The title compound features a polarized π-system due to resonance between the N—C(H)=C(H)—B and ionic N+=C(H)—C(H)=B− canonical forms.
Keywords: crystal structure, dioxaborolan-2-yl, resonance
Abstract
The title compound, C20H24BNO2, has a polarized π-system due to significant resonance between the N—C(H)=C(H)—B and ionic N+=C(H)—C(H)=B− canonical forms. The dihedral angles between the NC2B plane (r.m.s. deviation 0.0223 Å) and the C3N (r.m.s. deviation 0.0025 Å) and BCO2 (r.m.s. deviation 0.0044 Å) planes are 2.51 (12) and 3.09 (19)°, respectively. This indicates the lone pair of the nitrogen atom and a vacant p orbital of the boron atom are conjugated with the central C=C bond. In comparison with the carbazole analogue [Hatayama & Okuno (2012 ▸). Acta Cryst. E68, o84], the C—N and C—B bonds are shorter. The results are well explained by the increase in the contribution of the N+=C(H)—C(H)=B− canonical form in the title compound.
Structure description
The title compound, C20H24BNO2, has a hybrid π-conjugated system within the N—C(H)=C(H)—B fragment. The insertion of a π-conjugated system in the N—B bond affords a highly polarized π-system owing to the contribution of an ionic canonical structure, i.e. N+= C(H)—C(H)=B−. The contribution of the ionic canonical structure is small when p-phenylene is inserted into the N—B bond (Yuan et al., 2006 ▸). However, when a C≡C bond is inserted into the N—B bond (Onuma et al., 2015 ▸), a relatively large contribution of the ionic canonical structure is apparent. The structure of the C=C bond inserted system, namely 9-[(E)-2-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)ethenyl]-9H-carbazole has been reported (Hatayama & Okuno, 2012 ▸). In the title compound, the carbazole unit of the former is replaced by a diphenylamino residue (Fig. 1 ▸).
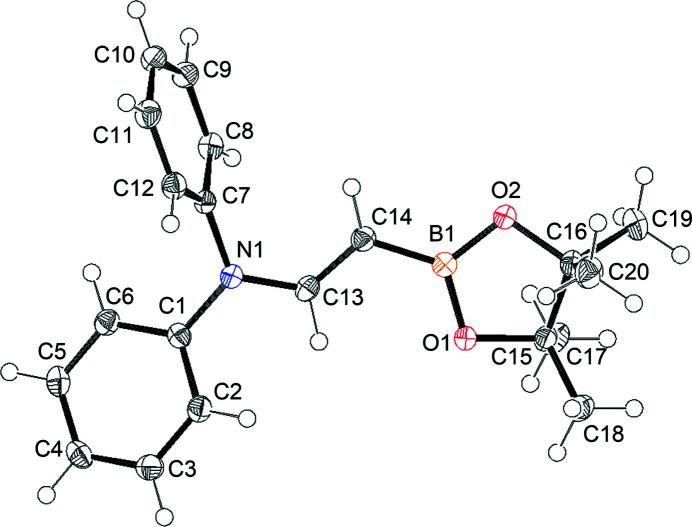
Figure 1.
The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level; H are atoms shown as small spheres.
The dihedral angles between the C13/C14/B1/N1 plane (r.m.s. deviation 0.0223 Å) and the N1/C1/C7/C13 (r.m.s. deviation 0.0025 Å) and B1/O1/O2/C14 (r.m.s. deviation 0.0044 Å) planes are 2.51 (12) and 3.09 (19)°, respectively, indicating the lone pair of the nitrogen atom and a vacant p orbital of the boron are conjugated with the central C=C bond. The C13—N1 [1.3824 (19) Å] and C14—B1 [1.532 (2) Å] bonds are shortened, compared with those in the carbazole analogue of 1.396 (3) Å and 1.537 (3) Å, respectively; the central C=C bond at 1.341 (2) Å is experimentally equivalent to that of 1.336 (4) Å in the carbazolyl derivative. The results are well explained by the increase in the contribution of the N+=C(H)—C(H)=B− canonical structure in the title compound. This is presumably because the nitrogen atom of diphenylamino group donates its lone pair to the π-system more effectively compared to that of the carbazolyl group, which leads to a decrease in the contribution of the N+=C(H)—C(H)=B− canonical structure in the latter.
Synthesis and crystallization
The title compound was obtained by hydroboration of N-ethynyl-N-phenylaniline (Tokutome & Okuno, 2013 ▸) with 4,4,5,5-tetramethyl-1,3,2-dioxaborolane in 16% yield. 1H NMR (CDCl3): δ 1.25 (s, 12H), 4.17 (d, J = 15.6 Hz, 1H), 7.07 (d, J = 7.7 Hz, 4H), 7.12 (t, J = 7.7 Hz, 2H), 7.31 (t, J = 7.7 Hz, 4H), 7.64 (d, J = 15.6 Hz, 1H).
Single crystals were obtained by recrystallization from hexane solution.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 1 ▸.
Table 1. Experimental details.
| Crystal data | |
| Chemical formula | C20H24BNO2 |
| M r | 321.23 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 93 |
| a, b, c (Å) | 32.071 (11), 6.011 (2), 22.219 (8) |
| β (°) | 122.590 (4) |
| V (Å3) | 3609 (2) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.07 |
| Crystal size (mm) | 0.13 × 0.11 × 0.05 |
| Data collection | |
| Diffractometer | Rigaku Saturn724+ |
| Absorption correction | Numerical (NUMABS; Rigaku, 1999 ▸) |
| T min, T max | 0.991, 0.996 |
| No. of measured, independent and observed [F 2 > 2.0σ(F 2)] reflections | 13892, 3869, 3041 |
| R int | 0.084 |
| (sin θ/λ)max (Å−1) | 0.639 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.052, 0.147, 1.09 |
| No. of reflections | 3869 |
| No. of parameters | 217 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.25, −0.27 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314622000839/tk4073sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622000839/tk4073Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622000839/tk4073Isup3.cml
CCDC reference: 2129837
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C20H24BNO2 | F(000) = 1376.00 |
| Mr = 321.23 | Dx = 1.182 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71075 Å |
| a = 32.071 (11) Å | Cell parameters from 5341 reflections |
| b = 6.011 (2) Å | θ = 1.5–31.1° |
| c = 22.219 (8) Å | µ = 0.07 mm−1 |
| β = 122.590 (4)° | T = 93 K |
| V = 3609 (2) Å3 | Prism, colourless |
| Z = 8 | 0.13 × 0.11 × 0.05 mm |
Data collection
| Rigaku Saturn724+ diffractometer | 3041 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 7.111 pixels mm-1 | Rint = 0.084 |
| ω scans | θmax = 27.0°, θmin = 1.5° |
| Absorption correction: numerical (NUMABS; Rigaku, 1999) | h = −33→40 |
| Tmin = 0.991, Tmax = 0.996 | k = −7→7 |
| 13892 measured reflections | l = −28→26 |
| 3869 independent reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.147 | H-atom parameters constrained |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0758P)2 + 0.8943P] where P = (Fo2 + 2Fc2)/3 |
| 3869 reflections | (Δ/σ)max < 0.001 |
| 217 parameters | Δρmax = 0.25 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt).The C-bound H atoms were placed at ideal positions and were refined as riding on their parent C atoms. The Uiso(H) values were set at 1.2Ueq(Csp2) and 1.5 Ueq(Csp3). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.34742 (4) | 0.98270 (18) | 0.51068 (5) | 0.0221 (3) | |
| O2 | 0.41736 (4) | 0.82168 (18) | 0.60407 (5) | 0.0218 (3) | |
| N1 | 0.36028 (5) | 0.6152 (2) | 0.35305 (7) | 0.0199 (3) | |
| C1 | 0.32337 (5) | 0.6584 (3) | 0.28043 (8) | 0.0190 (3) | |
| C2 | 0.29881 (6) | 0.8631 (3) | 0.25973 (8) | 0.0218 (3) | |
| H2 | 0.3090 | 0.9801 | 0.2936 | 0.026* | |
| C3 | 0.25961 (6) | 0.8955 (3) | 0.18984 (8) | 0.0242 (4) | |
| H3 | 0.2427 | 1.0342 | 0.1764 | 0.029* | |
| C4 | 0.24475 (6) | 0.7281 (3) | 0.13936 (8) | 0.0242 (4) | |
| H4 | 0.2173 | 0.7497 | 0.0919 | 0.029* | |
| C5 | 0.27059 (6) | 0.5283 (3) | 0.15912 (8) | 0.0239 (4) | |
| H5 | 0.2613 | 0.4146 | 0.1244 | 0.029* | |
| C6 | 0.30963 (6) | 0.4925 (3) | 0.22861 (8) | 0.0217 (3) | |
| H6 | 0.3271 | 0.3555 | 0.2412 | 0.026* | |
| C7 | 0.39973 (5) | 0.4638 (2) | 0.36945 (8) | 0.0186 (3) | |
| C8 | 0.40344 (6) | 0.2606 (3) | 0.40195 (8) | 0.0220 (3) | |
| H8 | 0.3798 | 0.2207 | 0.4133 | 0.026* | |
| C9 | 0.44182 (6) | 0.1166 (3) | 0.41769 (8) | 0.0237 (3) | |
| H9 | 0.4446 | −0.0221 | 0.4401 | 0.028* | |
| C10 | 0.47629 (6) | 0.1748 (3) | 0.40080 (8) | 0.0246 (4) | |
| H10 | 0.5025 | 0.0757 | 0.4115 | 0.030* | |
| C11 | 0.47247 (6) | 0.3772 (3) | 0.36832 (8) | 0.0248 (4) | |
| H11 | 0.4960 | 0.4166 | 0.3567 | 0.030* | |
| C12 | 0.43426 (6) | 0.5225 (3) | 0.35273 (8) | 0.0226 (3) | |
| H12 | 0.4317 | 0.6617 | 0.3307 | 0.027* | |
| C13 | 0.35865 (6) | 0.7131 (2) | 0.40803 (8) | 0.0194 (3) | |
| H13 | 0.3305 | 0.8029 | 0.3942 | 0.023* | |
| C14 | 0.39193 (6) | 0.6968 (3) | 0.47847 (8) | 0.0206 (3) | |
| H14 | 0.4197 | 0.6011 | 0.4957 | 0.025* | |
| C15 | 0.34973 (6) | 1.0522 (3) | 0.57530 (8) | 0.0210 (3) | |
| C16 | 0.40496 (6) | 1.0063 (3) | 0.63455 (8) | 0.0226 (3) | |
| C17 | 0.31353 (6) | 0.9064 (3) | 0.58176 (10) | 0.0296 (4) | |
| H17A | 0.3140 | 0.9481 | 0.6247 | 0.036* | |
| H17B | 0.2801 | 0.9272 | 0.5396 | 0.036* | |
| H17C | 0.3232 | 0.7500 | 0.5851 | 0.036* | |
| C18 | 0.33465 (6) | 1.2945 (3) | 0.56815 (9) | 0.0261 (4) | |
| H18A | 0.3361 | 1.3421 | 0.6115 | 0.031* | |
| H18B | 0.3573 | 1.3857 | 0.5616 | 0.031* | |
| H18C | 0.3008 | 1.3124 | 0.5267 | 0.031* | |
| C19 | 0.41396 (7) | 0.9327 (3) | 0.70589 (9) | 0.0344 (4) | |
| H19A | 0.4058 | 1.0548 | 0.7270 | 0.041* | |
| H19B | 0.3931 | 0.8038 | 0.6988 | 0.041* | |
| H19C | 0.4488 | 0.8920 | 0.7380 | 0.041* | |
| C20 | 0.43914 (6) | 1.1987 (3) | 0.64438 (10) | 0.0311 (4) | |
| H20A | 0.4321 | 1.3267 | 0.6648 | 0.037* | |
| H20B | 0.4737 | 1.1530 | 0.6767 | 0.037* | |
| H20C | 0.4336 | 1.2400 | 0.5980 | 0.037* | |
| B1 | 0.38529 (6) | 0.8323 (3) | 0.53116 (9) | 0.0188 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0242 (6) | 0.0246 (6) | 0.0168 (6) | 0.0045 (4) | 0.0105 (5) | −0.0009 (4) |
| O2 | 0.0246 (6) | 0.0227 (6) | 0.0180 (6) | 0.0047 (4) | 0.0114 (5) | −0.0001 (4) |
| N1 | 0.0220 (7) | 0.0207 (6) | 0.0176 (6) | 0.0024 (5) | 0.0110 (6) | −0.0006 (5) |
| C1 | 0.0176 (7) | 0.0236 (8) | 0.0171 (7) | −0.0014 (6) | 0.0101 (6) | 0.0001 (6) |
| C2 | 0.0258 (8) | 0.0213 (8) | 0.0214 (8) | −0.0001 (6) | 0.0149 (7) | −0.0009 (6) |
| C3 | 0.0248 (8) | 0.0283 (8) | 0.0242 (8) | 0.0046 (6) | 0.0164 (7) | 0.0051 (6) |
| C4 | 0.0211 (8) | 0.0338 (9) | 0.0172 (8) | −0.0005 (6) | 0.0101 (7) | 0.0024 (6) |
| C5 | 0.0247 (8) | 0.0298 (8) | 0.0201 (8) | −0.0048 (6) | 0.0139 (7) | −0.0042 (6) |
| C6 | 0.0239 (8) | 0.0219 (8) | 0.0233 (8) | 0.0005 (6) | 0.0154 (7) | −0.0004 (6) |
| C7 | 0.0178 (7) | 0.0201 (7) | 0.0159 (7) | 0.0012 (6) | 0.0077 (6) | −0.0024 (6) |
| C8 | 0.0215 (8) | 0.0241 (8) | 0.0224 (8) | −0.0025 (6) | 0.0132 (7) | −0.0023 (6) |
| C9 | 0.0256 (8) | 0.0209 (8) | 0.0231 (8) | 0.0005 (6) | 0.0121 (7) | −0.0002 (6) |
| C10 | 0.0215 (8) | 0.0267 (8) | 0.0226 (8) | 0.0024 (6) | 0.0099 (7) | −0.0038 (6) |
| C11 | 0.0216 (8) | 0.0312 (9) | 0.0244 (8) | −0.0018 (6) | 0.0143 (7) | −0.0035 (7) |
| C12 | 0.0253 (8) | 0.0236 (8) | 0.0206 (8) | −0.0007 (6) | 0.0135 (7) | −0.0002 (6) |
| C13 | 0.0210 (8) | 0.0183 (7) | 0.0241 (8) | −0.0015 (6) | 0.0155 (7) | −0.0021 (6) |
| C14 | 0.0202 (8) | 0.0212 (7) | 0.0216 (8) | 0.0012 (6) | 0.0121 (7) | −0.0011 (6) |
| C15 | 0.0252 (8) | 0.0207 (7) | 0.0203 (8) | 0.0021 (6) | 0.0143 (7) | −0.0009 (6) |
| C16 | 0.0272 (8) | 0.0227 (8) | 0.0195 (8) | 0.0014 (6) | 0.0136 (7) | −0.0011 (6) |
| C17 | 0.0329 (9) | 0.0244 (8) | 0.0404 (10) | −0.0022 (7) | 0.0256 (8) | −0.0032 (7) |
| C18 | 0.0313 (9) | 0.0214 (8) | 0.0286 (9) | 0.0041 (6) | 0.0182 (8) | 0.0009 (6) |
| C19 | 0.0430 (10) | 0.0406 (10) | 0.0193 (8) | 0.0060 (8) | 0.0166 (8) | 0.0008 (7) |
| C20 | 0.0280 (9) | 0.0308 (9) | 0.0300 (9) | −0.0045 (7) | 0.0127 (8) | −0.0091 (7) |
| B1 | 0.0193 (8) | 0.0187 (8) | 0.0203 (9) | −0.0020 (6) | 0.0118 (7) | −0.0007 (6) |
Geometric parameters (Å, º)
| O1—B1 | 1.380 (2) | C10—H10 | 0.9500 |
| O1—C15 | 1.4585 (18) | C11—C12 | 1.388 (2) |
| O2—B1 | 1.375 (2) | C11—H11 | 0.9500 |
| O2—C16 | 1.4623 (18) | C12—H12 | 0.9500 |
| N1—C13 | 1.3824 (19) | C13—C14 | 1.341 (2) |
| N1—C1 | 1.419 (2) | C13—H13 | 0.9500 |
| N1—C7 | 1.4369 (19) | C14—B1 | 1.532 (2) |
| C1—C2 | 1.398 (2) | C14—H14 | 0.9500 |
| C1—C6 | 1.403 (2) | C15—C18 | 1.516 (2) |
| C2—C3 | 1.388 (2) | C15—C17 | 1.522 (2) |
| C2—H2 | 0.9500 | C15—C16 | 1.560 (2) |
| C3—C4 | 1.386 (2) | C16—C19 | 1.516 (2) |
| C3—H3 | 0.9500 | C16—C20 | 1.526 (2) |
| C4—C5 | 1.390 (2) | C17—H17A | 0.9800 |
| C4—H4 | 0.9500 | C17—H17B | 0.9800 |
| C5—C6 | 1.384 (2) | C17—H17C | 0.9800 |
| C5—H5 | 0.9500 | C18—H18A | 0.9800 |
| C6—H6 | 0.9500 | C18—H18B | 0.9800 |
| C7—C12 | 1.390 (2) | C18—H18C | 0.9800 |
| C7—C8 | 1.391 (2) | C19—H19A | 0.9800 |
| C8—C9 | 1.387 (2) | C19—H19B | 0.9800 |
| C8—H8 | 0.9500 | C19—H19C | 0.9800 |
| C9—C10 | 1.390 (2) | C20—H20A | 0.9800 |
| C9—H9 | 0.9500 | C20—H20B | 0.9800 |
| C10—C11 | 1.386 (2) | C20—H20C | 0.9800 |
| B1—O1—C15 | 107.10 (11) | N1—C13—H13 | 116.0 |
| B1—O2—C16 | 107.02 (12) | C13—C14—B1 | 120.13 (14) |
| C13—N1—C1 | 121.41 (13) | C13—C14—H14 | 119.9 |
| C13—N1—C7 | 119.53 (13) | B1—C14—H14 | 119.9 |
| C1—N1—C7 | 119.05 (12) | O1—C15—C18 | 109.14 (12) |
| C2—C1—C6 | 118.90 (14) | O1—C15—C17 | 106.93 (13) |
| C2—C1—N1 | 120.80 (14) | C18—C15—C17 | 110.29 (13) |
| C6—C1—N1 | 120.25 (14) | O1—C15—C16 | 102.26 (12) |
| C3—C2—C1 | 120.12 (15) | C18—C15—C16 | 114.30 (13) |
| C3—C2—H2 | 119.9 | C17—C15—C16 | 113.29 (13) |
| C1—C2—H2 | 119.9 | O2—C16—C19 | 108.47 (13) |
| C4—C3—C2 | 120.84 (15) | O2—C16—C20 | 106.72 (13) |
| C4—C3—H3 | 119.6 | C19—C16—C20 | 110.72 (14) |
| C2—C3—H3 | 119.6 | O2—C16—C15 | 102.24 (12) |
| C3—C4—C5 | 119.03 (15) | C19—C16—C15 | 115.10 (14) |
| C3—C4—H4 | 120.5 | C20—C16—C15 | 112.83 (13) |
| C5—C4—H4 | 120.5 | C15—C17—H17A | 109.5 |
| C6—C5—C4 | 120.93 (15) | C15—C17—H17B | 109.5 |
| C6—C5—H5 | 119.5 | H17A—C17—H17B | 109.5 |
| C4—C5—H5 | 119.5 | C15—C17—H17C | 109.5 |
| C5—C6—C1 | 120.06 (15) | H17A—C17—H17C | 109.5 |
| C5—C6—H6 | 120.0 | H17B—C17—H17C | 109.5 |
| C1—C6—H6 | 120.0 | C15—C18—H18A | 109.5 |
| C12—C7—C8 | 120.28 (14) | C15—C18—H18B | 109.5 |
| C12—C7—N1 | 119.38 (14) | H18A—C18—H18B | 109.5 |
| C8—C7—N1 | 120.34 (13) | C15—C18—H18C | 109.5 |
| C9—C8—C7 | 119.68 (14) | H18A—C18—H18C | 109.5 |
| C9—C8—H8 | 120.2 | H18B—C18—H18C | 109.5 |
| C7—C8—H8 | 120.2 | C16—C19—H19A | 109.5 |
| C8—C9—C10 | 120.11 (15) | C16—C19—H19B | 109.5 |
| C8—C9—H9 | 119.9 | H19A—C19—H19B | 109.5 |
| C10—C9—H9 | 119.9 | C16—C19—H19C | 109.5 |
| C11—C10—C9 | 120.09 (15) | H19A—C19—H19C | 109.5 |
| C11—C10—H10 | 120.0 | H19B—C19—H19C | 109.5 |
| C9—C10—H10 | 120.0 | C16—C20—H20A | 109.5 |
| C10—C11—C12 | 120.06 (15) | C16—C20—H20B | 109.5 |
| C10—C11—H11 | 120.0 | H20A—C20—H20B | 109.5 |
| C12—C11—H11 | 120.0 | C16—C20—H20C | 109.5 |
| C11—C12—C7 | 119.77 (15) | H20A—C20—H20C | 109.5 |
| C11—C12—H12 | 120.1 | H20B—C20—H20C | 109.5 |
| C7—C12—H12 | 120.1 | O2—B1—O1 | 112.71 (13) |
| C14—C13—N1 | 127.95 (14) | O2—B1—C14 | 123.48 (14) |
| C14—C13—H13 | 116.0 | O1—B1—C14 | 123.79 (14) |
| C13—N1—C1—C2 | 30.2 (2) | C1—N1—C13—C14 | −176.36 (15) |
| C7—N1—C1—C2 | −150.72 (14) | C7—N1—C13—C14 | 4.5 (2) |
| C13—N1—C1—C6 | −147.37 (14) | N1—C13—C14—B1 | 175.57 (14) |
| C7—N1—C1—C6 | 31.7 (2) | B1—O1—C15—C18 | 145.27 (13) |
| C6—C1—C2—C3 | 3.7 (2) | B1—O1—C15—C17 | −95.43 (14) |
| N1—C1—C2—C3 | −173.86 (13) | B1—O1—C15—C16 | 23.86 (15) |
| C1—C2—C3—C4 | −1.1 (2) | B1—O2—C16—C19 | 146.11 (14) |
| C2—C3—C4—C5 | −1.8 (2) | B1—O2—C16—C20 | −94.56 (14) |
| C3—C4—C5—C6 | 2.1 (2) | B1—O2—C16—C15 | 24.11 (14) |
| C4—C5—C6—C1 | 0.6 (2) | O1—C15—C16—O2 | −28.89 (14) |
| C2—C1—C6—C5 | −3.4 (2) | C18—C15—C16—O2 | −146.69 (13) |
| N1—C1—C6—C5 | 174.15 (13) | C17—C15—C16—O2 | 85.81 (15) |
| C13—N1—C7—C12 | −113.40 (16) | O1—C15—C16—C19 | −146.25 (14) |
| C1—N1—C7—C12 | 67.48 (19) | C18—C15—C16—C19 | 95.96 (17) |
| C13—N1—C7—C8 | 66.41 (19) | C17—C15—C16—C19 | −31.54 (19) |
| C1—N1—C7—C8 | −112.71 (16) | O1—C15—C16—C20 | 85.36 (15) |
| C12—C7—C8—C9 | 0.0 (2) | C18—C15—C16—C20 | −32.44 (18) |
| N1—C7—C8—C9 | −179.76 (14) | C17—C15—C16—C20 | −159.93 (13) |
| C7—C8—C9—C10 | −0.2 (2) | C16—O2—B1—O1 | −10.22 (17) |
| C8—C9—C10—C11 | 0.2 (2) | C16—O2—B1—C14 | 168.01 (14) |
| C9—C10—C11—C12 | 0.1 (2) | C15—O1—B1—O2 | −9.76 (17) |
| C10—C11—C12—C7 | −0.3 (2) | C15—O1—B1—C14 | 172.01 (14) |
| C8—C7—C12—C11 | 0.3 (2) | C13—C14—B1—O2 | 178.88 (14) |
| N1—C7—C12—C11 | −179.94 (13) | C13—C14—B1—O1 | −3.1 (2) |
References
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Hatayama, Y. & Okuno, T. (2012). Acta Cryst. E68, o84. [DOI] [PMC free article] [PubMed]
- Onuma, K., Suzuki, K. & Yamashita, M. (2015). Chem. Lett. 44, 405–407.
- Rigaku (1999). NUMABS. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2008). CrystalClear. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2019). CrystalStructure. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tokutome, Y. & Okuno, T. (2013). J. Mol. Struct. 1047, 136–142.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yuan, Z., Entwistle, C. D., Collings, J. C., Albesa-Jové, D., Batsanov, A. S., Howard, J. A. K., Taylor, N. J., Kaiser, H. M., Kaufmann, D. E., Poon, S. Y., Wong, W. Y., Jardin, C., Fathallah, S., Boucekkine, A., Halet, J. F. & Marder, T. B. (2006). Chem. Eur. J. 12, 2758–2771. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314622000839/tk4073sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314622000839/tk4073Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314622000839/tk4073Isup3.cml
CCDC reference: 2129837
Additional supporting information: crystallographic information; 3D view; checkCIF report