Abstract
In a serotype 11A clone of erythromycin-resistant pneumococci isolated from young Greek carriers, we identified the nucleotide sequence of erm(A), a methylase gene previously described as erm(TR) in Streptococcus pyogenes. The erm(A) pneumococci were resistant to 14- and 15-member macrolides, inducibly resistant to clindamycin, and susceptible to streptogramin B. To our knowledge, this is the first identification of resistance to erythromycin in S. pneumoniae attributed solely to the carriage of the erm(A) gene.
Resistance of Streptococcus pneumoniae to erythromycin and the other macrolides is increasing in many parts of the world (1, 5, 7, 18). Strains resistant to erythromycin are also resistant to azithromycin, clarithromycin, and roxithromycin (25). Recently, it has been shown that pneumococci resistant to erythromycin have mainly one of two distinct resistance determinants, erm(B) or mef(A) (15, 17, 19, 20, 23; A. Tait-Kamradt, T. Davies, F. Brennan, F. Depardieu, P. Courvalin, J. Duignan, J. Petitpas, L. Wondrack, M. Jacobs, P. Appelbaum, and J. Sutcliffe, Addendum Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. LB-8, p. 15, 1999). mef(A) encodes an efflux pump that appears to be specific for 14- and 15-member macrolides. The remainder of the resistant strains carry an erm(B) methylase. In this case, an adenine residue in 23S rRNA is methylated, leading to reduced binding of 14-, 15-, and 16-member macrolides, lincosamides, and streptogramin B (MLSB) to their shared target site in the 50S ribosomal subunit. erm synthesis can be inducible or constitutive.
The nasopharynx is the main reservoir of antibiotic-resistant pneumococci in children, and carriage usually precedes infection (11). From 10 February 1997 to 10 February 1999, nasopharyngeal cultures for S. pneumoniae were performed for 2,448 Greek infants and toddlers who were enrolled in the Hellenic Antibiotic-Resistant Respiratory Pathogens Study. Children 2 to 23 months of age were enrolled from the outpatient clinics of four hospitals, as well as from the private offices of 14 practicing pediatricians in different areas of central and southern Greece (22). At the time the nasopharyngeal culture was obtained, the children were healthy and were brought to the pediatrician to be vaccinated or had signs and symptoms of an acute respiratory tract infection. Isolation, identification, susceptibility testing, and serotyping of the S. pneumoniae strains were performed as described previously (21, 22). Of a total of 781 pneumococcal isolates recovered from the 2,448 children studied, 137 (18%) were erythromycin resistant, with 67.9% of them carrying the erm(B) gene and 29.2% having mef(A) gene products (22). In 4 (2.9%) of the 137 erythromycin-resistant pneumococcal isolates, neither the erm(B) gene nor the mef(A) gene was identified. The present study was undertaken to investigate the phenotype, genotype, and mechanism of resistance of isolates carrying neither erm(B) nor mef(A).
The susceptibility of the four erythromycin-resistant S. pneumoniae isolates that carried neither erm(B) nor mef(A) to erythromycin, azithromycin, josamycin, streptogramin A and B, penicillin, and tetracycline was tested. MICs were determined in ambient air in microtiter trays with Mueller-Hinton broth supplemented with 2.5% lysed horse blood following recommendations by the National Committee for Clinical Laboratory Standards (12). All compounds were purchased from Sigma or made by published methods at Pfizer, Inc. Double disk diffusion analysis was performed as previously described (19). Induction was present when the zone of inhibition around the clindamycin or streptogramin B disk was blunted on the side next to the erythromycin disk.
Determination of erythromycin resistance mechanisms.
Primers for internal regions of erm(A), erm(B), erm(C), erm(TR), msr(A), mef(A), mph(A), mph(B), ere(A), and ere(B) have been described previously (20, 24). Primers designed from the S. pyogenes erm(TR) sequence (16) to amplify the entire class A gene were also used in this study: 5′-AAGATTAGTTCATTATAACC-3′ [−38 to −18 bp upstream of the start codon for erm(TR)] and 5′-TTATTGAAATAATTTGTAAC-3′ [anneals to the terminal 20 bases of erm(TR)]. Primers for mph(C) are based on the sequence of a putative macrolide phosphorylase from Staphylococcus aureus clinical strains (10; J. Cheng, T. Grebe, L. Wondrack, P. Courvalin, and J. Sutcliffe, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 837, p. 114, 1999) and are described in reference 24. Amplified PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and sequenced on an ABI 373XL automated sequencing apparatus with stretch upgrade (PE Biosystems, Foster City, Calif.) as described previously (24). Sequence comparisons were carried out with Vector NTI sequence analysis software (InforMax, Inc., North Bethesda, Md.).
Genotypic analysis of erythromycin-resistant S. pneumoniae.
Molecular analysis of the genotype of the four erythromycin-resistant S. pneumoniae isolates that carried neither erm(B) nor mef(A) was performed by pulsed-field gel electrophoresis (PFGE) as described previously (13).
Presence of the erm(A) gene in erythromycin-resistant pneumococci.
Genomic DNA from the four resistant isolates which possessed neither erm(B) nor mef(A) was isolated and subjected to PCR analysis with primers specific for macrolide esterases [ere(A) and ere(B)], phosphotransferases [mph(A), mph(B), and mph(C)], an ABC-binding transporter [msr(A)], and rRNA methylases [erm(TR), erm(A), and erm(C)] (16, 19, 20, 23, 24). Each isolate had a PCR product only when primers specific for the erm(TR) determinant were used. The use of primers encompassing the entire erm(TR) gene plus 38 bases upstream revealed that the nucleotide sequences from the four pneumococci were identical to the erm(TR) gene from a clinical strain of Streptococcus pyogenes (16). However, based on a recent classification of the MLSB resistance genes, erm(TR) has been assigned to class A as an erm(A) determinant (15).
The four S. pneumoniae isolates carrying the erm(A) gene were recovered from the nasopharynges of four children during an 11-month period (Table 1). These children were heavily colonized with pneumococcus, because colony counts revealed > 105 CFU/ml. The four children were living in unrelated parts of the city of Patras and its surroundings in southwestern Greece, and we were not able to identify any close contact among them.
TABLE 1.
Origin of the Streptococcus pneumoniae isolates carrying the erm(A) genea
| Strain no.b | Date of isolation | Age (mo) | No. of siblings (age in yr) | Clinical condition |
|---|---|---|---|---|
| 16 | 22 February 1997 | 11 | 2 (5 and 9) | Healthy |
| 96 | 12 April 1997 | 16 | 2 (16 and 19) | Acute otitis media |
| 215 | 31 October 1997 | 13 | 1 (4) | Acute otitis media |
| 357 | 28 January 1998 | 16 | None | Healthy |
Note that none of the children attended a day care center.
All strains were serotype 11A.
The MIC ranges of the antimicrobial agents tested were as follows: erythromycin, 0.78 to 3.12 μg/ml; azithromycin, 6.25 to 25 μg/ml; josamycin, 0.20 to 0.78 μg/ml; streptogramin A, 25 μg/ml; streptogramin B, 0.78 to 1.56 μg/ml; penicillin G, 0.1; and tetracycline, 6.25 μg/ml. The erm(A) pneumococcal isolates were inducibly resistant to clindamycin. Due to the large zone of inhibition around the erythromycin disk for the erm(A) strains, it was necessary to increase the spacing between disks beyond 12 to 16 mm to adequately identify blunting.
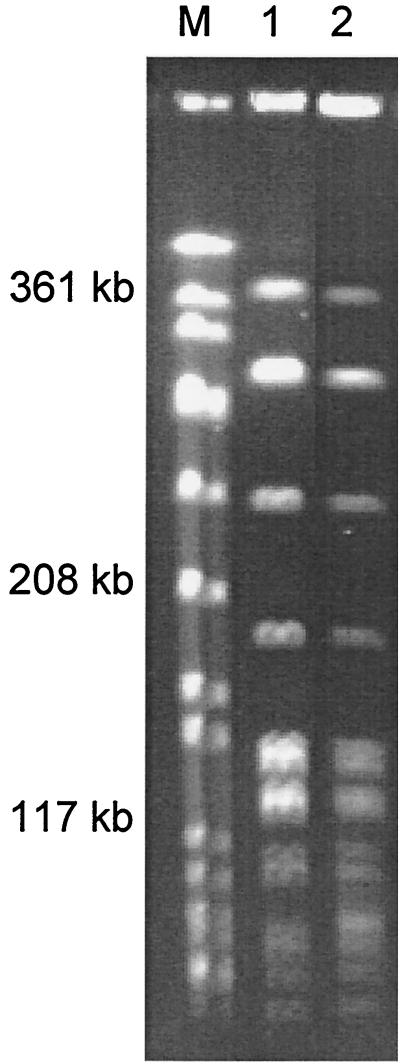
Molecular analysis by PFGE showed that the four serotype 11A erm(A) strains had a clonal relationship sharing an identical genotype. The PFGE patterns of two serotype 11A pneumococci are shown in Fig. 1.
FIG. 1.
SmaI PFGE patterns of erythromycin-resistant pneumococci carrying the erm(A) gene. Lane 1, strain 16; lane 2, strain 215. An SmaI digest of genomic DNA from S. aureus ATCC 8325 was used as the molecular weight standard (M).
To our knowledge, this is the first identification of resistance to erythromycin in S. pneumoniae attributed solely to carriage of the erm(A) gene. There has been one report of an erythromycin-resistant S. pneumoniae strain, which carried erm(A) [subclass erm(TR)] in addition to the erm(B) gene (2). erm(A) is an erythromycin resistance methylase gene which was recently described as erm(TR) in S. pyogenes strains in Finland (9, 16). Other studies have expanded the finding of erm(A)+ strains of S. pyogenes to Greece (our unpublished data), Italy (6), France (3), Spain (14), and Canada (4). In addition, the majority of group G, but not group C, streptococci, harbor erm(A) (8).
At the level of the clinical laboratory, data from the MIC and disk analysis of strains harboring erm(A) could possibly be interpreted as representing an M phenotype (macrolide resistant, but susceptible to clindamycin and streptomycin B), especially since streptogramin B is not routinely used in the disk analysis. The zone sizes for clindamycin in the erm(A) strains are intermediate, and the zones around the erythromycin disk can be intermediate. Because of the larger zones, it may be easy to miss the blunt that occurs between the erythromycin and clindamycin zones. The intermediate zones for the erm(A) strains translate to an equivocal result for clindamycin. However, given that the strain carries a methylase, it is highly likely these strains would be resistant to clindamycin therapy, unlike strains carrying mef(A).
REFERENCES
- 1.Baquero F, García-Rodríguez J A, García de Lomas J, Aguilar L The Spanish Surveillance Group for Respiratory Pathogens. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996–1997) multicenter surveillance study. Antimicrob Agents Chemother. 1999;43:357–359. doi: 10.1128/aac.43.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betriu C, Redondo M, Palau M L, Sánchez A, Gómez M, Culebras E, Boloix A, Picazo J J. Comparative in vitro activities of linezolid, quinupristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob Agents Chemother. 2000;44:1838–1841. doi: 10.1128/aac.44.7.1838-1841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen E, Fitoussi F, Doit C, Cohen R, Tanna A, George R, Loukil C, Brahimi N, Le Thomas I, Deforche D. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob Agents Chemother. 2000;44:1453–1457. doi: 10.1128/aac.44.6.1453-1457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Azavedo J C S, Yeung R H, Bast D J, Duncan C L, Borgia S B, Low D E. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob Agents Chemother. 1999;43:2144–2147. doi: 10.1128/aac.43.9.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doern G V, Pfaller M A, Kugler K, Freeman J, Jones R N. Prevalence of antimicrobial resistance among respiratory tract isolates of Streptococcus pneumoniae in North America: 1997 results from the SENTRY antimicrobial surveillance program. Clin Infect Dis. 1998;27:764–770. doi: 10.1086/514953. [DOI] [PubMed] [Google Scholar]
- 6.Giovanetti E, Montanari M P, Mingoia M, Varaldo P E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–1940. doi: 10.1128/aac.43.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs M R, Bajaksouzian S, Zilles A, Lin G, Pankuch G A, Appelbaum P C. Susceptibilities of Streptococcus pneumoniae and Haemophilus influenzae to 10 oral antimicrobial agents based on pharmacodynamic parameters: 1997 U.S. surveillance study. Antimicrob Agents Chemother. 1999;43:1901–1908. doi: 10.1128/aac.43.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kataja J, Seppälä H, Skurnik M, Sarkkinen H, Huovinen P. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob Agents Chemother. 1998;42:1493–1494. doi: 10.1128/aac.42.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataja J, Huovinen P, Skurnik M, Seppälä H The Finnish Study Group for Antimicrobial Resistance. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob Agents Chemother. 1999;43:48–52. doi: 10.1128/aac.43.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuoka M, Endou K, Kobayashi H, Inoue M, Nakajima Y. A plasmid that encodes three genes for resistance to macrolide antibiotics in Staphylococcus aureus. FEMS Microbiol Lett. 1998;167:221–227. doi: 10.1111/j.1574-6968.1998.tb13232.x. [DOI] [PubMed] [Google Scholar]
- 11.Musher D M. Streptococcus pneumoniae. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 5th ed. New York, N.Y: Churchill Livingstone; 2000. pp. 2128–2147. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement. M100-S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 13.Pankuch G A, Jueneman S A, Davies T A, Jacobs M R, Appelbaum P C. In vitro selection of resistance to four β-lactams and azithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2914–2918. doi: 10.1128/aac.42.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portillo A, Lantero M, Gastanares M J, Ruiz-Larrea F, Torres C. Macrolide resistance phenotypes and mechanisms of resistance in Streptococcus pyogenes in La Rioja, Spain. Int J Antimicrob Agents. 1999;13:137–140. doi: 10.1016/s0924-8579(99)00104-1. [DOI] [PubMed] [Google Scholar]
- 15.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seppälä H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shortridge V D, Flamm R K, Ramer N, Beyer J, Tanaka S K. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;26:73–78. doi: 10.1016/s0732-8893(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 18.Song J-H, Lee N Y, Ichiyama S, Yoshida R, Hirakata Y, Fu W, Chongthaleong A, Aswapokee N, Chiu C-H, Lalitha M K, Thomas K, Perera J, Yee T T, Jamal F, Warsa U C, Vinh B X, Jacobs M R, Appelbaum P C, Pai C H the ANSORP Study Group. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant pathogens (ANSORP) Study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 19.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syrogiannopoulos G A, Grivea I N, Katopodis G D, Geslin P, Jacobs M R, Beratis N G. Carriage of antibiotic-resistant Streptococcus pneumoniae in Greek infants and toddlers. Eur J Clin Microbiol Infect Dis. 2000;19:288–293. doi: 10.1007/s100960050477. [DOI] [PubMed] [Google Scholar]
- 22.Syrogiannopoulos G A, Grivea I N, Davies T A, Katopodis G D, Appelbaum P C, Beratis N G. Antimicrobial use and colonization with erythromycin-resistant Streptococcus pneumoniae in Greece during the first 2 years of life. Clin Infect Dis. 2000;31:887–893. doi: 10.1086/318118. [DOI] [PubMed] [Google Scholar]
- 23.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tait-Kamradt A, Davies T, Cronan M, Jacobs M R, Appelbaum P C, Sutcliffe J. Mutations in 23S rRNA and L4 ribosomal protein account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visalli M A, Jacobs M R, Appelbaum P C. Susceptibility of penicillin-susceptible and -resistant pneumococci to dirithromycin compared with susceptibilities to erythromycin, azithromycin, clarithromycin, roxithromycin, and clindamycin. Antimicrob Agents Chemother. 1997;41:1867–1870. doi: 10.1128/aac.41.9.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]