Abstract
ADP-ribosylation, a modification of proteins, nucleic acids, and metabolites, confers broad functions, including roles in stress responses elicited, for example, by DNA damage and viral infection and is involved in intra-and extracellular signaling, chromatin and transcriptional regulation, protein biosynthesis, and cell death. ADP-ribosylation is catalyzed by ADP-ribosyltransferases (ARTs), which transfer ADP-ribose from NAD+ onto substrates. The modification, which occurs as mono- or poly-ADP-ribosylation, is reversible due to the action of different ADP-ribosylhydrolases. Importantly, inhibitors of ARTs are approved or are being developed for clinical use. Moreover, ADP-ribosylhydrolases are being assessed as therapeutic targets, foremost as antiviral drugs and for oncological indications. Due to the development of novel reagents and major technological advances that allow the study of ADP-ribosylation in unprecedented detail, an increasing number of cellular processes and pathways are being identified that are regulated by ADP-ribosylation. In addition, characterization of biochemical and structural aspects of the ARTs and their catalytic activities have expanded our understanding of this protein family. This increased knowledge requires that a common nomenclature be used to describe the relevant enzymes. Therefore, in this viewpoint, we propose an updated and broadly supported nomenclature for mammalian ARTs that will facilitate future discussions when addressing the biochemistry and biology of ADP-ribosylation. This is combined with a brief description of the main functions of mammalian ARTs to illustrate the increasing diversity of mono- and poly-ADP-ribose mediated cellular processes.
Keywords: ADP-ribosylation, MARylation, PARP, PARylation, posttranslational modification
The ADP-ribosyltransferase superfamily
ADP-ribosylation, an ancient modification of proteins, nucleic acids, and metabolites, describes the reversible transfer of ADP-ribose (ADPr) from β-nicotinamide adenine dinucleotide (NAD+) onto a substrate. This modification is central to many cellular processes across all domains of life. The biochemical reaction is catalyzed primarily by enzymes of the ADP-ribosyltransferase (ART) clan or superfamily of proteins (Fig. 1A), which are found in all kingdoms of life, generating N-, O-, or S-glycosidic linkages at the 1" position of the nicotinamide ribose following a dissociative SN2 reaction scheme. Typically, the modification occurs as mono-ADP-ribosylation (MARylation), but some ARTs have the ability to synthesize polymers of ADPr (PAR chains or PARylation) (Fig. 1A). The information carried by both PAR and MAR can be disseminated by dedicated reader domains. Amino acids that are currently known to be ADP-ribosylated include Glu, Asp, Ser, Thr, Tyr, Arg, Lys, His, and Cys (Fig. 1B), while nucleic acids are typically modified at an exposed phosphate group. By targeting the glycosidic bonds, ADP-ribosylhydrolases are capable of either degrading PAR chains or removing the substrate proximal ADPr, or both in the case of ARH3 (ADP-ribosylhydrolase, ADP-ribosyl-acceptor hydrolases or ADP-ribosyl-glycohydrolases) (Fig. 1A). We refer the reader to recent reviews that summarize and expand on these observations [1–16].
Fig. 1.
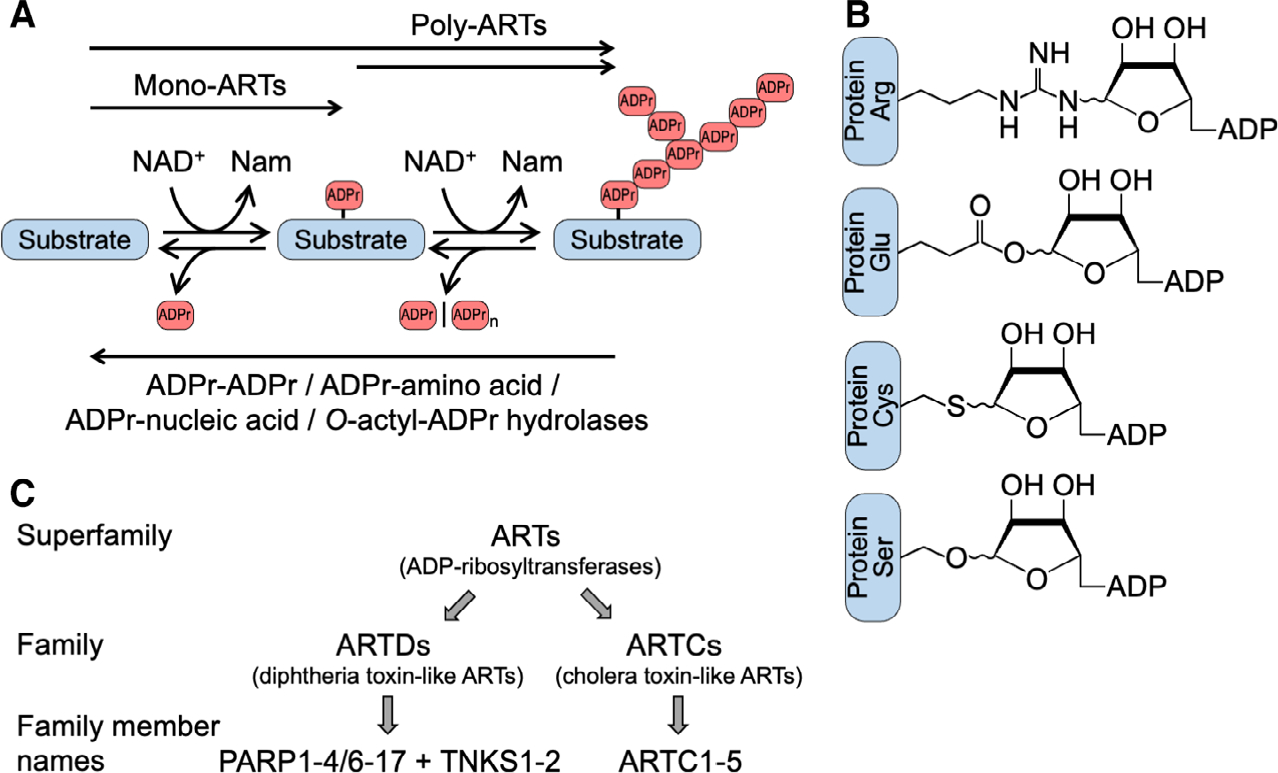
ADP-ribosylation is a reversible modification of proteins, nucleic acids, and metabolites. (A) Indicated are ADP-ribosyltransferases (ARTs), which catalyze mono- and poly-ADP-ribosylation (MARylation and PARylation, respectively). Poly-ARTs may modify naïive substrates generating PAR chains. Alternatively, poly-ARTs may use MARylated substrates and extend the modification to form polymers. The modification is removed by ADP-ribosylhydrolases. Some hydrolases degrade PAR chains (e.g., PARG and ARH3), and others cleave the glycosidic bond between the substrate and the proximal ADPr (e.g., MacroD1, MacroD2, TARG1, ARH1 and ARH3). Substrates include proteins, nucleic acids, and metabolites (the latter exemplified by O-acetyl-ADPr). The specificity of the enzymes is not indicated, some are highly selective, while others have broad specificity, including the modification of different classes of substrates. (B) Indicated are proteins with different ADPr acceptor amino acids and the corresponding N-, O-, or S-glycosidic linkages at the 1″ position of the nicotinamide ribose. Additional amino acids that have been detected to be modified include Asp, Asn, Lys, Thr, Tyr, His, Phospho-Ser, and diphthamide. Please note that for Ser-ADPr and Arg-ADPr, and probably for other linkages, the initial products are in the a configuration. Isomerization may occur, which is indicated. Additionally, migration to the 2″- and 3″-OH may happen for Glu-ADPr and Asp-ADPr. (C) Summary of the proposed nomenclature. ARTs represent the superfamily, which consists of 23 families, two being ARTD and ARTC [17,18]. Names that are commonly used for ARTD family members are PARP (the historic abbreviation for poly(ADP-ribose)polymerase) and TNKS (for tankyrase). Here, we propose PARP as a name on its own right, rather than an abbreviation.
The ART superfamily consists of 23 families when the Pfam database is interrogated [17,18]. ARTs are characterized by a comparable overall structure consisting of a split β-sheet and two helical regions, despite low conservation of primary amino acid sequences. ARTs can be divided into three clades, the H-Y-[EDQ] clade (clade 1), the R-[ST]-E clade (clade 2), and the H-H-h clade (not further discussed), containing the indicated amino acids in the active sites. These structural aspects have been widely discussed and will be summarized below [3,19,20]. The focus here will be on mammalian proteins that belong to clade 1 and clade 2.
Classification of mammalian ADP-ribosyltransferases
The majority of mammalian proteins that belong to clade 1 are referred to as PARPs and tankyrases (TNKS) or together as ARTDs (see below for definitions), which are located intracellularly [1,4,7,8,21]. The proteins expressed in mammals belonging to clade 2 are extracellular or secreted and designated ARTCs or ecto-ARTs [22–25]. The proposition to use the terminology ARTD and ARTC for two structurally related but distinct families of ARTs was made more than ten years ago [20]. It was based on the observation that the catalytic domains of diphtheria toxin and cholera toxin represent these two families with the above indicated specific configurations of amino acids, H-Y-[EDQ] and R-[ST]-E, in the catalytic cleft for NAD+ binding and catalysis, hence the addenda D and C to ART, respectively (Fig. 1C).
The ARTD/ARTC nomenclature as well as the proposed numbering of the members has been endorsed by some groups. However, most research groups have opted to continue to use the term PARP, in particular those working on PARP1 and PARP2 and on clinically relevant inhibitors of these enzymes as well as those who apply PARP inhibitors in patients. In contrast, ARTC has been well received and adopted more broadly. Here, we propose a revised nomenclature, taking into account the structural considerations, the biochemical specifications and the preferences for the names as used in the literature (Table 1). In brief, we suggest using ARTD and ARTC for the two structurally distinct families and PARP and TNKS with appropriate numbering as the names of the mammalian ARTD family members (Fig. 1C).
Table 1.
Names and properties of the family of diphtheria toxin-like ADP-ribosyltransferases.
| Preferred/suggested names for ARTs of the diphtheria toxin-like family (ARTD) | Alternatively/previously used names | Catalytic ADP-ribosylation activity |
|---|---|---|
|
| ||
| PARP1 | Poly-PARP, ARTD1, PARS, ADPRT | Poly |
| PARP2 | ARTD2 | Poly |
| PARP3 | ARTD3 | Mono |
| PARP4 | VPARP, ARTD4 | Mono |
| TNKS1 | Tankyrase 1, TNKS, PARP5a, ARTD5 | Poly |
| TNKS2 | Tankyrase 2, PARP5b ARTD6 | Poly |
| PARP6 | ARTD17 | Mono |
| PARP7 | TIPARP, ARTD14 | Mono |
| PARP8 | ARTD16 | Mono |
| PARP9 | BAL1, ARTD9 | Mono |
| PARP10 | ARTD10 | Mono |
| PARP11 | ARTD11 | Mono |
| PARP12 | ZC3HDC1, ARTD12 | Mono |
| PARP13 | ZAP, ZC3HAV1, ARTD13 | Inactive |
| PARP14 | BAL2, ARTD8 | Mono |
| PARP15 | BAL3, ARTD7 | Mono |
| PARP16 | ARTD15 | Mono |
It is worthwhile to briefly review how the naming of these enzymes evolved. Originally, the enzyme that synthesizes PAR chains was referred to as poly(ADP-ribose) synthetase and typically compared to DNA and RNA polymerases, also reflecting that PAR was initially thought to be a polyadenylic polymer [26]. In the 1980s and 1990s, the name poly(ADP-ribose) synthetase (PARS) as well as ADP-ribosyltransferase (ADPRT) were used, but then gradually replaced by poly(ADP-ribose) polymerase [27,28]. Subsequently, the abbreviation PARP was established, coined for an enzyme that synthesizes poly-ADP-ribose (a nucleic acid mimic molecule) from NAD+. This resulted in naming all mammalian clade 1 proteins with an H-Y-[EDQ] signature in the active center as poly(ADP-ribose) polymerases, poly-ADP-ribosepolymerases or PARP enzymes, and occasionally as poly-ADP-ribosyltransferases. Early on it was clear that PARP, now PARP1 and the founding member of the ART superfamily, transfers multiple ADP-ribose units in an iterative process directly onto protein substrates thus being biochemically speaking an ADP-ribosyltransferase that PARylates proteins. This is distinct from findings with bacterial toxins that MARylate their substrates and enzymes with this activity were initially referred to as NAD+:protein ADP-ribosyl transferases [26]. For many years, PARP1 was considered to be the only mammalian enzyme able to catalyze ADP-ribosylation. In the late 1990s, in part as a result of sequencing the human genome, an entire family of proteins was defined that is characterized by a sequence related to the catalytic domain of PARP1 and bacterial ARTs. The initial biochemical characterization of some of these proteins, for example, tankyrase (TNKS, for TRF1-interacting, ankyrin-related ADP-ribose polymerase [29]) and PARP2 [30] confirmed their ability to synthesize PAR, similar to PARP1 (Table 1, Fig. 2). It is important to note that these enzymes do not require a template for synthesizing PAR, different from both DNA- or RNA-synthesizing polymerases. Instead, they use an iterative transferase activity to polymerize PAR chains.
Fig. 2.
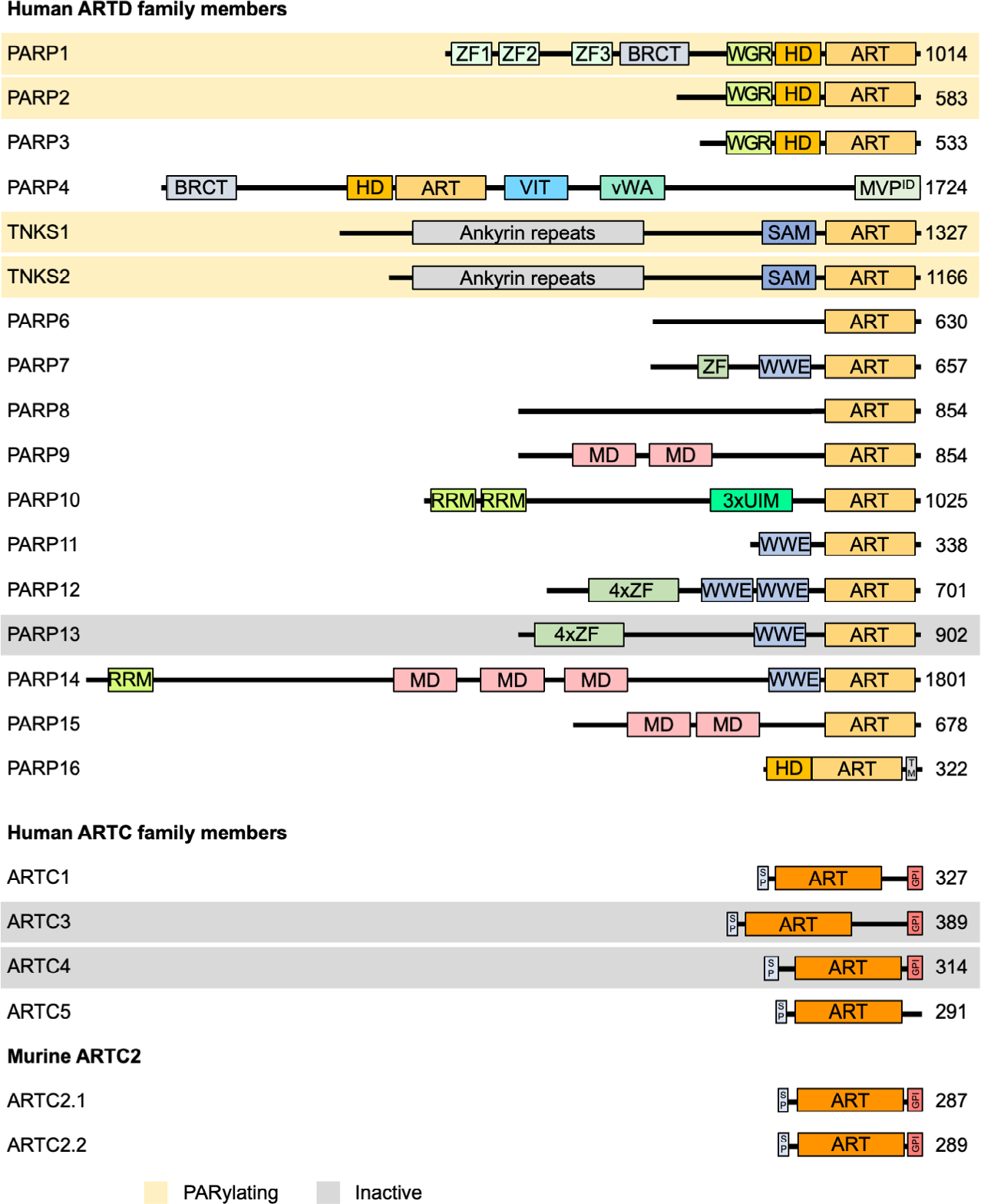
Domain architecture of ARTD and ARTC family members. Important domains of ARTDs and ARTCs family members are indicated. PARylating members as well as members that have been suggested to be catalytically inactive are highlighted in orange and gray, respectively. All other proteins possess MARylating activity. As discussed in the text, whether PARP9 is active has not been fully clarified. The potential coding region of human ARTC2 carries premature stop codons, displayed are the two proteins expressed from two closely related murine genes. ART, ADP-ribosyltransferase domain; BRCT, BRCA1 C terminus domain; GPI, glycosylphosphatidylinositol anchor; HD, helical domain; MD, macrodomain; MVPID, major vault protein interaction domain; RRM, RNA-recognition motif; SAM, sterile alpha motif; SP, signal peptide; TM, transmembrane motif; UIM, ubiquitininteraction motif; vWA, von Willebrand factor type A domain; WGR, conserved Trp-Gly-Arg motif domain; WWE, three conserved residues Trp-Trp-Glu motif domain; ZF, Zinc finger (light green, CCHC-type ZnF1 and ZnF2, CCCC-type ZnF3; purple, CCCH-type ZnF).
In contrast to these early findings, the more recent characterization of certain other ARTs, including PARP10, revealed that these enzymes only MARylate substrates and are unable to form PAR [31]. Sequence and structural considerations led to the hypothesis that the majority of the ARTD family members function as mono-ARTs rather than poly-ARTs. This original suggestion was based on the presence or absence of a catalytic glutamate or aspartate in the active center of clade 1 H-Y-[EDQ] proteins [31]. Furthermore, this was guided by the finding that mutating the corresponding glutamate in PARP1 resulted in an enzyme that was able to MARylate but could no longer PARylate [32]. Thus, at first sight the presence or absence of a glutamate in the active center specified catalytic activity. This left open the question how enzymes lacking this key acidic residue catalyze ADP-ribosylation. To overcome this conundrum, a model was proposed that suggests substrate-assisted catalysis as a mechanism to compensate for the lack of a catalytic glutamate [31]. Subsequent studies by different groups have provided evidence that most enzymes belonging to the ARTD family are mono-ARTs [33–36]. Moreover, two of the enzymes that possess a glutamate in the active center, PARP3 and PARP4, were also found to MARylate rather than PARylate their substrates [33,37], suggesting that additional determinants are important to distinguish between the ability to MARylate or PARylate substrates.
Revised nomenclature of mammalian ADP-ribosyltransferases
In eukaryotes, ARTD summarizes the intracellular ART family with PARP and TNKS as the names for individual proteins that possess a diphtheria toxin-like version of the ART fold, while ARTC describes the family of extracellular enzymes with a cholera toxin-like version of the ART fold (Fig. 2). The term poly-ADP-ribose-polymerase is widely used to define intracellular ARTs. In our view, this term is imprecise and should not be applied to describe the biochemical properties or the family of intracellular ARTs because most of the members do not synthesize PAR and those that can form PAR do so in a template-independent manner using their transferase activity (Table 2). Therefore, ADP-ribosyltransferase or ART as both a biochemical and family description is more accurate, and we suggest using these terms for all enzymes that can transfer ADP-ribose onto a substrate. ART defines the biochemical activity described by these enzymes, including PARPs, TNKSs, ecto-ARTs, and bacterial toxins, that is, the transfer of ADP-ribose. Included are also pseudo-enzymes without catalytic activity, such as PARP13, which possess a domain that exhibits high structural similarity to the catalytic domain of active ART members [38].
Table 2.
Revised recommended nomenclature of mammalian ADP-ribosyltransferases.
| ADP-ribosyltransferase (ART) | Description of the clan or superfamily of enzymes that can transfer an ADP-ribose group from NAD+ onto a substrate. It includes pseudo-enzymes |
| ARTD family | Defines one family of ARTs that is characterized by a catalytic domain structure related to diphtheria toxin-like bacterial ARTs. This includes the intracellular mammalian ART proteins. They are named: - PARPx (PARPx is the name of a particular ARTD family member; PARP is a name on its own right and it is proposed to not be used as abbreviation for poly-ADP-ribose-polymerase) - TNKSx (TNKSx is the name of a particular ARTD family member and the abbreviation for tankyrase) The previously defined numbering shall be used (see Table 1 and Fig. 2) |
| ARTC family | Defines one family of ARTs that is characterized by a catalytic domain fold related to cholera toxin-like bacterial ARTs. This includes the extracellular, membrane bound as well as secreted mammalian ART proteins (ecto-ARTs). These protein MARylate substrates, ARTC3 and ARTC4, are thought to be inactive |
| Poly-ARTs | ART superfamily members that have the capacity to iteratively transfer multiple ADP-ribose units onto a substrate, thereby forming PAR chains. Presently, they include the ARTD family members PARP1, PARP2, TNKS1, and TNKS2 |
| Mono-ARTs | ART superfamily members that are restricted to the transfer of one ADP-ribose unit, thereby producing MARylated substrates. This term should be used only when a distinction needs to be made to enzymes that can synthesize PAR. As the majority of ARTs MARylate substrates, the use of mono-ART is not necessary in most cases. This includes the following members of the ARTD family: PARP3, PARP4, PARP6-12, PARP14-16. PARP13 is inactive |
| Poly-ADP-ribose-polymerase/Poly(ADP-ribose)polymerase | We recommend that this term is no longer used. PARP should not be used as abbreviation for poly-ADP-ribose-polymerase but as a name in its own right (i.e., PARPx). Most of these proteins do not synthesize PAR, and those that can form PAR do so in a template-independent manner |
| Poly-PARP/mono-PARP | These terms should no longer be used |
To extend this logic, we recommend that PARP is not used as the abbreviation for poly-ADP-ribosepolymerase and the characterization of the intracellular ART family (Table 2). Instead, PARP should be defined as a term in its own right to describe distinct members of the ART family, for example, PARP1 and PARP7 denote a poly- and a mono-ART, respectively. In addition to the nomenclature in eukaryotes, the terms ARTD/ARTC can be used to discern ARTs in other kingdoms. Thus, a main aspect of classification is that it categorizes the enzymes according to their evolutionary history and structural properties, that is, whether they possess a fold with greater similarity to diphtheria toxin or cholera toxin. This nomenclature does not rely on the ability of enzymes to modify substrates by either MARylation or PARylation. Also, the term ART is the biochemically correct term for these enzymes.
Based on these considerations, we define here the names and acronyms and make recommendations for what and how these should be used (Table 2). PARPx and TNKSx stand for distinct members of the ARTD family (Figs 1C and 2, Table 1). ARTC constitutes another family of ARTs, which in mammals is congruent with ecto-ARTs (Figs 1C and 2). In our view, this revised use of PARPx and TNKSx as names of ARTD family members should make this nomenclature acceptable to both PARP- and ARTD-ologists. It also avoids conflicts with the widespread use of the name PARP particularly also in the clinical setting. As the ARTD family contains enzymes that can either MARylate or PARylate substrates, poly-ARTs and mono-ARTs could be used to clarify their biochemical reaction. We suggest omitting abbreviations such as pART and mART as these terms are frequently found with different meanings (e.g., in PubMed). The consequences of this line of thought are that names, which are intrinsically illogical and may lead to confusion, can be avoided. For example, the term mono-PARP, which can be found in the literature, should not be used any longer, because if taken as acronym for poly-ADP-ribose-polymerase, a mono-PARP would then refer to a mono-poly-ADP-ribose-polymerase. Thus, PARP1 is a poly-ART, while PARP10 is a mono-ART. Moreover, the term tankyrase has been well established, describing the two related ARTD family members TNKS1 and 2. We therefore recommend that these names are used rather than PARP5a and PARP5b or ARTD5 and ARTD6.
An additional concern lies in the numbering of the members of the ARTD family of ARTs (Fig. 1C, Table 1). As both PARPx and ARTDx are currently being used in parallel, the different numbers of the enzymes are a source of confusion. The original concept that was endorsed and published in 2010 attempted to order the intracellular ART family members according to sequence similarities in the catalytic domains and additional structural domains [20]. Due to the wider use of PARPx compared to ARTDx, we suggest that the original numbering be maintained in combination with PARP (Table 1, Fig. 2).
Functional properties of mammalian ARTD and ARTC ADP-ribosyltransferases
The proteins of the mammalian ART superfamily are associated with an increasing number of activities (summarized in Fig. 3). Moreover, due to advances in mass spectrometry, chemical genetics, proximity labeling and other methods, novel substrates, and interactors are being identified at a breathtaking rate (for recent findings see Ref. [9,34,39]). While exciting, this comes with some reservation as in many cases we only have limited functional information about the consequences of ADP-ribosylation, particularly relevant for MARylation. Defining functions is a major task and challenge for upcoming studies. At the same time, to be able to use a common nomenclature for these proteins will be highly valuable for future communication, avoiding confusion and misunderstandings.
Fig. 3.
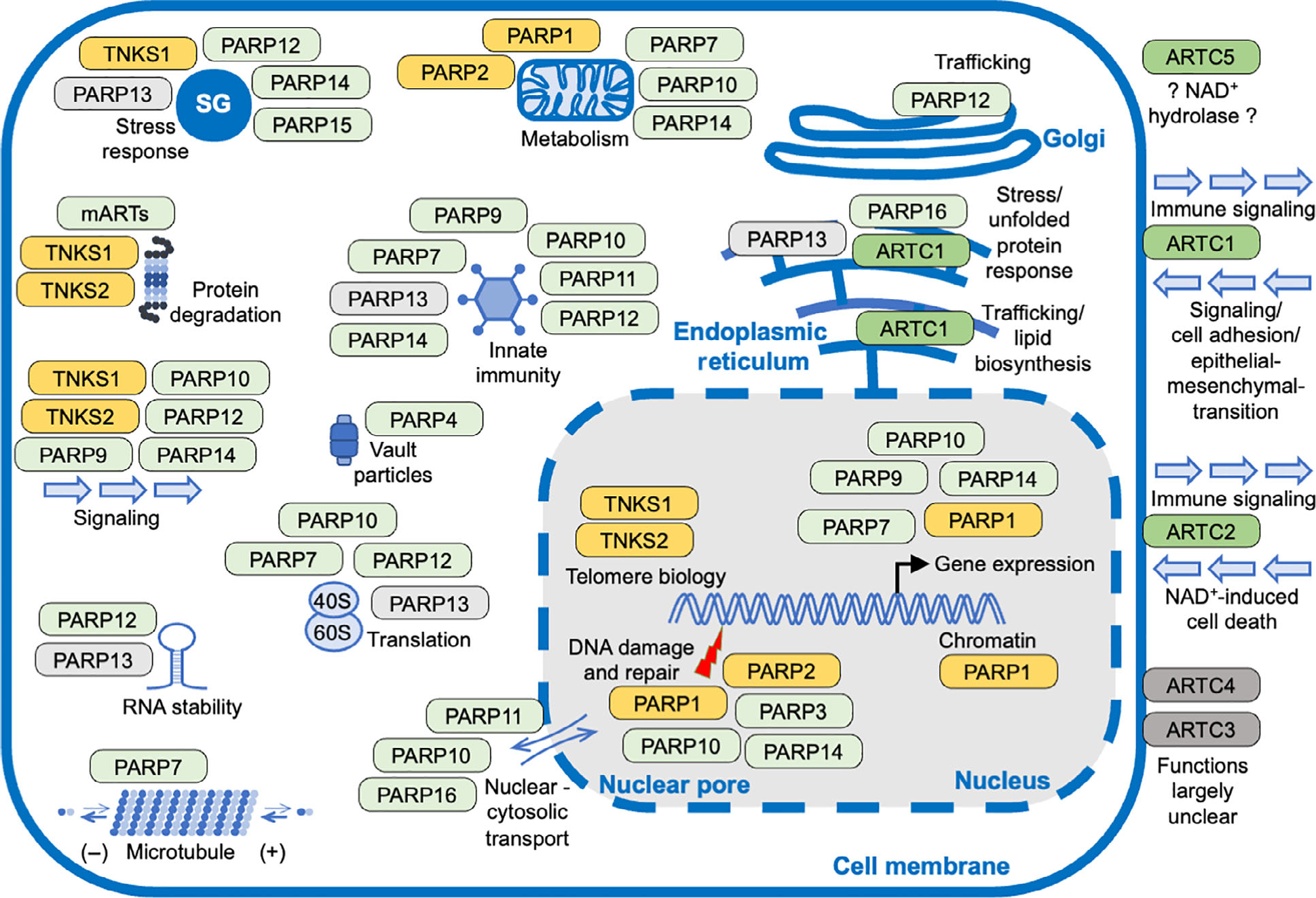
Summary of the functions of mammalian ART family members. The proteins belonging to the H-Y-[EDQ] clade (in lighter colors) and of the R-[ST]-E clade (in darker colors) are grouped together according to cellular processes in which they are proposed to be involved. The family members that are capable of synthesizing PAR chains are indicated in orange, those that MARylate substrates are in green, proteins for which so far no catalytic activity has been identified are in gray. While PARP and TNKS proteins are intracellular, ARTC proteins are translocated into the endoplasmic reticulum and transported to the plasma membrane. Intracellular and/or organelle associated functions are being discussed. ARTC1–4 are membrane associated through a GPI (glycosylphosphatidylinositol) anchor and ARTC5 is secreted. ARTC2 is not expressed in humans, but in other species. The figure gives an overview on important biological functions that appear to be regulated by ADP-ribosylation, it is not meant to be exhaustive. For details readers are referred to the cited, recent reviews. Please note that ARTC2 is not expressed in humans.
ARTD family members have been suggested to participate or to be associated with many different intracellular processes, located both in the nucleus as well as in the cytoplasm (Fig. 3). While numerous studies describe PARP1 as a key sensor that recognizes DNA damage and disseminates this information to regulate repair, cell cycle progression and cell death, among other consequences [40,41], the molecular functions of other family members are less well understood. Additional proteins have been identified that possess an ADP-ribosyltransferase domain classified as belonging to clade 1 [18]. One is tRNA 2’-phosphotransferase (TRPT1), an essential component of the fungal tRNA splicing machinery, while its role in mammalian cells is poorly defined [2]. Another clade 1 member is TASOR. So far, no catalytic activity has been associated with the ART domain of TASOR, which is part of the HUSH (human silencing hub) complex [42–44]. This complex is involved in methylating histone H3 lysine 9, a repressive chromatin mark [45]. Additional proteins include LRRC9, for which no catalytic activity has been reported so far, and NEURL4, which has been suggested to modify mitochondrial proteins [18,19,46]. Also of note is that PARP9, which was considered inactive, MARylates ubiquitin when in complex with DTX3L [47]. Recently, the RING-DTC domain of Deltex family E3 ubiquitin ligases was found to MARylate ubiquitin independently of PARP9 [48]. Finally, some sirtuins have been suggested to function as ADP-ribosyltransferases, although these activities have been debated [4]. The findings regarding DTX domain proteins and sirtuins suggest the existence of additional intracellular ART families that will need further exploration.
Members of the ARTC family or ecto-ARTs that are expressed in mammals are membrane bound facing the extracellular environment or are secreted [22–25]. Four family members are expressed in humans, six in mice. They regulate multiple activities including immune functions, signaling, and ER stress response (Fig. 3) [22,23]. Of note is that ARTC enzymes modify primarily arginines. While so far, no mammalian ARTD has been reported to possess a comparable target amino acid specificity, mass spectrometry studies indicate that arginine-ADP-ribosylation is abundant within cells [49–51], suggesting that one or more intracellular enzymes exist that modify arginine. ARTC1 appears to be predominantly located in the ER and may reach other membranous compartments in cells, possibly explaining the substantial arginine-ADP-ribosylation that can be detected intracellularly [22]. Further work is required to clarify the identity of the relevant enzyme(s).
Inhibitors of ADP-ribosyltransferases
Keeping with PARP as the name for most of the ARTD family members implies that PARP inhibitors (PARPi) are then substances that interfere with the functions of PARP enzymes. This is particularly relevant as in the clinical setting PARPi is an established term to describe inhibitors of PARP1, although the clinically approved inhibitors are not specific for PARP1 and also inhibit PARP2 and possibly other PARPs [52]. Importantly, PARP1/2 inhibitors are approved for the treatment of specific cancers, particularly those that are defective in homologous recombination-mediated repair, including tumors with mutations in the tumor suppressors BRCA1 or BRCA2, referred to as synthetic lethality [53–56]. Moreover, tankyrase inhibitors (TNKSi) are being developed as potential cancer therapeutics [57,58], defining an additional specific inhibitor entity associated with mammalian ARTD family members.
Recently, inhibitors of other PARP family members have been developed; these include inhibitors of PARP10 [59–61], PARP11 [62], and PARP14 [61,63,64]. Additionally, a selective inhibitor of PARP7 (RBN-2397) is in human clinical trials for treatment of patients with solid tumors (clinical trial: https://clinicaltrials.gov/ct2/show/NCT04053673). This is a significant development in the field because RBN-2397 is the first inhibitor of a MARylating enzyme in human clinical trials. While the terms PARPi and TNKSi will be helpful to define the two general subclasses of inhibitors, the recent disclosures of inhibitors of PARPs other than PARP1/2 and TNKS1/2 necessitate further specification for inhibitors. We suggest that the PARP family member name is used when describing an inhibitor with clear specificity; for example, a PARP7 selective inhibitor can be referred to as PARP7i.
As of now, the name PARPi implies inhibition of PARP1/2 but formally is not specific for these polymer forming enzymes. Also it does not imply how inhibitors impair protein function, be it via catalytic inhibition, via allosteric means or by interfering with protein-protein interactions, giving this term flexibility to also be used to describe entities such as PROTACs that could also eventually find their way into common experimental and/or clinical use [65–69].
Conclusions and consequences
Together, we suggest an updated, revised nomenclature of mammalian enzymes belonging to the ART superfamily. This nomenclature is based on the currently used names but also takes structural and biochemical functions of these proteins into consideration. In this, ARTD and ARTC define the two major families of the mammalian ART superfamily. The terms PARP and TNKS, which are widely used and very well established, stand for different ARTD members. In the context of the mammalian ART superfamily, we consider and thus propose PARP as a name on its own right, rather than an abbreviation for poly-ADP-ribose-polymerase. We are convinced that these modifications to the two existing versions of ART nomenclatures will unify and clarify how to describe and discuss the enzymes that ADP-ribosylate an ever-increasing number of substrates, and the processes that are controlled by this modification.
Acknowledgements
Funding for open access charge was supported by the German Research Foundation (LU466/16–2) and by core funding of the Institute of Biochemistry and Molecular Biology of RWTH Aachen University. JM was supported by the Intramural Research Program, NIH/NHLBI.
Abbreviations
- ADPr
ADP-ribose
- ARH
ADP-ribosylhydrolase/ADP-ribosyl-acceptor hydrolases/ADP-ribosyl-glycohydrolases
- ART
ADP-ribosyltransferase
- ARTC
ADP-ribosyltransferase cholera toxin-like
- ARTD
ADP-ribosyltransferase diphtheria toxin-like
- MAR
mono-ADP-ribose
- MARylation
mono-ADP-ribosylation
- NAD+
β-nicotinamide adenine dinucleotide
- PAR
poly-ADP-ribose
- PARP
suggested not to be used as abbreviation but rather as a term in its own right to describe distinct members of the ART family
- PARPi
PARP inhibitor
- PARylation
poly-ADP-ribosylation
- TNKS
tankyrase
Footnotes
Conflict of interest
AA is a co-founder of Tango Therapeutics, Azkarra Therapeutics, Ovibio Corporation; a consultant for SPARC, Bluestar, ProLynx, Earli, Cura, GenVivo, Ambagon, Phoenix Molecular Designs and GSK; a member of the SAB of Genentech, GLAdiator, Circle and Cambridge Science Corporation; receives grant/research support from SPARC and AstraZeneca; holds patents on the use of PARP inhibitors held jointly with AstraZeneca from which he has benefitted financially (and may do so in the future). PC is co-founder of Ribon Therapeutics, Inc. and ARase Therapeutics, Inc. WLK is a founder, consultant, and SAB member for Ribon Therapeutics, Inc. and ARase Therapeutics, Inc.
References
- 1.Kim DS, Challa S, Jones A & Kraus WL (2020) PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev 34, 302–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weixler L, Scharinger K, Momoh J, Luscher B, Feijs KLH & Zaja R (2021) ADP-ribosylation of RNA and DNA: from in vitro characterization to in vivo function. Nucleic Acids Res 49, 3634–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen MS & Chang P (2018) Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol 14, 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luscher B, Butepage M, Eckei L, Krieg S, Verheugd P & Shilton BH (2018) ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem Rev 118, 1092–1136. [DOI] [PubMed] [Google Scholar]
- 5.Szanto M & Bai P (2020) The role of ADP-ribose metabolism in metabolic regulation, adipose tissue differentiation, and metabolism. Genes Dev 34, 321–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abplanalp J & Hottiger MO (2017) Cell fate regulation by chromatin ADP-ribosylation. Semin Cell Dev Biol 63, 114–122. [DOI] [PubMed] [Google Scholar]
- 7.Hopp AK & Hottiger MO (2021) Uncovering the invisible: mono-ADP-ribosylation moved into the spotlight. Cells 10, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challa S, Stokes MS & Kraus WL (2021) MARTs and MARylation in the cytosol: biological functions, mechanisms of action, and therapeutic potential. Cells 10, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suskiewicz MJ, Palazzo L, Hughes R & Ahel I (2021) Progress and outlook in studying the substrate specificities of PARPs and related enzymes. FEBS J 288, 2131–2142. [DOI] [PubMed] [Google Scholar]
- 10.Rack JGM, Palazzo L & Ahel I (2020) (ADP-ribosyl) hydrolases: structure, function, and biology. Genes Dev 34, 263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Sullivan J, Tedim Ferreira M, Gagne JP, Sharma AK, Hendzel MJ, Masson JY & Poirier GG (2019) Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun 10, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teloni F & Altmeyer M (2016) Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res 44, 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupte R, Liu Z & Kraus WL (2017) PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31, 101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rack JG, Perina D & Ahel I (2016) Macrodomains: structure, function, evolution, and catalytic activities. Annu Rev Biochem 85, 431–454. [DOI] [PubMed] [Google Scholar]
- 15.Grimaldi G & Corda D (2019) ADP-ribosylation and intracellular traffic: an emerging role for PARP enzymes. Biochem Soc Trans 47, 357–370. [DOI] [PubMed] [Google Scholar]
- 16.Daniels CM, Ong SE & Leung AK (2015) The promise of proteomics for the study of ADP-ribosylation. Mol Cell 58, 911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyzewski Z, Gradowski M, Krysinska M, Dudkiewicz M & Pawlowski K (2021) A novel predicted ADP-ribosyltransferase-like family conserved in eukaryotic evolution. PeerJ 9, e11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L, Zhang D, de Souza RF, Anand S & Iyer LM (2015) The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr Top Microbiol Immunol 384, 3–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hottiger MO, Hassa PO, Luscher B, Schuler H & Koch-Nolte F (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35, 208–219. [DOI] [PubMed] [Google Scholar]
- 21.Gibson BA & Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13, 411–424. [DOI] [PubMed] [Google Scholar]
- 22.Di Girolamo M & Fabrizio G (2019) Overview of the mammalian ADP-ribosyl-transferases clostridia toxin-like (ARTCs) family. Biochem Pharmacol 167, 86–96. [DOI] [PubMed] [Google Scholar]
- 23.Rosado MM & Pioli C (2021) ADP-ribosylation in evasion, promotion and exacerbation of immune responses. Immunology. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seman M, Adriouch S, Haag F & Koch-Nolte F (2004) Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem 11, 857–872. [DOI] [PubMed] [Google Scholar]
- 25.Laing S, Unger M, Koch-Nolte F & Haag F (2011) ADP-ribosylation of arginine. Amino Acids 41, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayaishi O & Ueda K (1977) Poly(ADP-ribose) and ADP-ribosylation of proteins. Annu Rev Biochem 46, 95–116. [DOI] [PubMed] [Google Scholar]
- 27.Ueda K & Hayaishi O (1985) ADP-ribosylation. Annu Rev Biochem 54, 73–100. [DOI] [PubMed] [Google Scholar]
- 28.Hassa PO, Haenni SS, Elser M & Hottiger MO (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70, 789–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith S, Giriat I, Schmitt A & de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 30.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J & de Murcia G (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274, 17860–17868. [DOI] [PubMed] [Google Scholar]
- 31.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH & Luscher B (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32, 57–69. [DOI] [PubMed] [Google Scholar]
- 32.Marsischky GT, Wilson BA & Collier RJ (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem 270, 3247–3254. [DOI] [PubMed] [Google Scholar]
- 33.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I & Chang P (2014) Family-wide analysis of poly (ADP-ribose) polymerase activity. Nat Commun 5, 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez KM, Buch-Larsen SC, Kirby IT, Siordia IR, Hutin D, Rasmussen M, Grant DM, David LL, Matthews J, Nielsen ML et al. (2021) Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets. Elife 10, e60480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Paola S, Micaroni M, Di Tullio G, Buccione R & Di Girolamo M (2012) PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS One 7, e37352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catara G, Grimaldi G, Schembri L, Spano D, Turacchio G, Lo Monte M, Beccari AR, Valente C & Corda D (2017) PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci Rep 7, 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T & Helleday T (2010) PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem 285, 8054–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlberg T, Klepsch M, Thorsell AG, Andersson CD, Linusson A & Schuler H (2015) Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J Biol Chem 290, 7336–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buch-Larsen SC, Hendriks IA, Lodge JM, Rykaer M, Furtwangler B, Shishkova E, Westphall MS, Coon JJ & Nielsen ML (2020) Mapping physiological ADP-ribosylation using activated ion electron transfer dissociation. Cell Rep 32, 108176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisemann T & Pascal JM (2020) Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell Mol Life Sci 77, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Hernandez K, Rodriguez-Vargas JM, Schreiber V & Dantzer F (2016) Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin Cell Dev Biol 63, 92–101. [DOI] [PubMed] [Google Scholar]
- 42.Tchasovnikarova IA, Timms RT, Matheson NJ, Wals K, Antrobus R, Gottgens B, Dougan G, Dawson MA & Lehner PJ (2015) GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Wang GZ, Cingoz O & Goff SP (2018) NP220 mediates silencing of unintegrated retroviral DNA. Nature 564, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douse CH, Tchasovnikarova IA, Timms RT, Protasio AV, Seczynska M, Prigozhin DM, Albecka A, Wagstaff J, Williamson JC, Freund SMV et al. (2020) TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat Commun 11, 4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicetto D & Zaret KS (2019) Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr Opin Genet Dev 55, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardamone MD, Gao Y, Kwan J, Hayashi V, Sheeran M, Xu J, English J, Orofino J, Emili A & Perissi V (2020) ADP-ribosylation of mitochondrial proteins is mediated by neuralized-like protein 4 (NEURL4). bioRxiv 2020.12.28.424513 [PREPRINT]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang CS, Jividen K, Spencer A, Dworak N, Ni L, Oostdyk LT, Chatterjee M, Kusmider B, Reon B, Parlak M et al. (2017) Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol Cell 66, 503–516.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatrin C, Gabrielsen M, Buetow L, Nakasone MA, Ahmed SF, Sumpton D, Sibbet GJ, Smith BO & Huang DT (2020) Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases. Sci Adv 6, eabc0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leutert M, Menzel S, Braren R, Rissiek B, Hopp AK, Nowak K, Bisceglie L, Gehrig P, Li H, Zolkiewska A et al. (2018) Proteomic characterization of the heart and skeletal muscle reveals widespread arginine ADP-ribosylation by the ARTC1 ectoenzyme. Cell Rep 24, 1916–1929.e5. [DOI] [PubMed] [Google Scholar]
- 50.Hendriks IA, Larsen SC & Nielsen ML (2019) An advanced strategy for comprehensive profiling of ADP-ribosylation sites using mass spectrometry-based proteomics. Mol Cell Proteomics 18, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gehrig PM, Nowak K, Panse C, Leutert M, Grossmann J, Schlapbach R & Hottiger MO (2021) Gas-phase fragmentation of ADP-ribosylated peptides: arginine-specific side-chain losses and their implication in database searches. J Am Soc Mass Spectrom 32, 157–168. [DOI] [PubMed] [Google Scholar]
- 52.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D et al. (2012) Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol 30, 283–288. [DOI] [PubMed] [Google Scholar]
- 53.Faraoni I & Graziani G (2018) Role of BRCA mutations in cancer treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. Cancers (Basel) 10, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashworth A & Lord CJ (2018) Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol 15, 564–576. [DOI] [PubMed] [Google Scholar]
- 55.Lord CJ & Ashworth A (2017) PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrum AK, Vindigni A & Mosammaparast N (2019) Defining and modulating ‘BRCAness’. Trends Cell Biol 29, 740–751. [DOI] [PubMed] [Google Scholar]
- 57.Lehtio L, Chi NW & Krauss S (2013) Tankyrases as drug targets. FEBS J 280, 3576–3593. [DOI] [PubMed] [Google Scholar]
- 58.Mariotti L, Pollock K & Guettler S (2017) Regulation of Wnt/beta-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br J Pharmacol 174, 4611–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venkannagari H, Verheugd P, Koivunen J, Haikarainen T, Obaji E, Ashok Y, Narwal M, Pihlajaniemi T, Luscher B & Lehtio L (2016) Small-molecule chemical probe rescues cells from mono-ADP-ribosyltransferase ARTD10/PARP10-induced apoptosis and sensitizes cancer cells to DNA damage. Cell Chem Biol 23, 1251–1260. [DOI] [PubMed] [Google Scholar]
- 60.Morgan RK, Kirby IT, Vermehren-Schmaedick A, Rodriguez K & Cohen MS (2019) Rational design of cell-active inhibitors of PARP10. ACS Med Chem Lett 10, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holechek J, Lease R, Thorsell AG, Karlberg T, McCadden C, Grant R, Keen A, Callahan E, Schuler H & Ferraris D (2018) Design, synthesis and evaluation of potent and selective inhibitors of mono-(ADP-ribosyl)transferases PARP10 and PARP14. Bioorg Med Chem Lett 28, 2050–2054. [DOI] [PubMed] [Google Scholar]
- 62.Kirby IT, Kojic A, Arnold MR, Thorsell AG, Karlberg T, Vermehren-Schmaedick A, Sreenivasan R, Schultz C, Schuler H & Cohen MS (2018) A potent and selective PARP11 inhibitor suggests coupling between cellular localization and catalytic activity. Cell Chem Biol 25, 1547–1553.e12. [DOI] [PubMed] [Google Scholar]
- 63.Upton K, Meyers M, Thorsell AG, Karlberg T, Holechek J, Lease R, Schey G, Wolf E, Lucente A, Schuler H et al. (2017) Design and synthesis of potent inhibitors of the mono(ADP-ribosyl)transferase, PARP14. Bioorg Med Chem Lett 27, 2907–2911. [DOI] [PubMed] [Google Scholar]
- 64.Schenkel LB, Molina JR, Swinger KK, Abo R, Blackwell DJ, Lu AZ, Cheung AE, Church WD, Kunii K, Kuplast-Barr KG et al. (2021) A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants. Cell Chem Biol. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 65.Chamberlain PP & Hamann LG (2019) Development of targeted protein degradation therapeutics. Nat Chem Biol 15, 937–944. [DOI] [PubMed] [Google Scholar]
- 66.Wigle TJ, Ren Y, Molina JR, Blackwell DJ, Schenkel LB, Swinger KK, Kuplast-Barr K, Majer CR, Church WD, Lu AZ et al. (2021) Targeted degradation of PARP14 using a heterobifunctional small molecule. ChemBioChem 22, 2107–2110. [DOI] [PubMed] [Google Scholar]
- 67.Steffen JD, Tholey RM, Langelier MF, Planck JL, Schiewer MJ, Lal S, Bildzukewicz NA, Yeo CJ, Knudsen KE, Brody JR et al. (2014) Targeting PARP-1 allosteric regulation offers therapeutic potential against cancer. Cancer Res 74, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pollock K, Liu M, Zaleska M, Meniconi M, Pfuhl M, Collins I & Guettler S (2019) Fragment-based screening identifies molecules targeting the substrate-binding ankyrin repeat domains of tankyrase. Sci Rep 9, 19130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sowa ST, Vela-Rodriguez C, Galera-Prat A, Cazares-Olivera M, Prunskaite-Hyyrylainen R, Ignatev A & Lehtio L (2020) A FRET-based high-throughput screening platform for the discovery of chemical probes targeting the scaffolding functions of human tankyrases. Sci Rep 10, 12357. [DOI] [PMC free article] [PubMed] [Google Scholar]


