Abstract
Binding affinities of β-lactam antibiotics for the three penicillin binding proteins (PBPs) from Chlamydia trachomatis were determined in vitro and compared with their antichlamydial activities. Mecillinam selectively inhibited PBP1, with a 50% inhibitory concentration for PBP1 binding (0.2 μg/ml) similar to the MIC (0.1 μg/ml) and minimum bactericidal concentration (0.25 μg/ml). Although the other β-lactams inhibited a wider range of PBPs than mecillinam, their antichlamydial activities were inferior to that of mecillinam.
Chlamydiae are an important group of gram-negative intracellular pathogens which cause serious infections in humans (18). They have a complex development cycle involving infection by elementary bodies (EBs), transformation of EBs into reticulate bodies (RBs), RB division, differentiation of RBs back to EBs, and release of EBs following host cell lysis (18). Penicillin and other β-lactam antibiotics inhibit the growth of chlamydiae (1, 3, 8–10, 13–16) and have been used for the treatment of chlamydial infections, particularly when first-line agents are contraindicated (11, 14, 19, 24, 25). Exposure of chlamydiae to penicillin leads to the accumulation of large aberrant RBs, the so-called penicillin forms, which can resume normal development if penicillin is removed (15, 16). Consequently, it has been suggested that β-lactam antibiotics interfere with RB division (1, 3, 16).
The susceptibility of chlamydiae to β-lactams is consistent with the presence of penicillin binding proteins (PBPs) in these organisms (1, 3, 7, 12, 21). In other bacteria PBPs have roles in peptidoglycan (PG) metabolism through their activities as transpeptidases, carboxypeptidases, and endopeptidases (6, 23). Chlamydiae possess three PBPs, and although genomic analysis assigns transpeptidase activity to PBP1 and PBP2 (3, 7) and carboxypeptidase activity to PBP3 (3, 7), a major paradox of chlamydial biology is the inability to detect PG, or a PG-like polymer, in these organisms (1, 3, 7, 16). Furthermore, the apparent absence of genes encoding transglycosylase activity suggests that if a muramic acid-containing wall polymer is synthesized by chlamydiae, then it may exist only as a cross-linked disaccharide-peptide, i.e., as a glycanless wall polymer (7). In view of these observations, there is poor understanding of the mechanisms by which β-lactams prevent chlamydial growth, and it is not known, for instance, whether inhibition of all three PBPs is a requirement for antichlamydial activity or whether binding to individual PBPs is sufficient for growth inhibition. We have now addressed these issues by comparing the binding of various β-lactams to chlamydial PBPs in vitro with their antichlamydial activities.
We chose a panel of seven β-lactam antibiotics and determined their MICs, minimum bactericidal concentrations (MBCs), and affinities for PBPs in Chlamydia trachomatis 434 serotype L2 (Table 1). Benzylpenicillin, ampicillin, and cefotaxime were purchased from Sigma, and the other antibiotics were gifts from Merck Sharp & Dohme (imipenem, as Primaxin IV), Astra-Zeneca (meropenem), Leo Pharmaceuticals (mecillinam), and Biochemie GmBh (ceftriaxone).
TABLE 1.
Antichlamydial activities of selected penicillins, cephalosporins, and carbapenems and their competition for the PBPs of C. trachomatis
| Antibiotic | MIC (μg/ml) | MBC (μg/ml) | IC50 (μg/ml) for PBP binding
|
||
|---|---|---|---|---|---|
| PBP1 | PBP2 | PBP3 | |||
| Benzylpenicillin | 0.1 | 4.0 | 0.03 | 0.005 | 0.01 |
| Ampicillin | 0.1 | 2.0 | 0.01 | 0.005 | 0.5 |
| Mecillinam | 0.1 | 0.25 | 0.2 | 100 | 250 |
| Ceftriaxone | 2.0 | 16.0 | 0.1 | 0.01 | 1.0 |
| Cefotaxime | 32.0 | 250 | 0.1 | 0.04 | 5.0 |
| Imipenem | 64 | 512 | 0.01 | 0.2 | 0.005 |
| Meropenem | 16 | 64 | 0.01 | 0.1 | 0.005 |
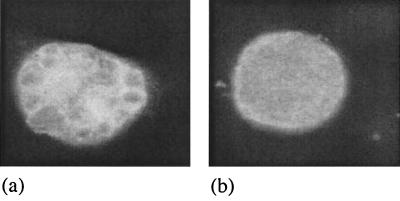
C. trachomatis was cultured in McCoy cell monolayers which were prepared on glass coverslips in flat-bottomed plastic vials. Confluent cells were infected with 103 inclusion-forming units of C. trachomatis in Dulbecco's modified Eagle medium (Gibco BRL, Paisley, United Kingdom) supplemented with 4% fetal calf serum and 1 μg of cycloheximide per ml (17). Cultures were centrifuged at 2,000 × g for 30 min at 30°C and then incubated at 37°C. At 1 h after infection, the medium was changed and the test antibiotic was added. For MBC determinations, the infected cells were scraped off the coverslips after 48 h and 0.1 ml was used to infect a second monolayer growing on antibiotic-free medium. After a further 48 h, the coverslips were fixed in methanol, stained with a fluorescein isothiocyanate-labeled monoclonal antibody to the chlamydial lipopolysaccharide (DAKO Ltd., Ely, United Kingdom), and then examined by microscopy at magnifications of ×400 and ×1,000. The MBC was defined as the lowest concentration of antibiotic in the first cycle of infection that resulted in no inclusions in the second cycle of infection. MICs were determined as described above, but after the first 48 h of incubation the cultures were fixed in methanol and stained. The MIC was defined as the lowest concentration of antibiotic that led to the formation of inclusions with abnormal morphology, as assessed by light microscopy (Fig. 1 illustrates mecillinam as an example).
FIG. 1.
Light micrograph of chlamydial inclusions growing in McCoy cells and stained with fluorescein isothiocyanate-labeled monoclonal antibody to the chlamydial lipopolysaccharide. Chlamydiae were treated with 0.25 μg of mecillinam per ml (a) or no antibiotic (PBS control) (b). The typical appearance of the β-lactam form can be seen in panel a, compared to a normal inclusion in panel b. Magnification, ×3,200.
PBP assays were performed essentially as previously described (2, 20), with the modifications described below. Chlamydiae were grown in McCoy cells in 150-cm2 tissue culture flasks with an inoculum that gave 95 to 100% infection without centrifugation. After 20 h, while the chlamydiae were still predominantly RBs, the medium was removed and the infected cells were scraped off into 10 ml of phosphate-buffered saline (PBS) per flask. The cells were sonicated for five 30-s cycles, and membranes were pelleted by centrifugation at 30,000 × g for 80 min at 4°C. The pellets were resuspended in PBS containing DNase I (25 μg/ml) and stored at −80°C. For PBP binding assays, 10 μl of membrane preparation (50 μg of protein) was added to 10 μl of β-lactam solution (various concentrations) in PBS. Samples were incubated at 35°C for 20 min, and then 2 μl of [3H]benzylpenicillin (Amersham Pharmacia Biotech, Amersham, United Kingdom) (37 mBq/ml; final concentration, 0.5 μg/ml) was added. After a further 20 min at 35°C, the reaction was stopped by the addition of 20 μl of sodium dodecyl sulfate (SDS) sample buffer (0.06M Tris-HCl, 1.4 M 2-mercaptoethanol, 4% [wt/vol] SDS, 20% [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue). Samples were boiled for 5 min and then centrifuged (30,000 × g, 10 min, 4°C). Proteins were separated by SDS-polyacrylamide gel electrophoresis, and the gels were analyzed as described previously (2) after autoradiography for 28 days using Amplify (Amersham Pharmacia) as the fluor. PBP binding assays were performed at least twice for each β-lactam antibiotic.
It is well documented that when C. trachomatis is cultured in the presence of β-lactam antibiotics, inclusions form with abnormal morphology (10). Since we wanted to determine the minimal concentration of antibiotic that affected chlamydial growth, we used morphological abnormality (Fig. 1) as a criterion for determining MICs, rather than complete inhibition of the development of inclusions, which has been used by others (8, 9). Thus, the MICs that we determined (Table 1) are not necessarily directly comparable with those reported in the literature. However, in agreement with others (8, 9), we observed that representative penicillins (benzylpenicillin, ampicillin, and mecillinam) were more active at both the MIC and MBC than cephalosporins (ceftriaxone and cefotaxime). The carbapenems imipenem and meropenem were also relatively inactive, in agreement with earlier findings on N-formimidoyl-thienamycin and imipenem (8, 9). Mecillinam was the most potent of the β-lactams we investigated, having an MBC and an MIC of 0.25 and 0.01 μg/ml, respectively (Table 1). Similar observations were made by Hammerschlag and Gleyzer (9), who found that mecillinam had the greatest antichlamydial activity among 12 β-lactams studied.
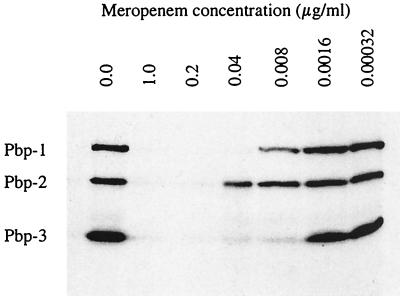
The PBP pattern of RBs from C. trachomatis is shown in Fig. 2, together with the results of a typical competition experiment, in this case using meropenem. On the basis of electrophoretic mobility, the PBPs had apparent molecular masses of 110 kDa (PBP1), 73 kDa (PBP2), and 40 kDa (PBP3). These molecular mass values differ from those reported by Barbour et al. (1) and those predicted by sequence analysis (12, 21). We have previously commented that these differences are probably explained by anomalous migration of the PBPs in SDS-polyacrylamide gel electrophoresis (22).
FIG. 2.
Fluorogram showing the competition of meropenem for the PBPs of C. trachomatis.
Data for competition of the seven β-lactam antibiotics with [3H]benzylpenicillin for the three PBPs are presented in Table 1. As the PBP assay is based on competitive binding, these data are expressed as 50% inhibitory concentrations (IC50s), i.e., the concentration of β-lactam required to inhibit subsequent binding of [3H]benzylpenicillin by 50% (2, 4, 5). The only quantitative PBP binding data previously reported for C. trachomatis RBs concern the binding of benzylpenicillin, where IC50s of 0.03 μg/ml (PBP1), 0.006 μg/ml (PBP2), and 0.003 μg/ml (PBP3) were reported (1). For benzylpenicillin we observed a similar range of IC50s, with no more than a sixfold difference in affinity between the individual PBPs.
Studies with other bacteria, particularly the Enterobacteriaceae and Pseudomonas aeruginosa, have indicated that the killing target or targets for a particular β-lactam are usually represented by those PBPs that are most readily saturated by the antibiotic (2, 4, 5, 23). On this basis, it appears that all three chlamydial PBPs may be targets for benzylpenicillin. The other β-lactams appear to exhibit some selectivity towards particular PBPs. The two carbapenems had lower affinities for PBP2 than for PBP1 and PBP3, and the two cephalosporins and ampicillin had lower affinities for PBP3 than for PBP1 and PBP2 (Table 1). With the exception of mecillinam, the MBC and sometimes also the MIC were significantly higher than the PBP binding values. This suggests that the antibiotics have some difficulty in crossing the various host cell and chlamydial permeability barriers that separate the PBPs in RBs from the external medium.
Compared with the other β-lactam antibiotics, mecillinam exhibited a different set of responses. This antibiotic displayed high selectivity for PBP1, and the IC50 for PBP1 binding (0.2 μg/ml) corresponded closely with both the MIC and MBC (Table 1). Thus, mecillinam appears to penetrate the various permeability barriers quite effectively and exerts antichlamydial activity by binding exclusively to PBP1. Furthermore, these data suggest that PBP1 is essential for chlamydial division and survival. In Escherichia coli mecillinam also has high affinity for a single PBP, in this case PBP2 (4–6, 23), which implies that chlamydial PBP1 and E. coli PBP2 may be functionally related. This is also supported by analysis of structural relatedness, since among the E. coli PBPs, chlamydial PBP1 has highest homology to E. coli PBP2 (7). Although PBP2 is required for the maintenance of cell shape in E. coli (4, 5, 23), the lethality of PBP2 inactivation has also been attributed to cell division inhibition (26). If chlamydial PBP1, by analogy, is involved in cell division, this would be consistent with our hypothesis (3) that in chlamydia PG, or a glycanless PG-like polymer, has a major role in RB division. Since mecillinam appears to specifically inhibit chlamydial PBP1, more detailed examination of the structure and morphology of mecillinam-inhibited cultures may provide a clue to the role of this PBP in the chlamydial cell cycle.
Finally, the results reported here support an earlier suggestion that in view of its potent antichlamydial activity, mecillinam may have a role in the treatment of chlamydial infection (9). Furthermore, our data now provide a molecular explanation for the potency of mecillinam, which can be attributed to both favorable cell penetration and high affinity for chlamydial PBP1.
REFERENCES
- 1.Barbour A G, Amano K-I, Hackstadt T, Perry L, Caldwell H D. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J Bacteriol. 1982;151:420–428. doi: 10.1128/jb.151.1.420-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton M G, Orr D C. Detection of bacterial penicillin-binding proteins and their role in the interpretation of the mode of action of β-lactam antibiotics. In: Russell A D, Quesnel L B, editors. Antibiotics: assessment of antimicrobial activity and resistance. Society for Applied Bacteriology Technical Series no. 18. London, United Kingdom: Academic Press; 1983. pp. 161–181. [Google Scholar]
- 3.Chopra I, Storey C, Falla T J, Pearce J H. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology. 1998;144:2673–2678. doi: 10.1099/00221287-144-10-2673. [DOI] [PubMed] [Google Scholar]
- 4.Curtis N A C, Orr D, Ross G W, Boulton M G. Competition of β-lactam antibiotics for the penicillin-binding proteins of Pseudomonas aeruginosa, Enterobacter cloacae, Klebsiella aerogenes, Proteus rettgeri, and Escherichia coli: comparison with antibacterial activity and effects upon bacterial morphology. Antimicrob Agents Chemother. 1979;16:325–328. doi: 10.1128/aac.16.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis N A C, Orr D, Ross G W, Boulton M G. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob Agents Chemother. 1979;16:533–539. doi: 10.1128/aac.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denome S A, Elf P K, Henderson T A, Nelson D E, Young K D. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghuysen J-M, Goffin C. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother. 1999;43:2339–2344. doi: 10.1128/aac.43.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gump D W. Antimicrobial susceptibility testing for some atypical microorganisms: chlamydiae, mycoplasmas, Rickettsia, and spirochetes. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: Williams and Wilkins; 1996. pp. 212–229. [Google Scholar]
- 9.Hammerschlag M R, Gleyzer A. In vitro activity of a group of broad-spectrum cephalosporins and other β-lactam antibiotics against Chlamydia trachomatis. Antimicrob Agents Chemother. 1983;23:493–494. doi: 10.1128/aac.23.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.How S J, Hobson D, Hart C A, Quayle E. A comparison of the in vitro activity of antimicrobials against Chlamydia trachomatis examined by Giemsa and a fluorescent antibody stain. J Antimicrob Chemother. 1985;15:399–404. doi: 10.1093/jac/15.4.399. [DOI] [PubMed] [Google Scholar]
- 11.Hueston W J, Lenhart J G. A decision analysis to guide antibiotic selection for Chlamydia infection during pregnancy. Arch Fam Med. 1997;6:551–555. doi: 10.1001/archfami.6.6.551. [DOI] [PubMed] [Google Scholar]
- 12.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomics of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 13.Kuo C-C, Grayston J T. In vitro drug susceptibility of Chlamydia sp. strain TWAR. Antimicrob Agents Chemother. 1988;32:257–258. doi: 10.1128/aac.32.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin D H, Pastorek J G, Faro S. In-vitro and in-vivo activity of parenterally administered β-lactam antibiotics against Chlamydia trachomatis. Sex Transm Dis. 1986;13:81–87. doi: 10.1097/00007435-198604000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto A, Manire G P. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol. 1970;101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moulder J W. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis. 1993;2:87–99. [PubMed] [Google Scholar]
- 17.Richmond S J, Bailey J M G, Bailey A S, Mearns G. Primary isolation of Chlamydia trachomatis and Chlamydia psittaci in in vitro cell culture. In: Collins C H, Grange J M, editors. Isolation and identification of micro-organisms of medical and veterinary importance. New York, N.Y: Academic Press; 1985. pp. 297–312. [Google Scholar]
- 18.Schachter J. Chlamydial infections. In: Gorbach S L, Bartlett J G, Blacklow N R, editors. Infectious diseases. W. B. Philadelphia, Pa: Saunders; 1992. pp. 817–822. [Google Scholar]
- 19.Silverman N S, Sullivan M, Hochman M, Womack M, Jungkind D L. A randomized, prospective trial comparing amoxicillin and erythromycin for the treatment of Chlamydia trachomatis in pregnancy. Am J Obstet Gynecol. 1994;170:829–832. doi: 10.1016/s0002-9378(94)70292-6. [DOI] [PubMed] [Google Scholar]
- 20.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 21.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 22.Storey C, Pearce J, Chopra I. Expression of penicillin binding protein 1 in Escherichia coli and yeast. In: Stephens R S, Byrne G I, Christiansen G, Clarke I N, Grayston J T, Bank R G, Ridgeway G L, Saikku P, Schachter J, Stamm W E, editors. Chlamydial infections. 1998. pp. 563–566. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, San Francisco, Calif. [Google Scholar]
- 23.Tomasz A. Mode of action of β-lactam antibiotics—a microbiologist's view. In: Demain A L, Solomon N A, editors. Antibiotics containing the beta-lactam structure, part 1. Handbook of experimental pharmacology. 67/1. Berlin, Germany: Springer-Verlag; 1983. pp. 15–96. [Google Scholar]
- 24.Toomey K E, Barnes R C. Treatment of Chlamydia trachomatis genital infection. Rev Infect Dis. 1990;12:S645–S655. doi: 10.1093/clinids/12.supplement_6.s645. [DOI] [PubMed] [Google Scholar]
- 25.Turrentine M A, Newton E R. Amoxicillin or erythromycin for the treatment of antenatal chlamydial infection: a meta analysis. Obstet Gynecol. 1995;86:1021–1025. doi: 10.1016/0029-7844(95)00296-4. [DOI] [PubMed] [Google Scholar]
- 26.Vinella D, Joseleau-Petit D, Thevenet D, Bouloc P, D'Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by ftsZ overexpression. J Bacteriol. 1993;175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]