA-Net), to identify infected regions from chest CT images automatically. In our SSA-Net, a self-attention mechanism is utilized to expand the receptive field and enhance the representation learning by distilling useful contextual information from deeper layers without extra training time, and spatial convolution is introduced to strengthen the network and accelerate the training convergence. Furthermore, to alleviate the insufficiency of labeled multi-class data and the long-tailed distribution of training data, we present a semi-supervised few-shot iterative segmentation framework based on re-weighting the loss and selecting prediction values with high confidence, which can accurately classify different kinds of infections with a small number of labeled image data. Experimental results show that SSA-Net outperforms state-of-the-art medical image segmentation networks and provides clinically interpretable saliency maps, which are useful for COVID-19 diagnosis and patient triage. Meanwhile, our semi-supervised iterative segmentation model can improve the learning ability in small and unbalanced training set and can achieve higher performance.
1. Introduction
Since the end of 2019, coronavirus disease 2019 (COVID-19), an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1 , has spread in the worldwide, which can cause acute respiratory illness and even lead to fatal acute respiratory distress syndrome (ARDS) (Chen et al., 2020). So far (Central European Time of December 20, 2021), the number of confirmed cases of COVID-19 has been more than 273.9 million, with more than 5.3 million deaths, according to the COVID-19 situation dashboard in the World Health Organization (WHO) website2 , and the number is continuing to increase. The health of human being all over the world are threatened and everyones life has been greatly affected due to the outbreak of the virus. Since it is highly contagious and we still lack appropriate treatment and vaccines, early detection of COVID-19 is essential to prevent spreading in time and to properly allocate limited medical resources. Among all virus detection methods, antigen testing is fast, but the sensitivity is also poor (Fang et al., 2020). Reverse transcription polymerase chain reaction (RT-PCR) has been considered as the gold standard for COVID-19 screening (Ai et al., 2020), which detects viral nucleic acid using nasopharyngeal and throat swabs (Bai et al., 2020). However, the results of RT-PCR testing are susceptible to low viral load or sampling errors, and result in high false negative rates (Xie et al., 2020). Meanwhile, the requirements for the testing laboratory environment are extremely strict and there is always a shortage of equipment under the epidemic (Liang et al., 2020), which would greatly limit and delay the diagnosis of suspected subjects. To find a fast and sufficiently accurate patient screening way becomes an unprecedented challenge to prevent the spread of the infection. Since most patients infected by COVID-19 are diagnosed with pneumonia, radiological examinations have also been used to diagnose and assess disease evolution as important complements to RT-PCR tests (Rubin et al., 2020). X-ray and computed tomography (CT) are two typical imaging methods for patients in the COVID-19 study (Shi et al., 2020). CT has a 3D view and the ribs in X-ray images may affect the lesion detection. The diagnostic accuracy of CT is much higher than that of X-ray in the early stage of the disease (Wong et al., 2020). Furthermore, chest CT screening on clinical patients has showed that its sensitivity outperforms that of RT-PCR (Fang et al., 2020) and it can even confirm COVID-19 infection in negative or weakly-positive RT-PCR cases (Xie et al., 2020). Therefore, in view of the particularity of prevention and control during the COVID-19 epidemic, it is suggested that CT should be the first choice for screening COVID-19 under the condition of limited nucleic acid detection (Huang, Wang, Li, Ren, Zhao, Hu, Zhang, Fan, Xu, Gu, et al., 2020, Chung, Bernheim, Mei, Zhang, Huang, Zeng, Cui, Xu, Yang, Fayad, et al., 2020, Lei, Li, Li, Qi, 2020). Although imaging features alone cannot make a definite diagnosis, combined with epidemiological history, clinical manifestations and imaging examinations, CT can greatly improve the accuracy of screening, especially for suspected patients and asymptomatic infections. This can help to effectively discover and isolate the source of infection as soon as possible and cut off the route of transmission, which has a positive effect on controlling the development of whole epidemic. In short, chest CT plays a key role in the diagnostic procedure for suspected patients and some recent reports have emphasized its performances (Dong et al., 2020). However, image reading in severe epidemic areas is a tedious and time-consuming task for radiologists, and the visual fatigue would increase the potential risk of missed diagnosis of some small lesions. In addition, radiologists’ judgement is usually influenced by personal bias and clinical experience. Thus, Artificial Intelligence (AI) technology is playing an increasingly important role in the struggle against COVID-19 (Shi et al., 2020).
In recent years, with the gradual deepening study of artificial intelligence technology, image segmentation has been developed rapidly, but it is still challenging to automatically segment the COVID-19 pneumonia lesions from CT scans, especially for multi-class pixel-level segmentation. First, the typical signs of infected lesions observed from CT slices have various complex and changeable appearances, irregular shapes and fuzzy borders. For example, as shown in Fig. 1 , the boundaries of ground-glass opacity (GGO) have low contrast and blurred appearance, and the blurring of boundaries also increases the difficulty of labeling. Second, the successful performance of popular deep convolutional neural networks (CNN), the core technology of the rising AI, is largely depended on the availability of large-scale, well-annotated data sets in the real world. However, it is quite difficult to collect sufficient training data from patients systematically due to the urgent nature of the pandemic, and high-quality annotations of multi-category infections are specially limited. Third, for screening, most of the pneumonia symptoms of collected patients are usually at the early stage, and the proportion of infected lesions in available image samples is small and uneven, which leads to the problem of long-tailed data distribution. In Table 1 , we can see that the number of annotated lesion pixels are far fewer than the background pixels, particularly the proportion of pulmonary consolidations in the data is quite small. In this paper, we deal with the above issues and propose a novel semi-supervised framework for COVID-19 lung infection segmentation from limited and incompletely annotated CT datasets.
Fig. 1.
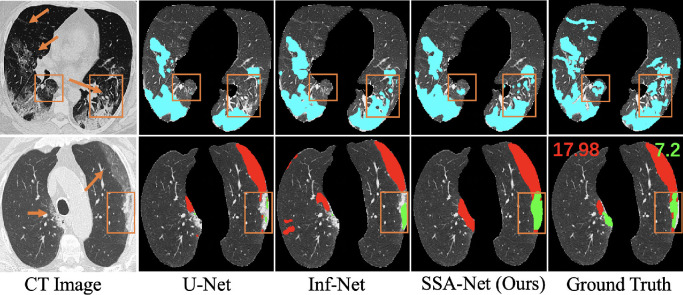
Two examples of COVID-19 positive CT scans from two different datasets and their corresponding segmentation results. The first row is a single-class lesion segmentation with lesions labeled in blue, and the second row is a multi-class lesion segmentation with ground-glass opacity (GGO) in red and consolidation in green. We can clearly see the fuzzy boundaries of the infected areas, highlighted with orange arrows. The red number and the green number marked in the last graph represent the proportions of GGO and consolidation respectively, which shows the issue of imbalanced class distribution. It can be seen that SSA-Net performs better in complicated lesion segmentation and the proposed semi-supervised few-shot learning framework outperforms other state-of-the-art algorithms in multi-class COVID-19 infection segmentation with limited training data, especially in regions labeled with orange boxes.
Table 1.
A summary of the Datasets in our experiments. denotes the total number of COVID-19 slices. denotes the number of labeled infection categories. , , , denote the percentage of pixels of lung area, GGO, consolidation, the total infection of GGO and consolidation, respectively. COVID-19 CT Segmentation dataset is available at https://medicalsegmentation.com/covid19/.
| Dataset | Lung | GGO | Con | G+C | ||
|---|---|---|---|---|---|---|
| COVID-19-CT-Seg (Jun et al., 2020) | 1848 | 1 | 15.46% | - | - | 1.84% |
| COVID-19 CT Segmentation dataset | 98 | 3 | 26.90% | 4.66% | 2.29% | 6.95% |
| COVID-19 CT Segmentation dataset | 370 | 2 | 21.46% | 1.97% | 0.51% | 2.48% |
The main contributions in our work are threefold:
(1) We present an encoder-decoder based deep neural network named spatial self-attention network (SSA-Net) for lesion segmentation. To take full advantage of the context information between the encoder layers, a self-attention distilling method is utilized, which can expand the receptive field and strengthen the self-learning without extra training time. For the sake of obtaining the low contrast and fuzzy boundary area effectively, spatial convolution is introduced for slicing the feature map and then convoluting slicer by slicer, so that the features can be effectively transferred in the direction of row and column.
(2) According to the long-tailed distribution of COVID-19 datasets and limited labeled data, we provide a semi-supervised few-shot iterative segmentation framework for multi-class infection segmentation, which leverages a large amount of unlabeled data to generate their corresponding pseudo labels, thereby augmenting the training dataset effectively. A re-weighting module is introduced to rebalance the category distribution based on the number of pixels for each category, and a trust module is added to select high confidence values and improve the credibility of pseudo labels.
(3) We conducted extensive experiments on two publicly available datasets. Ablation studies have demonstrated that both the spatial convolution and self-attention distilling are beneficial to improve the performance of infection segmentation. And comparative studies have revealed that SSA-Net with our semi-supervised few-shot learning strategy outperformed the state-of-the-art segmentation models and showed competitive performance compared with the state-of-the-art systems in COVID-19 challenge.
2. Related work
In this section, we mainly talk over three aspects of works closely related to our work, including context-enhanced deep learning for segmentation, few-shot learning and class balancing and COVID-19 pneumonia infection segmentation.
2.1. Context-enhanced deep learning for segmentation
In order to segment lesions in medical images, deep learning technology is widely used. U-Net is commonly used for lung region and lung lesion segmentation (Shi et al., 2020). U-Net is a full convolution network proposed by Ronneberger et al. (2015), which has a U-shaped architecture and symmetric encoding and decoding paths. Skip connections connect the layers of the same level in the two paths. Therefore, the network can learn more semantic information with limited data, and it is widely used in medical image segmentation. Thereafter, many variants of networks based on U-Net have been proposed, such as no-new-U-Net (nnU-Net) (Isensee et al., 2019), which is based on 2D and 3D vanilla U-Nets and can adapt the preprocessing strategy and network architecture automatically without manual tuning. Milletari et al., 2016 propose V-Net, which uses residual blocks as the basic convolution blocks for 3D medical images. He et al. (2016) put forward a new encoder-decoder network structure, ResNet, by introducing the residual blocks. Compared with the U-Net and other variants, ResNet can avoid the gradient vanishing and accelerate the network convergence, so we prefer to use ResNet as backbone. However, lesions in medical images are sometimes subtle and sparse, and the number of annotated lesion pixels is much fewer than the background pixels, which brings new challenges. Therefore, we need more contextual and spatial information to train deep models for the task.
Several schemes have been proposed to reinforce the representation ability of deep networks, e.g. some researches improve performance by deepening the network. The UNet++ (Zhou et al., 2018) inserts a nested convolutional structure between the encoding and decoding paths. In order to detect the ambiguous boundaries in medical images, Lee et al. (2020) present a structure boundary preserving segmentation framework, which uses a key point selection algorithm to predict the structure boundary of target. Indeed, the deeper the network is, the more information we can get. However, deepening the network is inefficient, and as the network deepens, it is easy to cause gradient explosion and gradient disappearance, and the optimization effect degrades. Meanwhile, these methods can greatly improve the performance of segmenting large and clustered objects, but they are easy to fail when encountering small and scattered lesions.
Another way is to exploit attention mechanism to optimize the deep learning. For example, Wang et al. (2020b) combine two 3D-ResNets with a prior-attention residual learning block to screen COVID-19 and classify the type of pneumonia. Ren et al. (2020) present a strategy with hard and soft attention modules. The hard-attention module generates coarse segmentation map, while the soft-attention module with position attention can capture context information precisely. Zhong et al. (2020) propose a squeeze-and-attention network which imposes pixel-group attention to conventional convolution to consider the spatial-channel interdependencies. However, the above methods need additional computation cost. Gao et al., 2020 propose a dual-branch combination network for COVID-19 classification and total lesion region segmentation simultaneously. A lesion attention module is used to combine classification features with corresponding segmentation features. Hou et al. (2019) propose a self-attention distillation (SAD) approach, which makes use of the networks own attention maps and perform top-down and layer-wise attention distillation within the network itself. Through the feature maps between the encoder layers, the model can learn from itself without extra labels and consumptions. The intuition of SAD is that useful contextual information can be distilled from the attention maps of successive layers through those of previous layers. The timepoint of training to add SAD to an existing network may affect the convergence time, and it is recommended to use SAD in a model pretrained to some extent. In this paper, we introduce the self-attention learning mechanism into a strengthened U-shaped segmentation network without pre-training. Then stronger labels will be generated from the feature maps of lower layers to guide the deeper layers for further representation learning. And our method is helpful to strengthen some obscure and scattered objects.
Many studies have confirmed that more information can be obtained at the encoder and the bottleneck of network. CE-Net (Gu et al., 2019) presents two modules at the bottleneck. One module uses multi-scale dilated convolution to extract rich features, while the other uses multi-scale pooling operation to further obtain context information. Besides, Shan et al. (2020) propose VB-Net, which is based on V-Net, to achieve more effective segmentation by adding bottleneck blocks by convolutions, but such models are computationally expensive. To utilize spatial information in neural networks, Pan et al. (2017) propose Spatial CNN (SCNN), in which slice-by-slice convolutions within feature maps are employed instead of traditional deep layer-by-layer convolutions, so that messages are transferred between pixels across rows and columns in the layer. In this paper, we attempt to introduce a spatial convolution block into the bottleneck of the encoder-decoder network by using a sequential message passing scheme similar to SCNN. This kind of message passing mechanism helps to propagate the information between neurons, avoid the influence of sparse and subtle supervision, and make better use of the contextual relationships of pixels. Therefore, the U-shaped neural network is strengthened and the training convergence of network can also be accelerated.
2.2. Few-shot learning and class balancing
Because manual labeling is time-consuming, laborious and expensive, many researchers have conducted studies in few-shot learning. Some researchers choose transfer learning (Raghu, Zhang, Kleinberg, Bengio, 2019, Minaee, Kafieh, Sonka, Yazdani, Jamalipour Soufi, 2020), which refers to applying the learned knowledge to other problems in different but related fields to solve new tasks. In addition, many studies augment the data through Generative Adversarial Networks (GAN) (Goodfellow et al., 2014) or its extensions (Mahapatra et al., 2018, Zhou, Gao, Cheng, Gu, Fu, Tu, Yang, Zhao, Liu, 2020), which create new images and corresponding masks, and then add the synthesized data to the training set to expand the training image. Mahapatra et al., 2018 propose a model to generate many synthetic disease images from real disease images by Conditional GAN. These algorithms are computationally intensive and may require additional annotation data. Apart from that, most advanced methods use class activation map (CAM) (Zhou et al., 2016) and gradient-weighted class activation map (Grad-CAM)(Selvaraju et al., 2017) for object localization and image-level weakly supervised semantic segmentation, which get results from feature heatmaps of the network. Sometimes, these methods are used as a basic step for semantic segmentation of large and clustered objects. For instance, Wang et al. (2020d) propose a self-supervised equivariant attention mechanism, in which CAM is combined with pixel correlation module to narrow the gap between full and weak supervisions. So that, for the segmentation of COVID-19 lesions, which are small and scattered, these methods are not ideal. Furthermore, Lee (2013) propose a semi-supervised framework to learn from limited data, which utilize the segmentation results with pseudo labels generated from the model to retrain the model. Then by continuous iterations, this strategy can use few labeled data and pseudo data to improve the performance of network, which is also confirmed in Fan et al., 2020. In this work, we build a similar iterative framework and add a trust module after each iteration to make the pseudo labels more reliable.
The issue of long-tailed training datasets has attracted a lot of attention in machine learning. Zhou et al. (2020b) propose a deep learning algorithm to solve the large-scene-small-object problem. In addition, Cui et al., 2019 present that as the sample number of a class increases, the penalty term of this class decreases significantly. Therefore, through theoretical derivation, they design a re-weighting scheme to re-balance the loss, so as to better achieve long-tailed classification. Kervadec et al. (2019) propose a boundary loss for highly unbalanced segmentation, which uses integrals over the interface between regions rather than using unbalanced integrals over regions. Wu et al. (2020) also present a new loss function called distribution-balanced loss for the multi-label recognition of long-tailed class distributions. This loss re-balances the weights considering the impact of label co-occurrence, and mitigates the over-suppression of negative labels. Different from these methods, we introduce a re-weighting module before the training of each iteration to balance the class distribution.
2.3. COVID-19 Pneumonia infection segmentation
Due to the lack of high-quality pixel-level annotation, a large number of AI-based studies are aimed at solving the issue of COVID-19 diagnosis (Kang et al., 2020) and lesion segmentation from the perspective of using limited training datasets. For example, Oh et al. (2020) provide a method of patch-based convolutional neural network, which has less trainable parameters for COVID-19 diagnosis. He et al. (2020) not only build a publicly-available dataset, but also propose a self-trans method to combine contrastive self-supervised learning with transfer learning to learn strong and unbiased feature representations. Wang et al. (2020c) propose a weakly-supervised deep learning framework for COVID-19 classification and lesion localization by using 3D CT volumes. The 3D deep neural network is used to predict the probability of infections, while the location of COVID-19 lesions is the overlap of the activation region in classification network and the unsupervised connected components. These works are much concerned about the detection of infectious locations and cannot obtain the shape and classification.
Certainly, many deep learning networks have been established to segment COVID-19 lesions. However, most of them are based on adequate data and supervised learning. Yan et al. (2020) introduce a deep CNN, which provides a feature variation block to adjust the global properties of features for the segmentation of COVID-19 lesions. Shan et al. (2020) use the human in the loop strategy for efficient annotation, which can help radiologists improve the automatic labeling of each case. In terms of public datasets, pixel-level annotations are often noisy. Wang et al. (2020a) present an adaptive mechanism to better deal with noisy labels. In their work, they propose a COVID-19 pneumonia lesion segmentation network and a noise-robust dice loss to better segment lesions of various scales and appearances as well. Although the adaptive mechanism can effectively obtain more high-quality annotations, it is very complicated to implement. Fan et al., 2020 propose Inf-Net to automatically segment infected area from CT images. A parallel partial decoder is used to aggregate high-level features and generate global features. Then, a reverse attention module and an edge attention module are used to enhance the representation of boundary. Meanwhile, a semi-supervised training strategy is also introduced.
Nevertheless, most research work ignores the imbalance of infection categories in datasets. In fact, whether it is GGO or consolidation, for doctors, better identification of the distribution of lesions in different stages is more conducive to understand patients condition and make treatment. Therefore, it is necessary to segment not only the total infected regions but also multi-class pneumonia infections with limited data.
3. Method
In this section, we first present the details of our proposed spatial self-attention network in terms of network architecture, self-attention learning, spatial convolution and loss function. We then present the semi-supervised few-shot learning framework for COVID-19 lesions segmentation based on the re-weighting module and the trust module.
3.1. Spatial self-attention network (SSA-Net)
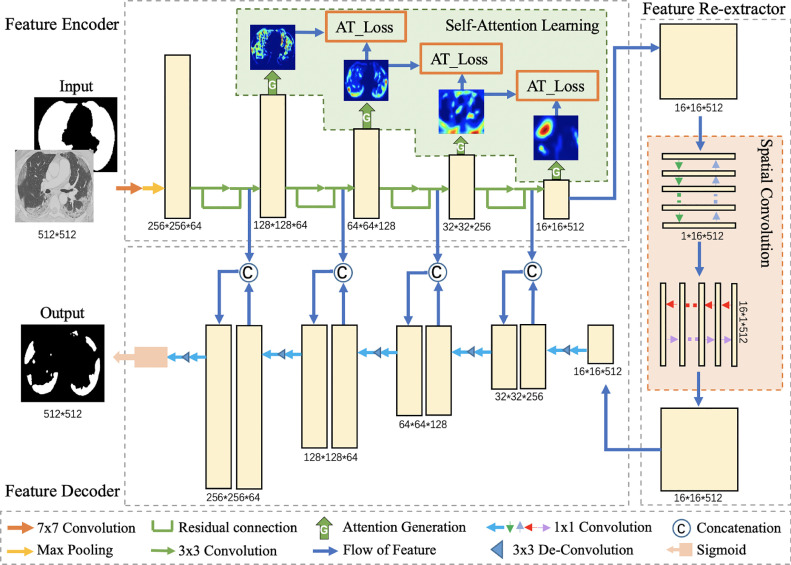
For the sake of obtaining more contextual and spatial information in the learning network and extracting the complex and obscure COVID-19 lesion areas effectively, we propose an encoder-decoder based deep neural network named Spatial Self-Attention network (SSA-Net) for lesion segmentation. As shown in Fig. 2 , the proposed SSA-Net consists of three major parts: a feature encoder with self-attention learning, a feature re-extractor with spatial convolution, and a feature decoder. Each CT slice is concatenated with its lung mask as the input of our proposed network to remove the background except the lungs. In this proposed method, we use ResNet34 (He et al., 2016) as the backbone approach in feature encoder module. Herein, a self-attention learning module is added after four residual blocks to enhance the representation learning by distilling layer-wise attention and useful contextual information from deeper layers. The feature map obtained from the fourth residual block is fed to perform spatial convolution in the feature re-extractor to transmit spatial information. Skip connections are used to concatenate the encoder and the decoder. Meanwhile, for the sake of improving the decoding performance, we use upscaling and deconvolution (Apostolopoulos et al., 2017) operations. Finally, after the sigmoid activation function, the result generated from the feature decoder has the same size as input.
Fig. 2.
The architecture of Spatial Self-Attention network (SSA-Net), which consists of three major parts: feature encoder, feature re-extractor and feature decoder. Each CT slice is concatenated with its lung mask as the input of network. In the feature encoder, a self-attention learning module is added after four residual blocks to enhance the representation learning by distilling layer-wise attention and useful contextual information from deeper layers.The feature map obtained from the fourth residual block is fed to perform spatial convolution in the feature re-extractor using a sequential scheme to transmit spatial information. Skip connections are used to concatenate the encoder layers with four decoder layers with upscaling and deconvolution operations. Finally, after a sigmoid activation function, the result is generated from the feature decoder.
3.1.1. Feature encoder
In this work, the feature encoder consists of four residual blocks for down-sampling operations, which is the same as the encoder of ResNet34. To strengthen the representation, we introduce a self-attention learning module after each residual block, and then the attention maps of previous layers can distil useful contextual information from those of successive layers, and the better representation learned at lower layers will in turn benefit the deeper layers. Through this kind of self-learning, the representation can be strengthened without extra training time and additional labels.
Self-Attention Learning: Several works (Hou, Ma, Liu, Loy, 2019, Ren, Huo, Xuan, Wei, Zhang, Wang, 2020, Zhong, Lin, Bidart, Hu, Daya, Li, Zheng, Li, Wong, 2020) have shown that attention mechanism can provide useful contextual information for segmentation. Thus, we introduce a self-attention learning mechanism to exploit attention maps derived from its own layers of network, without the need of additional labels and external supervisions. The attention maps used in this paper are activation-based attention maps. Specifically, is used to denote the output of -th residual blocks (), where denote the channel, height and width of output, respectively. The attention map is to map the three-dimensional feature of channel, height and width into a two-dimensional feature of height and width, namely . The distribution of spatial features is determined by considering the activated eigenvalues of each channel. The importance of each element on the final output depends on its absolute value in the map. Therefore, the attention map can be generated by a mapping function designed to calculate statistics of all the absolute values of elements across the channel dimension as follows:
| (1) |
where denotes the -th slice of in the channel dimension, and can be a natural number greater than 1. The larger the , the more attention will be paid to these highly activated regions. In our experiment, is set to 2, because it has been verified that this can maximize the performance improvement (Hou et al., 2019).
And then we perform spatial softmax operation () on . The size of attention map is different between two adjacent layers, so bilinear upsampling operation () is used to make the original feature and the target feature the same size. Formally, the whole process is represented by a function:
| (2) |
Finally, we use mean square difference loss () function to calculate the attention loss (, which is shown in Fig. 2) between the four adjacent features after each residual blocks. The formulation is:
| (3) |
So the total loss of self-attention learning is formulated as follow:
| (4) |
where is the number of samples, is the number of residual blocks, and is equal to 4 in this paper.
3.1.2. Feature re-extractor
The feature extractor is a newly spatial convolution module at the bottleneck of our encoder-decoder network. By using a sequential message passing scheme, this module is aimed to extract more spatial information between rows and columns in the feature map and strengthen the training.
Spatial Convolution: Several works (Gu, Cheng, Fu, Zhou, Hao, Zhao, Zhang, Gao, Liu, 2019, Pan, Shi, Luo, Wang, Tang, 2017) have made innovations at the bottleneck of encoder-decoder structures and achieved effective results. In order to improve the ability to explore spatial information of the network and better interpret the common low contrast and fuzzy boundary areas in COVID-19 CT images, we add a spatial convolution module to obtain the feature maps through channel wise convolutions with large kernels.
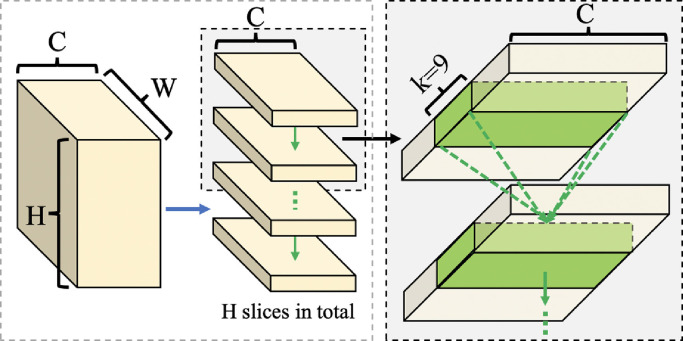
Specifically, the feature map obtained from the feature encoder is a 3D tensor with the size of , where , and is the number of channel, height and width respectively. As shown in Fig. 3 , taking the dimension as an example, that is, passing the message from top to bottom, the feature map would be cut into slices. denotes the kernel width. It represents that a pixel in the next slice can receive messages from pixels in the current slice. The first slice is convoluted by a convolution layer, and the output is added to the second slice, then the new output is then fed to the next convolution. This process is iterated for times to get the final output. The above operations are carried out in four directions, including downward, upward, leftward and rightward, to complete the spatial information transmission.
Fig. 3.
Detailed example of downward in spatial convolution.
Further, denotes the element of a 3D tensor , and , , represent the indexes of channel, height and width respectively. Thus, the spatial convolution function is
| (5) |
where denotes the update of the element, is the nonlinear activation function of ReLU. denotes the weight between an element in channel of the last slice and an element in channel of the current slice, with an offset of columns between the two elements.
3.1.3. Feature decoder
The feature decoder is designed for constructing the segmentation results from feature encoder and feature extractor. Through skip connections, the feature decoder can get more details from encoder to make up for the loss of information after pooling and convolutional operations. Each decoder layer includes a 1 1 convolution, a 3 3 transposed convolution and a 1 1 convolution. Based on skip connections and the concatenations of decoder layers, the output has the same size as input. In the end, we adopt the Sigmoid function as the activation function to generate the segmentation result.
3.1.4. Loss function
The total loss comprises of two terms. One term is a segmentation loss, and the other is a self-attention loss. The COVID-19 infected areas at an early stage, shown as GGO, are often scattered and occupy only a small region of image. When the proportion of foreground is too small, the Dice loss function proposed in Milletari et al., 2016 has been proved to be effective, so we prefer to consider Dice loss function as a segmentation loss in our task. All the networks for comparison are trained with the same loss function (Dice loss), so all the experiments were carried out under the same experimental settings. The Dice loss function is defined as follows:
| (6) |
where G denotes the ground truth and S represents the segmentation. The self-attention loss is mentioned in Eq. (4). Thus, as shown in Eq. (7), the sum of segmentation loss and self-attention loss is regarded as the total loss of the network.
| (7) |
where is the weight of self-attention learning loss to balance the influence of attention loss on the task, and set to 0.1 in our experiment.
3.2. Semi-supervised few-shot learning
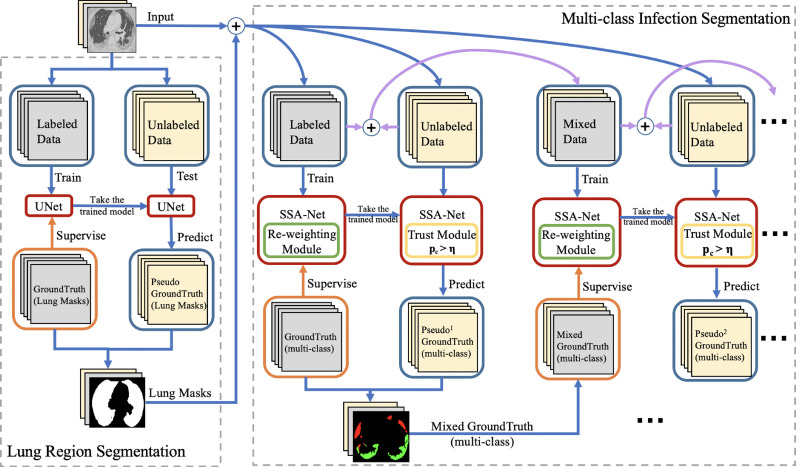
Due to the class unbalanced and limited labeled data of COVID-19 datasets, we propose a semi-supervised few-shot learning framework, which consists of two major parts: the lung region segmentation, and multi-class infection segmentation, as shown in Fig. 4 .
Fig. 4.
The architecture of the semi-supervised few-shot learning framework, which consists of two major parts: the lung region segmentation and iteration infection segmentation. A trained U-Net model is used to segment the lung region in each CT image as an initialization of multi-class infection segmentation. Then, each lung mask is concatenated with its CT image as the input of our multi-class infection segmentation. In this part, we firstly train the model with SSA-Net, and we introduce a re-weighting module to rebalance the class distribution. The unlabeled data are test by the pre-trained SSA model with a trust module to obtain more reliable pseudo labels. Secondly, we take the original data and generated pseudo data as new trainning dataset. Thirdly, we train a new SSA model using this dataset in the same way. Follow this method until all unlabeled images are predicted and the latest model is no longer improved.
3.2.1. Lung region segmentation
The lung region segmentation is an initial step of our COVID-19 lesion segmentation. First, we use a trained U-Net model provided by Hofmanninger et al. (2020) for the segmentation of lung region. Then, all unlabeled CT slices are segmented by the pre-trained U-Net to obtain all the boundaries of lung.
3.2.2. Multi-class infection segmentation
Because the manual labeling of professional doctors is not only time-consuming but also expensive, there are limited labeled public datasets, and fewer labels for multi-class infection areas. In this work, we present a semi-supervised few-shot learning strategy, which leverages a large number of unlabeled CT images to effectively augment the training dataset. Moreover, we introduce a re-weighting module and a trust module to balance the distribution of different lesion classes and to obtain more reliable pseudo labels.
An overview of our semi-supervised few-shot learning framework is shown in Fig. 4. Our framework is based on a random sampling strategy and uses unlabeled data to gradually expand the training dataset and generate pseudo labels. Each CT slice is concatenated with its lung mask generated by lung region segmentation as the input of our proposed SSA model. When training, we exploit a re-weighting module, which is a class re-balancing strategy based on the number of pixels for each class. And more reliable pseudo labels can be obtained from trust module by selecting high confidence values.
Specifically, the labeled dataset is divided into an original training set , a validation set and a test set . We firstly pretrain a SSA model with reweighting module using original labeled dataset . Meanwhile, we use the validation set with true labels to measure the performance of the new trained SSA model, and images in the unlabeled dataset are tested by the pre-trained with trust module to generate pseudo labels. Next, we randomly select generated pseudo samples, and add them into the original training set to make a strengthened training dataset. Then, we use this dataset to train a new SSA model in the same way and repeat this process. Therefore, the strengthened training set consists of the original training set and the pseudo-label training set . Once a new SSA model is generated, all the pseudo labels of images in will be renewed. If the number of unlabeled images in is reduced to be less than , we add all the remaining images in with their pseudo labels into the strengthened training set. If all the images of has been used, we will only update the pseudo labels of during the iteration. The iteration will stop when the DSC of validation set is no longer improved.
Re-weighting module: In this module, we introduce the cross entropy loss, which is more suitable for the condition of class imbalance in multi-class training tasks. However, because not only the number of consolidation samples in datasets is small, but the consolidation proportion of pixels in each image is also very small. In view of this kind of class imbalance, we calculate the pixel ratios of the two categories (GGO and consolidation) in all training data, and exploit the result to set their weights of the cross entropy loss. Therefore, the final loss function is defined as follow:
| (8) |
where denotes the number of total categories, and represents the pixel proportion of class in the training set, which consists of the original labeled training set and the current pseudo-label training set . The initial labeled training set is a multi-class data set. To guide the model to identify different types of lesions, we need to ensure that the original labeled training set contains samples of all categories. If the category is the same as the class of sample , is 1, otherwise it is 0. is the prediction probability of class of sample . In this way, it can ensure that the weight of a class with small proportion is more than 1, and the weight of a class with large proportion is less than 1, so as to achieve a balance of categories.
Trust module: Usually, we only pick up a class label which has the maximum predicted probability for each unlabeled sample. However, not all the predicted values are true values, and false values will guide the model to errors during the iterative process. The work in Lee (2013) has proved that the pseudo labels with high confidence are more effective. Hence, we add a trust model to re-evaluate the pseudo infection class labels obtained from current SSA model, by setting a threshold to select high confidence values, and the predicted pseudo label with credibility is defined as:
| (9) |
where denotes the final pseudo label after re-evaluation, represents the predicted infection category from the current SSA model, and denotes the maximum predicted probability of an unlabeled pixel. The pseudo label is set to 0 and the pixel is treated as uninfected lung region when the probability of predicted category is less than the threshold. The setting of is quite important. is the threshold for re-evaluating the pseudo infection class labels and selecting high confidence values. The higher is, the more confident of the pseudo label will be. Based on the experience, we try several values and is set to 0.95 in our experiments.
4. Experiments and results
4.1. COVID-19 pneumonia infection datasets
At present, many public datasets on COVID-19 are available for free. However, as mentioned above, due to the difficulty of manual labeling, most of the data only have image-wise labels for COVID-19 detection, and only a few datasets are labeled precisely for segmentation. Clinical CT scans collected from currently published COVID-19 CT datasets are used for our experiments.
One of the datasets is the COVID-19-CT-Seg dataset, which has been publicly available at here3 with CC BY-NC-SA license, and contains 20 public COVID-19 CT scans from the Coronacases Initiative and Radiopaedia. The corresponding annotations (Jun et al., 2020) including left lung, right lung, and infection can be freely downloaded at here4 . In Ma et al. (2020), we know that the last 10 cases in this dataset from Radiopaedia have been adjusted to lung window [-1250,250], and then normalized to [0,255]. While the other, the COVID-19 CT Segmentation dataset and its annotations are available at here5 , which includes 100 axial CT images from more than 40 patients with COVID-19 collected by the Italian Society of Medical and Interventional Radiology and 9 axial volumetric CT scans from Radiopaedia6 . In this dataset, the lung masks are contributed by Hofmanninger et al. (2020), and the images and volumes were segmented using three labels: ground-glass, consolidation and pleural effusion.
We use three datasets () for our experiments as shown in Table 2 . Firstly, the COVID-19-CT-Seg dataset consists of 1848 slices with lesion, which have been segmented by experienced radiologists. This dataset is used to demonstrate the effectiveness and stability of our proposed segmentation network. We consider these 1848 slices as . Same as the experiment in Ma et al. (2020), we split the twenty cases in into five groups randomly for 5-folder cross validation. Secondly, consists of 98 slices from the COVID-19 CT Segmentation dataset and we divide them into the same training set and validation set in the experiment of Fan et al., 2020. Finally, from the COVID-19 CT Segmentation dataset, we can obtain 468 slices with multi-class infection labels in total as which is used to confirm that our multi-class semi-supervised few-shot model is feasible and effective.
Table 2.
A summary of the Datasets in our experiments. denotes the total number of COVID-19 slices. denotes the number of lesion category.
| Dataset | From | ||
|---|---|---|---|
| COVID-19-CT-Seg | 1848 | 1 | |
| COVID-19 CT Segmentation dataset | 98 | 1 | |
| COVID-19 CT Segmentation dataset | 468 | 2 |
4.2. Experimental settings
Data preprocessing: In , in the light of the suggestions from instructions of the COVID-19-CT-Seg dataset7 , we preprocessed the image data, including adjusting the gray values to lung window [-1250,250], and then normalizing it to [0,255] for the previous ten groups of volumes. Besides, we cropped the last ten groups of images from 630 630 to 512 512, making them the same size as the previous ten groups. And we also performed the same operations in as well. The operating procedure of cropping is to calculate the center of gravity by using the lung label available in the corresponding dataset, and then calculate the cutting position by using the center of gravity.
Evaluation metrics: We used four metrics for quantitative evaluation between segmentation results and the ground truth , i.e., the Dice similarity coefficient (DSC), the 95-th percentile of Hausdoff Distance (HD), the Mean Absolute Error (MAE) and Normalized surface Dice (NSD). The first three measures are widely used in the evaluation of medical image processing, and the last one can better evaluate the situation of edge segmentation. For the measurements based on DSC and NSD (Nikolov et al., 2018), the higher the scores are, the better the segmentation performs. While on the contrary, for metrics of HD and the MAE, lower scores are supposed to be the better segmentation.
1) Dice Similarity Coefficient (DSC): This was first proposed in Milletari et al., 2016, and then widely used in medical image segmentation. The DSC is a similarity measure function, which is usually used to calculate the similarity of two samples. The formulation is as follows:
| (10) |
2) Hausdoff Distance (HD): This is also a commonly used measure to describe the similarity between segmentation result and the ground truth. DSC is sensitive to the inner filling of mask, while HD is sensitive to the boundary. HD is defined as follows:
| (11) |
The 95-th percentile of Hausdoff Distance () is the final value multiplied by 95% in order to eliminate the effect of a very small subset of outliers.
3) Mean Absolute Error (MAE): This is the average of absolute errors, which can better reflect the prediction error and it is defined as:
| (12) |
4)Normalized Surface Dice (NSD): Unlike the DSC, this measure assesses the overlap of the segmentation and ground truth surfaces with a specified tolerance () instead of the overlap of these two volumes. The surface here is represented by the boundary of mask. Then the segmentation surface and ground truth surface are expressed by G’ and S’ respectively, where G’ = and S’ = . And the border region of these two surfaces at tolerance are denoted by and , where , . The formula is:
| (13) |
where is set to 3 in our experiment, which is the same as Ma et al. mentioned in the Ma et al. (2020).
4.3. Ablation study
In this subsection, we evaluate different variants of the modules presented in Section 3 in order to prove the effectiveness of key components of our model, including the self-attention learning module and spatial convolution module in SSA-Net, and the re-weighting module and trust module in semi-supervised few-shot model.
4.3.1. Ablation experiments of SSA-Net
In order to investigate the importance of each component in SSA-Net, we combine spatial convolution (SC) and self-attention learning (SA) with backbone to get new models and use to train these models, which are devised as follows: backbone (), backbone+SA (), backbone+10episodes+SA (), backbone+20episodes+SA (), backbone+30episodes+SA (), backbone+40episodes+SA (), backbone+50episodes+SA (), backbone+60episodes+SA (), backbone+SC (), backbone+SC+SA ().
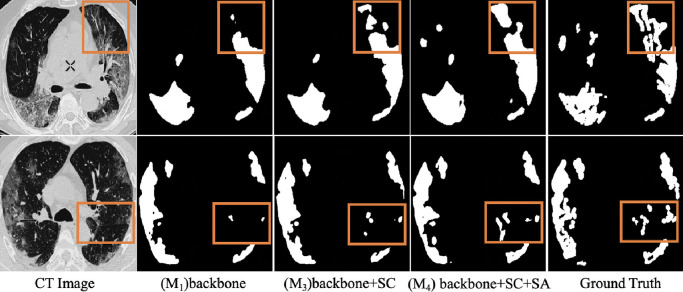
Effectiveness of self-attention learning: We compare and in Table 3 to evaluate the contribution of self-attention learning mechanism. The results clearly show that spatial convolution together with self-attention learning mechanism are useful to drive up performance. However, from model to model , by adding self-attention learning directly, we can also notice a drop in accuracy. As mentioned in Hou et al. (2019), the self-attention learning is assumed to be added to a half-trained model and the time to add the SA module has an effect on the convergence speed of the networks. Here, we also train the backbone by adding the single SA module at different timepoints (from 10 episodes to 50 episodes) and get new models ( - ) of . Table 3 displays the segmentation results in dataset 1 and all the networks are trained up to 150 episodes. The backbone with single SA module can achieve the best segmentation results when introducing the single SA started from the 40 episodes. It proves from one aspect that valuable self-attention contextual information can only be extracted from a model trained to a reasonable level. This accuracy decline reflects the effectiveness of the spatial convolution module as well, which strengthens the network and accelerates the training convergence. Fig. 5 displays two segmentation examples from . From the visual comparisons of and , we can obviously observe that the segmentation results, which is highlighted with orange boxes, show better performance in the model after introducing self-attention learning. It proves that the context information generated from self-attention learning is able to guide the network for better extracting more complex regions.
Table 3.
Ablation studies of our SSA-Net. denotes self-attention learning. denotes spatial covolution. The best results are highlighted in bold.
| Method | Dataset1 |
|||
|---|---|---|---|---|
| DSC | MAE | NSD | ||
| ()backbone | 0.6003 | 5.6866 | 0.0102 | 0.5126 |
| ()backbone+SA | 0.5498 | 6.5469 | 0.0263 | 0.4736 |
| ()backbone+10episodes+SA | 0.5529 | 6.3746 | 0.0219 | 0.4843 |
| ()backbone+20episodes+SA | 0.5878 | 6.0544 | 0.0182 | 0.5076 |
| ()backbone+30episodes+SA | 0.6069 | 5.8735 | 0.0147 | 0.5141 |
| ()backbone+40episodes+SA | 0.6144 | 5.7689 | 0.0121 | 0.5253 |
| ()backbone+50episodes+SA | 0.6100 | 5.7523 | 0.0139 | 0.5197 |
| ()backbone+60episodes+SA | 0.6032 | 5.8466 | 0.0156 | 0.5130 |
| ()backbone+SC | 0.6294 | 5.6036 | 0.0100 | 0.5375 |
| ()backbone+SC+SA | 0.6522 | 5.5260 | 0.0096 | 0.5643 |
Fig. 5.
Ablation studies of different modules for segmentation of COVID-19 pneumonia lesions. The model results show more details similar to ground truth after introducing spatial convolution, while after introducing self-attention learning, the contextual information generated is able to guide the network for better extracting more complex and scattered regions. The segmentation results highlighted with orange boxes show best performance in the model trained with both self-attention learning and spatial convolution.
Effectiveness of spatial convolution: From Tabel 3, all the metrics show that the models with spatial convolution make a better performance than models without this module. This clearly demonstrates that the use of spatial convolution can make the model segment the lesions more accurately. Furthermore, as shown in Fig. 5, we observe that the model shows more details similar to ground truth after introducing spatial convolution, especially the highlighted part in the orange box. Compared with the results of and , it also demonstrate that the spatial convolution module can not only help transfer the information between cows and columns in the backbone network, but also make better use of the context information to detect scattered and obscure lesions after introducing self-attention learning.
4.3.2. Ablation experiments of semi-supervised few-shot model
We further extend our SSA-Net to the segmentation of small samples multi-class lesions (GGO and consolidation). We use 98 slices in to train the semi-supervised models and the rest data is used for validation. The baselines we devised are as follows: SSA-Net with iteration (), SSA-Net based on re-weighting with iteration (), SSA-Net based on trust module with iteration () and SSA-Net based on re-weighting module and trust module with iteration ().
Effectiveness of re-weighting module: As shown in Table 4 , some evaluation metrics of reduce slightly compared with . The main reason is pseudo labels generated from the iteration model may contain more inaccurate results, so the re-weighting module can be affected and cannot work effectively in the following iterations. Therefore, we derive and based on trust module. The DSC of GGO and consolidation increase at the same time after introducing the re-weighting module. Although the and NSD of GGO have a faint decline, the average of most evaluation metrics have improved. The DSC and NSD raise to 0.5608 and 0.5128 respectively, while the HD95 descends to 0.0071.
Table 4.
Quantitative results of Different semi-supervised models trained with . denotes the model with iteration. denotes the model based on re-weighting module. denotes the model based on trust module. The best results are highlighted in bold.
| Method | Ground-glass opacity (GGO) |
Consolidation |
Average |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | MAE | NSD | DSC | MAE | NSD | DSC | MAE | NSD | ||||
| ()SSA-Net+ | 0.4058 | 7.3735 | 0.0237 | 0.3576 | 0.6251 | 3.1018 | 0.0025 | 0.5991 | 0.5155 | 5.2377 | 0.0131 | 0.4784 |
| ()SSA-Net+ | 0.4225 | 7.4845 | 0.0192 | 0.3605 | 0.6010 | 3.2384 | 0.0028 | 0.5661 | 0.5118 | 5.3615 | 0.0110 | 0.4633 |
| ()SSA-Net+ | 0.4622 | 5.5402 | 0.0117 | 0.4061 | 0.6343 | 3.0100 | 0.0026 | 0.5720 | 0.5483 | 4.2751 | 0.0072 | 0.4891 |
| ()SSA-Net+ | 0.4654 | 5.9266 | 0.0116 | 0.4016 | 0.6562 | 2.9541 | 0.0026 | 0.6239 | 0.5608 | 4.4404 | 0.0071 | 0.5128 |
Effectiveness of trust module: From these results of and in Table 4, it is evidential that trust module boosts the segmentation performance both in GGO and consolidation. Generally, we boost the performance by 3.28% and 1.07% in terms of the average DSC and average NSD, and reduce the average to 4.2751, the average MAE to 0.0072. Furthermore, we can observe from and that trust module is the basis of the re-weighting module. The re-weighting module can be effective under the condition of the trust module which is able to make pseudo labels more reliable.
4.4. Comparison of different deep learning networks
We compare our SSA-Net with two state-of-the-art deep learning networks, U-Net and nnU-Net, for semantic or medical image segmentation performance, and with Inf-Net, a COVID-19 infection segmentation network.
From the quantitative comparison shown in Table 5 , we can observe that nnU-Net, as an improved version of U-Net, has a better performance in segmentation tasks. This is mainly because nnU-Net has a more robust structure to adapt to a variety of datasets. Furthermore, the proposed SSA-Net is slightly better than nnU-Net in terms of DSC, and NSD in both and . Our SSA-Net improves the average DSC from 0.6447 to 0.6522, the average NSD from 0.5347 to 0.5643 and reduces the average from 5.7383 mm to 5.5260 mm in . While in , our SSA-Net improves the average DSC from 0.7500 to 0.7540, the average NSD from 0.5862 to 0.5876 and reduces the average from 7.1841 mm to 7.0464 mm. The improvements demonstrate that spatial convolution has the ability to obtain more information between rows and columns in images, and on this basis, the self-attention learning mechanism can offer more reliable context information. Compared with Inf-Net, the advantage of SSA-Net in is not obvious. In terms of DSC and NSD, our proposed SSA-Net outperforms by 1.14% and 1% respectively. But in , it is evident that all evaluation metrics of all networks increase significantly. However, our proposed SSA-Net has more advantages in this dataset. We observe that most patients represented by the CT images are in moderate or severe conditions, the lesion includes not only the fuzzy GGO in the early stage, but also the consolidation in the later stage in this small sample . Although the segmentation task in is more challenging than that in , our proposed SSA-Net can obtain more spatially complex information in a limited data set. And even if the lesions have a complex structure, it can perform better as well. The DSC, and NSD are better than others, reaching 0.7540, 7.0464 and 0.5876, respectively.
Table 5.
Quantitative evaluation of different networks for segmentation of single-class COVID-19 pneumonia lesions. The best results are highlighted in bold. The data marked with are inconsistent with that in Fan et al., 2020. The DSC and MAE of infnet here are better than the those in Fan et al., 2020 (0.682 and 0.082 respectively). The reason is that we are different in pre-processing. In Fan et al., 2020, they resize all the images to 352 352. But here, the size of images is adjusted to 512 512.
| Method |
|
|
||||||
|---|---|---|---|---|---|---|---|---|
| DSC | MAE | NSD | DSC | MAE | NSD | |||
| U-Net Ronneberger et al. (2015) | 0.5850 | 6.2653 | 0.0216 | 0.5151 | 0.6723 | 8.2343 | 0.1142 | 0.5489 |
| nnU-Net Isensee et al. (2019) | 0.6447 | 5.7383 | 0.0106 | 0.5347 | 0.7500 | 7.1841 | 0.0275 | 0.5862 |
| Inf-Net Fan et al., 2020 | 0.6408 | 5.5155 | 0.0092 | 0.5633 | 0.7236 | 7.0808 | 0.0311 | 0.5464 |
| SSA-Net(Ours) | 0.6522 | 5.5260 | 0.0096 | 0.5643 | 0.7540 | 7.0464 | 0.0305 | 0.5876 |
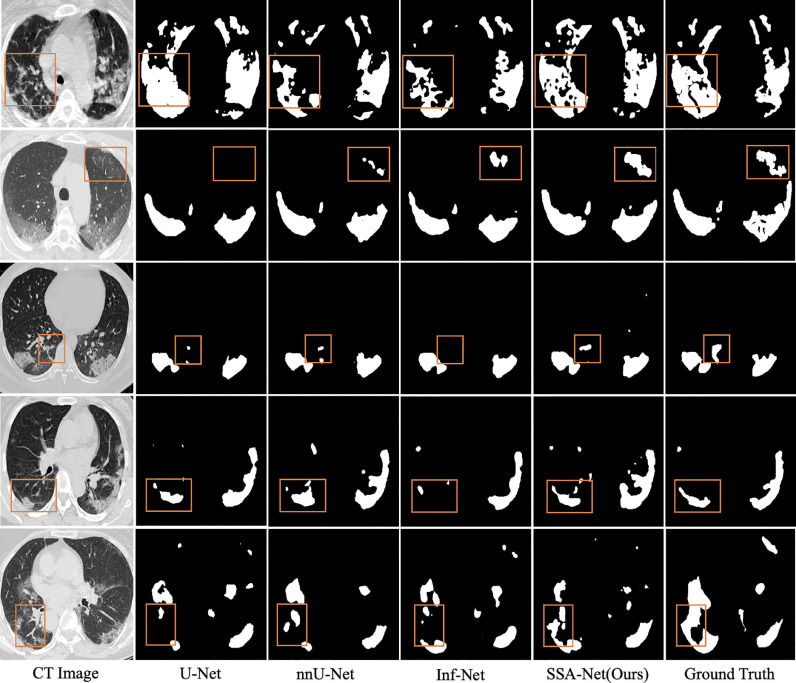
Fig. 6 shows a visual comparison of the results obtained from different networks in two different datasets. It can be observed that most of the current methods have improved the results, but they still perform poorly in the case of fuzzy areas and irregular shapes of COVID-19 lesions. However, our SSA-Net effectively alleviates this problem. Specifically, the segmentation results of SSA-Net are close to the ground truth, and there are fewer incorrectly segmented regions as well, especially for misty and scattered regions, which is attributed to the strengthened representation ability for fuzzy boundaries and irregular shapes of spatial convolution. Meanwhile, even in case of limited data in , SSA-Net can perform well, which is due to the role of self-attention learning to enable the model learn from itself, thereby further enhancing the ability of contextual expression.
Fig. 6.
Visual comparison of single-class infection segmentation results. The regions highlighted with orange boxes show the better performance of SSA-Net.
4.5. Results of semi-supervised few-shot learning
From Table 6 , our proposed SSA-Net has shown more competitive performance than other baseline methods. Besides, our proposed semi-supervised few-shot model (SSA-Net(I)) outperforms other algorithms in all evaluation metrics. By introducing the re-weighting module for class balancing and the trust module for generating more credible pseudo labels, our SSA-Net based semi-supervised learning framework enables the limited data to be utilized as much as possible. Compared with SSA-Net, in terms of GGO, SSA-Net(I) boost the performance by 5.02% in average DSC, 3.01% in NSD, and decrease the and MAE to 5.9266 and 0.0116 respectively. While in terms of consolidation, SSA-Net(I) still shows the best performance. The reason is that SSA-Net can obtain stronger receptive field and contextual information, which helps to detect scattered and complex lesions. In addition, the training of SSA-Net is a process of continuous reinforcement of spatial information, so SSA-Net can improve the self-learning ability of the network in the case of few training samples.
Table 6.
Quantitative evaluation of different models trained with for segmentation of multi-class COVID-19 pneumonia lesions. The best results are highlighted in bold.
| Method | ground-glass opacity (GGO) |
Consolidation |
Average |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC | MAE | NSD | DSC | MAE | NSD | DSC | MAE | NSD | ||||
| U-Net | 0.3596 | 7.3888 | 0.0320 | 0.3391 | 0.5277 | 3.6676 | 0.0030 | 0.4838 | 0.4437 | 5.5282 | 0.0175 | 0.4115 |
| nnU-Net | 0.4049 | 7.7792 | 0.0214 | 0.3395 | 0.4239 | 4.5909 | 0.0051 | 0.3697 | 0.4144 | 6.1851 | 0.0133 | 0.3546 |
| Inf-Net | 0.3021 | 8.9342 | 0.0448 | 0.3084 | 0.3987 | 4.8367 | 0.0054 | 0.2934 | 0.3504 | 6.8855 | 0.0251 | 0.3009 |
| SSA-Net | 0.4152 | 6.7788 | 0.0186 | 0.3713 | 0.4953 | 3.5529 | 0.0029 | 0.4529 | 0.4553 | 5.1659 | 0.0108 | 0.4121 |
| SSA-Net(I) | 0.4654 | 5.9266 | 0.0116 | 0.4016 | 0.6562 | 2.9541 | 0.0026 | 0.6239 | 0.5608 | 4.4404 | 0.0071 | 0.5128 |
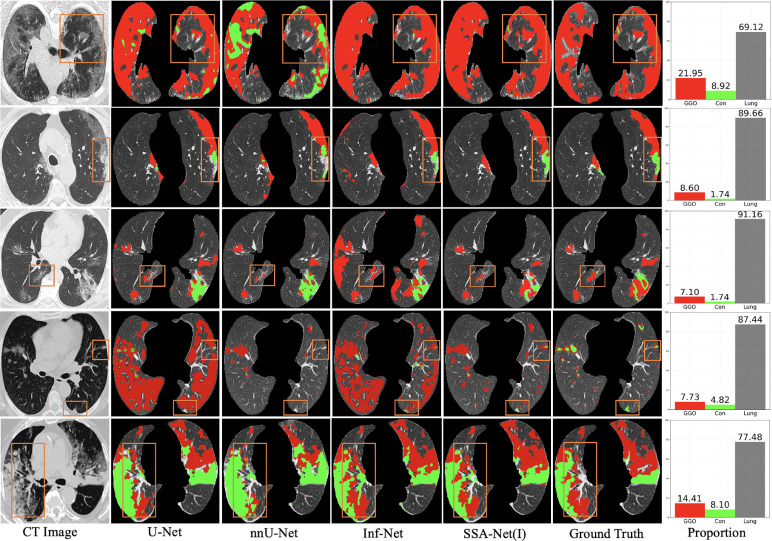
Fig. 7 shows the multi-class lesion segmentation results. Due to the small amount of training dataset, it is more prone to obtain wrong segmentations. Therefore, the baseline methods generate more incorrect results. On the contrary, the results of SSA-Net(I) are closer to the ground truth, because we set a threshold to get high confidence values and drop off the incorrect values. In addition, as can be observed in Fig. 7, the proportional distribution of classes in the last column shows that the data categories in dataset are unbalanced. Among them, lesions containing GGO and consolidation only account for a small proportion of the image, and the most part of images are uninfected lung regions. For small consolidations are quite difficult to segment correctly, but also easily affect the segmentation of GGO. However, our proposed small samples semi-supervised learning model based on SSA-Net can segment lesions more accurately, even if the lesions are small or the boundary is blurred. We can also draw the conclusion that our model can get the results more correctly, which is contributed to the effect of re-weighting module.
Fig. 7.
Visual comparison of multi-class infection segmentation results, where the red and green labels denote the GGO and consolidation, respectively. The first three examples are from , while the rest two are from . Besides, the bar charts in the last column are the proportional distributions of different categories, where the red, green and gray columns represent the GGO, consolidation and uninfected lung area, respectively.
5. Conclusion and future work
In this paper, we have proposed a novel COVID-19 pneumonia lesion segmentation learning network called Spatial Self-Attention network (SSA-Net), which exploits self-attention learning and spatial convolution to obtain more contextual information and can improve the performance in challenging segmentation task of COVID-19 infection areas. Furthermore, we have introduced our SSA-Net for multi-class lesion segmentation with small samples datasets. And we have presented a semi-supervised few-shot learning framework, in which a re-weighting module is utilized to re-balance the loss of different classes and solve the issue of long-tailed distribution of training data, and also a trust module is used to select high confidence values. Extensive experiments on public datasets have demonstrated that our proposed SSA-Net outperforms state-of-the-art medical image segmentation networks. At the same time, our semi-supervised iterative segmentation model also achieves higher performance by training limited data.
The proposed deep learning network can identify scattered and blurred lesions in complicated backgrounds, and which usually happens in medical images. In the future, we will apply it to other related tasks. In addition, due to the urgent nature of the COVID-19 global pandemic, it is difficult to systematically collect large datasets and annotations, especially multi-class annotations, for deep neural network training. Our few-shot multi-class semi-supervised training model only improves the model in process of getting more credible labels. In the near future, we plan to design a comprehensive system to detect, segment and analyze the COVID-19 pneumonia lesions automatically. Besides, we can get initial segmentation results to utilize class activation maps (Zhou, Khosla, Lapedriza, Oliva, Torralba, 2016, Selvaraju, Cogswell, Das, Vedantam, Parikh, Batra, 2017) generated from the feature maps of the network for data augmentation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the Natural Science Foundation of Zhejiang Province under grants LQ20H160052, LY22F030015, LY22F020016, LY21F020024, the National Natural Science Foundation of China under grant 11302195, 62002325, U1908210, and the Zhejiang Provincial Research Project on the Application of Public Welfare Technologies under grant LGF22F020023.
Footnotes
References
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos S., De Zanet S., Ciller C., Wolf S., Sznitman R. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2017. Pathological OCT retinal layer segmentation using branch residual U-shape networks; pp. 294–301. [Google Scholar]
- Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.B., Wang D.C., Mei J., et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020:200823. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Jia M., Lin T.Y., Song Y., Belongie S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2019. Class-balanced loss based on effective number of samples; pp. 9268–9277. [Google Scholar]
- Dong D., Tang Z., Wang S., Hui H., Gong L., Lu Y., Xue Z., Liao H., Chen F., Yang F., et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020 doi: 10.1101/2020.04.22.20074948. [DOI] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest ct for covid-19: comparison to rt-pcr. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 32073353
- Gao K., Su J., Jiang Z., Zeng L.L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y., et al. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2020;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Advances in Neural Information Processing Systems. 2014. Generative adversarial nets; pp. 2672–2680. [Google Scholar]
- Gu Z., Cheng J., Fu H., Zhou K., Hao H., Zhao Y., Zhang T., Gao S., Liu J. CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans. Med. Imaging. 2019;38(10):2281–2292. doi: 10.1109/TMI.2019.2903562. [DOI] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., Xie P. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medRxiv. 2020 doi: 10.1101/2020.04.13.20063941. [DOI] [Google Scholar]
- Hofmanninger, J., Prayer, F., Pan, J., Rohrich, S., Prosch, H., Langs, G., 2020. Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem. arXiv preprint arXiv:2001.11767. doi: 10.1186/s41747-020-00173-2. [DOI] [PMC free article] [PubMed]
- Hou Y., Ma Z., Liu C., Loy C.C. Proceedings of the IEEE International Conference on Computer Vision. 2019. Learning lightweight lane detection cnns by self attention distillation; pp. 1013–1021. [DOI] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee F., Jäger P.F., Kohl S.A., Petersen J., Maier-Hein K.H. Automated design of deep learning methods for biomedical image segmentation. arXiv preprint arXiv:1904.08128. 2019 [Google Scholar]
- Jun, M., Cheng, G., Yixin, W., Xingle, A., Jiantao, G., Ziqi, Y., Minqing, Z., Xin, L., Xueyuan, D., Shucheng, C., Hao, W., Sen, M., Xiaoyu, Y., Ziwei, N., Chen, L., Lu, T., Yuntao, Z., Qiongjie, Z., Guoqiang, D., Jian, H., 2020. COVID-19 CT Lung and Infection Segmentation Dataset. doi: 10.5281/zenodo.3757476. [DOI]
- Kang H., Xia L., Yan F., Wan Z., Shi F., Yuan H., Jiang H., Wu D., Sui H., Zhang C., et al. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- Kervadec H., Bouchtiba J., Desrosiers C., Granger E., Dolz J., Ayed I.B. International Conference on Medical Imaging with deep learning. PMLR; 2019. Boundary loss for highly unbalanced segmentation; pp. 285–296. [DOI] [Google Scholar]
- Lee D.H. Workshop on Challenges in Representation Learning. Vol. 3. ICML; 2013. Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks. [Google Scholar]
- Lee H.J., Kim J.U., Lee S., Kim H.G., Ro Y.M. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Structure boundary preserving segmentation for medical image with ambiguous boundary; pp. 4817–4826. [DOI] [Google Scholar]
- Lei J., Li J., Li X., Qi X. Ct imaging of the 2019 novel coronavirus (2019-ncov) pneumonia. Radiology. 2020;295(1) doi: 10.1148/radiol.2020200236. [DOI] [PMC free article] [PubMed] [Google Scholar]; 18–18
- Liang T., et al. Vol. 68. 2020. Handbook of covid-19 prevention and treatment. [Google Scholar]; The First Affiliated Hospital, Zhejiang University School of Medicine. Compiled According to Clinical Experience
- Ma J., Wang Y., An X., Ge C., Yu Z., Chen J., Zhu Q., Dong G., He J., He Z., et al. Towards efficient COVID-19 CT annotation: a benchmark for lung and infection segmentation. arXiv preprint arXiv:2004.12537. 2020 [Google Scholar]
- Mahapatra D., Bozorgtabar B., Thiran J.P., Reyes M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2018. Efficient active learning for image classification and segmentation using a sample selection and conditional generative adversarial network; pp. 580–588. [Google Scholar]
- Milletari F., Navab N., Ahmadi S.A. 2016 Fourth International Conference on 3D Vision (3DV) IEEE; 2016. V-Net: fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov S., Blackwell S., Mendes R., De Fauw J., Meyer C., Hughes C., Askham H., Romera-Paredes B., Karthikesalingam A., Chu C., et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. arXiv preprint arXiv:1809.04430. 2018 [Google Scholar]
- Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/tmi.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Pan X., Shi J., Luo P., Wang X., Tang X. Spatial as deep: spatial CNN for traffic scene understanding. arXiv preprint arXiv:1712.06080. 2017 [Google Scholar]
- Raghu M., Zhang C., Kleinberg J., Bengio S. Advances in Neural Information Processing Systems. 2019. Transfusion: Understanding transfer learning for medical imaging; pp. 3347–3357. [Google Scholar]
- Ren X., Huo J., Xuan K., Wei D., Zhang L., Wang Q. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) IEEE; 2020. Robust brain magnetic resonance image segmentation for hydrocephalus patients: Hard and soft attention; pp. 385–389. [DOI] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. International Conference on Medical image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., Schluger N.W., Volpi A., Yim J.J., Martin I.B., et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner society. Chest. 2020 doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [DOI] [Google Scholar]
- Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shi Y. Lung infection quantification of covid-19 in ct images with deep learning. arXiv preprint arXiv:2003.04655. 2020 doi: 10.1002/mp.14609. [DOI] [Google Scholar]
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/rbme.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/tmi.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Bao Y., Wen Y., Lu H., Luo H., Xiang Y., Li X., Liu C., Qian D. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/tmi.2020.2994908. [DOI] [PubMed] [Google Scholar]
- Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/tmi.2020.2995965. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang J., Kan M., Shan S., Chen X. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Self-supervised equivariant attention mechanism for weakly supervised semantic segmentation; pp. 12275–12284. [DOI] [Google Scholar]
- Wong H.Y.F., Lam H.Y.S., Fong A.H.T., Leung S.T., Chin T.W.-Y., Lo C.S.Y., Lui M.M.S., Lee J.C.Y., Chiu K.W.H., Chung T., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020:201160. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Huang Q., Liu Z., Wang Y., Lin D. European Conference on Computer Vision. Springer; 2020. Distribution-balanced loss for multi-label classification in long-tailed datasets; pp. 162–178. [DOI] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Wang B., Gong D., Luo C., Zhao W., Shen J., Shi Q., Jin S., Zhang L., You Z. COVID19 chest CT image segmentation–a deep convolutional neural network solution. arXiv preprint arXiv:2004.10987. 2020 [Google Scholar]
- Zhong Z., Lin Z.Q., Bidart R., Hu X., Daya I.B., Li Z., Zheng W.-S., Li J., Wong A. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Squeeze-and-attention networks for semantic segmentation; pp. 13065–13074. [DOI] [Google Scholar]
- Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [DOI] [Google Scholar]
- Zhou K., Gao S., Cheng J., Gu Z., Fu H., Tu Z., Yang J., Zhao Y., Liu J. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) IEEE; 2020. Sparse-GAN: sparsity-constrained generative adversarial network for anomaly detection in retinal OCT image; pp. 1227–1231. [Google Scholar]
- Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., et al. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/tmi.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. UNet++: a nested U-Net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]