Abstract
BACKGROUND:
Sleep staging is an important part of sleep research. Traditional automatic sleep staging based on machine learning requires extensive feature extraction and selection.
OBJECTIVE:
This paper proposed a deep learning algorithm without feature extraction based on one-dimensional convolutional neural network and long short-term memory.
METHODS:
The algorithm can automatically divide sleep into 5 phases including awake period, non-rapid eye movement sleep period (N1 N3) and rapid eye movement using the electroencephalogram signals. The raw signal was processed by the wavelet transform. Then, the processed signal was directly input into the deep learning algorithm to obtain the staging result.
RESULTS:
The accuracy of staging is 93.47% using the Fpz-Cz electroencephalogram signal. When using the Fpz-Cz and electroencephalogram signal, the algorithm can obtain the highest accuracy of 94.15%.
CONCLUSION:
These results show that this algorithm is suitable for different physiological signals and can realize end-to-end automatic sleep staging without any manual feature extraction.
Keywords: Sleep staging, deep learning, one-dimensional convolutional neural network, long short-term memory
1. Introduction
Sleep is an indispensable part of the body’s recovery and integration, and consolidation of memory. However, the increasing social pressure has caused sleep problems in most of the population, which may reduce the quality of life [1]. In the study of sleep quality and the diagnosis of sleep disorders, accurate sleep staging is the basis and also is a challenge.
The sleep process contains awake, non-rapid eye movement (NREM) and rapid eye movement (REM). Awake is a period before sleep. NREM consists of three stages: N1 N3 [2]. During the N1 period, the brain activity slows down with the relaxed muscles. The eye movement also slows down. Brain waves gradually become irregular. During the N2 period, the eye movements stop. The frequency of brain waves is only 1 2 Hz, but their amplitude is greatly increasing, showing a slowly changing curve in deep sleep stages. During REM period, the brain waves contain high-frequency and low-amplitude components with distinctive jagged waves. The eyes move rapidly with closed eyelids [3].
Sleep experts usually use polysomnography (PSG) to record electrical activities in different parts of the body [4]. They usually divide the signal into a series of small segments, each of which is 30 seconds long. By analyzing the characteristics of the segment, we could understand which sleep stage this segment belongs to. This process is called automatic sleep staging.
To improve the accuracy of automatic sleep staging, the researchers proposed various physiological signals to classify the sleep stages, such as EEG, ECG, EMG, EOG, blood oxygen, breathing, and so on [5, 6, 7, 8]. Zhang et al. chose fuzzy support vector machine method to extract temporal and spectral features of EEG, EMG and EOG, getting an average accuracy of 82.53% [9]. Gaiduk et al. derived an accuracy of 72% for wake, NREM, REM from three signals (respiratory, heart rate and movement) [10]. Noviyanto et al. applied both the respiratory parameters and ECG to sleep staging, achieving an accuracy of up to 79.78% [11]. In general, EEG signals are the most used, followed by ECG signals, then EOG signals [12, 13, 14, 15, 16].
The main steps of automatic sleep staging include signal preprocessing, feature extraction, feature selection, and classification. Traditionally, preprocessing methods have wavelet transform [17], principal component analysis (PCA) [18], independent component analysis (ICA) [19] and so on. Among them, wavelet transform is the most frequently used. After the preprocessing, many features are extracted including time domain, frequency domain and nonlinear features [20]. Because traditional methods fail to deal with the increasingly complex physiological signals, the criteria for feature selection and judgment in sleep staging are still unclear. Hence, more advanced signal-processing methods and machine learning algorithms have been employed [21, 22, 23].
To avoid the dependence on features, researchers chose deep learning for features classification. Moreover, deep learning has been employed on some challenging fields such as computer-based assessment of cardiac ECG data [24, 25, 26], detection of neurological disorders using EEG signals [27, 28]. Hassan et al. evaluated the classification effect of multiple classifiers, and concluded that these classifiers achieved an accuracy of 61.20% [29]. Längkvist et al. used deep brief network (DBN) to extract features from EEG, EMG and EOG with the accuracy of 67.4% [30]. Charbonnier et al. compared the staging effect of DBN and sparse auto-encoders (SAE) in automatic sleep staging, with the classification results of 72.2% and 77.7% respectively [31]. Phan et al. used convolutional neural network (CNN) to classify the time-frequency features which were manually extracted from the original signal EEG [32] Acharya et al. used 13 layers CNN to distinguish the class of EEG signals, and the accuracy was 88.7% [33]. Yildirim et al. extracted the features of EEG and EOG signals by 1D-CNN neural network, and obtained 91% accuracy [34]. Yulita et al. used the fast-convolutional method to extract features and softmax classifier for automatic sleep stage classification, with the accuracy of 84% [35]. Dong et al. used LSTM to perform automatic sleep staging on the EEG signals, with the accuracy of 78.94%–83.60% [36]. Michielli et al. proposed a cascaded LSTM architecture for automated sleep stage classification using single-channel EEG signals [37].
In this paper, to reduce the workload of feature processing and improve the accuracy of staging, a new deep learning algorithm based on 1D-CNN and LSTM is proposed for automatic sleep staging. This algorithm is an end-to-end structure. There are no hand-crafted features used for sleep staging. Besides, different physiological signals are tested. The accuracy of the algorithm is verified using a uniform preprocessing method. Multi-group test comparison is performed without changing any layer parameters of the deep learning algorithm to verify the generalization ability of the algorithm.
Based on the above research, different physiological signals are denoised by wavelet transform. Then, a single physiological signal and the combination of different physiological signals are used as data to be sent to the proposed deep learning model for classification. Finally, compared with a variety of deep learning models, the best combination of physiological signals is evaluated and the superiority of the algorithm is verified.
2. Materials and methods
2.1. Sleep datasets
The public sleep datasets named Sleep-EDF Database was used in this study. It includes 61 PSGs and scored recordings for 6 sleep stages [38, 39]. The records in this database can be used in two aspects of research. One is the study of the effects of age-related sleep on 79 healthy Caucasians between the ages of 25 and 101, taking no sleep-related drugs. The other is the study of temazepam effects on sleep in 22 Caucasian males and females who had mild difficulty falling asleep without other disease or medication. The EEG signal of the Fpz-Cz and Pz-Oz channels, and the EOG signal are recorded in this database. The sampling rate is 100 Hz.
In this study, EEG and EOG signals based on healthy subjects will be used for sleep staging. And, N3 and N4 are combined to produce new N3. Table 1 shows the detailed information of the used sleep data records for training and testing. Every sleep period is 30 seconds. A total of 17277 sleep periods were used in training set. Because the distribution of each period in the sleep record is unbalanced, the training set is composed of data of a unified period in the data records of 9 people. Each subject’s sleep data is longer than 13 hours, including a full night normal sleep cycle. Each testing group has more than 2700 sleep periods. The test data for each group contains a complete sleep data record more than 22 hours. Each night contains about 4 to 5 sleep cycles.
Table 1.
Sleep data distribution
| Sleep stages | Awake | N1 | N2 | N3 | REM | Total |
|---|---|---|---|---|---|---|
| Training data | 9313 | 859 | 4297 | 1174 | 1634 | 17277 |
| Testing data 1 | 1773 | 137 | 514 | 95 | 273 | 2792 |
| Testing data 2 | 2258 | 44 | 217 | 135 | 68 | 2722 |
| Testing data 3 | 1780 | 114 | 617 | 115 | 180 | 2806 |
| Testing data 4 | 2069 | 56 | 407 | 136 | 102 | 2770 |
| Testing data 5 | 2007 | 90 | 417 | 129 | 187 | 2830 |
2.2. Preprocessing
In this paper, the wavelet transform is used to denoise the original signals [40]. It can automatically adapt to the requirements of time-frequency signal analysis [41]. In practical applications, noise usually exists in the high frequency part, and the signal is mainly distributed in the low frequency part. Wavelet noise reduction is mainly done by decomposing the signal. The high-frequency signal is retained, and the low-frequency signal continues to decompose. Then threshold quantize the high-frequency coefficients of each layer of the signal. Finally, the signal is reconstructed. The db4 wavelet function was selected, and the number of the wavelet decomposition layer is 5. Stein unbiased estimation method was used to determine the threshold and the hard threshold function was selected to preprocess the original signals. The comparison between the original signals and the noise-reduced signals is shown in Fig. 1.
Figure 1.
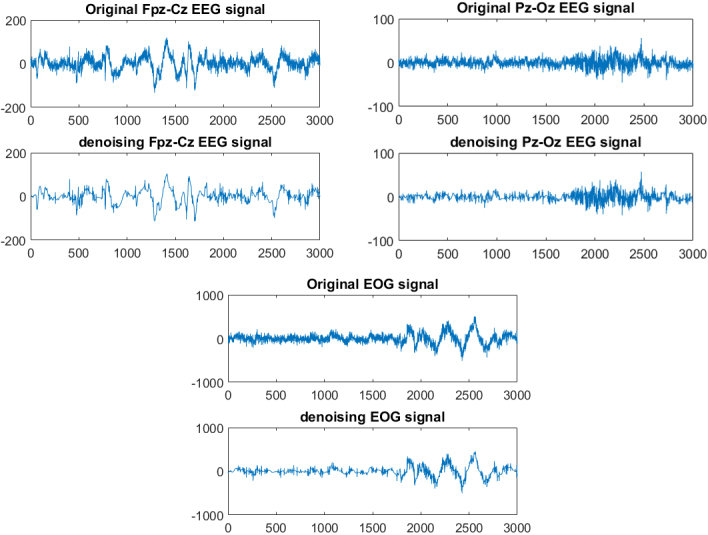
Filtering effects of different signals.
Compared with the original signals, the noise-reduced EEG signal of Fpz-Cz has an error(err) value of 39.02, a percentage(per) value of 0.923, and a signal noise ratio (snr) value of 7.599 dB. The noise-reduced EEG signal of Pz-Oz has an err value of 9.367, a per value of 0.946, and a snr value of 9.303 dB. Compared with the original signal, the EOG signal after noise reduction is 183.8, the value of per is 0.904, and the value of snr is 6.509 dB:
| (1) |
| (2) |
| (3) |
In these equations, is the noise-reduced signal, is the original signal and norm is the maximum singular value function. There is a certain difference between the noise-reduced signal and the original signal, and its energy loss is small. At the meantime, the local confounding on the original signal is more serious, and after the noise reduction processing, the confounding part is effectively eliminated.
2.3. Deep learning algorithm
2.3.1. CNN
CNN is frequently used to recognize two-dimensional images [42]. It combines the local perception, weight sharing, and down sampling. Local perception allows hidden units not to be full connections. Weight sharing enables different signals to share a convolution kernel. Down sampling refers to acquire the features after convolution. They can be expressed as Eqs (4) and (5).
| (4) |
| (5) |
Where , and represent weights, deviations, layers, regularization parameters, learning rate, the total number of training samples, momentum, update steps, and cost functions, respectively. For each layer of CNN, they update the weights and deviations by the above two equations.
1D-CNN is more suitable for one-dimensional input data, like biomedical signals [43]. For a 1D input signal and kernel , the convolution operation is defined in Eq. (6).
| (6) |
In this equation, the represents the discrete convolution operation. The convolution process outputs a feature map. means the elements in from to the dimension of . Thus, the output matrix can be represented by Eq. (7).
| (7) |
The structure of the CNN includes an input layer, a hidden layer, and an output layer. Convolution and pooling operations are usually done in the hidden layer. The final layer of the CNN model usually contains a layer of neural network, which performs the classification task.
2.3.2. LSTM
LSTM is a kind of time recurrent neural network. LSTM can process and predict important events with long intervals and delays in time series [44]. LSTM includes the input gates, the output gates, and the forget gates. Each input consists of two parts: the input state at time , and the output state at time . LSTM module is shown in Fig. 2.
Figure 2.
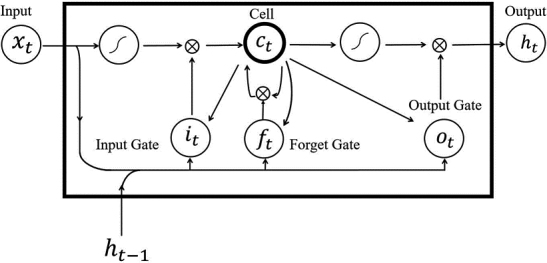
LSTM structure diagram.
The input gate can be represented as Eq. (8).
| (8) |
| (9) |
Forget gate controls what information will be removed from the cell memory.
| (10) |
Cell unit status updates information via forget gate and input gate.
| (11) |
Finally, the output result is obtained through the output gate.
| (12) |
| (13) |
In the above equation, is the state of the current input gate, is the state of the current forget gate, presents the input sequence, and is the output. and are the current and previous cell state respectively. And is the bias vector. is the separate weight vectors for each input, and is the logistic sigmoid function.
The key to LSTM is the state of the cell. In LSTM, the first step is to input the data into the cell through the forget gate which decides what information to discard, and then through the input gate which decides how much new information go to the cell state. Finally, the output gate determines the value of the output.
2.3.3. 1D CNN-LSTM
The proposed algorithm contains 13 layers to realize automatic sleep staging The algorithm model is shown in Fig. 3. There are 4 layers of 1D CNN and 3 layers of LSTM. The activation function for all convolutional layers is Relu. In addition, two dropout layers are added, which are used to avoid overfitting. The dropout rate is 0.5 and 0.4 respectively. The number of the output unit is 5, and the sleep is divided into 5 phases. The batch size is 200. The details of layers and parameters used in the proposed model are shown in Table 2.
Figure 3.
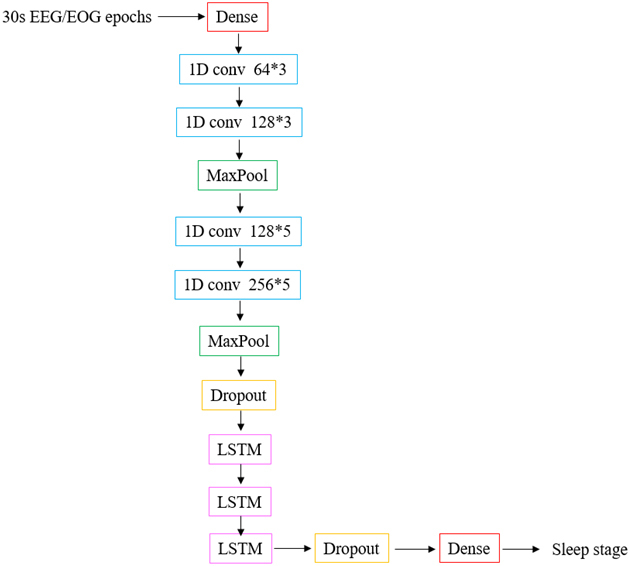
Deep learning classification model.
Table 2.
The details of layers and parameters
| No. | Layers | Filter*kernel size | Unit size | Parameters | Trainable parameters | Output size |
|---|---|---|---|---|---|---|
| 1 | Dense | – | 300 | Relu, strides 1 | 9300 | (N,200,300) |
| 2 | Conv1D | 64*3 | – | Relu, strides 1 | 57664 | (N,198,64) |
| 3 | Conv1D | 128*3 | – | Relu, strides 1 | 24704 | (N,196,128) |
| 4 | MaxPool | – | 2 | Strides 2 | 0 | (N,98,128) |
| 5 | Conv1D | 128*5 | – | Relu, strides 1 | 82048 | (N,94,128) |
| 6 | Conv1D | 256*5 | – | Relu, strides 1 | 164096 | (N,90,256) |
| 7 | MaxPool | – | 2 | Strides 2 | 0 | (N,45,256) |
| 8 | Dropout | – | – | Rate 0.5 | 0 | (N,45,256) |
| 9 | LSTM | – | 300 | – | 668400 | (N,45,300) |
| 10 | LSTM | – | 300 | – | 721200 | (N,45,300) |
| 11 | LSTM | – | 300 | – | 721200 | (N,300) |
| 12 | Dropout | – | 300 | Rate 0.4 | 0 | (N,300) |
| 13 | Dense | – | 5 | Softmax | 1806 | (N,5) |
As shown in Table 2, the parameters are set by inputting 30 s EEG and EOG signals, that is, 6000 datum per 30 s (data_dim 30, timesteps 200). If the single channel signal was used, that would be 100*30. Through the first one-dimensional convolutional layer, the size of the output data becomes 198*64. Then, MaxPool layer reduces the parameters and calculations, which can effectively prevent over-fitting. After all convolution and pooling, the output size of the data is 45*256. LSTM layer facilitates the final accurate classification. The final output with the highest probability is obtained by the softmax function. The total number of parameters is 2450418.
3. Results
In this paper, the 1D CNN-LSTM algorithm was used to perform automatic sleep staging of different physiological signals, and multiple test groups were selected for comparison. First, the single-channel signal was used for testing, and the results are shown in Table 3. The table shows that the Fpz-Cz EEG signal can realize better automatic sleep staging. The highest accuracy is 93.47%, with an average of 88.89%, all exceeding 85%. However, the experiments used Pz-Oz EEG signal shows very unstable results. The lowest accuracy is only 78%, and the highest accuracy is over 88%. Similarly, the sleep staging results based on EOG signals are also unstable. The accuracy of staging varies from 82.45% to 91.95%. Therefore, this algorithm is more suitable for the Fpz-Cz EEG signal. The relationship between accuracy and loss is shown in Fig. 4.
Table 3.
The results of sleep staging based on single-channel signal
| Testing number | Fpz-Cz | Pz-Oz | EOG |
|---|---|---|---|
| 1 | 85.42% | 82.88% | 82.45% |
| 2 | 88.59% | 85.93% | 88.91% |
| 3 | 86.67% | 78.23% | 84.75% |
| 4 | 93.47% | 88.16% | 91.95% |
| 5 | 90.32% | 86.93% | 84.84% |
| Average | 88.89% | 84.43% | 86.58% |
Figure 4.
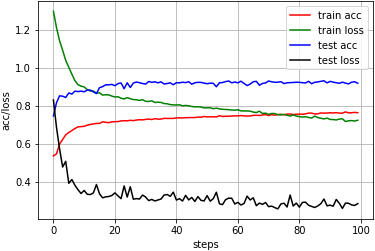
The accuracy and loss based on Fpz-Cz EEG.
It can be seen from Fig. 4 that the stability of the algorithm is high. During the testing, there are some small fluctuations in both accuracy and loss, but all remain balanced within an interval, indicating that the algorithm performs well and has credibility. Then, the next group experiments are based on multi-channel sleep physiological signals. The results are shown in Table 4. Compared with the single Pz-Oz signal, the accuracy of sleep staging based on the Pz-Oz and EOG signal has been improved. The accuracy of sleep staging was improved after the addition of EOG. The highest accuracy reached 94.15%. However, the combination of the two EEG signals did not improve the staging accuracy. According to the results, the more signals are selected, the stronger the interference may be for the deep learning algorithm. Therefore, the best choice is the Fpz-Cz+EOG.
Table 4.
The results of sleep staging based on multi-channel signals
| Testing number | Fpz-Cz+EOG | Pz-Oz+EOG | Fpz-Cz+Pz-Oz | Fpz-Cz+Pz-Oz+EOG |
|---|---|---|---|---|
| 1 | 87.39% | 84.60% | 82.38% | 84.60% |
| 2 | 89.31% | 90.37% | 91.70% | 88.28% |
| 3 | 89.45% | 87.53% | 84.43% | 87.17% |
| 4 | 94.15% | 90.14% | 91.30% | 89.64% |
| 5 | 89.58% | 90.39% | 89.33% | 88.41% |
| Average | 89.91% | 88.60% | 87.83% | 87.62% |
As can be seen from the confusion matrix in Table 5, the distribution of the data is not balanced. The highest accuracy is in the awake period. There are very few data during the N1 and N3 periods. Therefore, their accuracy is reduced accordingly. However, the algorithm does not have an over-fitting due to a large amount of data in the awake period. According to the confusion matrix, precision, recall, and f1-score can be calculated. Figure 5 illustrates the results of Test 4.
Table 5.
The results of test 4, shown here as an example
| Awake | N1 | N2 | N3 | REM | ||
|---|---|---|---|---|---|---|
| Expert Awake | 2007 | 23 | 2 | 5 | 32 | |
| N1 | 4 | 39 | 8 | 0 | 5 | |
| N2 | 1 | 2 | 363 | 25 | 16 | |
| N3 | 2 | 0 | 6 | 128 | 0 | |
| REM | 4 | 0 | 27 | 0 | 71 | |
| Total ACC | 94.15% |
Figure 5.
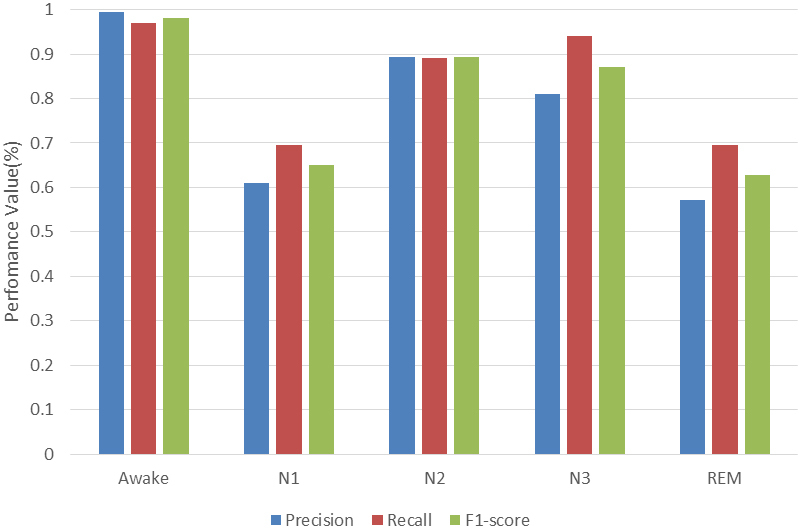
Performances obtained from test 4.
In the classification process, there are TP (the number of samples predicted to be positive in the positive category), TN (the number of samples predicted to be negative in the negative category), FP (the number of samples predicted to be positive in the negative category), FN (the number of samples predicted to be negative in the positive category).
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
represents the kappa coefficient. A kappa coefficient of 0 indicates that the classification accuracy is the opportunity level. And the kappa coefficient of 1 indicates the optimal classification effect. The kappa coefficient of this algorithm is 0.94.
ACC is classification accuracy. ACC0 1/Nr, where Nr represents the total amount of data. The accuracy of the algorithm in the awake period can reach 97%, and the accuracy of the N3 period is more than 80%. But the accuracy in the N1 and REM periods is low, due to the distribution and imbalance of the data set. For the N2 period and the REM period, which are usually difficult to distinguish, it can be seen from the confusion matrix that the confusion between the two periods is high. The amount of data in the REM period is small and will be confused in the N2 period. However, the accuracy of the staging that the algorithm exhibits in these two phases is also good. The relationship between accuracy and loss about this testing is shown in Fig. 6.
Figure 6.
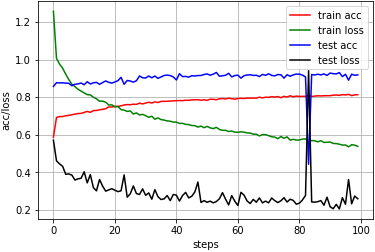
The accuracy and loss based on Fpz-Cz EEG and EOG.
As shown in Fig. 6, the algorithm shows good classification performance in this test. During the training process, the accuracy is constantly improved, and finally stabilized, and the loss is getting lower and lower. On the basis of training, the test accuracy becomes relatively higher and more stable. Post-accuracy basically maintained above 90%. An anomaly can be observed in test loss after more than 80 times iteration, then the anomaly returned to a stable level. Overall, the stability of the algorithm is high.
4. Discussion
Experts use a variety of physiological signal fusion sources to improve accuracy, although many studies on sleep staging are conducted using different datasets and classification algorithms. In data selection, there are two main points to be considered: one is which signals are selected, and the other is how many data are used. Existing public sleep data sets contain PSG records with multiple channels EEG, as well as EOG or EMG channels [45]. Studies on sleep staging are mainly based on EEG, supplemented by other physiological signals. In order to improve accuracy, experts prefer a variety of physiological signal fusion methods.
In terms of classification algorithm, very few researchers have used deep learning model. Common deep learning algorithms for sleep automatic staging include RBM, DBN, SAE, CNN, LSTM [34, 46]. Early studies have shown that the features extracted by deep learning are better than those of artificial extraction. Until the end-to-end approach was proposed, people realized that the deep learning algorithm could automatically extract features from the original signal [47]. With the deepening of research, the model of deep learning is more and more complex, and the number of layers of the network is increasing [48]. Those studies are summarized in Table 6.
Table 6.
Summary of automatic sleep staging classification
| Study | Channels/samples | Methods | Accuracy (%) |
|---|---|---|---|
| Ref. [29] | 1 EEG/– | Boosting | 61.20 |
| Ref. [30] | 2 EEG+2 EOG+1 EMG | DBN | 67.40 |
| Ref. [49] | 1 EEG+2 EOG+1 EMG/– | CNN+HMM+DBN | 69.78 |
| Ref. [50] | 1 EEG/15136 | NB | 71.8 |
| Ref. [51] | 1 EEG/37022 | CNN | 74.8 |
| Ref. [47] | 1 EEG/37022 | SAE | 78.9 |
| Ref. [32] | 2 EEG+2 EOG+1 EMG | DBN | 81.9 |
| Ref. [4] | 1 EEG/41950 | CNN-LSTM | 82 |
| Ref. [52] | 1 EEG+1 EOG/– | RF | 83 |
| Ref. [50] | 1 EEG/15136 | KNN | 83.56 |
| Ref. [36] | 20 EEG+2 EOG+1 EGC+3 EMG | LSTM | 83.60 |
| Ref. [31] | 4 EEG+1 EOG+1 EMG+1 ECG/66164 | MLP | 85.50 |
| Ref. [37] | 1 EEG/10280 | LSTM | 86.7 |
| Ref. [53] | 1 EEG/15188 | CEEMD+Bagging | 86.89 |
| Ref. [50] | 1 EEG/15136 | MLP | 87.13 |
| Ref. [54] | 1 EEG/14963 | VG-HVG+SVM | 87.5 |
| Ref. [33] | 1 EEG/– | CNN | 87.70 |
| Ref. [55] | 1 EEG/15188 | EEMD+RUSBoost | 88.07 |
| Ref. [56] | 1 EEG/– | CNN | 88.83 |
| Ref. [50] | 1 EEG/15136 | RF | 89.74 |
| Ref. [57] | 1 EEG/106376 | DWT+RF | 90.5 |
| Ref. [34] | 1 EEG+1 EOG/127512 | 1D CNN | 91.00 |
| Ref. [58] | 1 EOG/15188 | DWT+SVM | 91.70 |
| Proposed | 1 EEG/20047 | 1 D CNN+LSTM | 93.47 |
| 1 EEG+1 EOG/20047 | 94.15 |
Comments: NB (Naïve Bayes), KNN (k-nearest neighbor), MLP (Multi-layer perceptron), CEEMDAN (complete ensemble empirical mode decomposition), VG-HVG (visibility graph-horizontal VG), EEMD (ensemble empirical mode decomposition), DWT (discrete wavelet transform).
Table 6 summarizes different algorithms which perform automatic sleep staging based on different physiological signals. No matter whether single-channel signals or multi-channel signals are used, the accuracy is over 90%. In the meantime, different classification algorithms are used. Some classification algorithms like SVM or RF could achieve accurate sleep staging, but other deep learning algorithms like SAE, DBN or CNN used for sleep staging are not so good. Only a few of them have been over 90%. In general, the traditional machine learning algorithm is relatively mature in the automatic sleep staging. The deep learning algorithm in sleep staging is still in the exploration stage. Some studies use LSTM to achieve automatic sleep staging, but the results are not so ideal. Therefore, the combination of multiple methods is an effective way to improve the accuracy of staging. In this paper, the amount of data used is moderate, and the proposed algorithm has achieved high accuracy of over 90%, whether using a single signal or multiple signals.
In summary, the algorithm of 1D CNN-LSTM has the following advantages.
-
•
LSTM could overcome the problem of gradient disappearance in traditional neural network models, but the operation speed is slow. With the addition of CNN, the local receptive field and weight sharing characteristics can increase computing speed. In this study, using a single signal, the accuracy maintains above 80%, the average accuracy is above 85%, and the highest accuracy is 93.47%, indicating that the algorithm is suitable for automatic sleep staging of different physiological signals. The accuracy is improved by using a variety of physiological signals. The highest accuracy is 94.15%, which is result of the combination of the Fpz-Cz signal and the EOG signal. The proposed algorithm can achieve accurate sleep automatic staging, and also shows good classification performance in different tests, and can provide more accurate reference results in sleep research.
-
•
The classification algorithm has a high generalization ability and a strong learning ability. It is suitable for a variety of physiological signals without modifying any parameters. Moreover, it can learn useful information when the amount of data is not so huge, and can overcome the problem of data imbalance.
-
•
An end-to-end recognition structure for sleep staging is developed without any manual extraction of features.
However, there are still some shortcomings. The data source is single. There is room for improvement in the accuracy of staging. In future, the proposed model will be tested by different sleep databases.
5. Conclusion
In this study, a thirteen layers model was proposed for the classification of sleep stages. Performance comparison of different classification algorithms based on single-channel EEG, single-channel EOG, and a combination of EEG and EOG signals was presented. In addition, each group finished 5 test sets to make the results more convincing. The results showed that the model also had good classification performance for different physiological signals. When the EEG signal of Fpz-Cz channel was used, the accuracy was 93.47%. When the EEG signal of Fpz-Cz channel and the EOG signal were used, the accuracy of the staging reaches 94.15%. The algorithm had high accuracy and generalization ability. It could achieve more accurate sleep automatic staging without manual extraction features. In the future, this model can be used in sleep-related research. The fully automatic system could replace the traditional error-prone large-scale PSG signal manual expert inspection task. At the same time, deep learning itself is also an algorithm worthy of continuous research, especially for distinguishing sleep periods with high similarity.
Acknowledgments
This paper is sponsored by the National Natural Science Foundation of China (31700856), the Natural Science Foundation of Chongqing, China (cstc2018jcyjAX0163) and the special research project of philosophy and social science of Chongqing Medical University, China (Grant No. 201712).
Conflict of interest
No potential conflict of interest was reported by the authors.
References
- [1]. Wang Q, Zhao D, Wang Y, et al. Ensemble learning algorithm based on multi-parameters for sleep staging. Med Biol Eng Comput 2019; 57(8): 1693-1707. [DOI] [PubMed] [Google Scholar]
- [2]. Liang SF, Kuo CE, Hu YH, et al. A rule-based automatic sleep staging method. J Neurosci Methods 2012; 205(1): 169-176. [DOI] [PubMed] [Google Scholar]
- [3]. Sharma M, Goyal D, Achuth PV, et al. An accurate sleep stages classification system using a new class of optimally time-frequency localized three-band wavelet filter bank. Comput Biol Med 2018; 98: 58-75. [DOI] [PubMed] [Google Scholar]
- [4]. Supratak A, Dong H, Wu C, et al. DeepSleepNet: a model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans Neural Syst Rehabil Eng 2017; 25(11): 1998-2008. [DOI] [PubMed] [Google Scholar]
- [5]. Alickovic E, Subasi A. Ensemble SVM method for automatic sleep stage classification. IEEE Trans Instrum Meas 2018; 67(6): 1258-1265. [Google Scholar]
- [6]. Lajnef T, Chaibi S, Ruby P, et al. Learning machines and sleeping brains: automatic sleep stage classification using decision-tree multi-class support vector machines. J Neurosci Methods 2015; 250: 94-105. [DOI] [PubMed] [Google Scholar]
- [7]. Zafar R, Dass SC, Malik S. Electroencephalogram-based decoding cognitive states using convolutional neural network and likelihood ratio based score fusion. PLoS One 2017; 12(5): e0178410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Akin M, Kurt MB, Sezgin N, et al. Estimating vigilance level by using EEG and EMG signals. Neural Comput Appl 2018; 17(3): 227-236. [Google Scholar]
- [9]. Zhang Y, Zhang X, Liu W, et al. Automatic sleep staging using multi-dimensional feature extraction and multi-kernel fuzzy support vector machine. J Healthc Eng 2014; 5(4): 505-520. [DOI] [PubMed] [Google Scholar]
- [10]. Gaiduk M, Penzel T, Ortega JA, et al. Automatic sleep stages classification using respiratory, heart rate and movement signals. Physiol Meas 2018; 9(12): 124008. [DOI] [PubMed] [Google Scholar]
- [11]. Noviyanto A, Isa SM, Wasito I, et al. Selecting features of single lead ECG signal for automatic sleep stages classification using correlation-based feature subset selection. IJCSI International Journal of Computer Science Issues 2011; 8(5): 139-148. [Google Scholar]
- [12]. Jiang D, Yu MA, Yuanyuan W. Sleep stage classification using covariance features of multi-channel physiological signals on Riemannian manifolds. Comput Methods Programs Biomed 2019; 178: 19-30. [DOI] [PubMed] [Google Scholar]
- [13]. Movahedi F, Coyle JL, Sejdic E. Deep belief networks for electroencephalography: a review of recent contributions and future outlooks. IEEE J Biomed Health Inform 2018; 22(3): 642-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Jestrović I, Coyle JL, Sejdić E. Decoding human swallowing via electroencephalography: a state-of-the-art review. J Neural Eng 2015; 12(5): 051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Oliver Faust, Hajar Razaghi, Ragab Barika, et al. A review of automated sleep stage scoring based on physiological signals for the new millennia. Comput Methods Programs Biomed 2019; 176: 81-91. [DOI] [PubMed] [Google Scholar]
- [16]. U Rajendra Acharya, Shreya Bhat, Oliver Faust, et al. Nonlinear dynamics measures for automated EEG-based sleep stage detection. European Neurology 2015; 74(5-6): 268-287. [DOI] [PubMed] [Google Scholar]
- [17]. Thamarai P, Adalarasu K. Denoising of EEG, ECG and PPG signals using wavelet transform. J Pharm Sci 2018; 10(1): 156-161. [Google Scholar]
- [18]. Mateo J, Torres AM, García MA. Dynamic fuzzy neural network based learning algorithms for ocular artefact reduction in EEG recordings. Neural Processing Letters 2014; 39(1): 45-67. [Google Scholar]
- [19]. Zhao D, Wang Y, Wang Q, et al. Comparative analysis of different characteristics of automatic sleep stages. Comput Methods Programs Biomed 2019; 17553-72. [DOI] [PubMed] [Google Scholar]
- [20]. Mousavi S, Afghah F, Acharya UR. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PloS One 2019; 14(5): e0216456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Tzimourta KD, Tsilimbaris A, Tzioukalia K, et al. EEG-based automatic sleep stage classification. Biomed J 2018; 7(4): 1-6. [Google Scholar]
- [22]. Dereymaeker A, Pillay K, Vervisch J, et al. An automated quiet sleep detection approach in preterm infants as a gateway to assess brain maturation. Int J Neural Syst 2017; 27(6): 1750023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Pławiak P. Novel genetic ensembles of classifiers applied to myocardium dysfunction recognition based on ECG signals. Swarm Evol Comput 2018; 39: 192-208. [Google Scholar]
- [24]. Yildirim Ö. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput Biol Med 2018; 96: 189-202. [DOI] [PubMed] [Google Scholar]
- [25]. Acharya UR, Oh SL, Hagiwara Y, et al. A deep convolutional neural network model to classify heartbeats. Comput Biol Med 2017; 89: 389-396. [DOI] [PubMed] [Google Scholar]
- [26]. Yildirim Ö, Pławiak P, Tan RS, et al. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput Biol Med 2018; 102: 411-420. [DOI] [PubMed] [Google Scholar]
- [27]. Oh SL, Hagiwara Y, Raghavendra U, et al. A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput Appl. 2018.
- [28]. Antoniades A, Spyrou L, Martin-Lopez D, et al. Deep neural architectures for mapping scalp to intracranial EEG. Int J Neural Sys 2018; 28(8): 1850009. [DOI] [PubMed] [Google Scholar]
- [29]. Hassan AR, Subasi A. Automatic identification of epileptic seizures from EEG signals using linear programming boosting. Comput Methods Programs Biomed 2016; 136: 65-77. [DOI] [PubMed] [Google Scholar]
- [30]. Längkvist M, Karlsson L, Loutfi A. Sleep stage classification using unsupervised feature learning. Advances in Artificial Neural Systems 2012; 2012(5): 1-9. [Google Scholar]
- [31]. Charbonnier S, Zoubek L, Lesecq S, et al. Self-evaluated automatic classifier as a decision-support tool for sleep/wake staging. Computers in Biology & Medicine 2011; 41(6): 380-389. [DOI] [PubMed] [Google Scholar]
- [32]. Phan H, Andreotti F, Cooray N, et al. Joint classification and prediction CNN framework for automatic sleep stage classification. IEEE Trans Biomed Eng 2018; 66(5): 1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Acharya UR, Oh SL, Hagiwara Y, et al. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med 2018; 100: 270-278. [DOI] [PubMed] [Google Scholar]
- [34]. Yildirim Ö, Baloglu UB, Acharya UR. A deep learning model for automated sleep stages classification using psg signals. Int J Environ Res Public Health 2019; 16(4): 599(1-21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Yulita IN, Fanany MI, Arymurthy AM. Fast convolutional method for automatic sleep stage classification. Healthc. Inform. Res. 2018. [DOI] [PMC free article] [PubMed]
- [36]. Dong H, Supratak A, Pan W, et al. Mixed neural network approach for temporal sleep stage classification. IEEE Trans Neural Syst Rehabil Eng 2018; 26(2): 324-333. [DOI] [PubMed] [Google Scholar]
- [37]. Michielli N, Acharya UR, Molinari F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput Biol Med 2019; 106: 71-81. [DOI] [PubMed] [Google Scholar]
- [38]. http://www.physionet.org/physiobank/database/sleep-edfx/.
- [39]. Diykh M, Li Y, Wen P. EEG sleep stages classification based on time domain features and structural graph similarity. IEEE Trans Neural Syst Rehabil Eng 2016; 24(11): 1159-1168. [DOI] [PubMed] [Google Scholar]
- [40]. Manish Sharma, Sohamkumar Patel, Siddhant Choudhary, et al. Automated Detection of Sleep Stages Using Energy-Localized Orthogonal Wavelet Filter Banks. Arab J Sci Eng. 2019.
- [41]. Sharma, Manish, Dhere, Abhinav, et al. An Automatic Detection of Focal EEG Signals Using New Class of Time-Frequency Localized Orthogonal Wavelet Filter Banks. Knowl Based Syst. 2016: 118217-227.
- [42]. Russakovsky O, Deng J, Su H, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis 2015; 115(3): 211-252. [Google Scholar]
- [43]. Faust O, Hagiwara Y, Hong TJ, et al. Deep learning for healthcare applications based on physiological signals: a review. Comput Methods Programs Biomed 2018; 161: 1-13. [DOI] [PubMed] [Google Scholar]
- [44]. Baloglu UB, Yildirim Ö. Convolutional long-short term memory networks model for long duration EEG signal classification. J Mech Med Biol 2019; 19(1). [Google Scholar]
- [45]. O"Reilly C, Gosselin N, Carrier J, et al. Montreal Archive of Sleep Studies: an open-access resource for instrument benchmarking and exploratory research. J Sleep Res 2014; 23(6): 628-635. [DOI] [PubMed] [Google Scholar]
- [46]. Chambon S, Galtier MN, Arnal PJ, et al. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans Neural Syst Rehabil Eng 2018; 26(4): 758-769. [DOI] [PubMed] [Google Scholar]
- [47]. Tsinalis O, Matthews PM, Guo Y. Automatic sleep stage scoring using time-frequency analysis and stacked sparse autoencoders. Ann Biomed Eng 2016; 44(5): 1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Tripathy RK, Acharya UR. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern Biomed Eng 2018; 38(4): 890-902. [Google Scholar]
- [49]. Zhang J, Wu Y, Bai J, et al. Automatic sleep stage classification based on sparse deep belief net and combination of multiple classifiers. T I MEAS CONTROL 2016; 38(4): 435-451. [Google Scholar]
- [50]. Sharma R, Pachori RB, Upadhyay A. Automatic sleep stages classification based on iterative filtering of electroencephalogram signals. Neural Comput Appl 2017; 28(10(SI)): 2959-2978. [Google Scholar]
- [51]. Zhang J, Wu Y. A new method for automatic sleep stage classification. IEEE Trans Biomed Circuits Syst 2017; 11(5): 1097-1110. [DOI] [PubMed] [Google Scholar]
- [52]. Phan H, Andreotti F, Cooray N, et al. SeqSleepNet: end-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans Neural Syst Rehabil Eng 2019; 27(3): 400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Hassan AR, Bhuiyan MIH. Computer-aided sleep staging using complete ensemble empirical mode decomposition with adaptive noise and bootstrap aggregating. Biomed Signal Process Control 2016; 24: 1-10. [Google Scholar]
- [54]. Zhu G, Li Y, Wen PP. Analysis and classification of sleep stages based on difference visibility graphs from a single-channel EEG signal. IEEE J Biomed Health Inform 2014; 18(6): 1813-1821. [DOI] [PubMed] [Google Scholar]
- [55]. Hassan AR, Bhuiyan MIH. Automated identification of sleep states from EEG signals by means of ensemble empirical mode decomposition and random under sampling boosting. Comput Methods Programs Biomed 2017; 140: 201-210. [DOI] [PubMed] [Google Scholar]
- [56]. Malafeev A, Laptev D, Bauer S, et al. Automatic human sleep stage scoring using deep neural networks. Front Neurosci 2018; 12: 781(1-15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Da Silveira TLT, Kozakevicius AJ, Rodrigues CR. Single-channel EEG sleep stage classification based on a streamlined set of statistical features in wavelet domain. Med Biol Eng Comput 2017; 55(2): 343-352. [DOI] [PubMed] [Google Scholar]
- [58]. Rahman MM, Bhuiyan MIH, Hassan AR. Sleep stage classification using single-channel EOG. Comput Biol Med 2018; 102: 211-220. [DOI] [PubMed] [Google Scholar]


