Abstract
Genomic diversification of the mec regulator region mediated by IS431 was investigated for clinical isolates of methicillin-resistant staphylococci. A single rearranged form of the mecR1 gene due to IS431 insertion was detected in the three staphylococcal species, while another type of mecR1 truncation with IS431 and an IS431 located downstream of mecI were found only in Staphylococcus haemolyticus. Genetic differentiation of IS431 and staphylococcal isolates suggested transmission of mecDNA with IS431-mediated rearrangement among different staphylococcal species.
Methicillin resistance in staphylococci is defined by the presence of the mecA gene, which encodes PBP 2a, having low affinity to beta-lactam antibiotics (7, 20). The mecA gene in methicillin-resistant (MR) Staphylococcus aureus (MRSA) is located on a large genetic element designated mecDNA and is suggested to be transmitted from coagulase-negative staphylococci (CNS) (5, 9, 10).
Expression of mecA is originally controlled by the mec regulator proteins encoded by the mecR1 and mecI genes, which are located upstream of mecA (8), and methicillin resistance is induced by the presence of beta-lactams. That is, the mecI product (MecI) usually represses mecA expression (17), but this function is removed when the bacterial cells are exposed to beta-lactams (10). However, it is known that recent MRSA isolates are rendered constitutively resistant to beta-lactams through mutations generated in mec regulator regions and the resultant loss of the repression function of MecI. These mutations are nucleotide substitutions in the mecI or mecA promoter region or nucleotide deletion in mecI (9, 14, 21).
In addition to such genetic changes, truncation of mecR1 and deletion of mecI through insertion of IS1272 have been identified in some MR staphylococci (1, 16). IS1272 is prevalent primarily in Staphylococcus haemolyticus, but it is considered to have disseminated among other staphylococcal species and is associated with methicillin resistance of staphylococci (2). However, in our previous study (16), IS1272 was not found in some MR isolates with incomplete mec regulator genes, suggesting that the deletion of mec regulator regions was generated by a mechanism other than IS1272 insertion.
IS431, a well-known mobile genetic element in staphylococci, is 782 bp long (IS431mec) and contains an open reading frame (ORF) of a putative transposase gene and 14- to 22-bp terminal inverted repeats (3, 4). IS431 is implicated in transfer of a gene(s) or entire plasmid into other replicons or the chromosome, and particularly in transfer of antimicrobial resistance genes, because variable resistance genes are found to be flanked by IS431 (18, 19). In mecDNA, a pair of IS431 elements flanking a plasmid, pUB110, are located downstream of mecA in a prototype MRSA strain (N315) and other MRSA isolates (11).
In the present study, we investigated the rearrangement of the mec regulator region mediated by IS431 insertion. Previously, we examined a total of 118 clinical isolates of MR staphylococci with respect to the presence of mecR1 and mecI through PCR amplification of individual genes, and we found that 80 isolates possessed both mecR1 and mecI, while 23 isolates had an incomplete mecR1 truncated with IS1272 (16). However, neither mec regulator genes nor IS1272 was detected in 15 isolates (2 S. aureus, 1 Staphylococcus epidermidis, and 12 S. haemolyticus). These isolates were analyzed in the present study in regard to IS431 insertion into the mec regulator region. In addition to these, 16 staphylococcal isolates having both mecR1 and mecI were examined for the presence of IS431 downstream of mecA or mecI. S. aureus isolates were classified by coagulase type, coagulase gene type (13), and protein A type (15), and S. haemolyticus was discriminated by use of an arbitrarily primed PCR (AP-PCR) with ERIC2 and M13R primers (6, 22).
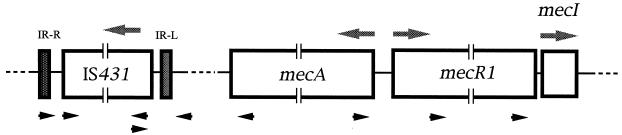
The presence of IS431 in mecDNA and its orientation were examined by PCR with primers with different directions complementary to mecA, mecR1, or IS431 sequences (Fig. 1). Extraction of bacterial DNA and PCR were performed as described previously (12), employing TaKaRa Ex Taq (Takara) as the Taq DNA polymerase. Nucleotide sequences around the insertion site and ORF of IS431 located at different sites were determined directly from PCR products by the dideoxynucleotide chain termination method using a Sequenase version 2 PCR Product Sequencing Kit (United States Biochemical, Cleveland, Ohio).
FIG. 1.
Schematic representation of mecA, mec regulator genes, and IS431 and locations of the primers used in this study. Arrowheads indicate primers, while arrows above ORFs of genes show directions of transcription.
The presence of IS431 upstream of mecA in a single isolate each of S. aureus and S. epidermidis and in 12 isolates of S. haemolyticus was confirmed. In one MRSA isolate, SH220, a PCR product suggesting the presence of IS431 upstream of mecA was not obtained. The IS431 downstream of mecI was detected only in the two S. haemolyticus isolates. The presence of an IS431 located downstream of mecA (IS431-A) was confirmed for all of the staphylococci examined in this study.
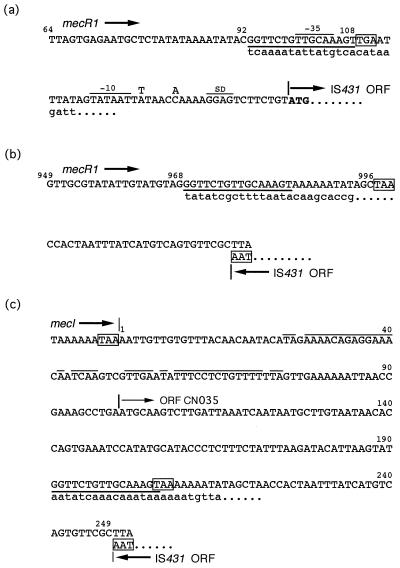
Two distinct types of IS431 insertion into mecR1 were clarified by nucleotide sequencing. In the first type (Fig. 2a), an IS431 (IS431-F) was linked with the 5′ 92 bp of the mecR1 gene (ΔmecR1a), and the transcription direction of its ORF was identical to that of mecR1. An ORF of the rearranged mecR1 gene contained the initial 16 bp of IS431 after the truncation site of mecR1 and was presumed to encode an extremely short peptide with 36 amino acids. This type of insertion, i.e., ΔmecR1a-IS431-F, was found in a single isolate each of S. aureus and S. epidermidis and in 10 S. haemolyticus isolates that were divided into at least three genetic groups (i, iii, and iv) by AP-PCR (data not shown) (Table 1). In the second type of insertion, which was detected only in S. haemolyticus, an IS431 (IS431-R) with the reverse orientation to that of IS431-F was integrated after the 5′ 968 bp of mecR1 (ΔmecR1b) (Fig. 2b). The rearranged ORF containing the partial IS431 sequence is suggested to encode a product of 332 amino acids. The two isolates having this mecDNA were classified into a single AP-PCR type (type ii) which was different from those found in other S. haemolyticus isolates with ΔmecR1a-IS431-F (Table 1). The IS431 located downstream of mecI (IS431-I) was inserted after nucleotide 190 from the termination codon of mecI (Fig. 2c). The transcription direction of the IS431-I ORF was opposite to that of mecR1, as seen for IS431-R.
FIG. 2.
(a and b) Nucleotide sequences of the 3′-end portions of ΔmecR1a (a) and ΔmecR1b (b), and partial sequences of IS431 [referred to as IS431-F (a) and IS431-R (b) in the text] integrated into mecR1, determined for staphylococcal isolates with incomplete mec regulator genes. Nucleotide numbers from the initial base of the mecR1 start codon are indicated on the sequences, and termination codons for the rearranged mecR1 sequence or IS431 are boxed. Underlining shows locations of the terminal inverted repeat (IR) of IS431 [IR-L (a) and IR-R (b)] (3). Lowercase letters indicate the mecR1 nucleotide sequence of a prototype MRSA strain, N315 (8). Nucleotides of IS431mec (4) which are different from those of IS431-F are indicated above the sequence. (c) Nucleotide sequence of a junction (numbered on the sequence from 1 to 249) between the 3′-end portion of mecI and IS431-I detected in S. haemolyticus isolates. Lowercase letters indicate the nucleotide sequence found in the prototype MRSA strain N315 (11) (GenBank accession no. D86934). Terminal codons of mecI, IS431, and the rearranged ORF of CN035 are boxed. IR-R of IS431 is shown by an underline, and inverted repeats downstream of mecI are shown by lines above the sequence.
TABLE 1.
Grouping of staphylococcal isolates based on genetic organization of IS431 and the mec regulator region
| Staphylococcal species | Isolate(s) (biological and/or genetic type)a | Position, orientation, and genotype of IS431 located near the mecA and mec regulator genesb |
|---|---|---|
| S. aureus | SH15 (IV,C,7), SH73 (IV,C,7), SH273 (VII,B,7), SH313 (VII,B,7), SH363 (VII,B,7), SH423 (II,A,13), SH443 (II,A,10), SH457 (II,A,10) | 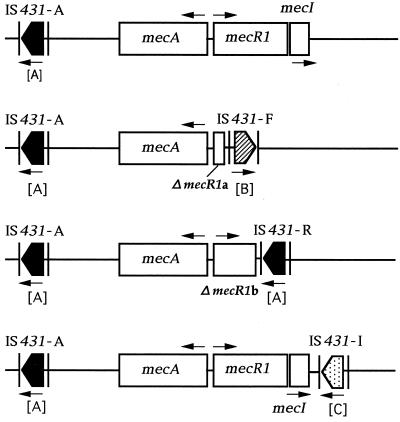 |
| S. epidermidis | SH350, SH214, SH366, SH390, SH418, SH520 | |
| S. aureus | SH20041 (II,E,10) | |
| S. epidermidis | SH513 | |
| S. haemolyticus | SH83 (i), SH84 (iii), SH335 (iv), SH426 (iii), SH514 (i), SH533 (i), SH534 (i), SH535 (i), SH536 (iii), SH538 (iii) | |
| S. haemolyticus | SH517 (ii), SH529 (ii) | |
| S. haemolyticus | SH339 (v), SH537 (vi) |
Coagulase types, coagulase gene types (13), and protein A types (15) (in this order) are shown for S. aureus, and genetic groups (i to vi) discriminated by AP-PCR fingerprinting are shown for S. haemolyticus.
Arrows indicate direction of transcription. IS431-A, IS431-F, IS431-R, and IS431-I were designated according to their positions relative to methicillin resistance genes. Genotypes (A, B, and C) of IS431 are given in brackets.
By comparison of the nucleotide sequences of the ORFs of the four IS431 elements (IS431-A, IS431-F, IS431-R, and IS431-I) located at different positions, three IS431 genotypes (A, B, and C) were discriminated (Fig. 3). Genotype A represents the one virtually identical to IS431mec, that was reported for MRSA strain BB270 (4). All of the IS431-A and IS431-R sequences were grouped into genotype A. IS431-F sequences were classified in genotype B, which showed sequence divergence of 16 to 20 nucleotides compared with the IS431mec sequence. Notably, the nucleotide substitution at position 144 (C to G) generates a new stop codon (Fig. 3); therefore, the IS431-F ORF is presumed to encode a short product. Genotype C included only IS431-I, which was detected in two S. haemolyticus isolates. In this genotype, a 17-bp sequence corresponding to nucleotides 29 to 45 of IS431mec was deleted, accompanied by substitution of several nucleotides in other regions (Fig. 3). The IS431-I ORF is suggested to be extremely short (69 bp) due to a frameshift caused by the sequence deletion. These findings suggested that IS431-R and IS431-A were derived from the same origin but were genetically distinct from IS431-F and IS431-I.
FIG. 3.
Alignment of partial nucleotide sequences of the three IS431 genotypes, A, B, and C, identified in this study. Sequences corresponding to ORF of IS431mec (4), numbered from the first nucleotide of the start codon, are shown. The sequence of genotype A is shown as the IS431mec sequence, and only substituted nucleotides found in some isolates are indicated above the sequence. In genotype B and C sequences, dots represent nucleotides identical to those of genotype A, while dashes denote gaps. Putative termination codons of genotypes B and C are boxed.
In addition to the genomic rearrangement of the mec regulator region via deletion and insertion with IS1272 (1, 16), our present study indicates that IS431 also played an important role in the genomic evolution of mecDNA and probably in modifying functions of mec regulator proteins. Considering the function of IS431 associated with gene transfer, it may be also possible that recombination between the IS431 copies occurs, leading to deletion of mecA and its transfer to other bacterial genomes, although no analysis of this was done in the present study.
In the present study, mecDNA with a rearranged form, ΔmecR1a-IS431-F, was detected in the three staphylococcal species, suggesting the transmission of this type of mecDNA among different staphylococcal species. However, other IS431-mediated genomic variations, ΔmecR1b-IS431-R and mecR1-mecI-IS431-I, were found only in S. haemolyticus. Taken together with our observation of a higher prevalence of IS431 in methicillin-susceptible CNS than in methicillin-susceptible S. aureus (unpublished data), it was assumed that IS431 had originally been prevalent in CNS, resulting in the occurrence of various forms of mecDNA with inserted IS431 in CNS, and that subsequently some of the mecDNAs having the rearranged mec regulator region, including those with ΔmecR1a-IS431-F, had been transmitted to S. aureus, as suggested for IS1272-integrated mecDNA (1). It is also noteworthy that the IS431 located 190 bases downstream of mecI, which was found in two S. haemolyticus isolates in the present study, had been observed in Staphylococcus sciuri subsp. rodentius strain K8 isolated from a rodent (23) (GenBank accession no. Y13096), suggesting the transmission of this type of mecDNA between these two staphylococcal species.
Since genomic diversity in the mec regulator region seems to be a good marker to discriminate mecDNA, further extensive studies to search for other rearranged forms of mec regulator genes from various staphylococcal species may be significant in understanding diverse routes of mecDNA dissemination among staphylococci.
REFERENCES
- 1.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberis-Maino L, Berger-Bachi B, Weber H, Beck W D, Kayser F H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59:107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- 4.Barberis-Maino L, Ryffel C, Kayser F H, Berger-Bachi B. Complete nucleotide sequence of IS431mec in Staphylococcus aureus. Nucleic Acids Res. 1990;18:5548. doi: 10.1093/nar/18.18.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couto I, de Lencastre H, Severina E, Kloos W, Webster J A, Hubner R J, Sanches I S, Tomasz A. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2:377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 6.Fang F C, McClelland M, Guiney D G, Jackson M M, Hartstein A I, Morthland V H, Davis C E, McPherson D C, Welsh J. Value of molecular epidemiologic analysis in a nosocomial methicillin-resistant Staphylococcus aureus outbreak. JAMA. 1993;270:1323–1328. [PubMed] [Google Scholar]
- 7.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) FEBS Lett. 1992;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 9.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 11.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect. 1994;113:259–266. doi: 10.1017/s0950268800051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi N, Taniguchi K, Kojima K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Analysis of methicillin-resistant and methicillin-susceptible Staphylococcus aureus by a molecular typing method based on coagulase gene polymorphisms. Epidemiol Infect. 1995;115:419–426. doi: 10.1017/s095026880005857x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi N, Taniguchi K, Urasawa S. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:717–720. doi: 10.1128/aac.42.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi N, Urasawa S, Uehara N, Watanabe N. Analysis of genomic diversity within the Xr-region of the protein A gene in clinical isolates of Staphylococcus aureus. Epidemiol Infect. 1999;122:241–249. doi: 10.1017/s0950268898001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi N, Urasawa S, Uehara N, Watanabe N. Distribution of insertion sequence-like element IS1272 and its position relative to methicillin resistance genes in clinically important staphylococci. Antimicrob Agents Chemother. 1999;43:2780–2782. doi: 10.1128/aac.43.11.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon B R, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morton T M, Johnston J L, Patterson J, Archer G L. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother. 1995;39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Belkum A, Bax R, Peerbooms P, Goessens W H F, van Leeuwen N, Quint W G V. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1993;31:798–803. doi: 10.1128/jcm.31.4.798-803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, de Lencastre H, Tomasz A. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J Bacteriol. 1998;180:236–242. doi: 10.1128/jb.180.2.236-242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]