Abstract
Autism spectrum disorder (ASD) is a prevalent neurodevelopmental disorder characterized by several alterations, including disorganized brain cytoarchitecture and excitatory/inhibitory (E/I) imbalance. We aimed to analyze aspects associated with the inhibitory components in ASD, using bioinformatics to develop notions about embryonic life and tissue analysis for postnatal life. We analyzed microarray and RNAseq datasets of embryos from different ASD models, demonstrating that regions involved in neuronal development are affected. We evaluated the effect of prenatal treatment with resveratrol (RSV) on the neuronal organization and quantity of parvalbumin-positive (PV+), somatostatin-positive (SOM+), and calbindin-positive (CB+) GABAergic interneurons, besides the levels of synaptic proteins and GABA receptors in the medial prefrontal cortex (mPFC) and hippocampus (HC) of the ASD model induced by valproic acid (VPA). VPA increased the total number of neurons in the mPFC, while it reduced the number of SOM+ neurons, as well as the proportion of SOM+, PV+, and CB+ neurons (subregion-specific manner), with preventive effects of RSV. In summary, metabolic alterations or gene expression impairments could be induced by VPA, leading to extensive damage in the late developmental stages. By contrast, due to its antioxidant, neuroprotective, and opposite action on histone properties, RSV may avoid damages induced by VPA.
Keywords: autism spectrum disorder, valproic acid, resveratrol, interneuron, synapse, GABA receptor
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder described as a behavioral dyad composed of (a) communication and social interaction impairments and (b) stereotyped or repetitive patterns of behavior [1]. Besides that, ASD presents a high prevalence (1:44 in children up to 8 years old in the USA) [2] and heterogeneity among individuals [3], resulting in a challenge for clinical diagnosis [4,5] and public health policies [6].
Epilepsy and other electrophysiological abnormalities are among the most prevalent ASD comorbidities, affecting up to 1/3 of the individuals with ASD [7,8,9]. This evidence leads to the most consolidated hypothesis regarding ASD pathophysiology—that the imbalance between excitation and inhibition is probably associated with impairments in the inhibitory component [10,11,12]. Interneurons are crucial for the inhibition of neural circuits [13]. Although they represent only 10–15% of the total neurons in the hippocampus (HC) [14] and 20–30% in the neocortex in humans [15], the high diversity of cellular shapes, populations, and functional properties highlight their importance in the brain [16,17]. Parvalbumin-positive (PV+) and somatostatin-positive (SOM+) interneurons comprise the majority of the GABAergic interneurons (40% and 30% in the cortex, respectively), followed by several smaller populations, including calbindin-positive (CB+) interneurons [16,18]. While SOM+ neurons contribute to the regulation of the local excitatory input integration in cortical regions [19], PV+ neurons are implicated in the integration among different regions [20] and between the hemispheres [21]. Moreover, changes in PV+ neuron inputs [22,23] and the intrinsic features of this subpopulation [24,25] are observed in animal models of ASD, while evidence regarding SOM+ is still incipient. In addition, dysfunctions in other inhibitory components of the E/I balance have already been described in ASD, such as decreased levels of GABA receptor subunits in the parietal cortex and cerebellum in postmortem analysis [26] and synaptic alterations (e.g., reduced pruning [27] and mutations in the genes of PSD-95, gephyrin, and neuroligins [28,29]).
Recently, neuroimmune aspects have emerged as important factors involved in triggering neurodevelopmental disorders. For example, maternal immune activation (MIA) induces ASD-like features, changes in the cytokine profile (especially IL-6), and imbalances in lymphocyte populations [30,31]. In addition, it is observed to alter the expression of genes associated with neurodevelopment, such as genes involved with migration, function, and placement of interneurons [32,33,34]. Similarly, prenatal exposure to valproic acid (VPA) in rodents, a well-established model of autism [35,36,37,38,39], induces interneuronal alterations in sensory areas [36] and HC [40,41]. Furthermore, those animals show alterations in the profile of brain and peripheral cytokines [42] and a reduction of T CD4 + lymphocytes in the lymph nodes [43], indicating a possible involvement of the neuroimmune axis in the VPA model.
Therefore, molecules that prenatally modulate the immune system may hold promise in preventing neurodevelopmental alterations. For example, MR-39, an agonist of the FRP2 receptor, modulates the expression of lipoxin A4 in hippocampal tissues of BTBR and VPA animals, also improving social behavior impairments [44]. Following this line, trans-resveratrol (RSV, 3,5,4’-trihydroxystilbene) has been studied in the context of schizophrenia [45], attention deficit hyperactivity disorder [46], and ASD due to its antioxidant, anti-inflammatory, and neuroprotective effects [47]. The mechanisms associated with the neuroprotective effects of polyphenols, in general, involve scavenging of reactive species of oxygen (and others), modulation of inflammatory cytokines, reduction of the aggregation of amyloid proteins, among several other effects [48]. Interestingly, prenatal treatment with RSV prevented behavioral and molecular impairments in the VPA model [35,36,38]. However, it remains unknown whether RSV exerts any preventive effect on the quantity of GABAergic interneurons and on the laminar organization in the cortex and HC. Thus, we aimed to evaluate RNA-Seq and microarray library datasets in order to identify altered biological pathways in the embryos from an ASD animal model. Subsequently, we aimed to verify which of these pathways could be modulated by RSV; another further goal of this study was to investigate the possible preventive effects of RSV in the VPA model related to GABAergic interneuron proportion and placement; synaptic proteins, and GABA receptor expression in the medial prefrontal cortex (mPFC) and HC from juvenile rats.
2. Results
2.1. Big Data Evaluation: Early Metabolic Alterations, Cell Cycle Dysfunctions, and Progressive Impairments in Embryos or Progenitor Cells from ASD-Associated Animal Models
In order to create insights regarding cortical embryonic alterations in the VPA model, we refined five datasets (DS) library repositories (Figure S2). The descriptions of the DS are summarized in Table 1.
Table 1.
Description of the datasets analyzed.
| Reference | Animal Model/Sample | Embryonic Day | Method |
|---|---|---|---|
| Balmer et al., 2014 (DS1) |
Neural differentiated hESC exposed to VPA | 6 h and 4 days after VPA exposure | Microarray |
| Canales et al., 2021 (DS2) |
MIA Poly(I:C), mouse cortex | E12.5, E14.5, and E17.5 | RNA-Seq |
| Cui et al., 2020 (DS3) |
Cortical organoids exposed to VPA | 5 days after exposure | RNA-Seq |
| Kalish et al., 2021 (DS4) |
MIA Poly(I:C), mouse brain | E14 and E18 | RNA-Seq (single-cell) |
| Oskvig et al., 2012 (DS5) |
MIA Poly(I:C), rat cortex | E15 | Microarray |
These analyses helped conduct the evaluation of the experimental results, enabling the creation of more grounded hypotheses about the changes identified in postnatal life. In DS2, we observed an enrichment of differentially expressed genes (DEGs) for pathways associated with carbohydrate metabolism, hypoxia response, and sensory organ development six hours after MIA induction at E12.5 (besides other expected alterations like sensory organ development represented by the eye). Interestingly, 1.83% of these DEGs had an ortholog described in the SFARI database. At E14.5, the lipid, purine, and mitochondrial metabolism were associated with upregulated genes, while the protein dynamics (including histone modification and ubiquitination), cell cycle, nucleic acid metabolism, and response to reactive oxygen species were associated with downregulated genes. Moreover, the cell adhesion, extracellular matrix, synapse, and GABA/glutamate pathways were associated with upregulated DEGs (5.52% of these genes had an ortholog described in the SFARI database). At E18.5, mitochondrial and purine metabolism were still associated with upregulated genes, together with cell adhesion, extracellular matrix, synapse, glutamate metabolism, MAP/ERK, and adenylyl cyclase/cAMP signaling. Protein dynamics (including histone modification and ubiquitination), cell cycle, and nucleic acid metabolism were still associated with downregulated genes, together with WNT, Notch, and Hippo signaling, GABAergic neuron differentiation, and neuronal migration. Of note, 7.6% of the DEGs had an ortholog described in the Simons Foundation Autism Research Initiative (SFARI) database.
In DS5, at E15, upregulated genes were associated with mitochondrial and nucleic acid metabolism, ubiquitination regulation, and cell cycle. On the other hand, downregulated genes were associated with RNA metabolism, gene expression, histone modifications, GABAergic neurons differentiation, and neuronal migration. Interestingly, 5.15% of the DEGs had an ortholog described in the SFARI database.
In DS4, at E14.5, the DEGs identified in different subregions, including cortical subplate, cortical layers, subventricular zone, ganglionic eminence, and neural cells such as interneurons and radial glia, pointed to enriched pathways associated with mitochondrial metabolism, nucleic acid metabolism, protein dynamics, and cell cycle. Of note, 4.45–9.93% of the DEGs had an ortholog described in the SFARI database, depending on the brain region and cell type. At E18.5, the same regions and cells, and also other cortical layers and oligodendrocytes, presented a higher restricted pattern of alterations, especially in the mitochondrial metabolism and protein translation and metabolism. Around 2.91–7.47% of the DEGs had an ortholog described in the SFARI database, except for one region (ganglionic eminence) and one cell (radial glia), which did not present any match with SFARI.
In DS1, the neural cells exposed to VPA demonstrated, after six hours, DEGs associated with nucleic acid metabolism, cell cycle, MAP/ERK, and adenylyl cyclase/cAMP signaling, and neuronal migration, with 5.84% of the DEGs presenting an ortholog described in the SFARI database. After four days, the DEGs were associated with the same pathways, and WNT and Notch signaling, extracellular matrix, cell adhesion, and response to hypoxia. Interestingly, 7.42% of the DEGs had an ortholog described in the SFARI database.
Finally, in DS3, the organoids exposed to VPA demonstrated upregulation of genes associated with carbohydrate and lipid metabolism, ion transport, and cell adhesion. The downregulated genes were associated with nucleic acid and protein metabolism, eye development, synapse, and the WNT pathway. Only 6.36% of the DEGs had an ortholog described in the SFARI database.
2.2. The RSV Treatment Prevented the Neuronal Number Alterations Induced by VPA in the mPFC
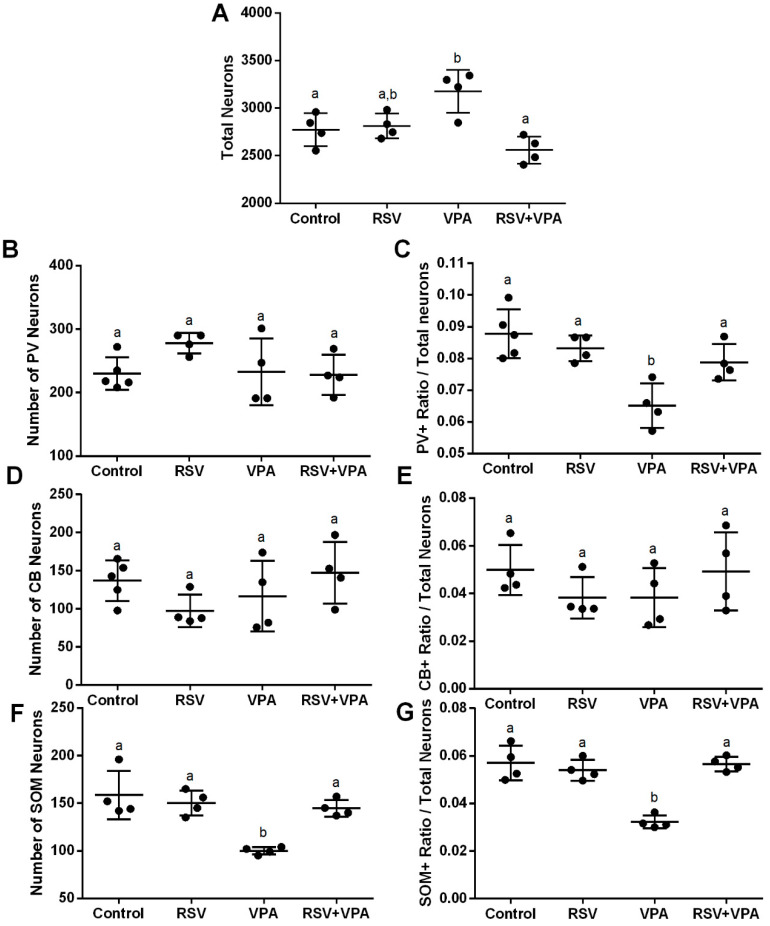
The absolute numbers of total neurons (NeuN + DAPI) and interneurons (CB+ NeuN + DAPI, PV+ NeuN + DAPI, and SOM+ NeuN + DAPI) were counted in each area. The ratio between the number of each interneuron and total neurons is a measurement of the proportion between the inhibitory (interneuron) and excitatory components (the majority of the total neurons). This is done in each subarea of the mPFC and in the mPFC as a whole. The RSV was able to prevent the increased number of total neurons induced by VPA (Figure 1A, interaction factor: F (1, 12) = 14.56, p = 0.0025; Cont-VPA ppost-hoc = 0.0361; RSV-VPA ppost-hoc = 0.0627; RSV + VPA-VPA = 0.0016); the decreased ratio of PV+ interneurons (Figure 1E, interaction factor: F (1, 13) = 9.314, p = 0.0093; Cont-VPA ppost-hoc = 0.006; RSV-VPA ppost-hoc = 0.0065; RSV + VPA-VPA = 0.0436); and the decreased number of SOM+ interneurons (Figure 1F, interaction factor: F (1, 12) = 12.39, p = 0.0042; Cont-VPA ppost-hoc = 0.0008; RSV-VPA ppost-hoc = 0.0030; RSV + VPA-VPA = 0.0074) as well as the SOM+ ratio (Figure 1G, interaction factor: F (1, 12) = 33.09, p < 0.0001; Cont-VPA ppost-hoc < 0.0001; RSV-VPA ppost-hoc = 0.0002; RSV + VPA-VPA < 0.0001). PV+ number (Figure 1B), CB+ number (Figure 1D) and CB+ ratio (Figure 1E) did not present significant differences among groups.
Figure 1.
RSV prevents the increase in the number of total neurons, the reduction in the PV+ ratio, and the reduction in the number and ratio of SOM+ induced by VPA in the whole mPFC. (A) Quantification of total neurons. (B) Quantification of CB+ interneurons. (C) Quantification of ratio of CB+ interneurons/total neurons. (D) Quantification of PV+ interneurons. (E) Quantification of ratio of PV+ interneurons/total neurons (F) Quantification of SOM+ interneurons. (G) Quantification of the ratio of SOM+ interneurons/total neurons. Values are shown as mean ± standard deviation. Statistical analysis: two-way ANOVA followed by Bonferroni, p < 0.05 was considered significant. NCON: 5, NRSV: 4, NVPA: 4, NRSV + VPA:4 CB+NeuN+DAPI, and PV+NeuN+DAPI; NCON: 4, NRSV: 4, NVPA: 4, NRSV + VPA:4 and SOM+NeuN+DAPI. Different letters indicate significant differences in the post-test when interaction was significant (p < 0.05).
2.3. The RSV Treatment Prevented the Increased Total Number of Neurons in the Deeper Layers and Whole PrL and IL
The data in Table 2 show that RSV was able to prevent the VPA-induced total neuron increase in deeper layers of PrL (Pre-Limbic Cortex), in the whole PrL, in deeper layers of IL (Infra Limbic Cortex), and in the whole IL. In the upper layers of PrL, a difference between the VPA and VPA-RSV groups was observed. In the deeper layers of aCC (anterior cingulate cortex) and whole aCC, RSV decreased the number of neurons. In the upper layers of IL and aCC, no significant differences were found.
Table 2.
Distribution profile of total neurons in the mPFC.
| Mean ± SD | F (DFn. DFd); p-Value | Pairwise Comparisons | ||
|---|---|---|---|---|
| Total Neurons aCC (II/III) |
CON: 336.00 ± 39.75 RSV: 321.00 ± 42.35 VPA: 371.25 ± 59.35 RSV + VPA: 318.75 ± 41.60 |
Interaction: F (1, 12) = 0.6523 p = 0.4350 VPA: F (1, 12) = 0.5052 p = 0.4908 RSV: F (1, 12) = 2.113 p = 0.1717 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.9108 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.8147 | |||
| Total Neurons aCC (IV/V) |
CON: 555.25 ± 76.01 RSV: 513.00 ± 13.71 VPA: 650.25 ± 49.85 RSV + VPA: 506.75 ± 39.78 |
Interaction: F (1, 12) = 4.086 p = 0.0661 # VPA: F (1, 12) = 3.140 p = 0.1018 RSV: F (1, 12) = 13.75 p = 0.0030 ** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.1197 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0132 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0096 ** | |||
| Total Neurons Whole aCC |
CON: 891.25 ± 104.45 RSV: 834.00 ± 51,153 VPA: 1021.00 ± 107.97 RSV + VPA: 825.50 ± 71.11 |
Interaction: F (1, 12) = 2.546 p = 0.1365 VPA: F (1, 12) = 1.960 p = 0.1868 RSV: F (1, 12) = 8.482 p = 0.0130 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.3342 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0606 # | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0469 * | |||
| Total Neurons PrL (II/III) |
CON: 368.25 ± 55.12 RSV: 363.50 ± 14.91 VPA: 404.75 ± 28.15 RSV + VPA: 325.75 ± 13.25 |
Interaction: F (1, 12) = 5.214 p = 0.0414 * VPA: F (1, 12) = 0.001478 p = 0.9700 RSV: F (1, 12) = 6.633 p = 0.0243 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.8305 | |||
| CON vs. RSV + VPA: | 0.5360 | |||
| RSV vs. VPA: | 0.5882 | |||
| RSV vs. RSV + VPA: | 0.7594 | |||
| VPA vs. RSV + VPA: | 0.0296 * | |||
| Total Neurons PrL (IV/V) |
CON: 572.50 ± 22.10 RSV: 611.25 ± 62.50 VPA: 693.00 ± 49.22 RSV + VPA: 536.00 ± 49.43 |
Interaction: F (1, 12) = 16.54 p = 0.0016 ** VPA: F (1, 12) = 0.8838 p = 0.3657 RSV: F (1, 12) = 6.036 p = 0.0302 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0244 * | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.2004 | |||
| RSV vs. RSV + VPA: | 0.2832 | |||
| VPA vs. RSV + VPA: | 0.0036 ** | |||
| Total Neurons Whole PrL |
CON: 940.75 ± 63.10 RSV: 974.75 ± 76.61 VPA: 1097.75 ± 61.32 RSV + VPA: 861.75 ± 60.10 |
Interaction: F (1, 12) = 16.93 p = 0.0014 ** F (1, 12) = 0.4497 p = 0.5152 F (1, 12) = 9.478 p = 0.0096 ** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0326 * | |||
| CON vs. RSV + VPA: | 0.6861 | |||
| RSV vs. VPA: | >0.1268 | |||
| RSV vs. RSV + VPA: | 0.1884 | |||
| VPA vs. RSV + VPA: | 0.0016 ** | |||
| Total Neurons IL (II/III) |
CON: 348.75 ± 47.98 RSV: 355.75 ± 26.98 VPA: 363.25 ± 20.85 RSV + VPA: 315.25 ± 17.41 |
Interaction: F (1, 12) =3.49 p = 0.0880 # VPA: F (1, 12) = 0.7707 p = 0.3972 RSV: F (1, 12) = 1.917 p =0.1914 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | 0.8139 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | 0.4623 | |||
| VPA vs. RSV + VPA: | 0.2447 | |||
| Total Neurons IL (IV/V) |
CON: 593.75 ± 39.22 RSV: 646.00 ± 26.24 VPA: 696.50 ± 51.39 RSV + VPA: 557.00 ± 54.16 |
Interaction: F (1, 12) = 18.75 p = 0.0010 ** VPA: F (1, 12) = 0.1041 p = 0.7525 RSV: F (1, 12) = 3.858 p = 0.0731 # |
CON vs. RSV: | 0.7209 |
| CON vs. VPA: | 0.0387 * | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.7910 | |||
| RSV vs. RSV + VPA: | 0.0904 # | |||
| VPA vs. RSV + VPA: | 0.0047 ** | |||
| Total Neurons Whole IL |
CON: 942.50 ± 27.47 RSV: 1001.75 ± 35.08 VPA: 1059.75 ± 66.74 RSV + VPA: 872.75 ± 53.72 |
Interaction: F (1, 12) = 26.01 p = 0.0003 *** VPA: F (1, 12) = 0.05921 p = 0.8119 RSV: F (1, 12) = 7.000 p = 0.0213 |
CON vs. RSV: | 0.6496 |
| CON vs. VPA: | 0.0297 * | |||
| CON vs. RSV + VPA: | 0.3820 | |||
| RSV vs. VPA: | 0.6908 | |||
| RSV vs. RSV + VPA: | 0.0158 * | |||
| VPA vs. RSV + VPA: | 0.0008 *** | |||
II/III, upper cortical layers; IV/V, deeper cortical layers; aCC, anterior cingulate cortex; IL, infralimbic cortex; mPFC, medial prefrontal cortex; PrL, prelimbic cortex; SD, standard deviation. p < 0.05 considered significant. * p < 0.05. ** p < 0.01, *** p < 0.001, # trend. Statistical analyses: two-way ANOVA parametric test followed by Bonferroni. NCON: 4. NRSV: 4. NVPA: 4. NRSV + VPA: 4.
2.4. The VPA Induced Alterations in PV+ Number and Ratio in Different Layers of the aCC and PrL
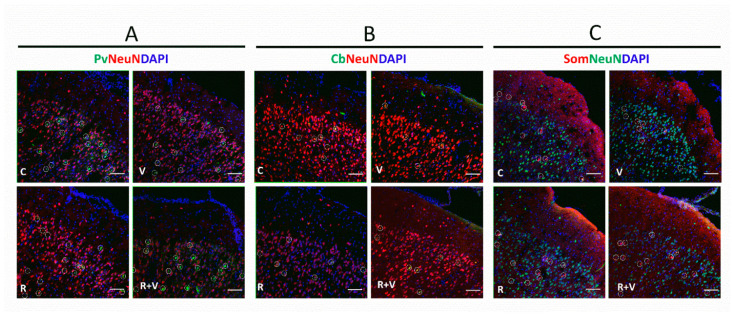
The data in Table 3 show that VPA decreased the number of PV+ neurons in the superficial layers of aCC, without RSV prevention. Interestingly, RSV prevented the VPA-induced decrease in PV+ ratio observed in the superficial layers of aCC. The PV+ ratio in the deeper layers of aCC was decreased by VPA, with partial prevention by RSV. When observing the whole aCC, the VPA decreased the PV+ ratio, which was prevented by RSV. In the superficial layers of PrL, the RSV prevented the VPA-induced increase in PV+ number. Regarding the ratio, a tendency was found in the interaction, and differences were identified in the isolated factors. In all the other regions, no differences were found among groups. Illustrative images of PV+ neurons are presented in Figure 2A.
Table 3.
Distribution profile of PV neurons in the mPFC.
| Mean ± SD | F (DFn. DFd); p-Value | Pairwise Comparisons | ||
|---|---|---|---|---|
| PV Total aCC (II/III) |
CON: 30.200 ± 2.863564 RSV: 28.750 ± 5.560276 VPA: 21.250 ± 7.274384 RSV + VPA: 25.000 ± 0.816 |
Interaction: F (1, 13) = 1.292 p = 0.2761 VPA: F (1, 13) = 7.709 p = 0.0157 * RSV: F (1, 13) = 0.2528 p = 0.6235 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0830 | |||
| CON vs. RSV + VPA: | 0.7351 | |||
| RSV vs. VPA: | 0.2497 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Ratio aCC (II/III) |
CON: 0.0804 ± 0.005 RSV: 0.0812 ± 0.017 VPA: 0.0506 ± 0.009 RSV + VPA: 0.0777 ± 0.005 |
Interaction: F (1, 13) = 6.862 p = 0.0212 * VPA: F (1, 13) = 11.06 p = 0.0055 ** RSV: F (1, 13) = 7.772 p = 0.0154 * |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.0050 ** | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.0061 ** | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.0152 * | |||
| PV Total aCC (IV/V) |
CON: 51.800 ± 8.55 RSV: 50.500 ± 13.89 VPA: 36.750 ± 15.37 RSV + VPA: 49.500 ± 10.96 |
Interaction: F (1, 13) = 1.391 p = 0.2593 VPA: F (1, 13) = 1.816 p = 0.2009 RSV: F (1, 13) = 0.9240 p = 0.3540 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.5361 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.8135 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9834 | |||
| PV Ratio aCC (IV/V) |
CON: 0.107 ± 0.0145 RSV: 0.0856 ± 0.01586 VPA: 0.0548 ± 0.0168 RSV + VPA: 0.0840 ± 0.019 |
Interaction: F (1, 13) = 9.992 p = 0.0075 ** VPA: F (1, 13) = 11.16 p = 0.0053 ** RSV: F (1, 13) = 0.2128 p = 0.6522 |
CON vs. RSV: | 0.4297 |
| CON vs. VPA: | 0.0024 | |||
| CON vs. RSV + VPA: | 0.3398 | |||
| RSV vs. VPA: | 0.1270 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.1606 | |||
| PV Total Whole aCC |
CON: 82.000 ± 11.25 RSV: 78.500 ± 16.60 VPA: 59.750 ± 23.60 RSV + VPA:74.500 ± 10.87 |
Interaction: F (1, 13) = 1.354 p = 0.2655 VPA: F (1, 13) = 2.801 p = 0.1181 RSV: F (1, 13) = 0.5144 p = 0.4859 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.3593 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.7401 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| PV Ratio Whole aCC |
CON: 0.094 ± 0.009 RSV: 0.084 ± 0.014 VPA: 0.053 ± 0.012 RSV + VPA 0.081 ± 0.011 |
Interaction: F (1, 13) = 11.45 p = 0.0049 ** VPA: F (1, 13) = 14.87 p = 0.0020 ** RSV: F (1, 13) = 2.065 p = 0.1744 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.0009 *** | |||
| CON vs. RSV + VPA: | 0.6141 | |||
| RSV vs. VPA: | 0.0177 * | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.0330 * | |||
| PV Total PrL (II/III) |
CON: 28.400 ± 2.88 RSV:24.500 ± 7.04 VPA: 38.750 ± 3.77 RSV + VPA:26.250 ± 6.94 |
Interaction: F (1, 13) = 4.159 p = 0.0483 * VPA: F (1, 13) = 7.359 p = 0.0178 * RSV: F (1, 13) = 7.675 p = 0.0159 * |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.0257 * | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.0138 * | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.0335 * | |||
| PV Ratio PrL (II/III) |
CON: 0.072 ± 0.004 RSV: 0.066 ± 0.009 VPA: 0.087 ± 0.003 RSV + VPA 0.070 ± 0.007 |
Interaction: F (1, 13) = 2.679 p = 0.0930 # VPA: F (1, 13) = 7.172 p = 0.0190 * RSV: F (1, 13) = 15.25 p = 0.0018 ** |
CON vs. RSV: | 0.7397 |
| CON vs. VPA: | 0.0474 * | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.0033 ** | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.0128 * | |||
| PV Total PrL (IV/V) |
CON: 49.200 ± 7.82 RSV: 56.000 ± 6.83 VPA: 50.750 ± 5.12 RSV + VPA: 48.750 ± 12.57 |
Interaction: F (1, 13) = 1.129 p = 0.3072 VPA: F (1, 13) = 0.4738 p = 0.5033 RSV: F (1, 13) = 0.3360 p = 0.5720 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Ratio PrL (IV/V) |
CON: 0.096 ± 0.020 RSV:0.084 ± 0.009 VPA: 0.068 ± 0.010 RSV + VPA:0.082 ± 0.020 |
Interaction: F (1, 13) = 2.460 p = 0.1408 VPA: F (1, 13) = 3.640 p = 0.0787 # RSV: F (1, 13) = 0.001322 p = 0.9715 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.1520 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| PV Total Whole PrL |
CON: 74.400 ± 8.82 RSV: 80.500 ± 13.52 VPA: 86.500 ± 9.000 RSV + VPA: 75.000 ± 17.92 |
Interaction: F (1, 13) = 2.051 p = 0.1758 VPA: F (1, 13) = 0.2884 p = 0.6003 RSV: F (1, 13) = 0.1930 p = 0.6676 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Ratio Whole PrL |
CON: 0.088 ± 0.013 RSV:0.078 ± 0.006 VPA: 0.072 ± 0.001 RSV + VPA:0.077 ± 0.014 |
Interaction: F (1, 13) = 2.101 p = 0.1709 VPA: F (1, 13) = 2.834 p = 0.1161 RSV: F (1, 13) = 0.3874 p = 0.5444 |
CON vs. RSV: | 0.9372 |
| CON vs. VPA: | 0.2425 | |||
| CON vs. RSV + VPA: | 0.7067 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| PV Total IL (II/III) |
CON: 28.800 ± 5.90 RSV: 29.250 ± 10.25 VPA: 33.750 ± 5.12 RSV + VPA: 25.000 ± 4.69 |
Interaction: F (1, 13) = 1.936 p = 0.1875 VPA: F (1, 13) = 0.01121 p = 0.9173 RSV: F (1, 13) = 1.576 p = 0.2315 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.5473 | |||
| PV Ratio IL (II/III) |
CON: 0.082 ± 0.010 RSV: 0.070 ± 0.015 VPA: 0.070 ± 0.010 RSV + VPA: 0.075 ± 0.008 |
Interaction: F (1, 13) = 2.644 p = 0.1279 VPA: F (1, 13) = 0.4062 p = 0.5350 RSV: F (1, 13) = 0.3412 p = 0.5691 |
CON vs. RSV: | 0.7940 |
| CON vs. VPA: | 0.7443 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Total IL (IV/V) |
CON: 45.40 ± 9.50 RSV: 56.00 ± 28.25 VPA: 48.00 ± 12.355 RSV + VPA: 53.50 ± 11.80 |
Interaction: F (1, 13) = 0.1646 p = 0.6915 VPA: F (1, 13) = 0.001360 p = 0.9711 RSV: F (1, 13) = 1.171 p = 0.2989 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Ratio IL (IV/V) |
CON:0.080 ± 0.020 RSV: 0.073 ± 0.031 VPA: 0.067 ± 0.016 RSV + VPA: 0.081 ± 0.020 |
Interaction: F (1, 13) = 1.046 p = 0.3251 VPA: F (1, 13) = 0.04663 p = 0.8324 RSV: F (1, 13) = 0.1055 p = 0.7504 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Total Whole IL |
CON: 74.60 ± 8.50 RSV: 80.50 ± 13.52 VPA: 81.75 ± 15.25 RSV + VPA: 77.50 ± 15.25 |
Interaction: F (1, 13) = 0.6249 p = 0.4434 VPA: F (1, 13) = 0.1045 p = 0.7517 RSV: F (1, 13) = 0.01651 p = 0.8997 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
| PV Ratio Whole IL |
CON: 0.082 ± 0.015 RSV: 0.072 ± 0.024 VPA: 0.068 ± 0.013 RSV + VPA: 0.079 ± 0.013 |
Interaction: F (1, 13) = 1.533 p = 0.2376 VPA: F (1, 13) = 0.1634 p = 0.6926 RSV: F (1, 13) = 0.006546 p = 0.9367 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | >0.9999 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | >0.9999 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | >0.9999 | |||
II/III, upper cortical layers; IV/V, deeper cortical layers; aCC, anterior cingulate cortex; IL, infralimbic cortex; mPFC, medial prefrontal cortex; PrL, prelimbic cortex; PV, parvalbumin-positive interneuron; SD, standard deviation. p < 0.05 was considered significant. * p < 0.05. ** p < 0.01, *** p < 0.001, # trend. Statistical analyses: two-way ANOVA parametric test followed by Bonferroni. NCON: 4. NRSV: 4. NVPA: 4. NRSV + VPA: 4.
Figure 2.
Representative immunofluorescence images of total neurons, PV+, CB+, and SOM+ in the mPFC. Representative images of the aCC, upper layers (II/III). (A) Pv, parvalbumin (green); NeuN (red); DAPI (blue). (B) Cb, calbindin (green); NeuN (red); DAPI (blue). (C) Som, somatostatin (red); NeuN (green); DAPI (blue). Scale bar: 50 µm. The respective interneurons are highlighted within white circles. aCC, anterior cingulate cortex; CB, calbindin-neurons; mPFC, medial frontal cortex; PV, parvalbumin-neurons; SOM, somatostatin-neurons.
2.5. The VPA Induced Alterations in CB+ Ratio in the Upper Layers of aCC, PrL, and IL
The data in Table 4 show that RSV prevented the decreased CB+ ratio induced by VPA in the superficial layers of aCC, but not in the superficial layers of PrL and IL. The differences found in the CB+ number in the superficial layers of aCC, ratio in the whole aCC, ratio in the whole PrL, and ratio in the IL were not associated with any specific factor after the post-hoc test. Illustrative images of PV+ neurons are presented in Figure 2B.
Table 4.
Distribution profile of CB neurons in the mPFC.
| Mean ± SD | F (DFn. DFd); p-Value | Pairwise Comparisons | ||
|---|---|---|---|---|
| CB Total aCC (II/III) |
CON: 19.20 ± 6.87 RSV: 16.75 ± 2.50 VPA: 13.25 ± 6.70 RSV + VPA: 21.75 ± 3.59 |
Interaction: F (1, 13) = 6.023 p = 0.0290 * VPA: F (1, 13) = 0.03959 p = 0.8454 RSV: F (1, 13) = 2.327 p = 0.1511 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.7519 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.5430 | |||
| VPA vs. RSV + VPA: | 0.1006 | |||
| CB Ratio aCC (II/III) |
CON: 0.057 ± 0.005 RSV: 0.050 ± 0.006 VPA: 0.034 ± 0.010 RSV + VPA: 0.064 ± 0.014 |
Interaction: F (1, 13) = 18.31 p = 0.0009 *** VPA: F (1, 13) = 1.081 p = 0.3175 RSV: F (1, 13) = 6.006 p = 0.0292 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.0117 * | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.1905 | |||
| RSV vs. RSV + VPA: | 0.2626 | |||
| VPA vs. RSV + VPA: | 0.0028 ** | |||
| CB Total aCC (IV/V) |
CON: 22.00 ± 7.25 RSV: 17.00 ± 4.45 VPA: 22.75 ± 10.00 RSV + VPA: 22.5 ± 9.95 |
Interaction: F (1, 13) = 0.4352 p = 0.5210 VPA: F (1, 13) = 0.7193 p = 0.4117 RSV: F (1, 13) = 0.5220 p = 0.4828 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio aCC (IV/V) |
CON: 0.059 ± 0.024 RSV: 0.037 ± 0.012 VPA: 0.039 ± 0.011 RSV + VPA: 0.042 ± 0.020 |
Interaction: F (1, 13) = 2.304 p = 0.1530 VPA:F (1, 13) = 0.7381 p = 0.4058 RSV:F (1, 13) = 1.381 p = 0.2610 |
CON vs. RSV: | 0.4334 |
| CON vs. VPA: | 0.6472 | |||
| CON vs. RSV + VPA: | 0.9794 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Total Whole aCC |
CON: 37.2 ± 13.92 RSV: 28.25 ± 8.15 VPA: 38.75 ± 19.77 RSV + VPA: 39.25 ± 17.40 |
Interaction: F (1, 13) = 0.4000 p = 0.5381 VPA: F (1, 13) = 0.7055 p = 0.4161 RSV: (1, 13) = 0.3198 p = 0.5813 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio Whole aCC |
CON: 0.058 ± 0.014 RSV: 0.042 ± 0.009 VPA: 0.038 ± 0.010 RSV + VPA 0.051 ± 0.017 |
Interaction: F (1, 13) = 5.701 p = 0.0328 * VPA: F (1, 13) = 0.9515 p = 0.3471 RSV: F (1, 13) = 0.03555 p = 0.8534 |
CON vs. RSV: | 0.5037 |
| CON vs. VPA: | 0.1775 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9213 | |||
| CB Total PrL (II/III) |
CON: 26.20 ± 7.35 RSV: 18.25 ± 1.70 VPA: 16.00 ± 9.75 RSV + VPA:19.25 ± 5.45 |
Interaction: F (1, 13) = 2.859 p = 0.1147 VPA: F (1, 13) = 1.929 p = 0.1882 RSV: F (1, 13) = 0.5036 p = 0.4905 |
CON vs. RSV: | 0.6284 |
| CON vs. VPA: | 0.2603 | |||
| CON vs. RSV + VPA: | 0.9078 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio PrL (II/III) |
CON: 0.079 ± 0.025 RSV: 0.059 ± 0.009 VPA: 0.034 ± 0.015 RSV + VPA: 0.057 ± 0.016 |
Interaction: F (1, 13) = 6.149 p = 0.0276 VPA: F (1, 13) = 7.593 p = 0.0164 RSV: F (1, 13) = 0.01238 p = 0.9131 |
CON vs. RSV: | 0.6541 |
| CON vs. VPA: | 0.0132 | |||
| CON vs. RSV + VPA: | 0.4615 | |||
| RSV vs. VPA: | 0.4187 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.5849 | |||
| CB Total PrL (IV/V) |
CON: 23.20 ± 4.35 RSV: 19.00 ± 4.95 VPA: 26.75 ± 14.05 RSV + VPA: 33.2 ± 7.65 |
Interaction: F (1, 13) = 1,748 p = 0.2089 VPA: F (1, 13) = 4.837 p = 0.0709 # RSV: F (1, 13) = 0.04947 p = 0.8275 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.6120 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.1830 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio PrL (IV/V) |
CON: 0.054 ± 0.018 RSV: 0.034 ± 0.009 VPA: 0.041 ± 0.017 RSV + VPA: 0.050 ± 0.014 |
Interaction: F (1, 13) = 3.755 p = 0.0747 VPA F (1, 13) = 0.04387 p = 0.8373 RSV: F (1, 13) = 0.6898 p = 0.4212 |
CON vs. RSV: | 0.3931 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9763 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Total Whole PrL |
CON: 51.00 ± 7.87 RSV: 36.25 ± 6.95 VPA: 42.75 ± 19.25 RSV + VPA:52.25 ± 7.90 |
interaction: F (1, 12) = 3.243 p = 0.0969 VPA: F (1, 12) = 0.3312 p = 0.5756 RSV: F (1, 12) = 0.1520 p = 0.7035 |
CON vs. RSV: | 0.8841 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.7124 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio Whole PrL |
CON: 0.062 ± 0.016 RSV: 0.043 ± 0.08 VPA: 0.038 ± 0.016 RSV + VPA: 0.052 ± 0.014 |
Interaction: F (1, 13) = 5.736 p = 0.0324 * VPA: F (1, 13) = 0.4439 p = 0.5169 RSV: F (1, 13) = 0.2759 p = 0.6082 |
CON vs. RSV: | 0.3221 |
| CON vs. VPA: | 0.2670 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Total IL (II/III) |
CON: 18.75 ± 5.50 RSV: 14.25 ± 4.70 VPA: 14.25 ± 7.00 RSV + VPA: 26.00 ± 11.5 |
Interaction: F (1, 11) = 3.947 p = 0.0662 # VPA: F (1, 11) = 7.953 p = 0.4461 RSV: F (1, 11) = 12.79 p = 0.4471 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.4421 | |||
| VPA vs. RSV + VPA: | 0.4421 | |||
| CB Ratio IL (II/III) |
CON: 0.069 ± 0.018 RSV: 0.043 ± 0.013 VPA: 0.031 ± 0.007 RSV + VPA: 0.060 ± 0.023 |
Interaction: F (1, 13) = 13.42 p = 0.0029 ** VPA: F (1, 13) = 1.476 p = 0.2460 RSV: F (1, 13) = 0.01159 p = 0.9159 |
CON vs. RSV: | 0.1364 |
| CON vs. VPA: | 0.0216 * | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.6924 | |||
| VPA vs. RSV + VPA: | 0.1324 | |||
| CB Total IL (IV/V) |
CON: 29.00 ± 10.90 RSV: 19.00 ± 6.13 VPA: 26.25.00 ± 6.18 RSV + VPA: 30.00 ± 8.25 |
Interaction: F (1, 13) = 3.990 p = 0.0672 # VPA: F (1, 13) = 1,271 p = 0.2800 RSV: F (1, 13) = 0.5051 p = 0.4898 |
CON vs. RSV: | 0.4251 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.3037 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Ratio IL (IV/V) |
CON: 0.045 ± 0.010 RSV: 0.035 ± 0.013 VPA: 0.040 ± 0.010 RSV + VPA: 0.048 ± 0.014 |
Interaction: F (1, 12) = 2.805 p = 0.1198 VPA: F (1, 12) = 0.3000 p = 0.5939 RSV: F (1, 12) = 0.07687 p = 0.7863 |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.8522 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
| CB Total Whole IL |
CON: 44.25 ± 9.45 RSV: 33.00 ± 9.70 VPA: 40.25 ± 12.00 RSV + VPA: 55.00 ± 18.65 |
Interaction: F (1, 13) = 5.378 p = 0.0597 # VPA: F (1, 13) = 0.7738 p = 0.3950 RSV: F (1, 13) = 0.01655 p = 0.8996 |
CON vs. RSV: | 0.5925 |
| CON vs. VPA: | 0.9999 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.2766 | |||
| VPA vs. RSV + VPA: | 0.9303 | |||
| CB Ratio Whole IL |
CON: 0.64 ± 0.027 RSV: 0.39 ± 0.011 VPA: 0.36 ± 0.008 RSV + VPA: 0.53 ± 0.018 |
Interaction: F (1, 13) = 5.736 p = 0.0324 * VPA: F (1, 13) = 0.4439 p = 0.5169 RSV: F (1, 13) = 0.2759 p = 0.6082 |
CON vs. RSV: | 0.3221 |
| CON vs. VPA: | 0.2670 | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.9999 | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.9999 | |||
II/III, upper cortical layers; IV/V, deeper cortical layers; aCC, anterior cingulate cortex; CB, calbindin-positive interneuron; IL, infralimbic cortex; mPFC, medial prefrontal cortex; PrL, prelimbic cortex; SD, standard deviation. p < 0.05 considered significant. * p < 0.05, ** p < 0.01, *** p < 0.001, # trend. Statistical analyses: two-way ANOVA parametric test followed by Bonferroni. NCON: 5. NRSV: 4. NVPA: 4. NRSV + VPA: 4.
2.6. The RSV Prevented the Widespread Impairments Induced by VPA in SOM+ Neurons
The data in Table 5 show that RSV prevented the VPA-induced decrease in SOM+ number and ratio in the superficial layers of aCC. The RSV was also able to prevent the decreased SOM+ number and ratio induced by VPA in the deeper layers of aCC. These results reflected a preventive effect of RSV in the whole aCC in both decreased SOM+ number and ratio. The RSV was also able to prevent the VPA-induced SOM+ ratio decrease in the superficial layers of PrL. In the deeper layers of PrL, RSV + VPA did not differ from any other group, but RSV prevented the reduction in the SOM+ ratio. In the whole PrL, RSV + VPA did not differ from any other group for the SOM+ number, but a prevention was observed in the ratio. In the superficial layers of IL, VPA decreased the numbers of SOM+ neurons. Regarding the ratio, a tendency was found in the interaction, and relevant differences were identified in the isolated factors. In the deeper layers of IL, RSV prevented the VPA-induced decrease of SOM+ neurons in both number and ratio. In the whole IL, RSV prevented the reductions in the SOM+ number and ratio. Illustrative images of PV+ neurons are presented in Figure 2C.
Table 5.
Distribution profile of SOM neurons in the mPFC.
| Mean ± SD | F (DFn. DFd); p-Value | Pairwise Comparisons | ||
|---|---|---|---|---|
| SOM Total aCC (II/III) |
CON: 18.5 ± 2.65 RSV: 17.5 ± 0.5 VPA: 12 ± 2.50 RSV + VPA: 18.75 ± 2.00 |
Interaction: F (1, 12) = 13.66 p = 0.0031 ** VPA: F (1, 12) = 6.270 p = 0.0277 * RSV: F (1, 12) = 7.521 p = 0.0178 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0053 ** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0179 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0040 ** | |||
| SOM Ratio aCC (II/III) |
CON: 0.060 ± 0.009 RSV: 0.059 ± 0.008 VPA: 0.034 ± 0.007 RSV + VPA: 0.068 ± 0.002 |
Interaction: F (1, 12) = 25.14 p = 0.0003 *** VPA: F (1, 12) = 6.460 p = 0.0259 * RSV: F (1, 12) = 22.93 p = 0.0004 *** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0011 ** | |||
| CON vs. RSV + VPA: | 0.8282 | |||
| RSV vs. VPA: | 0.0014 ** | |||
| RSV vs. RSV + VPA: | 0.6359 | |||
| VPA vs. RSV + VPA: | <0.0001 **** | |||
| SOM Total aCC (IV/V) |
CON: 33.25 ± 5.56 RSV: 30.50 ± 3.70 VPA: 21.00 ± 1.15 RSV + VPA: 31.25 ± 2.63 |
Interaction: F (1, 12) = 12.64 p = 0.0040 ** VPA: F (1, 12) = 9.888 p = 0.0085 ** RSV: F (1, 12) = 4.206 p = 0.0628 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0029 ** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0191 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0113 * | |||
| SOM Ratio aCC (IV/V) |
CON: 0.060 ± 0.010 RSV: 0.060 ± 0.008 VPA: 0.036 ± 0.007 RSV + VPA: 0.062 ± 0.005 |
Interaction: F (1, 12) = 15.49 p = 0.0020 ** VPA: F (1, 12) = 11.03 p = 0.0061 ** RSV: F (1, 12) = 13.84 p = 0.0029 ** |
CON vs. RSV: | 0.9999 |
| CON vs. VPA: | 0.0015 ** | |||
| CON vs. RSV + VPA: | 0.9999 | |||
| RSV vs. VPA: | 0.0019 ** | |||
| RSV vs. RSV + VPA: | 0.9999 | |||
| VPA vs. RSV + VPA: | 0.0009 *** | |||
| SOM Total Whole aCC |
CON: 51.75 ± 4.20 RSV: 48.00 ± 4.00 VPA: 33.00 ± 1.15 RSV + VPA: 50.00 ± 4.32 |
Interaction: F (1, 12) = 31.75 p = 0.0001 *** VPA: F (1, 12) = 20.69 p = 0.0007 *** RSV: F (1, 12) = 12.94 p = 0.0037 ** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | <0.0001 **** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0005 *** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0002 *** | |||
| SOM Ratio Whole aCC |
CON: 0.058 ± 0.006 RSV: 0.056 ± 0.007 VPA: 0.033 ± 0.005 RSV + VPA: 0.061 ± 0.005 |
Interaction: F (1, 12) = 23.64 p = 0.0004 *** VPA: F (1, 12) = 14.87 p = 0.0023 ** RSV: F (1, 12) = 21.35 p = 0.0006 *** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0003 *** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0004 *** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0001 *** | |||
| SOM Total PrL (II/III) |
CON: 19.75 ± 8.25 RSV: 18.25 ± 1.55 VPA: 12.50 ± 0.70 RSV + VPA: 17.25 ± 1.90 |
Interaction: F (1, 12) = 2.825 p = 0.1186 VPA: F (1, 12) = 2.825 p = 0.1186 RSV: F (1, 12) = 0.2721 p = 0.6114 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.2097 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.8721 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.8721 | |||
| SOM Ratio PrL (II/III) |
CON: 0.053 ± 0.016 RSV: 0.047 ± 0.003 VPA: 0.032 ± 0.004 RSV + VPA: 0.049 ± 0.007 |
Interaction: F (1, 12) = 8.612 p = 0.0125 * VPA: F (1, 12) = 2.893 p = 0.1147 RSV: F (1, 12) = 3.103 p = 0.1036 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0396 * | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.1841 | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0366 * | |||
| SOM Total PrL (IV/V) |
CON: 37.00 ± 5.90 RSV: 34.25 ± 5.75 VPA: 21.25 ± 3.86 RSV + VPA: 29.25 ± 2.63 |
Interaction: F (1, 12) = 5.170 p = 0.0422 ** VPA: F (1, 12) = 19.26 p = 0.0009 *** RSV: F (1, 12) = 1.233 p = 0.2886 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0030 ** | |||
| CON vs. RSV + VPA: | 0.2334 | |||
| RSV vs. VPA: | 0.0129 * | |||
| RSV vs. RSV + VPA: | 0.9636 | |||
| VPA vs. RSV + VPA: | 0.2037 | |||
| SOM Ratio PrL (IV/V) |
CON: 0.064 ± 0.008 RSV: 0.056 ± 0.008 VPA: 0.032 ± 0.007 RSV + VPA: 0.055 ± 0.005 |
Interaction: F (1, 12) = 18.82 p = 0.0010 VPA: F (1, 12) = 22.16 p = 0.0005 *** RSV: F (1, 12) = 4.211 p = 0.0626 # |
CON vs. RSV: | 0.7919 |
| CON vs. VPA: | 0.0002 *** | |||
| CON vs. RSV + VPA: | 0.5095 | |||
| RSV vs. VPA: | 0.0027** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0042 ** | |||
| SOM Total Whole PrL |
CON: 56.75 ± 12.55 RSV: 51.50 ± 7.25 VPA: 33.75 ± 3.40 RSV + VPA: 46.50 ± 2.00 |
Interaction: F (1, 12) = 5.739 p = 0.0338 * VPA: F (1, 12) = 13.89 p = 0.0029 ** RSV: F (1, 12) = 0.9963 p = 0.3379 |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0059 ** | |||
| CON vs. RSV + VPA: | 0.4661 | |||
| RSV vs. VPA: | 0.0353 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.2012 | |||
| SOM Ratio Whole PrL |
CON: 0.060 ± 0.011 RSV: 0.053 ± 0.006 VPA: 0.031 ± 0.004 RSV + VPA: 0.054 ± 0.005 |
Interaction: F (1, 12) = 18.37 p = 0.0011 ** VPA: F (1, 12) = 15.21 p = 0.0021 ** RSV: F (1, 12) = 4.905 p = 0.0469 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0005 *** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0059 ** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0037 ** | |||
| SOM Total IL (II/III) |
CON: 15.50 ± 1.75 RSV: 17.00 ± 2.45 VPA: 11.50 ± 2.50 RSV + VPA: 15.25 ± 1.50 |
Interaction: F (1, 12) = 1,152 p = 0.3043 VPA: F (1, 12) = 7.521 p = 0.0178 * RSV: F (1, 12) = 6.270 p = 0.0277 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.1163 | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0179 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.1587 | |||
| SOM Ratio IL (II/III) |
CON: 0.045 ± 0.007 RSV: 0.048 ± 0.008 VPA: 0.032 ± 0.006 RSV + VPA: 0.048 ± 0.004 |
Interaction: F (1, 12) = 4.503 p = 0.0553 # VPA: F (1, 12) = 3.938 p = 0.0706 # RSV: F (1, 12) = 9.410 p = 0.0098 ** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0794 # | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0230 * | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0193 * | |||
| SOM Total IL (IV/V) |
CON: 34.50 ± 7.15 RSV: 33.75 ± 1.00 VPA: 21.75 ± 3.00 RSV + VPA: 33.00 ± 1.45 |
Interaction: F (1, 12) = 9.167 p = 0.0105 * VPA: F (1, 12) = 11,60 p = 0.0052 ** RSV: F (1, 12) = 7.019 p = 0.0212 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0040 ** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0064 ** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0103* | |||
| SOM Ratio IL (IV/V) |
CON: 0.058 ± 0.011 RSV: 0.052 ± 0.002 VPA: 0.032 ± 0.003 RSV + VPA: 0.059 ± 0.005 |
Interaction: F (1, 12) = 29.00 p = 0.0002 *** VPA: F (1, 12) = 9.617 p = 0.0092 ** RSV: F (1, 12) = 12.21 p = 0.0044 ** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0004 *** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0033 ** | |||
| RSV vs. RSV + VPA: | 0.7939 | |||
| VPA vs. RSV + VPA: | 0.0002 *** | |||
| SOM Total Whole IL |
CON: 50.00 ± 8.70 RSV: 50.75 ± 3.30 VPA: 33.25 ± 4.71 RSV + VPA: 48.25 ± 2.63 |
Interaction: F (1, 12) = 6.840 p = 0.0226 * VPA: F (1, 12) = 12.48 p = 0.0041 ** RSV: F (1, 12) = 8.356 p = 0.0136 * |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0057 ** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0041 ** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | 0.0128 * | |||
| SOM Ratio Whole IL |
CON: 0.053 ± 0.008 RSV: 0.051 ± 0.003 VPA: 0.032 ± 0.003 RSV + VPA: 0.055 ± 0.001 |
Interaction: F (1, 12) = 31.96 p = 0.0001 *** VPA: F (1, 12) = 12.93 p = 0.0037 ** RSV: F (1, 12) = 21.75 p = 0.0005 *** |
CON vs. RSV: | >0.9999 |
| CON vs. VPA: | 0.0002 *** | |||
| CON vs. RSV + VPA: | >0.9999 | |||
| RSV vs. VPA: | 0.0005 *** | |||
| RSV vs. RSV + VPA: | >0.9999 | |||
| VPA vs. RSV + VPA: | <0.0001 **** | |||
II/III, upper cortical layers; IV/V, deeper cortical layers; aCC, anterior cingulate cortex; IL, infralimbic cortex; mPFC, medial prefrontal cortex; PrL, prelimbic cortex; SD, standard deviation; SOM, somatostatin-positive interneuron. p < 0.05 was considered significant. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, # trend. Statistical analyses: two-way ANOVA parametric test followed by Bonferroni. NCON: 4. NRSV: 4. NVPA: 4. NRSV + VPA: 4.
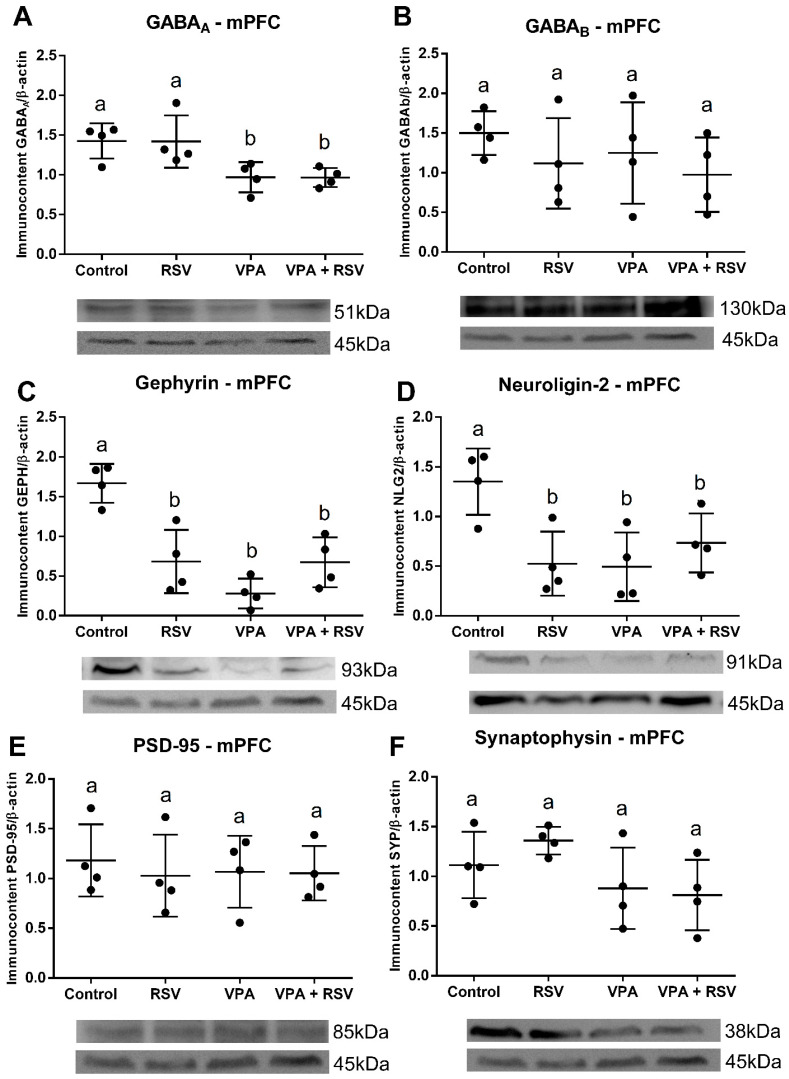
2.7. Both VPA and RSV Changed the Levels of Synaptic Proteins, whereas the Level of GABAA Was Affected Only by VPA
The protein quantification shows that VPA decreased GABAA, without RSV prevention (VPA: F (1, 12) = 16.00, p = 0.0018) (Figure 3A). Both RSV and VPA decreased gephyrin (Interaction: F (1, 12) = 21.56, p = 0.006; Cont-RSV ppost-hoc = 0.0031; Cont-VPAppost-hoc = 0.0001; Cont-RSV + VPAppost-hoc = 0.0029) (Figure 3C) and neuroligin-2 (Interaction: F (1, 12) = 10.77, p = 0.0066; Cont-RSV ppost-hoc = 0.0220, Cont-VPAppost-hoc = 0.0172; Cont-RSV + VPA ppost-hoc = 0.1128) (Figure 3D). No differences were observed among groups for GABAB (Figure 3B), PSD-95 (Figure 3E), and synaptophysin (Figure 3F).
Figure 3.
VPA induced a reduction in the immunocontent of GABAA, gephyrin, and neuroligin-2 (the last two also reduced by RSV) in the mPFC. The immunocontent of GABA receptors and synaptic proteins was normalized by the β-actin loading control. Values are shown as mean ± standard deviation. (A) GABAA immunocontent. (B) GABAB immunocontent. (C) Gephyrin immunocontent. (D) Neuroligin-2 immunocontent. (E) PSD-95 immunocontent. (F) Synaptophysin immunocontent. Statistical analysis: two-way ANOVA followed by Bonferroni, p < 0.05 was considered significant. NCON: 4, NRSV: 4, NVPA: 4, NRSV + VPA:4. Different letters indicate significant differences in the post-test when interaction was significant (p < 0.05).
2.8. The VPA Decreased the Number of Total Neurons and Altered the Ratio of Interneurons in the DG, without Full Prevention by RSV
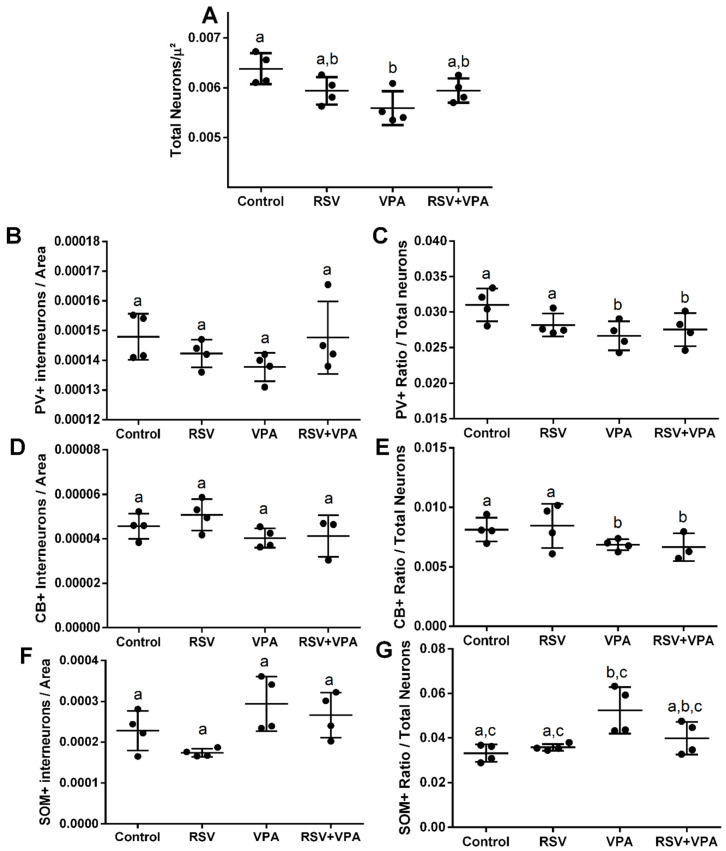
The VPA decreased the number of total neurons in DG (Interaction: F (1, 12) = 7.441, p = 0.0183; Cont-VPAppost-hoc = 0.0166) (Figure 4A), in PV+ ratio (VPA: F (1, 12) = 5.732, p = 0.0339) (Figure 4C), and in CB+ ratio (VPA: F (1, 11) = 5.709, p = 0.0359) (Figure 4E), and increased the SOM+ ratio (Interaction: F (1, 12) = 4.840, p = 0.0481; Cont-VPA ppost-hoc = 0.0023; RSV-VPA ppost-hoc= 0.0024; RSV + VPA-VPA ppost-hoc = 0.054) (Figure 4G). No differences were observed in the number of PV+ (Figure 4B), CB+ (Figure 4D), and SOM+ neurons (Figure 4F) among groups. Illustrative images of total neurons, PV+, CB+, and SOM+ are presented in Figure 5A–C.
Figure 4.
VPA reduced total neurons, CB+ ratio, and PV+ ratio while increasing the SOM+ ratio in the DG without prevention by RSV. (A) Quantification of total neurons. (B) Quantification of CB+ interneurons. (C) Quantification of the ratio of CB+ interneurons/total neurons. (D) Quantification of PV+ interneurons. (E) Quantification of the ratio of PV+ interneurons/total neurons. (F) Quantification of SOM+ interneurons. (G) Quantification of the ratio of SOM+ interneurons/total neurons. Values are shown as mean ± standard deviation. Statistical analysis: two-way ANOVA followed by Bonferroni, p < 0.05 was considered significant. NCON: 4, NRSV: 4, NVPA: 4, NRSV+VPA:4 CB+NeuN+DAPI, and PV+NeuN+DAPI; NCON: 4, NRSV: 4, NVPA: 4, NRSV+VPA:3 and SOM+NeuN+DAPI. Different letters indicate significant differences in the post-test when interaction was significant (p < 0.05).
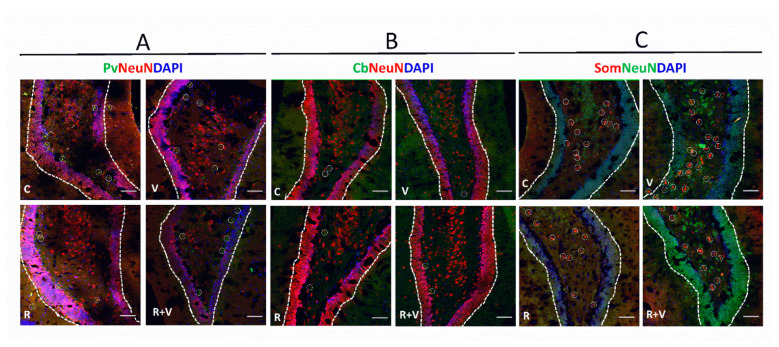
Figure 5.
Representative image of total neurons, PV+, CB+, and SOM+ immunofluorescence in the HC. Representative images of the SG, upper layers (II/III). (A) Pv, parvalbumin (green); NeuN (red); DAPI (blue). (B) Cb, calbindin (green); NeuN (red); DAPI (blue). (C) Som, somatostatin (red); NeuN (green); DAPI (blue). Scale bar: 50 µm. The respective interneurons are highlighted within white circles. aCC, anterior cingulate cortex; CB, calbindin-neurons; DG, dentate gyrus; HC, hippocampus; PV, parvalbumin-neurons; SOM, somatostatin-neurons.
2.9. The VPA Altered the Interneuronal Composition in CA1, CA2, CA3, and RSV Presented a per se Effect in CA3
The VPA group decreased the CB+ number in CA1, CA2, and CA3, following a decreased ratio of these neurons in CA2 and CA3 (Supplementary Materials Table S3). The RSV had a per se effect in CA3, decreasing PV+ numbers, but not altering the ratio (Supplementary Materials Table S2). The VPA increased the SOM+ number and ratio in the CA2, while RSV had a per se effect in CA3, increasing the number of SOM+ neurons without affecting the ratio (Supplementary Materials Table S4). For the total neurons, a significant difference was only found between VPA and RSV + VPA groups in CA2 (Supplementary Materials Table S1). No differences were found in other parameters.
2.10. The Immunocontent of the Analyzed Proteins Did Not Differ among Groups in the Hippocampus
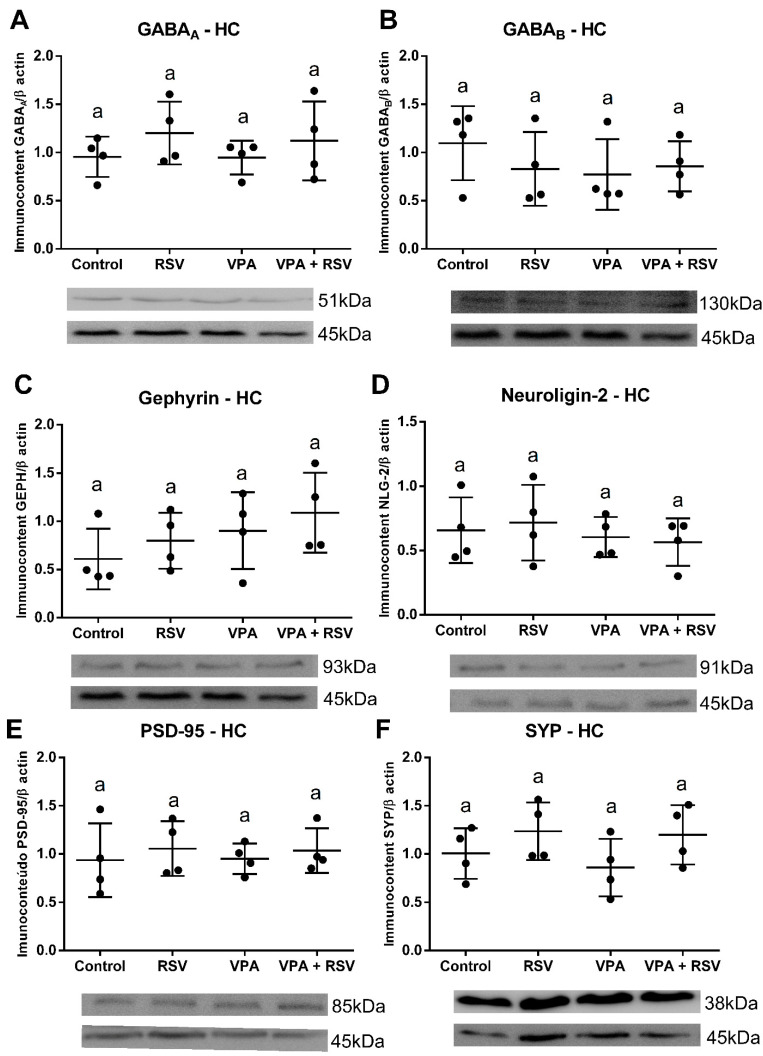
In the HC, no significant differences were found for all parameters evaluated. GABAA (Interaction: F (1, 12) = 0.06436 p = 0.8040. VPA: F (1, 12) = 0.09175 p= 0.7672. RSV: F (1, 12) = 2.034 p = 0.1793) (Figure 6A), GABAB (Interaction: F (1, 12) = 0.9989 p = 0.3373. VPA: F (1, 12) = 0.7182 p = 0.4133. RSV: F (1, 12) = 0.2652 p = 0.6159) (Figure 6B), gephyrin (Interaction: F (1, 12) = 8.221 × 10−5 p = 0.9929. VPA: F (1, 12) = 2.657 p = 0.1291. RSV: F (1, 12) = 1.099 p = 0.3152) (Figure 6C), neuroligin-2 (Interaction: F (1, 12) = 0.1844 p = 0.6753. VPA: F (1, 12) = 0.8125 p = 0.3851. RSV: F (1, 12) = 0.007832 p = 0.9309) (Figure 6D), PSD-95 (Interaction: F (1, 12) = 0.01751 p = 0.8969. VPA: F (1, 12) = 0.001065 p = 0.9745. RSV: F (1, 12) = 0.5443 p = 0.4748) (Figure 6E), and synaptophysin (Interaction: F (1, 12) = 0.1378 p = 0.7169. VPA: F (1, 12) = 0.3949 p = 0.5415. RSV: F (1, 12) = 3.797 p = 0.0751) (Figure 6F).
Figure 6.
There were no significant differences in the expression of synaptic proteins and GABA receptors in the HC. The immunocontent of GABA receptors and synaptic proteins was normalized by the β-actin loading control. Values are shown as mean ± standard deviation. (A) GABAA immunocontent. (B) GABAB immunocontent. (C) Gephyrin immunocontent. (D) Neuroligin-2 immunocontent. (E) PSD-95 immunocontent. (F) Synaptophysin immunocontent. Statistical analysis: two-way ANOVA followed by Bonferroni, p < 0.05 was considered significant. NCON: 4, NRSV: 4, NVPA: 4, NRSV + VPA:4.
3. Discussion
Changes in the organization of brain cytoarchitecture directly impact not only the local circuits but also the integration among different brain regions. In the autistic brain, cortical disorganization [49,50] and both high local connectivity and low long-range connectivity have already been described [51]. Here, we first studied microarray/RNA-Seq repository datasets of embryos from ASD animal models in order to investigate enriched pathways for the DEGs identified in them.
Firstly, the carbohydrate metabolic imbalance observed in E12.5 in DS2 was also observed in the organoids exposed to VPA (DS3), indicating that this may be the starting point of several subsequent alterations. The proliferation of neuronal progenitors relies mostly on aerobic glycolysis as the energetic source [52]; thus, an alteration in this metabolic pathway may induce early proliferative issues. In the subsequent days (E14.5 and E17.5), the pathways appear to induce a general condition of acceleration of neuronal differentiation, with upregulation of adhesion, neurotransmitter, and synaptic pathways to the detriment of the cell-cycle, gene expression, and protein dynamics regulation. Many of these features are also observed in brain organoids exposed to VPA (DS3), probably impacting the final disposition and organization of the neurons in different brain regions.
VPA has already demonstrated an influence in carbohydrate metabolism and mitochondrial function [53], increasing the production of reactive oxygen species [54]. RSV is a known antioxidant and anti-inflammatory molecule, and, thus, the early treatment (starting in E6.5) may attenuate a possible metabolic alteration induced by VPA. Moreover, VPA is a known inhibitor of histone deacetylases [55,56], while RSV is an activator of sirtuins [57], which may counteract the alterations in gene expression and cell-cycle. Thus, RSV may create a neuroprotective background, preventing alterations caused by VPA and expansion of initial damage throughout embryonic life, resulting in the maintenance of the neuronal composition in the mPFC and HC (to a lesser extent) in postnatal life.
Considering these data and our previous data from adult animals of the VPA animal model (P120) [41], which presented alterations in the neuronal composition of the HC, including disturbances in PV+, CB+, and SOM+, we studied here the same structure and also expanded it for mPFC in young animals (P30). Now, we demonstrate a substantial disorganization in the mPFC and HC neuronal cytoarchitecture in the VPA group, as well as important preventive effects of prenatal treatment with RSV, especially in the mPFC.
The VPA group showed an increased number of total neurons, while the interneurons presented either a reduced ratio or number in the mPFC, depending on the subpopulation. This numerical increase (even not significant in some subregions) is relevant because the ratio of interneurons/total neurons can be influenced by subtle alterations. We demonstrated that the deeper layers of PrL and IL presented the most significant increase in the number of total neurons. Postmortem analysis of ASD patients already demonstrated an increased number of neurons in the mini-columns of the frontal and parietal cortex [49] and patches of disorganization in the cortical layers, especially in the deeper layers [50]; moreover, an increase in the number of total neurons was observed in the dorsolateral cortex, the homologous region to the mPFC [58] in humans.
Next, we explored the gene expression datasets to identify potential mechanisms that could underlie the increased number of neurons in the cortex of VPA mice. The majority of the cortical neurons are excitatory pyramidal cells (about 75–80%) [59]. These neurons are generated in the ventricular zone in the early stages of embryogenesis (around E10 in rodents) [60] and reach the cortex through radial migration from the cortical subplate. VPA animals display an increased number of non-GABAergic neurons and thickness of the cortical layers, concomitant with changes in the expression of cell cycle proteins, suggesting maintenance of the proliferative phase for a longer time [61]. When we observed DS4 gene expression data, we noticed that the cortical subplate, the migrating cells from the subventricular zone, and even the radial glia (directly associated with migration guidance) displayed DEGs associated with cell cycle and gene expression in E14.
Next, we investigated the distribution of specific interneuron subpopulations in the mPFC. Prenatal exposure to VPA reduced the number of GABAergic SOM+ interneurons and the proportion of SOM+ and PV+ interneurons, with no general effect on CB+ (only specific alterations in the subregions). PV+, SOM+, and CB+ are mostly generated in different segments of the GE, developing a migration route that starts around E12.5 in rodents (the same day as the prenatal exposure to VPA) [18].
The SOM+ neurons originate in the medial portion of the GE (MGE) through an initial signaling system based on the increase in SHH expression followed by the expression of the NKX2.1 factor [62]. Since this interneuron population was the only one whose absolute number changed and considering that their migration starts earlier, it is possible that the drastic damage induced by VPA may occur when these cells are still in the proliferative stages. Interestingly, prenatal exposure to VPA at E9.5 reduced the SHH expression in E11.5 embryos [63], which could explain the SOM+ impairments.
On the other hand, the absence of changes in the absolute number (already described in the mPFC of the VPA model [64]) along with the reduction in the ratio of PV+ may suggest a subtle change potentially associated with migration processes, as seen in the anomalous pattern of distribution throughout the subregions. Indeed, while the aCC showed a reduction in number and proportion, the upper layers of PrL showed a completely opposite pattern. Previously, we observed that VPA animals showed an increased proportion of PV+ neurons in the upper layers of the somatosensory area, which was prevented by RSV [36]. CB+ presented a similar pattern to PV+ in relation to the ratio, and these subtle alterations may be associated with the small percentage of this interneuron population.
Interestingly, in DS4, the GE and emerging interneurons at E14 presented major alterations in cell-cycle, gene expression, and protein dynamics, which could result in alterations in the interneuronal proliferation and migration since they are strictly regulated by a sequence of transcription factors, including SHH, NKX2.1, DLX, LHX, SOX.
In addition to the changes in the number and proportion of GABAergic neurons, prenatal exposure to VPA induced a reduction in the immunocontent of the GABAA receptor, a finding already observed in postmortem analysis of ASD patients in the aCC [65] and in the frontal and parietal cortices [26]. Moreover, this alteration possibly contributes to the histological changes observed because this receptor plays an important role in neuronal migration throughout development [66]. Finally, the similar effect of VPA and RSV in reducing the immunocontent of gephyrin and neuroligin-2, two major constituents of inhibitory synapses, may point to an involvement of the Notch pathway, a signaling route highlighted in the DS2 as an altered pathway in late embryonic life, which involved the modulation of synapses [67,68] and is capable of being modulated by both VPA and RSV [69]. However, RSV alone did not cause major histological or behavioral alterations, similar to what was shown in previous studies from our research group [35,36,38].
In the HC, it was possible to observe that prenatal exposure to VPA mainly induced alterations in the DG. The reduction in the total neurons and the alterations in the interneurons, especially SOM+, may induce circuit imbalances with other regions, especially the mPFC, given the important role of SOM+ in integrating the HC and mPFC [70]. VPA is known to reduce neurogenesis in the HC [71] and induce the misplacement of neurons through a pathway mediated by the CXCL12 chemokine and its receptor, CXCR4 [72], which also plays a role in the migration of interneurons. Alterations in CB+ are present in several regions; however, the relatively low abundance of these cells in the HC may hinder accurate quantification. RSV has already demonstrated effects on the modulation of HC interneurons in adults [73]. Thus, prenatal treatment with RSV may cause alterations in the fate of these cells in specific situations. The absence of alterations in the synaptic proteins and GABA receptors in the HC suggests that VPA effects in this region may be restricted to modulation of neuronal populations and organization of brain cytoarchitecture.
4. Materials and Methods
4.1. Animals
Wistar rats from the Center for Reproduction and Experimentation of Laboratory Animals (CREAL) were housed in the bioterium of the Department of Biochemistry at UFRGS and maintained under a standard 12/12 h light/dark cycle at a constant temperature of 22 ± 2 °C with food and water ad libitum. The Ethics Commission of the Federal University of Rio Grande do Sul approved this project (CEUA-UFRGS #35733). The animals were euthanized by an anesthetic overdose of ketamine (300 mg/kg) and xylazine (40 mg/kg) (concentrations three times higher than the concentration required to obtain an anesthetic-surgical plan). All experimental procedures were performed in accordance with ethical principles in accordance with the Euthanasia Practice Guidelines of the National Council for Animal Experimentation Control (CONCEA) (Normative Resolution N. 13, 2013), NIH Guide for the Care and Use of Laboratory Animals, as well as Brazilian Arouca Law (11,794 of 8 October 2008).
4.2. Drugs and Prenatal Treatments
Wistar rats were mated overnight, and pregnancy was confirmed the next morning through the presence of spermatozoa in the female’s vaginal smear; when the pregnancy was confirmed, that day was considered the embryonic day 0.5 (E0.5). From E6.5 to E18.5, the pregnant rats received a daily subcutaneous injection of RSV (Fluxome, Stenløse, Denmark) at 3.6 mg/kg or equivalent volume of vehicle (dimethyl sulfoxide P.A. (DMSO)), as previously described [35,36]. At E12.5, pregnant rats received a single intraperitoneal injection of either VPA at 600 mg/kg (Acros Organics, Morris Plains, Morris County, NJ, USA) or vehicle (saline solution 0.9%). The four experimental groups, according to the treatment received, were the following: Control (vehicles), RSV, VPA, and RSV + VPA. Pregnant rats were singly housed at E18 for parturition. We considered the day of birth to be postnatal day 0 (P0). The female pups were euthanized at postnatal day (P) P21, and only males were used in this work. After weaning at P21, the male offspring were kept until P30. The total number of animals used in the study was nine control, eight RSV, eight VPA, and eight RSV + VPA divided randomly in experiments, generated from five control dams, four RSV, nine VPA, and nine RSV + VPA (the excedent offspring was destined to other projects, ensuring full use of the biological material). Loss rate for the VPA groups was approximately 50% in this protocol. The sample size used in each experiment is described in the corresponding figures and/or tables.
4.3. Immunofluorescence
The tissues were fixed and cryopreserved in OCT® and cut in a Leica® cryostat (−20 °C). The slices (25 µm) corresponding to the mPFC and HC were placed on histological slides covered with poly-L-lysine and post-fixed with 4% paraformaldehyde. The brain coordinates were the following: bregma 3.72/3.24 (mPFC and subregions: anterior cingulate [aCC], prelimbic [PrL], and infralimbic [IL]) and −2.92/−3.00 (HC and subregions: dentate gyrus [DG], CA1, CA2, and CA3) according to Paxinos Atlas (5th edition) [74]. Three slices were alternately placed in each histological slide, stained with specific primary antibodies for NeuN combined with PV, SOM, or CB, in addition to corresponding secondary antibodies associated with a fluorophore and nuclear DAPI dye according to the protocol described by Fontes-Dutra et al. [36]. Technical information and concentrations of the reagents used in the immunofluorescence assays are summarized in Supplementary Materials Table S1. The images were obtained using the Olympus FV1000® confocal microscope at the Center for Microscopy and Microanalysis (CMM-UFRGS) (Supplementary Figure S1 demonstrates the subdivisions established for the analyzed regions). Each coronal section was photographed in stacks by the confocal microscope (8, on average; dimensions: 635.9 × 635.9 microns). The analyses were performed manually by two trained researchers who were blinded to the experimental groups using the Cell Counter plug-in in the ImageJ® software [75]. Quantification was conducted by counting the cells in 8 stacks of at least 2 slices per animal (all stacks were counted individually and with the overlapping image).
The results are shown as the absolute number of total neurons (NeuN+DAPI) and interneurons (CB+NeuN+DAPI, PV+NeuN+DAPI, and SOM+NeuN+DAPI) normalized by area and as the ratio between the number of interneurons and total neurons to obtain a proportion between the inhibitory (interneuron) and excitatory components (the majority of the total neurons) according to the following formula: (CB+, PV+ or SOM+) Interneurons/Total neurons (based on Fontes-Dutra et al. [36]. This ratio was made separately for each interneuron evaluated. The mPFC was subdivided into three subregions, named aCC, PrL, and IL. Each of these regions were subdivided into upper layers (II/III) and deeper layers (IV/V). The total number of neurons, the number of each interneuron (PV+, CB+, and SOM+), and the ratio (interneuron/total neurons) were evaluated in each subfield (i.e., upper layers of aCC, deeper layers of aCC, upper layers of PrL, deeper layers of PrL, upper layers of IL, deeper layers of IL). The amount observed in the deeper + upper layers of a subregion represents the whole subregion (i.e., deeper layers of aCC + upper layers of aCC = whole aCC). The amount observed in whole aCC + whole PrL + whole IL represents the whole mPFC. The HC was subdivided into four subregions: DG, CA1, CA2, and CA3. In each of them, the total number of neurons, the number of each interneuron (PV+, CB+, and SOM+), and the ratio (interneuron/total neurons) were evaluated.
4.4. Western Blotting
Samples from mPFC and HC were homogenized and prepared in a buffer containing 10% SDS, 100 mM EDTA, 500 mM TRIS/HCl buffer (pH 8), and protease inhibitors. The supernatant was collected after centrifugation at 14.000× g for 20 min at 4 °C. Total proteins were quantified by the Lowry method [76], and the samples were prepared in a buffer containing glycerol, bromophenol blue, 500 mM TRIS/HCl buffer, and β-mercaptoethanol. Equal amounts of protein (40 µg) were applied to 10% polyacrylamide gels, separated by unidimensional electrophoresis, and transferred to nitrocellulose membranes to detect the immunocontent of GABAA, GABAB, gephyrin, neuroligin-2, PSD-95 and synaptophysin proteins using specific primary antibodies according to the protocol adapted from Deckmann et al., [77]. Technical information and concentrations of the reagents used in the Western Blotting assays are summarized in Supplementary Materials Table S1. After incubation with corresponding secondary peroxidase-associated antibodies (HRP), the chemiluminescent signal was detected using the ImageQuant™ LAS 4000 system (GE HealthCare Life Sciences®, Chicago, IL, USA). The quantification of the relative protein content was performed with the ImageJ® software, and the data were normalized by the endogenous marker β-actin.
4.5. Transcriptomic Analysis
To provide insights into the embryonic processes that could lead to the alterations observed in the postnatal brain of ASD models, we selected five RNA-Seq and microarray datasets [32,78,79,80,81] from MIA animal models, VPA-exposed cell cultures, and cortical organoids (Table 1) since databases of VPA-induced animal models are not available yet. The differentially expressed genes (DEGs) of each dataset were analyzed with Cytoscape® [82] using the BiNGO® plug-in [83] to evaluate Gene Ontology (GO) enrichments in a determined set of genes, providing tables with the statistically significant most representative GO terms. We also compared the DEGs observed in each dataset with the Simons Foundation Autism Research Initiative (SFARI) gene database [84] to observe the percentage of DEGs that have an ortholog already described as altered in ASD.
4.6. Statistical Analysis
All the analyses were performed using the GraphPad Prism 6 software (GraphPad Software, La Jolla, CA, USA). Kolmogorov–Smirnov and Shapiro–Wilk tests of normality were applied to determine the data distribution. As the data presented a normal distribution, we chose a parametric test (two-way ANOVA) followed by a Bonferroni post-test. When there was an interaction effect, pairwise comparison was analyzed in the post-hoc; when there was no effect, the effect of exposure to factors (VPA or RSV) was analyzed.
5. Conclusions
The present study demonstrated that the prenatal treatment with RSV was able to prevent important alterations in the neuronal composition of the mPFC induced by prenatal exposure to VPA, probably improving parameters associated with the E/I balance. These findings are in accordance with several other studies that have already demonstrated the neuroprotective effects of RSV in psychiatric disorders, not only in animal models but also in humans, highlighting the translational value of the study. The transcriptomic analysis allowed the establishment of hypotheses to explain the developmental context of these interventions, highlighting the pathways such as WNT, NOTCH, and others in which VPA and RSV may act. Next, we demonstrated that prenatal exposure to VPA alters the neuronal profile in the mPFC and HC, impacting the number and proportion of interneurons, indicating a possible E/I imbalance. Moreover, VPA also induced alterations in the immunocontent of a GABA receptor and synaptic proteins in the mPFC, adding another layer of evidence to comprehend the alterations in the circuitry of this region. In summary, prenatal treatment with RSV was able to prevent neuronal alterations in the mPFC. In addition, our analyses suggest that the investigation of mechanisms involved in the development of interneurons, brain cytoarchitecture, and synaptic content can be a promising strategy to expand the understanding of the pathophysiology of ASD.
Acknowledgments
We would like to thank all of the funding agencies, including CNPq, CAPES, INCT-NIM, and FIPE-HCPA. We also would like to thank Vitória Cassola de Lemos (Edinburgh, Scotland) for the English writing revision.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms23084075/s1.
Author Contributions
J.S.-T., I.D., G.C.-C., G.D.-F.N., G.B.-N., G.B.S., M.F.-D., R.R. and C.G., experimental design, sample processing, and intellectual contribution. R.R. and C.G., acquisition of financial resources. J.S.-T., I.D., G.C.-C. and M.F.-D., immunofluorescence analyses. J.S.-T., I.D. and GDFN, Western blotting analyses. J.S.-T., I.D., G.C.-C., G.D.-F.N., G.B.-N., G.B.S., M.F.-D. and C.G., data discussion and manuscript preparation. All authors have read and agreed to the published version of the manuscript.
Funding
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES); Instituto Nacional de Ciência e Tecnologia em Neuroimunomodulação (INCT-NIM) (Project number 465489/2014-1); Fundo de Incentivo à Pesquisa e Eventos do Hospital de Clínicas de Porto Alegre (FIPE-HCPA).
Institutional Review Board Statement
All procedures were approved by the local Ethics Commission on the Use of Animals (CEUA-UFRGS 35733) and performed according to ethical principles in accordance with the NIH Guide for the Care and Use of Laboratory Animals, as well as Brazilian Arouca Law (11,794, of 8 October 2008).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (DSM-5) American Psychiatric Association; Virginia, VA, USA: 2013. American Psychiatric DSM 5. [Google Scholar]
- 2.Maenner M.J., Shaw K.A., Baio J., Washington A., Patrick M., DiRienzo M., Christensen D.L., Wiggins L.D., Pettygrove S., Andrews J.G., et al. Prevalence of autism spectrum disorder among children aged 8 Years-Autism and developmental disabilities monitoring network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020;69:1–12. doi: 10.15585/mmwr.ss6904a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masi A., DeMayo M.M., Glozier N., Guastella A.J. An Overview of Autism Spectrum Disorder, Heterogeneity and Treatment Options. Neurosci. Bull. 2017;33:183–193. doi: 10.1007/s12264-017-0100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zwaigenbaum L., Penner M. Autism spectrum disorder: Advances in diagnosis and evaluation. BMJ. 2018;361:k1674. doi: 10.1136/bmj.k1674. [DOI] [PubMed] [Google Scholar]
- 5.Shulman C., Esler A., Morrier M.J., Rice C.E. Diagnosis of Autism Spectrum Disorder Across the Lifespan. Child Adolesc. Psychiatr. Clin. N. Am. 2020;29:253–273. doi: 10.1016/j.chc.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Baxter A.J., Brugha T.S., Erskine H.E., Scheurer R.W., Vos T., Scott J.G. The epidemiology and global burden of autism spectrum disorders. Psychol. Med. 2015;45:601–613. doi: 10.1017/S003329171400172X. [DOI] [PubMed] [Google Scholar]
- 7.Spence S.J., Schneider M.T. The Role of Epilepsy and Epileptiform EEGs in Autism Spectrum Disorders. Pediatr. Res. 2009;65:599. doi: 10.1203/PDR.0b013e31819e7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton P.F., Carcani-Rathwell I., Hutton J., Goode S., Howlin P., Rutter M. Epilepsy in autism: Features and correlates. Br. J. Psychiatry. 2011;198:289–294. doi: 10.1192/bjp.bp.109.076877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukmanji S., Manji S.A., Kadhim S., Sauro K.M., Wirrell E.C., Kwon C.S., Jetté N. The co-occurrence of epilepsy and autism: A systematic review. Epilepsy Behav. 2019;98:238–248. doi: 10.1016/j.yebeh.2019.07.037. [DOI] [PubMed] [Google Scholar]
- 10.Nelson S.B., Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozzi Y., Provenzano G., Casarosa S. Neurobiological bases of autism-epilepsy comorbidity: A focus on excitation/inhibition imbalance. Eur. J. Neurosci. 2018;47:534–548. doi: 10.1111/ejn.13595. [DOI] [PubMed] [Google Scholar]
- 12.Selten M., Bokhoven H., van Kasri N.N. Inhibitory control of the excitatory/inhibitory balance in psychiatric disorders. F1000Research. 2018;7:23. doi: 10.12688/f1000research.12155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux L., Buzsáki G. Tasks for inhibitory interneurons in intact brain circuits. Neuropharmacology. 2015;88:10–23. doi: 10.1016/j.neuropharm.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelkey K.A., Chittajallu R., Craig M.T., Tricoire L., Wester J.C., McBain C.J. Hippocampal GABAergic Inhibitory Interneurons. Physiol. Rev. 2017;97:1619–1647. doi: 10.1152/physrev.00007.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markram H., Toledo-Rodriguez M., Wang Y., Gupta A., Silberberg G., Wu C. Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 16.Tremblay R., Lee S., Rudy B. GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron. 2016;91:260–292. doi: 10.1016/j.neuron.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defelipe J., López-Cruz P.L., Benavides-Piccione R., Bielza C., Larrañaga P., Anderson S., Burkhalter A., Cauli B., Fairén A., Feldmeyer D., et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 2013;14:202. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelsom C., Lu W. Development and specification of GABAergic cortical interneurons. Cell Biosci. 2013;3:19. doi: 10.1186/2045-3701-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentet L.J., Kremer Y., Taniguchi H., Huang Z.J., Staiger J.F., Petersen C.C.H. Unique functional properties of somatostatin-expressing GABAergic neurons in mouse barrel cortex. Nat. Neurosci. 2012;15:607–612. doi: 10.1038/nn.3051. [DOI] [PubMed] [Google Scholar]
- 20.Xia F., Richards B.A., Tran M.M., Josselyn S.A., Takehara-Nishiuchi K., Frankland P.W. Parvalbumin-positive interneurons mediate neocortical-hippocampal interactions that are necessary for memory consolidation. eLife. 2017;6:e27868. doi: 10.7554/eLife.27868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho K.K.A., Davidson T.J., Bouvier G., Marshall J.D., Schnitzer M.J., Sohal V.S. Cross-hemispheric gamma synchrony between prefrontal parvalbumin interneurons supports behavioral adaptation during rule shift learning. Nat. Neurosci. 2020;23:892–902. doi: 10.1038/s41593-020-0647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson J.R., Bartley A.F., Hays S.A., Huber K.M. Imbalance of Neocortical Excitation and Inhibition and Altered UP States Reflect Network Hyperexcitability in the Mouse Model of Fragile X Syndrome. J. Neurophysiol. 2008;100:2615. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polepalli J.S., Wu H., Goswami D., Halpern C.H., Südhof T.C., Malenka R.C. Modulation of excitation on parvalbumin interneurons by neuroligin-3 regulates the hippocampal network. Nat. Neurosci. 2017;20:219–229. doi: 10.1038/nn.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L.J., Liu N., Cheng T.L., Chen X.J., Li Y.D., Shu Y.S., Qiu Z.L., Zhang X.H. Conditional deletion of Mecp2 in parvalbumin-expressing GABAergic cells results in the absence of critical period plasticity. Nat. Commun. 2014;5:5036. doi: 10.1038/ncomms6036. [DOI] [PubMed] [Google Scholar]
- 25.Yu F.H., Mantegazza M., Westenbroek R.E., Robbins C.A., Kalume F., Burton K.A., Spain W.J., McKnight G.S., Scheuer T., Catterall W.A. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat. Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- 26.Fatemi S.H., Reutiman T.J., Folsom T.D., Thuras P.D. GABAAreceptor downregulation in brains of subjects with autism. J. Autism Dev. Disord. 2009;39:223–230. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang G., Gudsnuk K., Kuo S.H., Cotrina M.L., Rosoklija G., Sosunov A., Sonders M.S., Kanter E., Castagna C., Yamamoto A., et al. Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parente D.J., Garriga C., Baskin B., Douglas G., Cho M.T., Araujo G.C., Shinawi M. Neuroligin 2 nonsense variant associated with anxiety, autism, intellectual disability, hyperphagia, and obesity. Am. J. Med. Genet. A. 2017;173:213–216. doi: 10.1002/ajmg.a.37977. [DOI] [PubMed] [Google Scholar]
- 29.Guang S., Pang N., Deng X., Yang L., He F., Wu L., Chen C., Yin F., Peng J. Synaptopathology involved in autism spectrum disorder. Front. Cell. Neurosci. 2018;12:471. doi: 10.3389/fncel.2018.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass R., Norton S., Fox N., Kusnecov A.W. Maternal immune activation with staphylococcal enterotoxin A produces unique behavioral changes in C57BL/6 mouse offspring. Brain. Behav. Immun. 2019;75:12–25. doi: 10.1016/j.bbi.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Boulanger-Bertolus J., Pancaro C., Mashour G.A. Increasing role of maternal immune activation in neurodevelopmental disorders. Front. Behav. Neurosci. 2018;12:230. doi: 10.3389/fnbeh.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oskvig D.B., Elkahloun A.G., Johnson K.R., Phillips T.M., Herkenham M. Maternal immune activation by LPS selectively alters specific gene expression profiles of interneuron migration and oxidative stress in the fetus without triggering a fetal immune response. Brain. Behav. Immun. 2012;26:623–634. doi: 10.1016/j.bbi.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergdolt L., Dunaevsky A. Brain changes in a maternal immune activation model of neurodevelopmental brain disorders. Prog. Neurobiol. 2019;175:1–19. doi: 10.1016/j.pneurobio.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontes-Dutra M., Rabelo B., Santos-Terra J., Deckmann I., Schwingel G.B., Gottfried C. Perinatal Inflammation and Adult Psychopathology. Springer; Cham, Switzerland: 2020. Maternal Immune Activation and Neuropsychiatric Disorders: The Intricate Puzzle of Autism Spectrum Disorder. [Google Scholar]
- 35.Bambini-Junior V., Zanatta G., Della Flora Nunes G., Mueller de Melo G., Michels M., Fontes-Dutra M., Nogueira Freire V., Riesgo R., Gottfried C. Resveratrol prevents social deficts in animal model of autism induced by valproic acid. Neurosci. Lett. 2014;583:176–181. doi: 10.1016/j.neulet.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 36.Fontes-Dutra M., Santos-Terra J., Deckmann I., Brum Schwingel G., Della-Flora Nunes G., Hirsch M.M., Bauer-Negrini G., Hedin-Pereira C., Bambini-Junior V., Riesgo R.S., et al. Resveratrol prevents cellular and behavioral sensory alterations in the animal model of autism induced by valproic acid. Front. Synaptic Neurosci. 2018;10:9. doi: 10.3389/fnsyn.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontes-Dutra M., Della-Flora Nunes G., Santos-Terra J., Souza-Nunes W., Bauer-Negrini G., Hirsch M.M., Green L., Riesgo R., Gottfried C., Bambini-Junior V. Abnormal empathy-like pro-social behaviour in the valproic acid model of autism spectrum disorder. Behav. Brain Res. 2019;364:11–18. doi: 10.1016/j.bbr.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 38.Hirsch M.M., Deckmann I., Fontes-Dutra M., Bauer-Negrini G., Della-Flora Nunes G., Nunes W., Rabelo B., Riesgo R., Margis R., Bambini-Junior V., et al. Behavioral alterations in autism model induced by valproic acid and translational analysis of circulating microRNA. Food Chem. Toxicol. 2018;115:336–343. doi: 10.1016/j.fct.2018.02.061. [DOI] [PubMed] [Google Scholar]
- 39.Gottfried C., Bambini-Junior V., Baronio D., Zanatta G., Bristot R., Vaccaro T., Riesgo R. Recent Advances in Autism Spectrum Disorders. Volume I IntechOpen; London, UK: 2013. Valproic Acid in Autism Spectrum Disorder: From an Environmental Risk Factor to a Reliable Animal Model. [Google Scholar]
- 40.Watanabe Y., Murakami T., Kawashima M., Hasegawa-Baba Y., Mizukami S., Imatanaka N., Akahori Y., Yoshida T., Shibutani M. Maternal Exposure to Valproic Acid Primarily Targets Interneurons Followed by Late Effects on Neurogenesis in the Hippocampal Dentate Gyrus in Rat Offspring. Neurotox. Res. 2017;31:46–62. doi: 10.1007/s12640-016-9660-2. [DOI] [PubMed] [Google Scholar]
- 41.Santos-Terra J., Deckmann I., Schwingel G.B., Paz A.V.C., Gama C.S., Bambini-Junior V., Fontes-Dutra M., Gottfried C. Resveratrol prevents long-term structural hippocampal alterations and modulates interneuron organization in an animal model of ASD. Brain Res. 2021;1768:147593. doi: 10.1016/j.brainres.2021.147593. [DOI] [PubMed] [Google Scholar]
- 42.Deckmann I., Schwingel G.B., Fontes-Dutra M., Bambini-Junior V., Gottfried C. Neuroimmune Alterations in Autism: A Translational Analysis Focusing on the Animal Model of Autism Induced by Prenatal Exposure to Valproic Acid. Neuroimmunomodulation. 2018;25:285–299. doi: 10.1159/000492113. [DOI] [PubMed] [Google Scholar]
- 43.Baronio D., Bauer-Negrini G., Castro K., Della-Flora Nunes G., Riesgo R., Mendes-Da-Cruz D.A., Savino W., Gottfried C., Bambini-Junior V. Reduced CD4 T Lymphocytes in Lymph Nodes of the Mouse Model of Autism Induced by Valproic Acid. Neuroimmunomodulation. 2018;25:280–284. doi: 10.1159/000491395. [DOI] [PubMed] [Google Scholar]
- 44.Cristiano C., Volpicelli F., Crispino M., Lacivita E., Russo R., Leopoldo M., Calignano A., Perrone-Capano C. Behavioral, Anti-Inflammatory, and Neuroprotective Effects of a Novel FPR2 Agonist in Two Mouse Models of Autism. Pharmaceuticals. 2022;15:161. doi: 10.3390/ph15020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magaji M.G., Iniaghe L.O., Abolarin M., Abdullahi O.I., Magaji R.A. Neurobehavioural evaluation of resveratrol in murine models of anxiety and schizophrenia. Metab. Brain Dis. 2017;32:437–442. doi: 10.1007/s11011-016-9927-6. [DOI] [PubMed] [Google Scholar]
- 46.Rafeiy-Torghabeh M., Ashraf-Ganjouei A., Moradi K., Bagheri S., Mohammadi M.R., Akhondzadeh S. Resveratrol adjunct to methylphenidate improves symptoms of attention-deficit/hyperactivity disorder: A randomized, double-blinded, placebo-controlled clinical trial. Eur. Child Adolesc. Psychiatry. 2020;30:799–807. doi: 10.1007/s00787-020-01562-z. [DOI] [PubMed] [Google Scholar]
- 47.Quincozes-Santos A., Gottfried C. Resveratrol modulates astroglial functions: Neuroprotective hypothesis. Ann. N. Y. Acad. Sci. 2011;1215:72–78. doi: 10.1111/j.1749-6632.2010.05857.x. [DOI] [PubMed] [Google Scholar]
- 48.Caruso G., Torrisi S.A., Mogavero M.P., Currenti W., Castellano S., Godos J., Ferri R., Galvano F., Leggio G.M., Grosso G., et al. Polyphenols and neuroprotection: Therapeutic implications for cognitive decline. Pharmacol. Ther. 2022;232:108013. doi: 10.1016/j.pharmthera.2021.108013. [DOI] [PubMed] [Google Scholar]
- 49.Casanova M.F., van Kooten I.A.J., Switala A.E., van Engeland H., Heinsen H., Steinbusch H.W.M., Hof P.R., Trippe J., Stone J., Schmitz C. Minicolumnar abnormalities in autism. Acta Neuropathol. 2006;112:287–303. doi: 10.1007/s00401-006-0085-5. [DOI] [PubMed] [Google Scholar]
- 50.Stoner R., Chow M.L., Boyle M.P., Sunkin S.M., Mouton P.R., Roy S., Wynshaw-Boris A., Colamarino S.A., Lein E.S., Courchesne E. Patches of Disorganization in the Neocortex of Children with Autism. N. Engl. J. Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belmonte M.K., Allen G., Beckel-Mitchener A., Boulanger L.M., Carper R.A., Webb S.J. Autism and abnormal development of brain connectivity. J. Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salsaa M., Pereira B., Liu J., Yu W., Jadhav S., Hüttemann M., Greenberg M.L. Valproate inhibits mitochondrial bioenergetics and increases glycolysis in Saccharomyces cerevisiae. Sci. Rep. 2020;10:11785. doi: 10.1038/s41598-020-68725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tung E.W.Y., Winn L.M. Valproic Acid Increases Formation of Reactive Oxygen Species and Induces Apoptosis in Postimplantation Embryos: A Role for Oxidative Stress in Valproic Acid-Induced Neural Tube Defects. Mol. Pharmacol. 2011;80:979–987. doi: 10.1124/mol.111.072314. [DOI] [PubMed] [Google Scholar]
- 55.Göttlicher M., Minucci S., Zhu P., Krämer O.H., Schimpf A., Giavara S., Sleeman J.P., Lo Coco F., Nervi C., Pelicci P.G., et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phiel C.J., Zhang F., Huang E.Y., Guenther M.G., Lazar M.A., Klein P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 57.Borra M.T., Smith B.C., Denu J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 58.Courchesne E., Mouton P.R., Calhoun M.E., Semendeferi K., Ahrens-Barbeau C., Hallet M.J., Barnes C.C., Pierce K. Neuron Number and Size in Prefrontal Cortex of Children With Autism. JAMA. 2011;306:2001. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 59.Kanari L., Ramaswamy S., Shi Y., Morand S., Meystre J., Perin R., Abdellah M., Wang Y., Hess K., Markram H. Objective Morphological Classification of Neocortical Pyramidal Cells. Cereb. Cortex. 2019;29:1719–1735. doi: 10.1093/cercor/bhy339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y.K., Fujishima K., Kengaku M. Differentiation of Apical and Basal Dendrites in Pyramidal Cells and Granule Cells in Dissociated Hippocampal Cultures. PLoS ONE. 2015;10:e0118482. doi: 10.1371/journal.pone.0118482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimura K., Mitsuhashi T., Shibata S., Shimozato S., Takahashi T. In Utero Exposure to Valproic Acid Induces Neocortical Dysgenesis via Dysregulation of Neural Progenitor Cell Proliferation/Differentiation. J. Neurosci. 2016;36:10908–10919. doi: 10.1523/JNEUROSCI.0229-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butt S.J.B., Fuccillo M., Nery S., Noctor S., Kriegstein A., Corbin J.G., Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 63.Oyabu A., Narita M., Tashiro Y. The effects of prenatal exposure to valproic acid on the initial development of serotonergic neurons. Int. J. Dev. Neurosci. 2013;31:202–208. doi: 10.1016/j.ijdevneu.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Lauber E., Filice F., Schwaller B. Prenatal Valproate Exposure Differentially Affects Parvalbumin-Expressing Neurons and Related Circuits in the Cortex and Striatum of Mice. Front. Mol. Neurosci. 2016;9:150. doi: 10.3389/fnmol.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oblak A., Gibbs T.T., Blatt G.J. Decreased GABA A receptors and benzodiazepine binding sites in the anterior cingulate cortex in autism. Autism Res. 2009;2:205–219. doi: 10.1002/aur.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bortone D., Polleux F. KCC2 Expression Promotes the Termination of Cortical Interneuron Migration in a Voltage-Sensitive Calcium-Dependent Manner. Neuron. 2009;62:53–71. doi: 10.1016/j.neuron.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giniger E. Notch signaling and neural connectivity. Curr. Opin. Genet. Dev. 2012;22:339–346. doi: 10.1016/j.gde.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salama-Cohen P., Arevalo M.-A., Grantyn R., Rodriguez-Tebar A. Notch and NGF/p75NTR control dendrite morphology and the balance of excitatory/inhibitory synaptic input to hippocampal neurones through Neurogenin 3. J. Neurochem. 2006;97:1269–1278. doi: 10.1111/j.1471-4159.2006.03783.x. [DOI] [PubMed] [Google Scholar]
- 69.Ji Y., Ke Y., Gao S. Intermittent activation of notch signaling promotes bone formation. Am. J. Transl. Res. 2017;9:2933–2944. [PMC free article] [PubMed] [Google Scholar]
- 70.Abbas A.I., Sundiang M.J.M., Henoch B., Morton M.P., Bolkan S.S., Park A.J., Harris A.Z., Kellendonk C., Gordon J.A. Somatostatin Interneurons Facilitate Hippocampal-Prefrontal Synchrony and Prefrontal Spatial Encoding. Neuron. 2018;100:926–939.e3. doi: 10.1016/j.neuron.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juliandi B., Tanemura K., Igarashi K., Tominaga T., Furukawa Y., Otsuka M., Moriyama N., Ikegami D., Abematsu M., Sanosaka T., et al. Reduced Adult Hippocampal Neurogenesis and Cognitive Impairments following Prenatal Treatment of the Antiepileptic Drug Valproic Acid. Stem Cell Rep. 2015;5:996–1009. doi: 10.1016/j.stemcr.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Danzer S.C. Valproic Acid Leads New Neurons Down the Wrong Path. Epilepsy Curr. 2019;19:132–133. doi: 10.1177/1535759719835366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra V., Shuai B., Kodali M., Shetty G.A., Hattiangady B., Rao X., Shetty A.K. Resveratrol Treatment after Status Epilepticus Restrains Neurodegeneration and Abnormal Neurogenesis with Suppression of Oxidative Stress and Inflammation. Sci. Rep. 2015;5:17807. doi: 10.1038/srep17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paxinos G., Watson C. The Rat Brain in Stereotaxic Coordinates—The New Coronal Set. 5th ed. Volume 1. Academic Press; Cambridge, MA, USA: 2004. [Google Scholar]
- 75.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 77.Deckmann I., Santos-Terra J., Fontes-Dutra M., Körbes-Rockenbach M., Bauer-Negrini G., Schwingel G.B., Riesgo R., Bambini-Junior V., Gottfried C. Resveratrol prevents brain edema, blood–brain barrier permeability, and altered aquaporin profile in autism animal model. Int. J. Dev. Neurosci. 2021;81:579–604. doi: 10.1002/jdn.10137. [DOI] [PubMed] [Google Scholar]
- 78.Balmer N.V., Klima S., Rempel E., Ivanova V.N., Kolde R., Weng M.K., Meganathan K., Henry M., Sachinidis A., Berthold M.R., et al. From transient transcriptome responses to disturbed neurodevelopment: Role of histone acetylation and methylation as epigenetic switch between reversible and irreversible drug effects. Arch. Toxicol. 2014;88:1451–1468. doi: 10.1007/s00204-014-1279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cui K., Wang Y., Zhu Y., Tao T., Yin F., Guo Y., Liu H., Li F., Wang P., Chen Y., et al. Neurodevelopmental impairment induced by prenatal valproic acid exposure shown with the human cortical organoid-on-a-chip model. Microsyst. Nanoeng. 2020;6:49. doi: 10.1038/s41378-020-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalish B.T., Kim E., Finander B., Duffy E.E., Kim H., Gilman C.K., Yim Y.S., Tong L., Kaufman R.J., Griffith E.C., et al. Maternal immune activation in mice disrupts proteostasis in the fetal brain. Nat. Neurosci. 2020;24:204–213. doi: 10.1038/s41593-020-00762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Canales C.P., Estes M.L., Cichewicz K., Angara K., Aboubechara J.P., Cameron S., Prendergast K., Su-Feher L., Zdilar I., Kreun E.J., et al. Sequential perturbations to mouse corticogenesis following in utero maternal immune activation. eLife. 2021;10:e60100. doi: 10.7554/eLife.60100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maere S., Heymans K., Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 84.Banerjee-Basu S., Packer A. SFARI Gene: An evolving database for the autism research community. Dis. Model. Mech. 2010;3:133–135. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or Supplementary Material.