Abstract
Over several decades, the disease pattern of intractable disease has changed from acute infection to chronic disease accompanied by immune and metabolic dysfunction. In addition, scientific evidence has shown that humans are holobionts; of the DNA in humans, 1% is derived from the human genome, and 99% is derived from microbial genomes (the microbiome). Extracellular vesicles (EVs) are lipid bilayer-delimited nanoparticles and key messengers in cell-to-cell communication. Many publications indicate that microbial EVs are both positively and negatively involved in the pathogenesis of various intractable diseases, including inflammatory diseases, metabolic disorders, and cancers. Microbial EVs in feces, blood, and urine show significant differences in their profiles between patients with a particular disease and healthy subjects, demonstrating the potential of microbial EVs as biomarkers for disease diagnosis, especially for assessing disease risk. Furthermore, microbial EV therapy offers a variety of advantages over live biotherapeutics and human cell EV (or exosome) therapy for the treatment of intractable diseases. In summary, microbial EVs are a new tool in medicine, and microbial EV technology might provide us with innovative diagnostic and therapeutic solutions in precision medicine.
Subject terms: Biomarkers, Biological therapy
Microbiome: Hacking microbial communication networks for clinical applications
The tiny membrane-bound vesicles containing various biomolecules that the organisms comprising our microbiome release could offer a powerful tool for precision medicine. Our bodies are home to trillions of microbes, which interact closely with our tissues to maintain a healthy physiological environment. Yoon-Keun Kim of the Institute of MD Healthcare, Seoul, South Korea, and colleagues have reviewed current research into the extracellular vesicles that these microbes use to communicate with other microbes and their human hosts. The authors note that these vesicles affect tissues throughout the body, and their activities have been linked to various disorders including asthma, Crohn’s disease and cancer. A deeper understanding of how these vesicles prevent or accelerate various conditions in different individuals could yield useful new diagnostic biomarkers and provide the foundation for interventions that are optimized for each patient.
Introduction
The approaches to precision medicine defined by the Precision Medicine Initiative1 and National Research Council2 allow doctors and researchers to predict more accurately which treatments and preventive strategies will work in patients with a particular disease. These definitions indicate that precision medicine can guide the most effective health care decisions for a given patient and thus provide the best quality of therapy while reducing unnecessary medical interventions3. Precision medicine enables the individualization of patient care from diagnosis to therapy selection and monitoring using a patient’s own data in engineering technology to identify individualized treatment strategies using population-wide data4.
With the genomic revolution of the 21st century came the ability to rapidly sequence entire genomes in a matter of days at increasingly more affordable costs. In addition, artificial intelligence (AI) platforms have been used to optimize combination therapy in preclinical and clinical trials4. Thus, the prospect of deciphering an individual’s unique genome as a roadmap for precision medicine has become a tantalizing goal for scientists and physicians around the world. However, several challenges have been encountered when applying human genomic strategies in precision medicine and in translation from the bench to the clinic. To address these issues, technological improvements, including long-read sequencing methods, algorithms for indel and structural variants, graph reference approaches, and standardization of nomenclature, have been suggested4. The variety of next-generation methods through which precision medicine solutions have been crafted include genetic sequencing, proteomic analysis, and AI strategies. The genomic analysis ultimately remains the core foundation of most precision medicine applications.
The human genome and human microbiome projects have revealed that microbial protein-encoding genes are 360 times more abundant than human genes5. In 1991, Lynn and Fester6 defined the term “holobiont”, which is derived from Ancient Greek hólos (“whole”) and the word biont for a unit of life. In 1994, Jefferson7 defined the term “hologenome” when he introduced the hologenome theory of evolution at a plenary lecture at Cold Spring Harbor Laboratory. The hologenome theory of evolution reboots elements of Lamarckian evolution and recasts the individual human as a community or a holobiont that is the host plus all of its symbiotic microbiota species8,9. Variation in the hologenome may encode phenotypic plasticity of the holobiont and can be subject to evolutionary changes caused by selection and drift if portions of the hologenome are transmitted between generations with reasonable fidelity. One of the important outcomes of recasting the individual as a holobiont subject to evolutionary forces is that genetic variation in the hologenome can be brought about by changes in the host genome and microbiome, including new acquisitions of microbes, horizontal gene transfers, and changes in microbial abundance within hosts. Thus, humans might be holobionts, with 99% of their DNA attributed to microbial genomes (the microbiome) and 1% of their DNA attributed to the human genome, according to the hologenome theory of evolution.
Extracellular vesicles (EVs) are lipid bilayer-delimited particles that are naturally released from almost all types of cells and cannot replicate. The vast majority of EVs are smaller than 200 nm. EVs, according to the biogenesis route, are divided into exosomes, ectosomes, and apoptotic bodies10. EVs carry cargoes of proteins, nucleic acids, lipids, metabolites, and even organelles from parent cells. Most cells that have been studied to date are thought to release EVs, including archaeal, bacterial, fungal, plant, and human cells11. Microbial EVs (MEVs) appear to be key messengers in host cell−microbiota communication. Recent experimental evidence shows that MEVs are key messengers in the communication between host cells and the microbiota. In this review, we will address MEVs as a new factor in precision medicine, focusing on 1) MEV biogenesis and functions, 2) the importance of MEVs in disease pathogenesis, and 3) the application of MEVs in the development of diagnostic and therapeutic solutions for precision medicine.
Microbial EV biogenesis
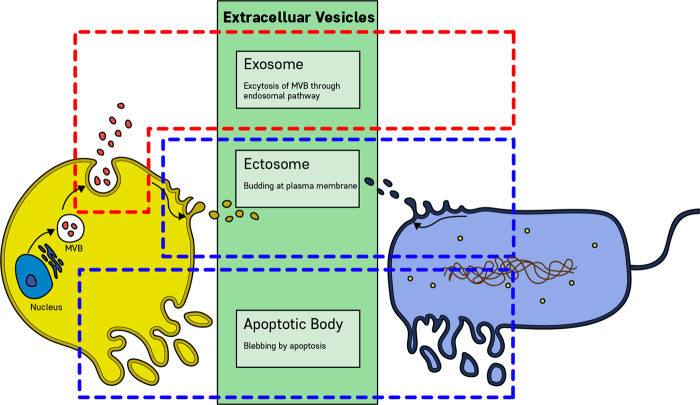
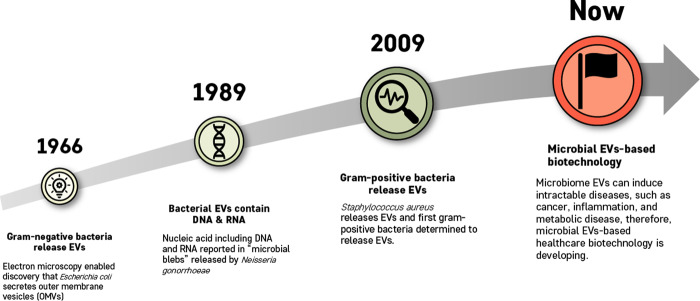
The biogenesis of EVs is a very tightly regulated process governed by multiple signaling molecules and begins with receptor activation that is unique for each cell type12. Eukaryotic EV biogenesis is well characterized, whereas MEV biogenesis has only recently been elucidated13. According to the method of biogenesis, eukaryotic cell-derived EVs are classified as exosomes, ectosomes (or shedding vesicles), and apoptotic bodies14,15, while prokaryotic cell-derived EVs are classified as ectosomes and apoptotic bodies16 (Fig. 1). Both gram-negative and gram-positive bacteria produce ectosomes, known as outer membrane (OM) vesicles (OMVs) and membrane vesicles (MVs), respectively16. The key milestones of bacterial EVs are as follows (Fig. 2): (1) gram-negative bacteria-derived OMVs were first discovered in 196617, (2) the fact that bacterial EVs contain nucleic acids was reported in 198918, (3) gram-positive bacteria-derived EVs were first included in a publication in 200919, and (4) since 2010, much evidence has shown that both gram-negative and gram-positive bacteria release EVs that are positively or negatively involved in disease pathogenesis.
Fig. 1. Biogenesis of extracellular vesicles (EVs) derived from eukaryotic cells and prokaryotic cells.
Extracellular vesicles can be classified into three main classes: Exosome, exosomes are formed within the endosomal network and released upon fusion of multivesicular bodies (MVBs) with the plasma membrane. Ectosomes, ectosomes are produced by outward budding and fission of the plasma membrane. Apoptotic bodies, apoptotic bodies are released as blebs of cells by apoptosis. Eukaryotic cells can release exosomes, ectosomes, and apoptotic bodies, whereas prokaryotic cells are able to secrete ectosomes and apoptotic bodies.
Fig. 2. Key milestones of microbial EV research.
The main findings of such studies are that gram-negative bacteria release EVs, bacterial EVs contain DNA and RNA, and gram-positive bacteria release EVs. Currently, microbial EV-based biotechnology is developing dramatically.
When OMVs are released from gram-negative bacteria during their proliferation, the OM bulges outwardly, and the connection between the OM and peptidoglycans is broken. The next steps are an accumulation of peptidoglycan fragments and misfolded proteins in the periplasmic space and enrichment of species-specific membrane curvature-inducing molecules16,20. However, this mechanism cannot sufficiently explain OMV biogenesis. Thus, new mechanisms for OMV formation have been reported. Several studies have shown that overexpressed and misfolded periplasmic proteins can cause budding, and specific proteins have been found to be enriched in or excluded from OMVs, suggesting a more sophisticated biogenesis pathway. The accumulation of periplasmic proteins, as well as the accumulation of curvature-inducing OM proteins, induces additional budding events21,22. Another study showed that the VacJ/Yrb ATP-binding cassette transport system was involved in OMV formation. In that report, a decrease in or deletion of the VacJ or Yrb genes was found to result in phospholipid accumulation in the outer leaflet of the OM. This asymmetric expansion of the outer leaflet then caused an outward bulging of the OM. Then, further enrichment of the positive and negative curvature-inducing phospholipids in both leaflets supported the budding of the OM. Ultimately, the released OMV was enriched in phospholipids incorporated into the outer leaflet of the vesicle membrane20,23.
Gram-positive bacteria lack an OM and have a much thicker peptidoglycan cell wall. Thus, until 2009, it was assumed that gram-positive bacteria could not release EVs. However, recent experimental evidence has shown that gram-positive bacteria release MVs19,24,25. The biogenesis mechanisms of MVs during the proliferation of gram-positive bacteria have not been as clearly elucidated compared to those of gram-negative bacterial ectosome biogenesis. One hypothesis is that MVs may be forced through the cell wall by turgor pressure after their release from the plasma membrane26. Another hypothesis is that cell walls loosened by enzymes may enable the release of MVs. For example, Staphylococcus aureus (S. aureus)-derived MVs carry peptidoglycan-degrading enzymes, which can modify thick gram-positive peptidoglycan cell walls19. Thus, membrane fluidity and cell wall integrity, affected by peptidoglycan cross-linking, might be critical for the release of MVs25. The third hypothesis is that EVs may pass through channels such as protein channels or structural cables from the intracellular to the extracellular environment26.
Regarding the biogenesis of apoptotic bodies, much evidence has revealed that bacterial cell apoptosis is induced in response to environmental stress, biofilm formation, and genetic transformation for the benefit of the entire colony rather than the individual cell27. Once the programmed cell death signaling pathways are activated, DNA damage and fragmentation are induced, and subsequently, cell membrane blebs, are induced, resulting in cell fragmentation and the release of apoptotic bodies15.
Microbial EV components and physiological functions
Nanometer-sized MEV particles contain a variety of functional components, including lipids, proteins, nucleic acids, and metabolites, within a spherical phospholipid bilayer and play a key role in cell-to-cell communication10,16,21,26.
Membrane components: lipids, carbohydrates, and proteins
Lipids in MEVs can play roles as drug delivery vehicles. The bilayer lipids of OMVs mainly contain lipopolysaccharide (LPS) and OM lipids28. Liposomal vesicles can be incorporated into cultured mammalian cells with no cytotoxic effects. The variety of glycolipids and glycoproteins in liposomal vesicles suggests the potential to modify the cellular composition and introduce biologically active molecules into cells16,29. In addition, the asymmetric distribution of EV lipids suggests that EVs could be synthesized in lipid domains, which could potentiate a lipoprotein-dependent effect on cargo loading25. For example, lipid domains, such as functional membrane microdomains, were found to exist in the membranes of Bacillus subtilis30,31. Therefore, EVs have been considered advantageous as pharmacological vehicles.
Lipids, carbohydrates, and proteins in the MEV membrane are crucial for specific targeting. In particular, the physical state of the lipids in EVs is important for determining the pathways of EVs16. The EV lipid bilayer is adapted to different target environment conditions, such as pH, for optimal function32. The carbohydrates in the cell membrane include receptors for glycoproteins, suggesting that the tissue distribution and cellular uptake of phospholipid EVs could be controlled by carbohydrate determinants on the EV surface33,34. Modification of the EV surfaces with specific synthetic glycolipids significantly affected the tissue distribution and stability of the EVs in mice35,36. Furthermore, the recognition systems of cell membrane carbohydrates include receptors for galactose-terminated glycoproteins in hepatocytes, 6-phosphomannosyl residue-containing glycoproteins in fibroblasts, and mannose-terminated glycoproteins in macrophages16,37–39. These membrane glycoprotein receptors are essential for specific targeting. Generally, membrane proteins are essential components for EV functions, such as immunity, targeting, and pathogenicity, and EV proteins can play significant roles in regulatory processes, cellular responses, host−microbe interactions, and targeting32. OM proteins have been shown to participate in flagellum assembly, pore formation, transport of specific substrates, OM stabilization, antibiotic resistance, proteolysis, and host bacteria interaction28.
Luminal components: proteins, nucleic acids, and metabolites
Proteomics studies have shown that there are a number of proteins in the membrane or luminal space of MEVs, which demonstrate complex protein organization19,25,40,41. Most EV membrane proteins originate from the cytoplasmic membrane, while EV luminal proteins are cytoplasmic proteins packaged during vesiculogenesis25. For example, S. aureus EVs (SaEVs) were found to have more than 200 proteins associated with EVs, and 160 of these proteins were cytoplasmic proteins42. Furthermore, α-hemolysin is localized in the lumen of SaEVs43. α-Hemolysin in EVs induces necrotic cell death via toxin entry into the cytoplasm of keratinocytes, whereas soluble α-hemolysin induces keratinocyte death. Thus, α-hemolysin in the EV lumen enhances keratinocyte death and the evasion of host immune defenses, illustrating the clinical significance of EVs in luminal protein delivery43.
Previous studies have shown that MEVs carry DNA, rRNA, tRNA, mRNA, and sRNA16,25. The majority of RNAs in EVs are sRNAs and are noncoding16,44. In addition, a large proportion of sRNAs in EVs are from uncharacterized intergenic regions and may also modulate gene expression in target cells44,45. However, there is much less evidence regarding the properties and functions of the nucleic acids in MEVs44,46. Further studies using next-generation sequencing (NGS) and transcriptome sequencing are needed to ascertain the content and function of MEV nucleic acids25. Previous studies suggest that the nucleic acids in MEVs could interfere with the host transcriptome and inhibit protein synthesis25.
The nucleic acids in EVs mediate horizontal gene transfer and immunomodulatory effects16. To affect gene expression in target cells directly, RNAs in EVs enter the cytoplasm. Once in the cytoplasm, microbial RNAs might interact with or be modified by target cell factors. Complementarity between sRNAs in EVs and RNAs in target cells might induce the attenuation or enhancement of gene expression via transcription, translation, or mRNA processing. The mechanisms involved likely vary greatly depending on the specific RNAs in EVs and the presence of specific target cell factors. mRNAs in EVs might be translated to produce microbial proteins in target cells44. Microbial nucleic acids are sensed by either endosomal or cytoplasmic receptors in host cells47. Several MEVs trigger pattern recognition receptor signaling to promote protective immunity, a property that has been harnessed for vaccine development44.
Recently, it has been reported that EVs are metabolically active16. Although emerging evidence suggests that EVs can act as metabolic regulators, the involvement of EVs in metabolic activity and the existence and function of metabolites in MEVs have not been completely characterized16. Thus, it is necessary to further investigate the metabolites in MEVs to demonstrate their potential metabolic functions.
Microbial EVs in pathogenesis
Previous studies elucidated the role of MEVs in disease development and the pathogenesis of diseases, including skin, lung, and gut diseases, metabolic diseases, and cancers (Table 1).
Table 1.
Role of microbial EVs in pathogenesis.
| Organ | Disease | EV species of origin | Pathogenic effects | Ref. |
|---|---|---|---|---|
| Skin | Atopic dermatitis | Staphylococcus aureus (S. aureus) | The application of S. aureus EVs induced activation of dermal fibroblasts, increasing the production of pro-inflammatory mediators such as IL-6, thymic stromal lymphopoietin, macrophage inflammatory protein-1a, and eotaxin. In addition, the application of S. aureus EVs caused epidermal thickening with infiltration of the dermis by mast cells and eosinophils, which are associated with the enhanced cutaneous production of IL-4, IL-5, IFN-c, and IL-17. | Hong et al.121 |
| α-Hemolysin in S. aureus EVs was cytotoxic to HaCaT keratinocytes and induced necrosis. In addition, α-hemolysin in S. aureus EVs induced skin barrier disruption and epidermal hyperplasia and caused dermal inflammation. | Hong et al.48 | |||
| Acne vulgaris | Propionibacterium acnes (P. acnes) |
P. acnes EVs significantly increased the secretion of cytokines, such as IL-8, GM-CSF, CXCL-1, and CXCL-5, promoting the infiltration of neutrophils. In addition, P. acnes EVs induce dysregulated epidermal differentiation of human keratinocytes via TLR2-mediated signaling pathways. P. acnes EVs may play pivotal roles in the early stages of the development of acne vulgaris by initiating inflammatory cytokine cascades in keratinocytes. |
Choi et al.50 | |
| Lung | Asthma or chronic obstructive pulmonary disease (COPD) | Staphylococcus aureus (S. aureus) |
After stimulation with S. aureus EVs, alveolar macrophages induced both TNF-a and IL-6 production, and airway epithelial cells induced only IL-6 production. Repeated airway exposure to S. aureus EVs induced both Th1 and Th17 cell responses and neutrophilic pulmonary inflammation, mainly via a TLR2-dependent mechanism. In terms of adjuvant effects, airway sensitization with S. aureus EVs and OVA resulted in neutrophilic pulmonary inflammation after OVA challenge alone. This phenotype was partly reversed by the absence of IFN-γ or IL-17. Neutrophilic inflammation led to airway hyperreactivity and fibrosis that contributed to asthma development, and the combination of increased elastase production and fibrosis caused COPD. |
Kim et al.55 |
| Pulmonary inflammation | Pseudomonas aeruginosa (P. aeruginosa) | P. aeruginosa EVs increased the concentrations of several chemokines and cytokines in the lungs and alveolar macrophages. The inflammatory responses to P. aeruginosa EVs were partly regulated by the TLR2 and TLR4 pathways. P. aeruginosa EVs cause pulmonary inflammation, which was only partly controlled by TLR2 and TLR4. Furthermore, P. aeruginosa EVs caused dose- and time-dependent pulmonary cellular inflammation. | Park et al.57 | |
| Emphysema | Escherichia coli (E. coli) | Airway exposure to E. coli EVs increased the production of pro-inflammatory cytokines, such as TNF-a and IL-6. In addition, the repeated inhalation of E. coli EVs induced neutrophilic inflammation and emphysema, and elastase activity was enhanced. Emphysema and elastase activity were reversed by the absence of the IFN-γ or IL-17A genes. In addition, during the early period, lung inflammation was dependent on IL-17A and TNF-a. Moreover, the production of IFN-γ was eliminated by the absence of IL-17A, whereas IL-17A production was not abolished by IFN-γ absence. In summary, E. coli EVs induced IL-17A-dependent neutrophilic inflammation and thereby emphysema via upregulation of elastase activity. | Kim et al.54 | |
| Lung fibrosis |
Bacteroides ovatus (B. ovatus), Bacteroides stercoris (B. stercoris), Prevotella melaninogenica (P. melaninogenica) |
The EVs of three bacterial species (B. ovatus, B. stercoris, and P. melaninogenica) induced IL-17B expression and consequently promoted Th17 cell development via TLR-Myd88 adaptor signaling. The expanded lung microbiota induced IL-17B expression and consequently Th17 cell expansion to promote lung fibrosis. | Yang et al.58 | |
| Chronic inflammatory airway diseases | Staphylococcus aureus (S. aureus), Escherichia coli (E. coli) | E. coli EVs and S. aureus EVs induced MUC5AC expression via the ERK1/2 and p38 MAPK signaling pathways in human airway epithelial cells. E. coli EVs and S. aureus EVs significantly activated phosphorylation of ERK1/2 MAPK and p38 MAPK. An ERK1/2 MAPK inhibitor, a p38 MAPK inhibitor, an ERK1/2 MAPK siRNA, and a p38 MAPK siRNA significantly blocked the MUC5AC mRNA expression induced by E. coli EVs and S. aureus EVs. | Bae et al.56 | |
| Gastrointestinal | Gastric disease | Helicobacter pylori (H. pylori) |
H. pylori and H. pylori EVs induced the expression of IL-8 mRNA and protein. IL-8 expression was induced to different levels in response to H. pylori EVs from hosts with different gastric diseases. Exposure to H. pylori EVs increased the phosphorylation and reduced the degradation of inhibitor of NF-κB alpha. H. pylori EVs may aid the development of various gastric diseases by inducing IL-8 and NF-κB expression. |
Choi et al.69 |
| Inflammatory bowel disease | Vibrio cholera (V. cholerae) | V. cholerae EVs activated NOD1 and NOD2, resulting in NF-κB signaling and a pro-inflammatory immune response. NOD1 and NOD2 activation by EVs is of particular interest for inflammatory diseases such as Crohn’s disease, in which targeting NOD signaling may be an effective therapeutic strategy. | Bielig et al.66 | |
| Escherichia coli (E. coli) | E. coli EVs activated NOD1 signaling pathways in intestinal epithelial cells. NOD1 silencing and RIP2 inhibition significantly abolished EV-mediated activation of NF-κB and subsequent IL-6 and IL-8 expression. In addition, endocytosed EVs colocalized with NOD1, triggered the formation of NOD1 aggregates, and promoted NOD1 association with early endosomes. | Canas et al.67 | ||
| Metabolic | Type 2 diabetes (T2D) | Pseudomonas panacis (P. panacis) |
P. panacis EVs blocked the insulin signaling pathway in both skeletal muscle and adipose tissue, thereby promoting glucose intolerance in skeletal muscle. Moreover, P. panacis EVs induced typical diabetic phenotype characteristics, such as glucose intolerance after glucose administration or systemic insulin injection. P. panacis EVs induced T2D via the induction of insulin resistance in insulin-responsive organs. |
Choi et al.71 |
| Diabetes mellitus | Porphyromonas gingivalis (P. gingivalis) | P. gingivalis EVs equipped with gingipain proteases were translocated to the liver. In addition, the hepatic glycogen synthesis in response to insulin was decreased, and thus, high blood glucose levels were maintained. P. gingivalis EVs also attenuated insulin-induced Akt/GSK-3β signaling in a gingipain-dependent fashion in hepatic HepG2 cells. The delivery of gingipains mediated by P. gingivalis EV elicited changes in glucose metabolism in the liver and contributed to the progression of diabetes mellitus. | Seyama et al.72 | |
| Cancer | Stomach cancer | Helicobacter pylori (H. pylori) | H. pylori EVs contained CagA and VacA. They induced the production of TNF-α, IL-6 and IL-1β by macrophages and IL-8 by gastric epithelial cells. Additionally, H. pylori EVs induced the expression of IFN-γ, IL-17, and H. pylori EV-specific IgG1. H. pylori EVs infiltrated and remained in the stomach for an extended time. H. pylori EVs, which are abundant in the gastric juices of gastric cancer patients, can induce inflammation and possibly cancer in the stomach, mainly via the production of inflammatory mediators from gastric epithelial cells after selective uptake by the cells. | Choi et al.77 |
| Lung cancer | Staphylococcus aureus (S. aureus) Escherichia coli (E. coli) | S. aureus and E. coli EVs triggered the Th17 response. Generally, S. aureus and E. coli EVs induced IL-17 production through polarized Th17 cells and thus caused neutrophilic inflammation. The neutrophilic inflammatory response induces epithelial cell dysplasia and MMP expression, which can lead to lung cancer. | Yang et al.53 | |
| Central nervous system | Alzheimer’s disease | Paenalcaligenes hominis (P. hominis) |
Oral administration of P. hominis EVs caused cognitive impairment in mice. P. hominis EV treatment increased NF-κB+/Iba1+, LPS+/Iba1+, and IL-1R+ cell counts in the hippocampus. P. hominis EV treatment reduced BDNF expression in the hippocampus while increasing IL-1β expression in the blood. Vagotomy significantly reduced the occurrence of cognitive impairment caused by P. hominis EV gavage. Vagotomy inhibited EV-induced changes in NF-κB+/Iba1+, LPS+/Iba1+, and IL-1R+ cell populations in the hippocampus. The occurrence of cognitive impairment induced by P. hominis EVs was significant. Fluorescein isothiocyanate (FITC)-conjugated EVs were detected in the pyramidal region of the hippocampus. However, vagotomy significantly reduced the FITC-conjugated EV-containing CD11c+ cell population. Furthermore, oral gavage of P. hominis EVs increased bacterial 16S rDNA levels in the hippocampus. The translocation of P. hominis EVs through the vagus nerve resulted in cognitive impairment induced by brain inflammation. |
Lee et al.81 |
| Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans) | A. actinomycetemcomitans EVs administered via intracardiac injection into mice were successfully delivered to the brain after crossing the blood–brain barrier, and the extracellular RNA cargos increased the expression of TNF-α through the TLR-8 and NF-κB signaling pathways in the mice. Thus, host gene regulation by microRNAs originating from A. actinomycetemcomitans EVs is a novel mechanism for host gene regulation, and transfer of A. actinomycetemcomitans EV extracellular RNAs to the brain may cause neuroinflammatory diseases like Alzheimer’s disease. | Han et al.79 |
GM-CSF granulocyte−macrophage colony-stimulating factor, ERK extracellular signal-regulated kinase, MAPK mitogen-activated protein kinase, NOD nucleotide oligomerization domain, NLR NOD-like receptor, MMP matrix metalloproteinase.
Skin diseases
Atopic dermatitis (AD) is a chronic inflammatory disease of the skin and is defined as eczematous lesions with pruritus and xerosis. AD skin lesions show distinct features, such as abnormal skin barrier function with epidermal hyperplasia and S. aureus colonization. Abnormal skin barrier function induced by the death of keratinocytes is one of the major causative factors of AD43. Skin inflammation has been associated with SaEVs43,48. SaEVs increase IL-6, IL-17, serum IgG1, IgE, thymic stromal lymphopoietin, macrophage inflammatory protein-1α, and eotaxin levels and recruit mast cells and eosinophils in the skin48. A previous study on the metagenomics of MEVs showed that Staphylococcus accounted for the highest proportion of bacterial genera in AD patients, and the proportion of Staphylococcus significantly decreased after treatment49.
If the skin barrier is disrupted, pathogens can penetrate the human body. Therefore, skin barrier disruption is a major cause of AD exacerbation43. α-Hemolysin can induce keratinocyte cell death, consequently enhancing skin penetration of high-molecular-weight allergens. SaEV-associated α-hemolysin was found to induce keratinocyte necrosis and induce epidermal thickening and eosinophilic inflammation in the dermis. SaEVs were internalized into the cytoplasm of keratinocytes, and EVs efficiently delivered α-hemolysin to the cytoplasm. These findings suggest that EV-associated toxins are key molecules in disease pathogenesis43. Therefore, SaEVs are useful in the development of in vivo models of AD49. Propionibacterium acnes (P. acnes) EVs induce dysregulated epidermal differentiation in human keratinocytes via TLR2-mediated signaling pathways. P. acnes EVs may play pivotal roles in the early stages of the development of acne vulgaris50.
Lung diseases
The main causative factor of chronic inflammatory diseases in the lung has been suggested to be exposure to chemical substances such as cigarette smoke and air pollution51,52 because of the inherent pharmacological or toxic effects of such chemical substances. However, the etiology of a large proportion of chronic inflammatory diseases is not related to these factors. Biological factors, including allergens, viruses, and bacterial factors, can cause severe inflammatory reactions52.
Airway exposure to MEVs triggers two main pathophysiological mechanisms, Th17, and Th1 responses, based on whether the parent bacterial cell is extracellular or intracellular, respectively. Specifically, extracellular bacteria-derived EVs generally cause neutrophilic inflammation through IL-17 release by polarized Th17 cells, whereas intracellular MEVs induce mononuclear inflammation through IFN-γ produced by polarized Th1 cells53. The pathogenic MEV-induced neutrophilic inflammatory response leads to airway hyperreactivity and fibrosis that contribute to asthma development. The combination of increased elastase production and fibrosis induced by neutrophilic inflammation can cause emphysema, which is a key pathology of chronic obstructive pulmonary disease (COPD). Furthermore, intracellular MEVs lead to Th1 polarization and subsequent IFN-γ-induced mononuclear inflammation, leading to increased elastase production in the alveoli and causing pulmonary tuberculosis-associated emphysema53.
In several in vivo tests, MEVs caused severe airway inflammation, which resulted in severe asthma and emphysema52. Previous studies have shown that repeated exposure to Escherichia coli (E. coli) EVs (EcEVs) in the airways led to IL-17-dependent neutrophilic inflammation and emphysema in conjunction with elastase upregulation54. Repeated airway exposure to SaEVs induced both Th1 and Th17 immune responses with IFN-γ and IL-17, respectively, and neutrophilic pulmonary inflammation was increased primarily through TLR2 engagement55. EcEVs and SaEVs induced MUC5AC expression in human airway epithelial cells. MUC5AC, a major mucin, is overexpressed in chronic inflammatory airway diseases and causes airway obstruction, increases susceptibility to infection, and decreases pulmonary function56. In addition, Pseudomonas aeruginosa EVs increase pulmonary inflammation through activation of the TLR2 and TLR4 pathways through dose-dependent increases in chemokines (CXCL1 and CCL2) and cytokines (IL-1β, TNF-α, IL-6, and IFN-γ), as well as increases in neutrophils and macrophages57. In addition, EVs from Bacteroides ovatus, Bacteroides stercoris, and Prevotella melaninogenica induce IL-17 expression and consequently promote Th17 cell development associated with lung fibrosis58.
Indoor dust containing MEVs induces both Th1 and Th17 responses associated with neutrophilic pulmonary inflammation59,60 and over 50% of the patients with childhood asthma were found to be sensitized to MEVs in indoor dust, suggesting that MEVs in indoor dust could be an important causative factor in the pathogenesis of childhood asthma59. Moreover, the anti-indoor dust EV IgG antibody was found to be significantly associated with the risk of asthma, COPD, and lung cancer (LC)61. These results suggest the potential of MEVs as a major risk factor in the development of inflammatory diseases and cancer in the lung.
Gastrointestinal disease
Inflammatory bowel disease (IBD) is characterized by chronic inflammation of the gastrointestinal tract. Although the exact etiology of IBD is under investigation, disruption of the normal intestinal flora and normal intestinal function have been linked to the development of IBD. Alteration of the commensal gut microbial ecosystem, which interacts with gut epithelial cells and plays significant roles in the maintenance of the mucosal barrier, induces dysfunctional immunomodulation and metabolic activity in the gut62. Such disruptions lead to reductions in mucus barrier functionality, including decreased tight junctions in the intestinal epithelial lining. Since the integrity of the mucosal barrier is significantly reduced, inflammatory agents such as microbial components in the intestinal lumen can induce mucosal inflammation associated with IBD63,64.
The NOD-like receptor (NLR) family, which is a family of cytoplasmic pattern recognition receptors, initiates inflammation by activating signaling pathways, including NF-κB signaling, followed by induction of caspase-1-mediated cleavage of IL-1β/IL-18 and production of gasdermin, which causes pyroptotic cell death via the formation of pores in the plasma membrane. NLRs or NLR-associated proteins have been found to be genetically associated with susceptibility to IBD65. Vibrio cholerae EVs and EcEVs activate NOD1 signaling66,67, resulting in NF-κB signaling and a pro-inflammatory immune response. NOD1 activation by MEVs can lead to IBDs, such as Crohn’s disease68.
Helicobacter pylori (H. pylori) EVs (HpEVs) may play a significant role in the development of various gastric diseases through IL-8 production and NF-κB activation. HpEVs may play a crucial role in the pathogenesis of gastric disease associated with H. pylori69.
Metabolic diseases
Commensal bacteria in the gastrointestinal tract play an important role in nutrient absorption and the fermentation of dietary fibers. In addition, the gut microbiota provides a critical function in the production of short-chain fatty acids (SCFAs) for energy sources and epigenetic regulation via histone deacetylase inhibition70. Gut microbiota dysbiosis resulting in irregular SCFA production can lead to dysregulated host metabolic functionality. Moreover, MEVs can participate in the development of metabolic diseases. Pseudomonas panacis EVs promoted by a high-sucrose/fat diet can induce type 2 diabetes (T2D) via the induction of insulin resistance in insulin-responsive organs. Pseudomonas panacis EVs blocked the insulin signaling pathway in both skeletal muscle and adipose tissue, thereby promoting glucose intolerance in skeletal muscle, while these EVs induced typical diabetic phenotype characteristics, such as glucose intolerance after glucose administration or systemic insulin injection71.
In one study, Porphyromonas gingivalis EVs equipped with proteases gingipains were translocated to the liver. Subsequently, hepatic glycogen synthesis in response to insulin was decreased, and high blood glucose levels were maintained. Porphyromonas gingivalis EVs also attenuated insulin-induced Akt/GSK-3β signaling in a gingipain-dependent fashion in hepatic HepG2 cells. The delivery of gingipains mediated by Porphyromonas gingivalis EVs elicited changes in glucose metabolism in the liver and contributed to the progression of T2D72. These results suggest a close relationship between MEVs and metabolic disease.
Cancers
MEVs are known to be associated with tumors, and cancer patients show different MEVs than healthy people. Previous studies have elucidated that Th17 cells play an important role in pathogen clearance during host defense reactions but induce tissue inflammation in the pathogenesis of autoimmune disease73. Recent studies reported that Th17 cells played an important role in LC pathogenesis74,75, and the immune effector T cells in small-cell LC patients included a large proportion of Th17 cells76. Airway exposure to SaEVs and EcEVs triggers the Th17 response. Generally, OMVs induce IL-17 production through polarized Th17 cells and thus cause neutrophilic inflammation. The neutrophilic inflammatory response induces epithelial cell dysplasia and the induction of matrix metalloproteinase expression, possibly leading to LC. COPD also increases the risk of LC53.
The abundances of H. pylori and HpEVs in gastric cancer patients were significantly higher than those in healthy people. HpEVs contain CagA and VacA proteins that can induce the production of TNF-α, IL-6, IL-1β, and IL-8. In addition, HpEVs induce the expression of IFN-γ, IL-17, and EV-specific IgGs in mice. In summary, HpEVs might be involved in the pathogenesis of gastric cancer, mainly via the induction of the production of inflammatory mediators by gastric epithelial cells after selective uptake by the cells77.
Central nervous system (CNS) diseases
MEVs can penetrate the blood–brain barrier (BBB) through three possible mechanisms: (1) receptor-mediated transcytosis, (2) paracellular passage in disease states, or (3) via EVs in infected immune cells. Once MEVs are inside brain tissues after penetrating the BBB, the components and cargo in EVs act as ligands of innate immune receptors, such as TLRs, and the NALP3 inflammasome, and activate the inflammatory immune response78.
A previous study showed that Aggregatibacter actinomycetemcomitans EVs can cross the mouse BBB and are present in the brain 24 h after intracardiac injection, inducing TNF-α production via extracellular RNAs through the TLR-8 and NF-κB signaling pathways79. In the mouse brain, increased TNF-α may cause neuroinflammatory diseases, such as Alzheimer’s disease79. Similarly, Aggregatibacter actinomycetemcomitans EVs can successfully deliver extracellular RNAs to brain monocytes/microglial cells to cause neuroinflammation associated with the upregulation of IL-6 through NF-κB activation80. Oral administration of Paenalcaligenes hominis EVs increased bacterial 16 S rDNA levels in the hippocampus, causing cognitive impairment in mice, whereas oral administration of LPS caused more severe cognitive impairment. Paenalcaligenes hominis EV treatment increased NF-κB+/Iba1+, LPS+/Iba1+, and IL-1R+ cell counts and reduced BDNF expression in the hippocampus while increasing IL-1β expression in the blood81. These findings provide evidence that some MEVs are important in the pathogenesis of neuroinflammatory diseases.
Recently, we performed microbiome analysis of EVs in blood and brain tissue samples from brain tumor (BT) patients and healthy controls (HCs)82. Blood EV microbiome analysis at the phylum level showed that the abundance of Firmicutes was significantly higher in BT patients than in HCs, whereas those of Proteobacteria and Actinobacteria were significantly lower. In addition, at the genus level, the proportions of EVs from Ruminococcaceae UCG-014, Lachnospiraceae NK4A136, Lactobacillus, Ruminococcaceae UCG-013, Ruminiclostridium 6, and Peptoclostridium were significantly higher but those from Escherichia-Shigella, Blautia, Bifidobacterium, Streptococcus, and Sphingomonas were significantly lower in BT patients than in HCs. Moreover, brain tissue microbiome analysis at the genus level showed that the abundance of EVs from Bacteroides and Erysipelatoclostridium was significantly higher but those from Chloroplast(c), Prevotella 9, and Saccharibacteria(p) were significantly lower in BT patients than in HCs82. These findings suggest that MEVs might be closely linked to the pathogenesis of CNS diseases, although further research is needed.
Microbial EVs as biomarkers
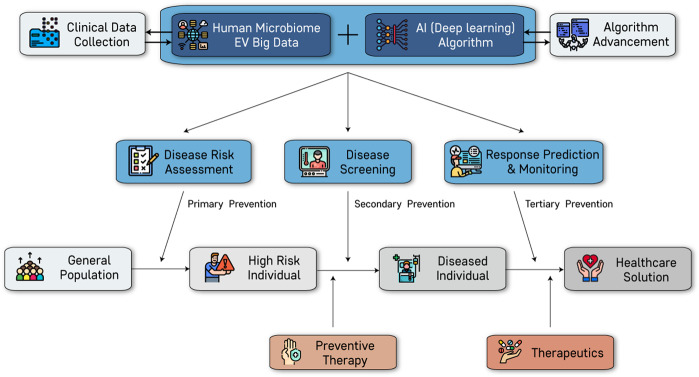
Liquid biopsy, also known as fluid biopsy or fluid phase biopsy, is the sampling and analysis of nonsolid biological tissue. MEVs are a newly emerging biomarker that can be attested for by liquid biopsy. Moreover, AI-based analysis of clinical and MEV data enables the development of a more accurate prediction of disease status (Fig. 3).
Fig. 3. Application of microbial EVs for precision medicine.
Precision medicine goals, including risk assessment, screening, prediction, and monitoring, are based on analysis using an artificial intelligence (AI) algorithm through human microbial EV big data and clinical data. High-risk individuals and diseased individuals can be identified by risk assessment, and preventive therapy can reduce disease risk. Diseased individuals identified by screening and diagnosis are suggested therapeutics based on the results of response prediction and monitoring. These processes can realize microbial EV-based health care solutions.
Commensal microbes secrete MEVs for cell-to-cell communication. MEVs penetrate the mucosal barrier, circulate throughout the body, and then are excreted via feces, urine, and exhaled air in their intact forms, unlike live microbes, which are restricted to the mucosal lumen or skin surface71. Certain MEVs act as etiological agents of various diseases, while some MEVs have a protective role in disease pathogenesis. Thus, MEVs circulating in our body provide us with good information for health and disease status.
An organism’s evolutionary history is documented in its genome. rDNA encoding rRNA changes slowly and is useful for investigating relationships between taxa that diverged hundreds of millions of years ago. In contrast, mitochondrial DNA evolves rapidly and is more useful for exploring more recent evolutionary events. MEVs harbor 16S rDNA, which provides taxonomy information83. However, there are some issues in comparing published microbiome data. First, the regions targeted during the investigation of the microbiota are different for the 16S rDNA and NGS platforms. The diversity and abundance of the bacterial mock community are different between the V1-V3 region and V3-V4 region of the 16S rRNA or MiSeq platform (Illumina, USA) and the Roche 454 platform (Roche, Switzerland)84. Second, microbiome information differs among the databases used for taxonomic assignments. There are several public databases, such as SILVA, RDP, Greengenes, EzTaxon, and NCBI, and these databases have different taxonomic classifications85. Third, these public databases are updated continuously, and unassigned taxa have the potential to be assigned in the future.
Disease diagnosis and prediction goals, such as risk assessment, early diagnosis, treatment response prediction, and disease monitoring, are important for the reduction of mortality and the increase of quality of life86,87. MEVs are novel biomarkers for in vitro diagnostics. Several 16S rDNA amplicon studies using serum, urine, feces, and skin-fluid samples have shown significant differences in MEV composition between healthy and patient groups (Table 2). The clinical performance of the diagnostic models as evaluated by area under the curve (AUC) analysis using external validation set was 0.97, 1.00, 0.95, 0.93, 0.87, 0.82, 1.00, and 1.00 for asthma88, AD89, colorectal cancer90, BT82, hepatocellular carcinoma91, gastric cancer92, pancreatic cancer93, and biliary tract cancer94, respectively. Furthermore, diagnostic models were developed to predict the prognosis of hepatic diseases91. MEV-based models, in combination with microbiome and metabolome data, provided the best performance; the performance of colorectal cancer models using both metagenomic and metabolomic data was better than that of models using microbiome or metabolome data alone90. Moreover, EV microbiome data provided better performance when combined with tumor markers; the diagnostic performance in ovarian cancer was better using both MEV data and the tumor marker CA-125 than either biomarker alone95.
Table 2.
Diagnostic models using microbiome data derived from EVs.
| Patient group | Sample type | Genera | Diagnostics | Ref. | ||
|---|---|---|---|---|---|---|
| Enriched in case | Enriched in control | Method | Performance | |||
| Colorectal cancer | Feces | Faecalibacterium, Eubacterium, Ruminococcus, Bifidobacterium | Escherichia-Shigella, Pseudomonas, Methylobacterium, Molicutes, Proteus | Logistic regression using age, sex and metagenomic biomarkers selected by statistical analysis | AUC: 0.95, Sen: 0.90, Spe: 1.00, Acc: 0.93 | Kim et al.90 |
| Logistic regression using age, sex and metagenomic and metabolomic biomarkers selected by statistical analysis | AUC: 1.00, Sen: 1.00, Spe: 1.00, Acc: 1.00 | Kim et al.90 | ||||
| Atopic dermatitis | Blood | Escherichia-Shigella, Enterococcus, Alistipes, Klebsiella, Veillonella, Bifidobacterium, Akkermansia, Bacteroides | Acinetobacter, Pseudomonas, Parabacteroides, Proteus, Prevotella, Dialister, Rhizobium, Sphingomonas, Staphylococcus | Logistic regression using biomarkers selected by LEfSe | AUC: 1.00, Sen: 1.00, Spe: 1.00, Acc: 1.00 | Yang et al.89 |
| Urine | Pseudomonas, Alicyclobacillus, Propionibacterium, Corynebacterium | Lactobacillus, Leuconostoc, Lactococcus, Bradyrhizobium | – | – | Kim et al.107 | |
| Skin washing fluid | Staphylococcus, Pseudomonas, Streptococcus, Acinetobacter | Alcaligenaceae, Sediminibacterium, Lactococcus, Phaeospirillum, Rhodococcus, Lactobacillus, Methylobacterium | – | – | Kim et al.122 | |
| Asthma | Blood | Klebsiella, Bacteroides, Alistipes, Subdoligranulum, Bifidobacterium, Faecalibacterium, Veilonella, Eubacterium, Parabacteroides, Prevotella | Pseudomonas, Akkermansia, Citrobacter, Staphylococcus, Micrococcus, Acinetobacter, Lactobacillus, Corynebacterium, Sphingomonas, Propionibacterium, Cupriavidus, Streptococcus | Logistic regression using biomarkers selected by LEfSe with age and sex as covariates | AUC: 0.97, Sen: 0.92, Spe: 0.93, Acc: 0.92 | Lee et al.88 |
| – | – | Logistic regression using antibacterial EV IgG, IgG1, and IgG4 with smoking status as a covariate | AUC: 0.78, Sen: 0.65, Spe: 0.88, Acc: 0.71 | Yang et al.96 | ||
| Autism | Urine | Halomonas, Streptococcus, Rhodococcus, Bacteroidales S24-7, Akkermansia, | Pseudomonas, Sphingomonas, Agrobacterium, Achromobacter | – | – | Lee et al.123 |
| Bipolar depressive disorder | Blood | Faecalibacterium, Dialister, Klebsiella, Bacteroidales S24-7, Escherichia-Shigella, Ruminococcus, Alistipes, Prevotella | – | – | Rhee et al.124 | |
| Major depressive disorder | Blood | Dialister, Faecalibacterium, Prevotella, Alistipes, Bacteroidales S24-7, Corynebacteriaceae, Escherichia-Shigella | Pseudomonas | – | – | Rhee et al.124 |
| Brain tumor | Blood | Turicibacter, Lactococcus, Lactobacillus, Staphylococcus, Peptoclostridium, Diaphorobacter, Klebsiella, Propionibacterium, Acinetobacter, Salmonella | Stenotrophomonas, Sphingomonas, Actinomyces, Streptococcus, Bifidobacterium, Knoellia, Pseudomonas, Corynebacterium, Veillonella | Logistic regression using biomarkers selected by LEfSe | AUC: 0.97, Sen: 0.93, Spe: 0.90, Acc: 0.91 | Yang et al.82 |
| Machine learning algorithm based on the gradient boosting machine (GBM) model | AUC: 0.99, Sen: 1.00, Spe: 0.94 | Yang et al.82 | ||||
| Tissue | Bacteroides, Erysipelatoclostridium | Bactroidales S24-7, Prevotella | – | – | Yang et al.82 | |
| Chronic rhinitis | Urine | Propionibacterium, Methylobacterium, Enhydrobacter | Achromobacter, Enterobacteriaceae | – | – | Samra et al.125 |
| Allergic rhinitis | Urine | Methylobacterium | Agrobacterium, Achromobacter, Enterobacteriaceae | – | – | Samra et al.125 |
| Atopic asthma | Urine | Methylobacterium, Sphingomonadaceae | Enterobacteriaceae | – | – | Samra et al.125 |
| Hepatocellular carcinoma | Blood | Staphylococcus, Acinetobacter | Pseudomonas, Streptococcus | Logistic regression using age, sex and biomarkers selected by statistical analysis | AUC: 0.88, Sen: 0.73, Spe: 0.85, Acc: 0.82 | Cho et al.91 |
| Biliary tract cancer | Blood | Ralstonia | Corynebacterium, Comamonas | Logistic regression using stepwise selection with age and sex as covariates | AUC: 1.00, Sen: 1.00, Spe: 1.00, Acc: 1.00 | Lee et al.94 |
| Preterm birth | Blood | Bacteroides, Lactobacillus, Sphingomonas, Rhizobium | Delftia, Pseudomonas, Stenotrophomonas | – | – | You et al.126 |
| Alcoholic hepatitis | Feces | Veillonella, Lactobacillales | Eubacterium, Oscillibacter, Christensenellaceae | – | – | Kim et al.127 |
| COPD | Blood | – | – | Logistic regression using antibacterial EV IgG, IgG1, and IgG4 with smoking status as a covariate | AUC: 0.79, Sen: 0.90, Spe: 0.61, Acc: 0.81 | Yang et al.96 |
| Gastric cancer | Urine | Corynebacterium 1, Neisseria, Fusobacterium, Diaphorobacter, Actinomyces, Porphyromonas, Cloacibacterium, Peptoniphilus | Acinetobacter, Staphylococcus, Bifidobacterium, Sphingomonas | Logistic regression using metagenomic biomarkers selected by statistical analysis | AUC: 0.82, Sen: 0.68, Spe: 0.85, Acc: 0.76 | Park et al.92 |
| Lung cancer | Blood | – | – | Logistic regression using antibacterial EV IgG, IgG1, and IgG4 with smoking status as a covariate | Auc: 0.81, Sen: 0.85, Spe: 0.61, Acc: 0.80 | Yang et al.96 |
| Lung cancer (from COPD) | Blood | – | – | Logistic regression using antibacterial EV IgG, IgG1, and IgG4 with smoking status as a covariate | AUC: 0.74, Sen 0.69, Spe: 0.69, Acc: 0.69 | Yang et al.96 |
| Ovarian cancer (from benign ovarian tumor) | Blood | Acinetobacter | Logistic regression using biomarkers, age, serum CA-125 levels, and Acinetobacter EVs selected by statistical analysis | AUC: 0.85, Sen: 0.82, Spe: 0.68 | Kim et al.95 | |
| Pancreatic cancer | Blood | Ruminococcaceae UCG-014, Lachnospiraceae NK4A136 group, Akkermansia, Turicibacter, Ruminiclostridium, Lachnospiraceae UCG-001 | Stenotrophomonas, Sphingomonas, Propionibacterium, Corynebacterium 1 | Logistic regression using age, sex and biomarkers selected by statistical analysis | AUC: 1.00, Sen: 1.00, Spe: 0.92 | Kim et al.93 |
AUC area under curve, Sen sensitivity, Spe specificity, Acc accuracy.
The presence of antibodies against MEVs can also provide good information for the diagnosis of disease. The presence of IgG antibodies against EVs in indoor dust is closely associated with asthma, COPD, and LC61. The levels of IgGs against pathogenic MEVs, such as EVs from S. aureus, Acinetobacter baumannii, Enterobacter cloacae, and Pseudomonas aeruginosa abundant in indoor dust, showed good AUC values: 0.78 for asthma, 0.79 for COPD, and 0.81 for LC. Thus, MEV-based diagnostic technologies, including MEV composition assessment and immunoassays to assess MEVs, can provide information on exposure to etiological agents96.
Therapeutic potential of microbial EVs
That commensal bacteria play an essential role in human health is beyond doubt, and it is now widely accepted that humans are holobionts or supra-organisms whose collective metabolic potential exceeds the sum of the individual eukaryotic and prokaryotic components. Probiotics or live biotherapeutics (LBPs) are live microorganisms that, when administered in adequate amounts, confer a health benefit on the host and are widely used in many countries in clinical practice. Paraprobiotics are currently referred to as “modified probiotics”, “inactivated probiotics”, “nonviable probiotics”, or “ghost probiotics”97–99. Paraprobiotics are a newly emerging modality, an immobilized version of probiotics that has gained traction in recent years due to concerns about the possibility of low tolerance of probiotics or LBPs, especially in pediatric populations and severely ill or immunocompromised patients100. The mechanism of action (MoA) of LBPs and parabiotics is not well understood, although the possible MoAs include immune system regulation and interference with pathogen attachment to host cells. Limited research has found that immobilized parabiotics release key bacterial components, including lipoteichoic acids, peptidoglycans, and exopolysaccharides, that exhibit key immunomodulatory effects and antagonizing effects against pathogens100.
Postbiotics, also known as metabiotics, biogenics, or simply metabolites, are soluble factors secreted by live microbes or released after microbial lysis, and they provide physiological benefits to the host101. Postbiotics are a newly emerging modality in therapy. Cell-to-cell communication is tightly regulated, and its disruption contributes to disease development. Intercellular communication can occur either via cell-to-cell contact or via soluble factors102. Soluble factors include proteins and small molecules that bind specific receptors on target cells, which then trigger a signaling cascade that will affect the cell phenotype. However, more recently, another process also involved in this type of communication has been highlighted. In addition to producing specific factors, microbial cells can send a package of information that is enclosed by a cell membrane, also called MEVs. Moreover, recent scientific evidence has shown that certain MEVs have protective effects against disease development or progression16,103–105. Thus, MEVs belong to the category of postbiotics and are a new modality for next-generation therapy to complement current therapeutics, such as small molecules, proteins, monoclonal antibodies, and cell therapeutics (Table 3).
Table 3.
Therapeutic mechanisms of microbial EVs.
| Species | Indication | Mechanism | Ref. |
|---|---|---|---|
| Akkermansia muciniphila (A. muciniphila) | Colitis | In vitro pretreatment with A. muciniphila EVs induced secretion of the proinflammatory cytokine IL-6 from colon epithelial cells. Additionally, oral application of A. muciniphila EVs protected against IBD characteristics, such as bodyweight loss, decreased colon length, and inflammatory cell infiltration of colon wall. | Kang et al.104 |
| Metabolic disease | A. muciniphila EV administration enhanced tight junction function, reduced bodyweight gain, and improved glucose tolerance in diabetic mice. Additionally, A. muciniphila EVs decreased the gut permeability of LPS-treated Caco-2 cells. The expression of occludin was increased by A. muciniphila EV treatment. | Chelakkot et al.106 | |
| Bacteroides flagilis (B. flagilis) | Colitis | PSA in B. flagilis EVs induced immunomodulatory effects and prevented experimental colitis. DCs sensed B. flagilis EV-associated PSA through TLR2, resulting in enhancement of regulatory T cells and anti-inflammatory cytokine production. DCs treated with B. flagilis EVs containing PSA prevented experimental colitis. | Shen et al.128 |
| Bifidobacterium longum (B. longum) | Food allergy | B. longum EVs bound specifically to mast cells and induced apoptosis without affecting T cell immune responses. Furthermore, injection of B. longum EVs carrying extracellular solute-binding protein into mice markedly reduced the occurrence of diarrhea in a mouse food allergy model. | Kim et al.111 |
| Bifidobacterium bifidum (B. bifidum) | Allergen-specific immunotherapy | DCs stimulated by B. bifidum EVs strongly promoted differentiation of functional CD25highFOXP3highCD127−/low Treg cells and induced the higher levels of IL-10 than of the proinflammatory cytokines IFN-γ, TNF-α and IL-17. B. bifidum EVs can be potentially used as an effective adjuvant for immunotherapy. | Lopez et al.129 |
| Bacteroides vulgatus (B. vulgatus) | Systemic disease | BMDCs stimulated by B. vulgatus EVs led to the induction of a tolerant semimature phenotype. Thereby, the MAMPs delivered by B. vulgatus EVs were crucial for the interaction and the resulting maturation of immune cells. In addition to molecules binding to host TLR4, a yet unknown ligand of TLR2 is indispensable for the conversion of immature BMDCs into a semimature state. Thus, by crossing the epithelial mucus layer and directly contacting host cells, B. vulgatus EVs mediated cross-tolerance via the transport of various Toll-like receptor antigens. | Maerz et al.130 |
| Escherichia coli (E. coli) | Cancer | Administered E. coli EVs specifically targeted and accumulated in the tumor tissue and subsequently induced the production of the antitumor cytokines CXCL10 and IFN-γ. | Kim et al.131 |
| Klebsiella pneumonia (K. pneumoniae) | Breast cancer | K. pneumoniae EVs enhanced the antihormonal effects of tamoxifen in MCF7 cells via Cyclin E2 and p-ERK. | An et al.132 |
| Lactobacillus plantarum (L. plantarum) | Atopic dermatitis | In vitro, IL-6 secretion from keratinocytes and macrophages was decreased and cell viability was restored with L. plantarum EV treatment. Additionally, in mouse AD models, L. plantarum EV administration reduced epidermal thickening and the IL-4 level. | Kim et al.107 |
| Skin inflammation | L. plantarum EVs promoted differentiation of human monocytic THP1 cells toward an anti-inflammatory M2 phenotype, specifically, the M2b phenotype, by inducing biased expression of cell surface markers and cytokines associated with M2 macrophages. Pretreatment or posttreatment with L. plantarum EVs under inflammatory M1 macrophage-favoring conditions, induced by LPS and IFN-γ, inhibited M1-associated surface marker (HLA-DRα) expression. Moreover, L. plantarum EVs treatment significantly induced IL-1β, GM-CSF and IL-10 expression in human skin organ cultures. Hence, L. plantarum EVs can trigger M2 macrophage polarization in vitro, and induce an anti-inflammatory phenomenon in human skin. | Kim et al.108 | |
| Depression | L. plantarum EVs could have a role in regulating neuronal function and stress-induced depressive-like behaviors. HT22 cells treated with the stress hormone glucocorticoid had reduced expression of BDNF and Sirt1, whereas L. plantarum EVs treatment reversed glucocorticoid-induced decreases in the expression of BDNF and Sirt1. siRNA-mediated knockdown of Sirt1 in HT22 cells decreased BDNF4, a splicing variant of BDNF, and Creb expression, suggesting that Sirt1 plays a role in the L. plantarum EV-induced increase in BDNF and Creb expression. L. plantarum EV treatment in mice rescued the reduced expression of BDNF, and blocked stress-induced depressive-like behaviors. Furthermore, L. plantarum EV injection inhibited the reduced expression of BDNF and Nt4/5 and stress-induced depressive-like behaviors. These results suggest that L. plantarum EVs can change the expression of neurotropic factors in the hippocampus and induce antidepressant-like effects in mice with stress-induced depression. | Choi et al.117 | |
| Lactobacillus paracasei (L. paracasei) | Inflammatory bowel disease |
In in vitro experiments, L. paracasei EVs reduced the expression of the LPS-induced pro-inflammatory cytokines (IL-1α, IL-1β, IL-2, and TNF-α), LPS-induced inflammation in HT29 cells, and the activation of inflammation-associated proteins (COX-2, iNOS, NF-κB, and NO) but increased the expression of the anti-inflammatory cytokines IL-10 and TGF-β. In in vivo mouse experiments, oral administration of L. paracasei EVs also protected against DSS-induced colitis by reducing weight loss, maintaining colon length, and decreasing the disease activity index. In addition, L. paracasei EVs induced the expression of endoplasmic reticulum stress-associated proteins, while the inhibition of these proteins blocked the anti-inflammatory effects of L. paracasei EVs in LPS-treated HT29 cells, restoring the pro-inflammatory effects of LPS. L. paracasei EVs attenuated LPS-induced inflammation in the intestine through endoplasmic reticulum stress activation. |
Choi et al.105 |
| Lactobacillus kefir (L. kefir), Lactobacillus kefiranofaciens (L. kefiranofaciens), Lactobacillus kefirgranum (L. kefirgranum) | Inflammatory bowel disease | Treatment of TNF-α-stimulated Caco-2 cells with each kefir-derived Lactobacillus EV type (L. kefir, L. kefiranofaciens, and L. kefirgranum) reduced both mRNA expression and the IL-8 level in association with inhibition of TNF-α signaling by reducing the phosphorylation of p65, a subunit of NF-κB. Subsequent administration of kefir-derived Lactobacillus EVs into IBD mice alleviated bodyweight loss and rectal bleeding and enhanced stool consistency. Histological examination showed that kefir-derived Lactobacillus EVs substantially reduced the infiltration of transmural leukocytes and loss of goblet cells within the colon, and the serum level of myeloperoxidase was significantly lower in the EV-treated group than control group. | Seo et al.109 |
| Lactobacillus sakei (L. sakei) | Immune system disease | IgA production in Peyer’s patch cells was enhanced by L. sakei EVs. In addition, stimulation of BMDCs with L. sakei EVs increased gene expression of inducible NO synthase, retinaldehyde dehydrogenase 2, and several inflammatory cytokines via TLR2. | Miyoshi et al.110 |
| Lactobacillus rhamnosus (L. rhamnosus) | Enteric nervous system disease | Ingested labeled L. rhamnosus EVs were detected in murine Peyer’s patch DCs within 18 h. After 3 days, Peyer’s patch and mesenteric lymph node DCs assumed a regulatory phenotype and increased functional regulatory CD4+25+Foxp3+ T cells. L. rhamnosus EVs similarly induced phenotypic changes in cocultured DCs via multiple pathways including C-type lectin receptors specific intercellular adhesion molecule-3 grabbing nonintegrin-related 1 and Dectin-1, as well as TLR-2 and TLR-9. L. rhamnosus EVs also decreased the amplitude of neuronally dependent MMCs in an ex vivo model of peristalsis. Gut epithelial application of L. rhamnosus EVs caused an increase in the number of action potentials recorded in adjacent patch-clamped sensory neurons. | AI-Nedawi et al.133 |
PSA polysaccharide A, DC dendritic cell, BMDC bone marrow-derived dendritic cell, ERK extracellular signal-regulated kinase, MAMP microbe-associated molecular pattern.
Therapy for immune and inflammatory diseases
Akkermansia muciniphila (A. muciniphila) is considered a beneficial anaerobic bacterium living in the large bowel. A previous study104 showed that A. muciniphila EVs (AmEVs) were decreased in dextran-induced colitis mice vs. healthy mice. In vitro pretreatment with AmEVs improved the production of the proinflammatory cytokine IL-6 by EcEV-stimulated intestinal epithelial cells. In addition, AmEVs protected against dextran-induced IBD phenotypes, including bodyweight loss, decreased colon length, and inflammatory cell infiltration of the colon wall. Oral therapy with AmEVs protected against LPS-induced gut barrier disruption, and this beneficial effect was mediated by the AMPK signaling pathway106.
Lactobacillus EVs have the possibility to be developed as a medicine for chronic inflammatory diseases such as IBD and AD105,107. Lactobacillus plantarum (L. plantarum) EVs (LpEVs) have anti-inflammatory effects and prevent SaEV-induced cell death107. Oral administration of LpEVs has a therapeutic effect against SaEV-induced atopic dermatitis107. LpEVs promoted the differentiation of human monocytic THP1 cells toward an anti-inflammatory M2 phenotype108. Treatment of LpEVs under inflammatory M1 macrophage-favoring conditions with LPS and IFN-γ inhibited M1 macrophage-associated HLA-DRα expression108. Furthermore, LpEV treatment significantly induced IL-1β, GM-CSF, and IL-10 expression in human skin organ cultures108. In the case of IBD, the administration of Lactobacillus paracasei EVs reduced weight loss, maintained colon length, decreased the disease activity index, and protected against dextran-induced IBD105. Lactobacillus paracasei EVs inhibited signaling via NF-kB, which is a major transcription factor inducing expression of the inflammatory cytokines IL-6 and TNF-α, and the expression of inducible nitric oxide synthase and cyclooxygenase-2, which is mediated by the unfolded protein response of the endoplasmic reticulum (ER) (UPRER) against ER stress105. Moreover, EVs derived from Lactobacillus kefir, Lactobacillus kefiranofaciens, and Lactobacillus kefirgranum inhibited the production of IL-8 from TNF-α-activated Caco-2 cells, reduced rectal bleeding, and improved stool consistency in IBD mice109. In addition, it was reported that Lactobacillus sakei EVs play crucial roles in the immune system through enhanced IgA production in Peyer’s patch cells110.
Bifidobacterium longum EVs were reported to alleviate food allergy symptoms mainly by inducing apoptosis of mast cells in the small intestine111. In terms of the components in Bifidobacterium longum EVs participating in mast cell apoptosis, extracellular solute-binding protein in the EVs specifically reduced the number of mast cells and alleviated diarrhea in mice with food allergies.
Therapy for metabolic diseases
A. muciniphila LBP and parabiotics (or lysates) have been shown to be beneficial in reducing fat mass and glycemia in T2D patients. The benefits involved the interaction of the A. muciniphila OM protein Amuc_1100 with host cell TLR2112. Interestingly, it was shown that AmEVs were decreased in T2D patients vs. HCs113. Moreover, oral therapy with AmEVs had a therapeutic effect against metabolic changes, such as the obesity and glucose intolerance induced by a high-fat diet. These metabolic benefits are likely linked to the beneficial effect of AmEVs on gut barrier integrity, which is mainly mediated by AMPK signlaing113. Taken together, the beneficial effects of A. muciniphila on metabolic phenotypes might be mediated by AmEVs.
Immunomodulatory therapy for malignant diseases
Cancer immunotherapies such as immune checkpoint inhibitors and chimeric antigen receptor therapies are now being recognized as a promising approach to overcome various malignant diseases114. MEVs are nanosized particles that can easily penetrate the gut mucosal barrier and readily interact with immune cells in the gut, giving them great therapeutic potential in the immunology−oncology area. A previous study reported that systemic administration of genetically modified EcEVs significantly and effectively induced antitumor immune responses and fully eradicated established tumors without notable adverse effects115. Furthermore, that study showed that systematically administered EcEVs targeted and accumulated in the tumor tissue and subsequently induced the production of the antitumor cytokines CXCL10 and IFN-γ. Moreover, the study found that IFN-γ-deficient mice did not induce such an EV-mediated immune response, suggesting that this antitumor effect is dependent on IFN-γ.
Therapy for CNS diseases
Recent evidence indicates that the gut microbiota plays a key role in regulating mental disorders, but the mechanisms by which the gut microbiota regulates brain function are controversial. L. plantarum is a gram-positive bacterium present in dairy, fermented vegetables, and the gut116. Recent studies have reported the beneficial effects of L. plantarum on stress-induced behavioral dysfunction117; for example, it improves cognitive deficits118, attenuates anxiety behavior119, and reduces oxidative stress markers120. In addition, we investigated whether LpEVs have a therapeutic effect against stress-induced depression117. The study showed that LpEVs protected against and reversed the depressive behaviors induced by psychological stress. The beneficial effect of LpEVs was linked to the overexpression of neurotrophic factor BDNF and the Nt4/5 and Sirt1 genes, which are decreased by the stress hormone glucocorticoid117. Collectively, these findings suggest that secreted L. plantarum EVs show highly beneficial effects and LpEVs can be used to develop novel therapies that penetrate the BBB and modulate CNS function.
Concluding remarks
We reviewed MEVs as a new tool for precision medicine, especially focusing on the role of MEVs in disease pathogenesis and as biomarkers and MEV therapeutic potential. The pathogenesis of currently intractable diseases is related to cellular aging and increased reactive oxygen species (ROS). Intracellular ROS cause loss of proteostasis, mitochondrial dysfunction, genome instability, and telomerase exhaustion, thereby leading to aging-related diseases, such as immune, metabolic, and neurodegenerative diseases and cancers. According to changes in disease patterns, medical needs have shifted from caring for sick patients to promoting health, from invasive to noninvasive diagnosis methods, and from toxic or expensive drugs to safe drugs with reasonable costs. To meet medical needs, advances in biological data and AI technologies that enable disease prediction and tailored therapy are needed in the near future.
Despite intense microbiome research, there is still much to learn about the intrinsic mechanisms of microbes, especially those of effector microbial products exchanged between the microbiota and host. An exact understanding of microbiota-to-host communication is essential for developing translation strategies employing microbial products for human health. In this context, the study of MEVs is receiving increasing interest. Evidence from nonclinical studies provides us with an opportunity to develop novel first-in-class therapeutic modalities: microbiome-related nanotherapeutics.
MEVs are commensal nonliving materials that act on the human immune system to induce protective responses. Studies have shown that MEVs are absorbed into and distributed in various organs after oral administration, unlike exosomes (human cell-derived EVs). Furthermore, MEVs have a unique ability to target distant organs and effectively penetrate cellular organelles compared with LBPs (live microbes), which are generally restricted to the mucosal or skin surface. Moreover, MEVs can be conveniently administered orally or topically, as opposed to biologics requiring intravenous infusion or subcutaneous injection. LBPs, exosomes, and MEVs are considered to be promising next-generation therapies to complement or replace current biologics, such as protein, monoclonal antibody, cell, and gene therapies. Among the candidates for next-generation therapies, MEVs offer a variety of advantages over LBPs and exosomes in terms of pharmacology (pharmacokinetics and pharmacodynamics), safety, and manufacturing, as summarized in Table 4.
Table 4.
Comparison of microbial EV therapy vs. live biotherapeutics (LBP) and human cell EV (exosome) therapies.
| Microbial EVs | Live biotherapeutics (LBP) | Human cell EVs (Exosomes) | |
|---|---|---|---|
| Pharmacology (PK/PD) |
• Multimodal mechanism of action (MoA) • Nanosized EVs enter systemic circulation • Independent of viability • Targeting of specific organs & cellular organelles • Targeting of distant organs (esp. brain) • Efficacious oral application possible |
• No penetration of host cells • Generally restricted to the GI tract • Dependent on viability • Targeting of specific organs unfeasible |
• Targeting of distant organs unfeasible • Oral administration unfeasible |
| Safety |
• No proliferation • Minimal safety concerns (commensal derived) • Low potential for epigenetic modification |
• Uncontrollable proliferation • Concerns for immune-compromised individuals |
• Potential for epigenetic modification |
| CMC |
• Minimal storage & stability issues • High yield • Low cost |
• Storage & stability hurdles |
• Low yield • High cost |
PK pharmacokinetic, PD pharmacodynamic, CMC chemistry, manufacturing, and control.
The study of MEVs is rapidly evolving. In the near future, it is expected that research efforts in this area will contribute to the understanding of the role of MEVs in disease pathogenesis, which will provide an essential, solid basis for precision medicine solutions, such as in vitro diagnostics based on circulating MEV profiling and therapeutics based on EVs from beneficial bacteria as postbiotics.
Acknowledgements
This research was supported by the Korean Health Technology R&D Project Grant of the Korean Health Industry Development Institute (KHIDI) funded by the Korean Ministry of Health & Welfare (HR14C0001 and HR16C0001) and the Technology Innovation Program (20013712) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
Author contributions
All authors involved in data investigation. J.Y. and T.S.S. wrote and analysed this manuscript. Y.K.J. and Y.K.K.contributed substantially to discussion of the content to review and edit the manuscript before submission. Y.K.K supervised the project.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jinho Yang, Tae-Seop Shin.
References
- 1.National Library of Medicine. What is precision medicine? MedlinePlushttps://ghr.nlm.nih.gov/primer/precisionmedicine/definition (2018).
- 2.National Research Council. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (National Academies Press, 2011). [PubMed]
- 3.Ginsburg GS, Phillips KA. Precision Medicine: From Science to Value. Health Aff. 2018;37:694–701. doi: 10.1377/hlthaff.2017.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho D, et al. Enabling technologies for personalized and precision medicine. Trends Biotechnol. 2020;38:497–518. doi: 10.1016/j.tibtech.2019.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health. NIH Human Microbiome Project defines normal bacterial makeup of the body. NIH Newshttps://www.nih.gov/news-events/news-releases/nih-human-microbiome-project-defines-normal-bacterial-makeup-body (2012).
- 6.Margulis, L. & Fester, R. (eds) Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. (Mit Press, 1991). [PubMed]
- 7.Jefferson, R. Agriculture, Environment and the Developing World: A Future of PCR. Part 4: The Hologenome Plenary lecture at Cold Spring Harbor Laboratoryhttps://www.youtube.com/watch?v=pgL3rmZL9P0 (1994).
- 8.Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. MBio. 2016;7:e01395–15. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, et al. The function and clinical application of extracellular vesicles in innate immune regulation. Cell Mol. Immunol. 2020;17:323–334. doi: 10.1038/s41423-020-0391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocucci E, Meldolesi J. Ectosome and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;6:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Gustafson D, Veitch S, Fish JE. Extracellular vesicles as protagonists of diabetic cardiovascular pathology. Front. Cardiovasc. Med. 2017;4:71. doi: 10.3389/fcvm.2017.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Kim EK, McDowell A, Kim YK. Microbe-derived extracellular vesicles as a smart drug delivery system. Transl. Clin. Pharmacol. 2018;26:103–110. doi: 10.12793/tcp.2018.26.3.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Work E, Knox KW, Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann. N. Y. Acad. Sci. 1966;133:438–449. doi: 10.1111/j.1749-6632.1966.tb52382.x. [DOI] [PubMed] [Google Scholar]
- 18.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee EY, et al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 20.Roier S, Zingl FG, Cakar F, Schild S. Bacterial outer membrane vesicle biogenesis: a new mechanism and its implications. Microb. Cell. 2016;3:257–259. doi: 10.15698/mic2016.06.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roier S, et al. A noverl mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016;7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera J, et al. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl Acad. Sci. USA. 2010;107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briaud P, Carroll RK. Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect. Immune. 2020;88:e00433–20. doi: 10.1128/IAI.00433-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allocati N, Masulli M, Di Ilio C, De Laurenzi V. Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis. 2015;6:e1609. doi: 10.1038/cddis.2014.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee EY, Choi DS, Kim KP, Gho YS. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 2018;27:535–555. doi: 10.1002/mas.20175. [DOI] [PubMed] [Google Scholar]
- 29.Poste G, Papahadjopoulos D. Lipid vesicles as carriers for introducing materials into cultured cells: influence of vesicle lipid composition on mechanism(s) of vesicle incorporation into cells. Proc. Natl Acad. Sci. USA. 1976;73:1603–1607. doi: 10.1073/pnas.73.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strahl H, Errington J. Bacterial membranes: structure, domains, and function. Annu. Rev. Microbiol. 2017;71:519–538. doi: 10.1146/annurev-micro-102215-095630. [DOI] [PubMed] [Google Scholar]
- 31.Lopez D, Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 2017;36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int. J. Nanomed. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldeschwieler JD. Phospholipid vesicle targeting using synthetic glycolipid and other determinants. Ann. N. Y. Acad. Sci. 1985;446:349–367. doi: 10.1111/j.1749-6632.1985.tb18413.x. [DOI] [PubMed] [Google Scholar]
- 34.Bussian RW, Wriston JC., Jr Influence of incorporated cerebrosides on the interaction of liposomes with HeLa cells. Biochim. Biophys. Acta. 1977;471:336. doi: 10.1016/0005-2736(77)90261-9. [DOI] [PubMed] [Google Scholar]
- 35.Mauk MR, Gamble RC, Baldeschwieler JD. Vesicle targeting: timed release and specificity for leukocytes in mice by subcutaneous injection. Science. 1980;207:309–311. doi: 10.1126/science.7350660. [DOI] [PubMed] [Google Scholar]
- 36.Mauk MR, Gamble RC, Baldeschwieler JD. Targeting of lipid vesicles: specificity of carbohydrate receptor analogues for leukocytes in mice. Proc. Natl Acad. Sci. USA. 1980;77:4430–4434. doi: 10.1073/pnas.77.8.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashwell G, Morell AG. The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv. Enzymol. Relat. Areas Mol. Biol. 1974;41:99–128. doi: 10.1002/9780470122860.ch3. [DOI] [PubMed] [Google Scholar]
- 38.Achord DT, Brot FE, Sly WS. Inhibition of the rat clearance system for agalacto-orosomucoid by yeast mannans and by mannose. Biochem. Biophys. Res. Commun. 1977;77:409–415. doi: 10.1016/S0006-291X(77)80213-1. [DOI] [PubMed] [Google Scholar]
- 39.Stahl PD, Rodman JS, Miller MJ, Schlesinger PH. Evidence for receptormediated binding of glycoproteins, glycoconjugates, and lysosomal glycosidases by alveolar macrophages. Proc. Natl Acad. Sci. USA. 1978;75:1399–1403. doi: 10.1073/pnas.75.3.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi DS, et al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11:3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 41.Jeon J, et al. Proteomic analysis of extracellular vesicles derived from Propionibacterium acnes. Proteom. Clin. Appl. 2017;11:1600040. doi: 10.1002/prca.201600040. [DOI] [PubMed] [Google Scholar]
- 42.Tartaglia NR, et al. Staphylococcus aureus extracellular vesicles elicit an immunostimulatory response in vivo on the murine mammary gland. Front Cell Infect. Microbiol. 2018;8:277. doi: 10.3389/fcimb.2018.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong SW, et al. An important role of alpha-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS ONE. 2014;9:e100499. doi: 10.1371/journal.pone.0100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E. Extracellular vesicle RNA: a universal mediator of microbial communication? Trends Microbiol. 2018;26:401–410. doi: 10.1016/j.tim.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Sjostrom AE, Sandblad L, Uhlin BE, Wai SN. Membrane vesicle-mediated release of bacterial RNA. Sci. Rep. 2015;5:15329. doi: 10.1038/srep15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitto NJ, et al. Bacteria membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017;7:7072. doi: 10.1038/s41598-017-07288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eigenbrod T, Dalpkem AH. Bacterial RNA: An underestimated stimulus for innate immune responses. J. Immunol. 2015;195:411–418. doi: 10.4049/jimmunol.1500530. [DOI] [PubMed] [Google Scholar]
- 48.Hong SW, et al. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011;66:351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim MH, et al. A metagenomic analysis provides a culture-independent pathogen detection for atopic dermatitis. Allergy Asthma Immunol. Res. 2017;9:453–461. doi: 10.4168/aair.2017.9.5.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi EJ, et al. Propionibacterium acnes-derived extracellular vesicles promote Acne-like phenotypes in human epidermis. J. Invest. Dermatol. 2018;138:1371–1379. doi: 10.1016/j.jid.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Brandt EB, et al. Exposure to allergen and diesel exhaust particles potentiates secondary allergen-specific memory responses, promoting asthma susceptibility. J. Allergy Clin. Immunol. 2015;136:295–303. doi: 10.1016/j.jaci.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Kim YK, Kang TS, Jee YK, Kim YY. Importance of indoor dust biological ultrafine particles in the pathogenesis of chronic inflammatory pulmonary disease. Environ. Health Toxicol. 2017;32:e2017021. doi: 10.5620/eht.e2017021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Kim EK, Park HJ, McDowell A, Kim YK. The impact of bacteria-derived ultrafine dust particles on pulmonary diseases. Exp. Mol. Med. 2020;52:338–347. doi: 10.1038/s12276-019-0367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YS, et al. Extracellular vesicles derived from Gram-negative bacteria, such as Escherichia coli, induce emphysema mainly via IL-17A-mediated neutrophilic inflammation. J. Immunol. 2015;194:3361–3368. doi: 10.4049/jimmunol.1402268. [DOI] [PubMed] [Google Scholar]
- 55.Kim MR, et al. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy. 2012;67:1271–1281. doi: 10.1111/all.12001. [DOI] [PubMed] [Google Scholar]
- 56.Bae CH, Choi YS, Song SY, Kim YK, Kim YD. Escherichia coli–derived and Staphylococcus aureus–derived extracellular vesicles induce MUC5AC expression via extracellular signal related kinase 1/2 and p38 mitogen-activated protein kinase in human airway epithelial cells. Int. Forum Allergy Rhinol. 2017;7:91–98. doi: 10.1002/alr.21844. [DOI] [PubMed] [Google Scholar]
- 57.Park KS, et al. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 2013;49:637–645. doi: 10.1165/rcmb.2012-0370OC. [DOI] [PubMed] [Google Scholar]
- 58.Yang D, et al. Dysregulated lung commensal bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity. 2019;50:692–706. doi: 10.1016/j.immuni.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Kim YS, et al. Extracellular vesicles, especially derived from Gram-negative bacteria, in indoor dust induce neutrophilic pulmonary inflammation associated with both Th1 and Th17 cell responses. Clin. Exp. Allergy. 2013;43:443–454. doi: 10.1111/cea.12085. [DOI] [PubMed] [Google Scholar]
- 60.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim YS, et al. IgG sensitization to extracellular vesicles in indoor dust is closely associated with the prevalence of non-eosinophilic asthma, COPD, and lung cancer. Allergy Asthma Immunol. Res. 2016;8:198–205. doi: 10.4168/aair.2016.8.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin TS, Kim EK, McDowell A, Kim YK. Impact of gut microbiota extracellular vesicles on inflammatory and metabolic disease. J. Clin. Nutr. Food Sci. 2019;2:73–78. [Google Scholar]
- 63.Tlaskalová-Hogenová H, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michielan A, D’Incà R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rubino SJ, Selvanantham T, Girardin SE, Philpott DJ. Nod-like receptors in the control of intestinal inflammation. Curr. Opin. Immunol. 2012;24:398–404. doi: 10.1016/j.coi.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 66.Bielig H, et al. NOD-like receptor activation by outer membrane vesicles from Vibrio cholerae non-O1 nonO1390 strains is modulated by the quorum-sensing regulator HapR. Infect. Immune. 2011;79:1418–1427. doi: 10.1128/IAI.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Canas MA, Fabrega MJ, Gimenez R, Badia J, Baldoma L. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front. Microbiol. 2018;9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilmore WJ, Johnston EL, Zavan L, Bitto NJ, Kaparakis-Liaskos M. Immunomodulatory roles and novel applications of bacterial membrane vesicles. Mol. Immunnol. 2021;134:72–85. doi: 10.1016/j.molimm.2021.02.027. [DOI] [PubMed] [Google Scholar]
- 69.Choi MS, Ze EY, Park JY, Shin TS, Kim JG. Helicobacter pylori-derived outer membrane vesicles stimulate interleukin 8 secretion through nuclear factor kappa B activation. Korean J. Intern. Med. 2021;36:857–867. doi: 10.3904/kjim.2019.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7:2839–2849. doi: 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi Y, et al. Gut microbe-derived extracellular vesicles induce insulin resistance, thereby impairing glucose metabolism in skeletal muscle. Sci. Rep. 2015;5:15878. doi: 10.1038/srep15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seyama M, et al. Outer membrane vesicles of Porphyromonas gingivalis attenuate insulin sensitivity by delivering gingipains to the liver. Biochim. Biophys. Acta. 2020;1866:165731. doi: 10.1016/j.bbadis.2020.165731. [DOI] [PubMed] [Google Scholar]
- 73.Duan MC, Zhong XN, Liu GN, Wei JR. The Treg/Th17 paradigm in lung cancer. J. Immunol. Res. 2014;2014:730380. doi: 10.1155/2014/730380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reppert S, et al. A role for T-bet-mediated tumour immune surveillance in anti-IL-17A treatment of lung cancer. Nat. Commun. 2011;2:600. doi: 10.1038/ncomms1609. [DOI] [PubMed] [Google Scholar]
- 75.Li Y, et al. Effects of IL-17A on the occurrence of lung adenocarcinoma. Cancer Biol. Ther. 2011;12:610–616. doi: 10.4161/cbt.12.7.16302. [DOI] [PubMed] [Google Scholar]
- 76.Koyama K, et al. Reciprocal CD4+ T-cell balance of effector CD62Llow CD4+ and CD62LhighCD25+ CD4+ regulatory T cells in small cell lung cancer reflects disease stage. Clin. Cancer Res. 2008;14:6770–6779. doi: 10.1158/1078-0432.CCR-08-1156. [DOI] [PubMed] [Google Scholar]
- 77.Choi HI, et al. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp. Mol. Med. 2017;49:e330. doi: 10.1038/emm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cuesta CM, Guerri C, Urena J, Pascual M. Role of microbiota-derived extracellular vesicles in gut-brain communication. Int. J. Mol. Sci. 2021;22:4235. doi: 10.3390/ijms22084235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han EC, et al. Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. FASEB J. 2019;33:13412–13422. doi: 10.1096/fj.201901575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ha JY, Choi SY, Lee JH, Hong SH, Lee HJ. Delivery of periodontopathogenic extracellular vesicles to brain monocytes and microglial IL-6 promotion by RNA cargo. Front. Mol. Biosci. 2020;7:596366. doi: 10.3389/fmolb.2020.596366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee KE, et al. The extracellular vesicle of gut microbial Paenalcaligenes hominis is a risk factor for vagus nerve-mediated cognitive impairment. Microbiome. 2020;8:107. doi: 10.1186/s40168-020-00881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J, et al. Brain tumor diagnostic model and dietary effect based on extracellular vesicle microbiome data in serum. Exp. Mol. Med. 2020;52:1602–1613. doi: 10.1038/s12276-020-00501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi EB, et al. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy. 2014;69:517–526. doi: 10.1111/all.12374. [DOI] [PubMed] [Google Scholar]
- 84.Castelino M, et al. Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 2017;17:23. doi: 10.1186/s12866-017-0927-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balvociute M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics. 2017;18:114. doi: 10.1186/s12864-017-3501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hendriksen JMT, Geersing GJ, Moons KGM, de Groot JAH. Diagnostic and prognostic prediction models. J. Thromb. Haemost. 2013;11:129–141. doi: 10.1111/jth.12262. [DOI] [PubMed] [Google Scholar]
- 87.Seo JH, Lee JW, Cho D. The market trend analysis and prospects of cancer molecular diagnostics kits. Biomater. Res. 2018;22:2. doi: 10.1186/s40824-017-0111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee JH, et al. Metagenome analysis using serum extracellular vesicles identified distinct microbiota in asthmatics. Sci. Rep. 2020;10:15125. doi: 10.1038/s41598-020-72242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J, et al. Diagnostic models for atopic dermatitis based on serum microbial extracellular vesicle metagenomic analysis: a pilot study. Allergy Asthma Immunol. Res. 2020;12:792–805. doi: 10.4168/aair.2020.12.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim DJ, et al. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci. Rep. 2020;10:2860. doi: 10.1038/s41598-020-59529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cho EJ, et al. Circulating microbiota-based metagenomic signature for detection of hepatocellular carcinoma. Sci. Rep. 2019;9:7536. doi: 10.1038/s41598-019-44012-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park JY, et al. Bacteria-derived extracellular vesicles in urine as a novel biomarker for gastric cancer: integration of liquid biopsy and metagenome analysis. Cancers. 2021;13:4687. doi: 10.3390/cancers13184687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim JR, et al. Microbiome markers of pancreatic cancer based on bacteria-derived extracellular vesicles acquired from blood samples: a retrospective propensity score matching analysis. Biology. 2021;10:219. doi: 10.3390/biology10030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee H, Lee HK, Min SK, Lee WH. 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer. World J. Surg. Oncol. 2020;18:19. doi: 10.1186/s12957-020-1793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim SI, et al. Metagenomic analysis of serum microbe-derived extracellular vesicles and diagnostic models to differentiate ovarian cancer and benign ovarian tumor. Cancers. 2020;12:1309. doi: 10.3390/cancers12051309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang J, et al. Lung disease diagnostic model through IgG sensitization to microbial extracellular vesicles. Allergy Asthma Immunol. Res. 2020;12:669–683. doi: 10.4168/aair.2020.12.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akter S, Park JH, Jung HK. Potential health-promoting benefits of paraprobiotics, inactivated probiotic cells. J. Microbiol. Biotechnol. 2020;30:477–481. doi: 10.4014/jmb.1911.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taverniti V, Guglielmetti S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: proposal of paraprobiotic concept) Genes Nutr. 2011;6:261–274. doi: 10.1007/s12263-011-0218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Almada CN, Almada CN, Martinez CRR, Sant’Ana AS. Paraprobiotics: evidences on their ability to modify biological responses, inactivation methods and perspectives on their application in foods. Trends Food Sci. Technol. 2016;58:96–114. doi: 10.1016/j.tifs.2016.09.011. [DOI] [Google Scholar]
- 100.Pique N, Berlanga M, Minana-Galbis D. Health benefits of heat-killed (tyndallized) probiotics: an overview. Int. J. Mol. Sci. 2019;20:2534. doi: 10.3390/ijms20102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aguilar-Toala JE, et al. Postbiotics: an evolving term within the functional foods field. Trends Food Sci. Technol. 2018;75:105–114. doi: 10.1016/j.tifs.2018.03.009. [DOI] [Google Scholar]
- 102.Fafian-Labora JA, O’Loghlen A. Classical and nonclassical intercellular communication in senescence and ageing. Trends Cell Biol. 2020;30:628–639. doi: 10.1016/j.tcb.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Liu Y, Defourny KAY, Smid EJ, Abee T. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 2018;9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang CS, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi JH, et al. Lactobacillus paracasei-derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Exp. Mol. Med. 2020;52:423–437. doi: 10.1038/s12276-019-0359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chelakkot C, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018;50:e450. doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim MH, et al. Lactobacillus plantarum-derived extracellular vesicles protect atopic dermatitis induced by Staphylococcus aureus-derived extracellular vesicles. Allergy Asthma Immunol. Res. 2018;10:516–532. doi: 10.4168/aair.2018.10.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim W, et al. Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro. J. Extracell. Vesicles. 2020;9:1793514. doi: 10.1080/20013078.2020.1793514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seo MK, Park EJ, Ko SY, Choi EW, Kim S. Therapeutic effects of kefir grain Lactobacillus-derived extracellular vesicles in mice with 2,4,6-trinitrobenzene sulfonic acid-induced inflammatory bowel disease. J. Dairy Sci. 2018;101:8662–8671. doi: 10.3168/jds.2018-15014. [DOI] [PubMed] [Google Scholar]
- 110.Miyoshi Y, et al. Mechanisms underlying enhanced IgA production in Peyer’s patch cells by membrane vesicles derived from Lactobacillus sakei. Biosci. Biotechnol. Biochem. 2021;85:1536–1545. doi: 10.1093/bbb/zbab065. [DOI] [PubMed] [Google Scholar]
- 111.Kim JH, et al. Extracellular vesicle–derived protein from Bifidobacterium longum alleviates food allergy through mast cell suppression. J. Allergy Clin. Immunol. 2016;137:507–516. doi: 10.1016/j.jaci.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 112.Wang J, et al. The outer membrane protein Amuc_1100 of Akkermansia muciniphila promotes intestinal 5-HT biosynthesis and extracellular availability through TLR2 signalling. Food Funct. 2021;12:3597–3610. doi: 10.1039/D1FO00115A. [DOI] [PubMed] [Google Scholar]
- 113.Poste G, Papahadjopoulos D, Vail WJ. Lipid vesicles as carriers for introducing biologically active materials into cells. Methods Cell Biol. 1976;14:33–71. doi: 10.1016/S0091-679X(08)60468-9. [DOI] [PubMed] [Google Scholar]
- 114.Sambi M, Bagheri L, Szewczuk MR. Current challenges in cancer immunotherapy: multimodal approaches to improve efficacy and patient response rates. J. Oncol. 2019;2019:4508794. doi: 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim OY, et al. Bacterial outer membrane vesicles suppress tumor by interferon- γ-mediated antitumor response. Nat. Commun. 2017;8:626. doi: 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Evanovich E, de Souza Mendonca Mattos PJ, Guerreiro JF. Comparative genomic analysis of Lactobacillus plantarum: an overview. Int. J. Genomics. 2019;2019:4973214. doi: 10.1155/2019/4973214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choi J, Kim YK, Han PL. Extracellular vesicles derived from Lactobacillus plantarum increase BDNF expression in cultured hippocampal neurons and produce antidepressant-like effects in mice. Exp. Neurobiol. 2019;28:158–171. doi: 10.5607/en.2019.28.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tian F, et al. The therapeutic protection of a living and dead Lactobacillus strain against aluminum-nduced brain and liver injuries in C57BL/6 mice. PLoS ONE. 2017;12:e0175398. doi: 10.1371/journal.pone.0175398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis DJ, et al. Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish. Sci. Rep. 2016;6:33726. doi: 10.1038/srep33726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dhaliwal J, et al. Lactobacillus plantarum MTCC 9510 supplementation protects from chronic unpredictable and sleep deprivation-induced behaviour, biochemical and selected gut microbial aberrations in mice. J. Appl. Microbiol. 2018;125:257–269. doi: 10.1111/jam.13765. [DOI] [PubMed] [Google Scholar]
- 121.Bin L, et al. Staphylococcus aureus alpha-toxin modulates skin host response to viral infection. J. Allergy Clin. Immunol. 2012;130:683–691. doi: 10.1016/j.jaci.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim MH, et al. A metagenomics analysis provides a culture-independent pathogen detection for atopic dermatitis. Allergy Asthma Immunol. Res. 2017;9:453–461. doi: 10.4168/aair.2017.9.5.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee Y, et al. Rapid assessment of microbiota changes in individuals with autism spectrum disorder using bacteria-derived membrane vesicles in urine. Exp. Neurobiol. 2017;26:307–317. doi: 10.5607/en.2017.26.5.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rhee SJ, et al. Comparison of serum microbiome composition in bipolar and major depressive disorders. J. Psychiatr. Res. 2020;123:31–38. doi: 10.1016/j.jpsychires.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 125.Samra M, et al. Urine bacteria-derived extracellular vesicles and allergic airway diseases in children. Int. Arch. Allergy Immunol. 2019;178:150–158. doi: 10.1159/000492677. [DOI] [PubMed] [Google Scholar]
- 126.You YA, Yoo JY, Kwon EJ, Kim YJ. Blood microbial communities during pregnancy are associated with preterm birth. Front. Microbiol. 2019;10:1122. doi: 10.3389/fmicb.2019.01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim SS, et al. Microbiome as a potential diagnostic and predictive biomarker in severe alcoholic hepatitis. Aliment Pharmacol. Ther. 2021;53:540–551. doi: 10.1111/apt.16200. [DOI] [PubMed] [Google Scholar]
- 128.Shen Y, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2021;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lopez P, et al. Treg-inducing membrane vesicles from Bifidobacterium bifidum LMG13195 as potential adjuvants in immunotherapy. Vaccine. 2012;30:825–829. doi: 10.1016/j.vaccine.2011.11.115. [DOI] [PubMed] [Google Scholar]
- 130.Maerz JK, et al. Outer membrane vesicles blebbing contributes to B. vulgatus mpk-mediated immune response silencing. Gut Microbes. 2018;9:1–12. doi: 10.1080/19490976.2017.1344810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim OY, et al. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017;8:626. doi: 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.An J, et al. Bacterial extracellular vesicles affect endocrine therapy in MCF7 cells. Medicine. 2021;100:e25835. doi: 10.1097/MD.0000000000025835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.AI-Nedawi K, et al. Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB J. 2014;29:684–695. doi: 10.1096/fj.14-259721. [DOI] [PubMed] [Google Scholar]