Abstract
Cellular membranes are highly dynamic in shape. They can rapidly and precisely regulate their shape to perform various cellular functions. The protein’s ability to sense membrane curvature is essential in various biological events such as cell signaling and membrane trafficking. As they are bound, these curvature-sensing proteins may also change the local membrane shape by one or more curvature driving mechanisms. Established curvature-sensing/driving mechanisms rely on proteins with specific structural features such as amphipathic helices and intrinsically curved shapes. However, the recent discovery and characterization of many proteins have shattered the protein structure–function paradigm, believing that the protein functions require a unique structural feature. Typically, such structure-independent functions are carried either entirely by intrinsically disordered proteins or hybrid proteins containing disordered regions and structured domains. It is becoming more apparent that disordered proteins and regions can be potent sensors/inducers of membrane curvatures. In this article, we outline the basic features of disordered proteins and regions, the motifs in such proteins that encode the function, membrane remodeling by disordered proteins and regions, and assays that may be employed to investigate curvature sensing and generation by ordered/disordered proteins.
Graphical Abstract
Keywords: Disordered proteins, Protein–membrane interactions, Membrane curvature, Entropic effect
Introduction
Biological membranes are the essential structures in all kingdoms of life on earth as they compartmentalize distinct intracellular organelles [such as the Golgi apparatus, endoplasmic reticulum (ER), and nucleus] and define the cell’s outer boundary [i.e., plasma membrane (PM)] (Alberts et al. 2002). The membranes are composed of phospholipid bilayers, and their primary function is to serve as a physical barrier that not only separates distinct intracellular compartments but also separates the cell interior from the external environment (extracellular space) (Fakhree et al. 2019a). Far more than just a passive interface, membranes can adopt a variety of intricate and beautiful shapes as they are highly dynamic (Simunovic et al. 2019). The majority of which are thought to have evolved for a specific cellular function. A striking example of how membrane shape and cellular function are intimately interconnected is found in membrane trafficking pathways (Zeno et al. 2019a).
Most membrane-bound intracellular organelles have highly complex shapes. For example, ER is an interconnected network of tubules and flat sheets. Golgi apparatus contains a stack of perforated sheets. The mitochondrial inner membranes are divided by cristae, which are thin sheet-like structures with dimensions similar to ER sheets and Golgi cisternae (Shibata et al. 2009; Kozlov et al. 2014). Across all these structures, there is a large membrane curvature in their cross-sections. The radii of these curvatures, varying between 20 and 50 nm (Cui et al. 2013), are just a few times greater than the membrane thickness (3–5 nm) (Tsai et al. 2021). On the other hand, PM is usually a large, relatively flat structure but contains numerous fine micro-membrane structures that aid large curvature to the flat PM. The local and dynamic deformations of a limited area of the PM result in the generation of protrusions like filopodia and lamellipodia, as well as invaginations like caveolae and endocytic clathrin-coated pits (CCPs) (Suetsugu et al. 2014).
The examples mentioned above indicate that the deformation of the membrane such that they adopt large curvatures is imperative for various cellular events. A membrane, nonetheless, is naturally resistant to deformation (Helfrich 1973). Therefore, to generate (induce or drive) membrane curvature, proteins are employed by cells (Zimmerberg and Kozlov 2006; Stachowiak et al. 2013). It is well known that a variety of protein-driven pathways are involved in generating large membrane curvatures in a highly orchestrated fashion (Shibata et al. 2009; Baumgart et al. 2011; Has and Das 2021; Tsai et al. 2021). In addition, proteins also have the potency to sense the membrane curvature and bind to specific geometric cues, and then utilize the curvature as molecular information to organize many cellular processes spatiotemporally (Has and Das 2021). Note that proteins bound or embedded to the membrane are generally required for communication with the outside world and response to external stimuli (signal transduction pathways) (Cho and Stahelin 2005; Grecco et al. 2011). In short, proteins play a crucial role in precise cellular function because of their capability to transport molecules and chemical signals in/out of distinct cell compartments.
Proteins interact with a membrane by introducing their hydrophobic domains into the hydrophobic region of bilayers and attracting their hydrophilic domains to the bilayer surface via physical forces such as electrostatic interactions and hydrogen bonding (Israelachvili 2015). Until recently, it was thought that a protein’s function depended on its ability to form a single, well-defined three-dimensional (3D) molecular structure. This structure–function paradigm can be observed for many proteins or protein domains (White and Wimley 1999). In recent decades, the discovery of disordered (or unstructured or unfolded) proteins and protein regions (discussed in the next section) shattered the structure–function paradigm. Similar to structured proteins, recent studies have shown that disordered proteins and protein regions are also capable to sense/induce membrane curvature (Zeno et al. 2019a; Busch et al. 2015; Snead et al. 2017; Zeno et al. 2019b). After a quick overview of disordered proteins and protein regions in this review, we will focus on how proteins recognize their binding targets and facilitate interaction with them using conserved sequence motifs. We then explore how membrane-bound disordered proteins and protein regions remodel membrane characteristics, which in turn determines membrane trafficking processes. We next go through many experimental assays and computational approaches for curvature sensing and generation by proteins utilized for quantitative and qualitative analysis. Although most of the assays have been employed for structured proteins, they may be tried for unstructured ones to study their curvature sensitivity and driving capacity. At the end of our overview, we highlight open research areas that need to be addressed in the future. We believe that this detailed analysis will help biophysicists to understand the role of many disordered proteins in curvature sensing and generation of the membrane.
Abundance of Intrinsically Disordered Proteins (IDPs) and Protein Regions (IDPRs) in Membranes
In the human genome, around 5500–7500 genes have been predicted to translate into membrane-interacting proteins, which indicates that approximately 26–36% of the human genome’s protein-coding genes are translated into membrane proteins (Fagerberg et al. 2010). The protein–lipid mass ratio in a PM is found to be around 1:1, whereas it is greater than 3:1 for mitochondrial membrane (Müller et al. 2008). The average center-to-center gap between proteins in the PM, based on a 1:1 protein–lipid ratio, is estimated around 10 nm (Phillips et al. 2009), which is comparable to the space between proteins in cytoplasm (Ellis 2001). Given the importance of membrane proteins in intra/intercellular communication, chemical gradient maintenance, and membrane remodeling activities, it is no wonder that they are the key participants in human physiology, disease pathology, and drug development (Marinko et al. 2019). Understanding the molecular functions of membrane proteins requires determining their structures. In general, proteins are made up of single or many domains, each of which can perform different molecular functions. These domains are structured (or ordered or folded) domains that fold independently and make precise tertiary interactions. Such domain-containing proteins are called structured proteins and they exhibit small-scale undulations under physiological conditions. Their function has long been thought to be inextricably related to a well-defined 3D structure, which is determined by the protein’s primary amino acid (AA) sequence (polypeptide chains). This close relationship between shape and function has been studied for many membrane proteins (White and Wimley 1999).
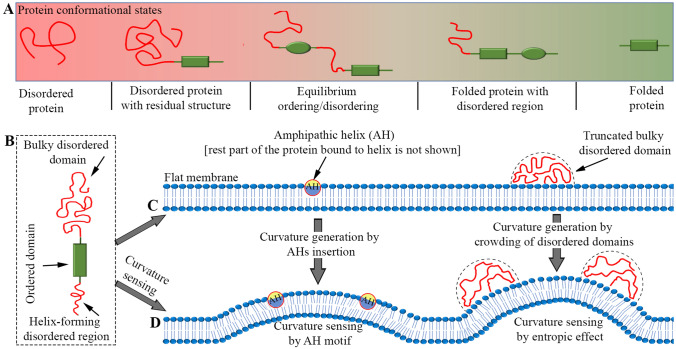
Unlike many structured proteins which must have a rigid 3D structure to fulfill their function, a substantial fraction of eukaryotic proteome consists of AA sequences that do not form a stable 3D structure but remain disordered in physiological conditions (Wright and Dyson 1999; Uversky 2002, 2003; Dyson and Wright 2005; Tompa 2005; Uversky and Dunker 2010; van der Lee et al. 2014). Proteins can have a structural disorder at short stretches (i.e., folded proteins with small disordered segments), long regions (i.e., disordered proteins with residual folded domains), or even the entire structure (Fig. 1A–D). It is essential to note that the majority of proteins do not have a completely disordered segment but are composed of some secondary structure with local disordered loops (see Fig. 1B), and some are even composed of structural domains connected by disordered linkers (Fukuchi et al. 2006). According to Protein Data Bank (PDB), only a slight fraction of crystal structures are entirely devoid of unstructured regions (Obradovic et al. 2003). Almost all human proteins contain unstructured regions within their N/C-terminus regions. Approximately 97% of proteins are predicted to have disordered regions in the first or last five AAs (Pentony and Jones 2010). The protein regions that demonstrate intrinsic disorder are called intrinsically disordered protein regions (IDPRs) (van der Lee et al. 2014; Habchi et al. 2014; Cornish et al. 2020), for instance, Epsin1 and CALM/AP180 proteins, as well as Bin/Amphiphysin/Rvs (BAR) domain proteins such as Amphiphysin (Amph) and Endophilin (Endo) (Pietrosemoli et al. 2013). Proteins, such as ubiquitin (Robustelli et al. 2018) and SEC22A/SEC22C (Pietrosemoli et al. 2013), without IDPRs are called ordered proteins. In addition, proteins [such as alpha-synuclein (S) (Jensen et al. 2011; Robustelli et al. 2018)] with entirely disordered AA sequences that lack any tertiary structure are referred to as intrinsically disordered proteins (IDPs) (van der Lee et al. 2014).
Fig. 1.
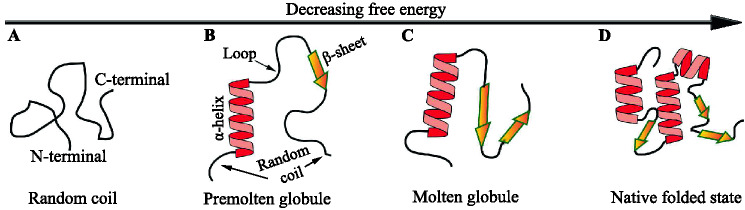
The dynamic ensemble of IDPs/IDPRs. Proteins can be characterized as either coil-like structures (non-compact unfolded chains, called IDPs) with maybe little or no secondary structures throughout the entire AA sequences (A), or as having a module with both IDPRs and folded domains as shown in B and C, or as native folded structures (D). The folded domains in B and C are the secondary structures composed of -helices and/or -sheets. While secondary structures fluctuate around their native position in premolten globules (B), molten globules possess almost native-like secondary structures and folding patterns with no close packing of their side chains (C). The disordered loop shown in B connects two distinct secondary structural elements. Schematic reproduced from Uversky and Finkelstein (2019)
The IDPs/IDPRs behave more like random polymers than stably ordered domains (Pietrosemoli et al. 2013). Consequently, protein’s intrinsic disorder represents a highly dynamic ensemble of conformations (Uversky 2002; Uversky et al. 2012). According to decreasing free energy and increasing compaction (from left to right, see Fig. 1A–D), conformations of a protein can be unfolded (random coil), premolten globule (PMG), molten globule (MG), and native folded states. Proteins disorder can be split into two classes based on structural features. Despite their flexibility, members of the first group are compact and have a well-defined secondary structure but lack a stable 3D shape, i.e., they have the characteristics of typical MGs. Proteins from another group may behave almost as random coils or as PMGs (significant secondary structure but less than that found in MGs) (Uversky 2002). Random coils can be considered as IDPs (Fig. 1A). The structure of IDPR-containing proteins is similar to that of PMG (Fig. 1B) or MG (Fig. 1C), which retain the structural elements of a native secondary structure and the approximate location of the folded state, but are flexible at the loops and ends. Both IDPs and IDPRs have now been identified as essential components of numerous proteins involved in membrane remodeling and coated-vesicle formation (Owen et al. 2004; Dafforn and Smith 2004; Pietrosemoli et al. 2013).
In eukaryotes, the total protein fractions disordered in entire sequence are predicted to be around 5–15%, and around 35–52% have at least one long IDPR () (Ward et al. 2004; Orosz and Ovádi 2011). In contrast, the long IDPRs in prokaryotic proteins are (Ward et al. 2004; Tompa 2005). As a large portion of the human proteome, membrane proteins also possess IDPs and IDPRs. A considerable fraction of the transmembrane (TM) proteins () involved in signaling have IDPRs of substantial length () (Iakoucheva et al. 2002), with a somewhat lower total prevalence in all TM proteins (41%) (Minezaki et al. 2007). Specifically, IDPRs are enriched in the regions extending from membrane-attached proteins into the cytoplasm, and cells use these cytoplasmic tails to transmit external signals across the membrane (Minezaki et al. 2007). An array of TM proteins has been found to have membrane-bound tails (Cornish et al. 2020).
In humans, IDPRs length profile follows a power law, with many-short disordered regions and a significant incidence of longer ones (Tompa and Kalmar 2010). Both structured and unstructured regions are necessary for the repertoire of the functions that proteins can have in many cellular processes, including transcription, translation, membrane trafficking pathways, and signal transduction (Iakoucheva et al. 2002; Tompa 2005; Dyson and Wright 2005; Snead and Eliezer 2019). According to both experimental and computational investigations, IDPRs have been observed in many types of proteins, i.e., globular, fibrous, and membrane proteins (Andreeva et al. 2014). Primary structural features of IDPs/IDPRs have been investigated mainly using small-angle X-ray scattering (SAXS), gel filtration, analytical ultracentrifugation, NMR, and various other spectroscopic methods (Kalthoff et al. 2002b; Dyson and Ewright 2002; Dyson and Wright 2005; Meier et al. 2008). These features include their molecular size, level of structural heterogeneity, the role of transient structure in coupled folding and binding events, aggregation tendencies, and presence of persistent structure (Dyson and Wright 2002, 2005; Meier et al. 2008).
The inability of IDPs/IDPRs to fold into a stably folded 3D structure is imprinted in the biased AA sequences, which are depleted in the hydrophobic regions that usually drive protein folding. Moreover, they are enriched in hydrophilic regions (charged and polar side chains) that prefer to remain in contact with water (Kotta-Loizou et al. 2013; van der Lee et al. 2014). The high degree of conformational entropy, high net charge, and a paucity of hydrophobic segments are all characteristics of IDPs/IDPRs (Cornish et al. 2020). As a result, IDPs/IDPRs assume a disordered and extended structural ensemble that is highly adaptable and compatible with particular functional modes (Varadi et al. 2014).
Intrinsically Disordered Yet Functional Motifs
Structured proteins achieve a global lowest energy state by folding, making the structure stable and functional. The energy minimization during the folding process leads to the burying of hydrophobic AAs inside the protein core instead of exposing them in the aqueous milieu. Under physiological conditions, structured proteins show small-scale fluctuations. In contrast to structured proteins, most IDPs/IDPRs are rich in AAs with charged or polar side chains that are not energetically penalized by exposure to the aqueous environment. Therefore, they do not need to be hidden inside the protein structure (Awile et al. 2010; Babu 2016). IDPs/IDPRs are thus unstructured due to a high net charge and low hydrophobicity. Albeit the unstructured regions have only a few hydrophobic AAs, those that do exist have their side chains exposed to an aqueous environment and are thus primed to be buried.
The energy landscape of IDPs/IDPRs is characterized by a large number of local minima with comparatively low transition barriers between different conformations [see “Abundance of intrinsically disordered proteins (IDPs) and protein regions (IDPRs) in membranes” section]. Such a wide range of interconverting conformations enable IDPs/IDPRs to interact with various binding partners. Indeed, one IDP/IDPR can act as diverse binders, interacting not only with numerous proteins (which can be other IDPs/IDPRs or folded proteins/domains) but also with lipid membranes, nucleic acids, as well as inorganic ions (Uversky and Dunker 2010). Conversely, some IDPRs do not interact with any biological targets but rather serve as flexible linkers between domains that keep them moving throughout functional activities or as flexible tails that regulate ordered domains (Uversky 2002; Tompa 2005). While IDPs/IDPRs-protein binding reactions have been extensively explored, IDPs/IDPRs–membrane interactions are still underinvestigated.
The IDPs/IDPRs are frequently involved in high specificity but low affinity ( M range) interactions, allowing for fast binding site interchange between several interacting partners. Interactions usually involve the folding of many IDPs/IDPRs upon their binding to the targets. The folding might take place for the entire unstructured protein, substantial portions, or only a few brief segments (Mittag et al. 2010). While some IDPs/IDPRs undergo disorder-to-order transitions only upon binding to a specific partner, many others form the so-called disordered, dynamic, or fuzzy complexes in the case when they stay disordered in bound state (Tompa and Fuxreiter 2008; Sharma et al. 2015). The majority of coupled folding and binding processes feature amphipathic motifs that are embedded inside more extensive disordered sequences (Dyson and Wright 2002, 2005). IDPs/IDPRs, like structured proteins, have unique motifs in their AAs that allow them to interact with other binding partners and become functional. The existence of one or more of the three structural elements listed below determines how an unfolded protein functions (van der Lee et al. 2014).
Short Linear Motifs (SLiMs)
SLiMs are usually 3–10 contiguous AA residues (Fakhree et al. 2019a) found mostly in IDPRs (over 80%) of a protein, and they serve as docking sites for protein–protein interactions. A SLiM interaction with a protein domain is sometimes called domain–SLiM interactions (Hugo et al. 2010). They form well-defined structures when they bind with their partners (Fuxreiter et al. 2007; Strome et al. 2018). SLiMs have low affinity for their binding targets [i.e., interaction dissociation constant ( is in 1–150 μM range (Diella et al. 2008)], allowing them to participate in reversible and short-lived interactions (Wright and Dyson 2009). Multiple SLiMs in the same protein normally compensate for the low binding affinity of individual SLiM-mediated interactions (Fig. 2) with the target proteins. Consequently, the apparent affinity due to multiple SLiMs dramatically increases (Davey et al. 2012). Due to their small size and low binding affinity, the functional importance of the SLiMs is inherently difficult to determine by bioinformatic approaches (Davey et al. 2012), and much information about these SLiMs, as well as the confirmation of their biological activities, are procured from experimentation (Neduva and Russell 2005).
Fig. 2.
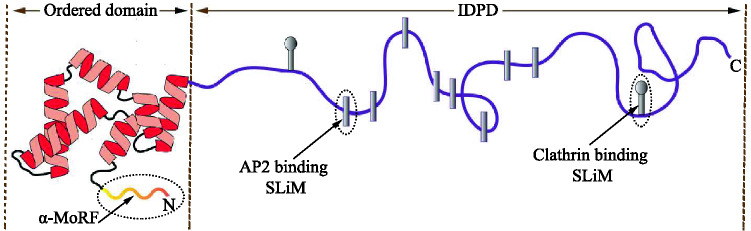
Motif/domain structures of Epsin1. This protein contains an ordered domain (i.e., ENTH domain) and a bulky disordered region (i.e., IDPD). The ENTH domain comprises a small disordered segment (i.e., MoRF motif), which acquires an -helical secondary structure upon binding to the membrane. The IDPD, which remains unstructured in solution, contains multiple SLiMs that attach to the N-terminal domain of clathrin and -appendage domain of AP2 protein. Reproduced from Kalthoff et al. (2002a)
Many dynamic networks rely on reversible yet high-avidity SLiM-mediated interactions, such as those dictating cell signaling, where several multi-protein complexes quickly assemble and disintegrate (Diella et al. 2008). A prominent example of SLiMs in IDPRs is the recognition sites for diverse post-translational modifications (PTMs) (Babu 2016; Lin et al. 2017) like phosphorylation and ubiquitylation (Woodsmith et al. 2013). Recently, researchers have identified a set of SLiM candidates in AEC2 and integrin TM proteins, likely to act in the host cell entry system of the novel coronavirus 2 (SARS-COV-2) (Mészáros et al. 2021; Kliche et al. 2021). In another example, motif PXXP that mediates interaction with the SH3 domain can be considered a SLiM (Diella et al. 2008). SLiMs are also involved in protein targeting to particular cellular compartments. A couple of examples are C-terminally located motifs such as KDEL and KKXX. As the KDEL motif causes the disordered protein to reside within the ER lumen, the KKXX promotes cytoplasmic or TM localization (Stornaiuolo et al. 2003).
Molecular Recognition Features (MoRFs)
MoRFs are longer interaction motif than SLiMs (usually 10–70 AAs), and they are also located within IDPs/IDPRs (van der Lee et al. 2014). In contrast to SLiMs, MoRFs are not described based on AA sequences but as interaction-prone unfolded regions that can generate secondary structure when bound. They can be divided into three categories, based on the secondary structure that they adopt when they bind to cellular targets, i.e., -MoRF (Fig. 2), -MoRF, and i-MoRFs, which represent alpha helices, beta strands, and irregular structures, respectively (van der Lee et al. 2014). Alpha helices are the most common membrane-interacting structure adopted upon binding, undergoing a disorder-to-order transition. As MoRFs name implies, they are thought to play a role in molecular recognition, the first step in interactions. Further, studies reveal that the unique AA composition, structural and physicochemical characteristics of these interfaces play a central role in the interaction between MoRFs and their binding partners (Vacic et al. 2007). Moreover, similar to SLiMs, MoRFs usually engage with their molecular targets with a modest affinity but high specificity. Many IDPs/IDPRs go through coupled folding and binding steps after recognizing their binding partners, resulting in stable secondary structures (Dyson and Wright 2002). According to MoRFs functional analysis, around 20% of MoRF-containing proteins are TM (Mohan et al. 2006). A typical example of a MoRF binding domain is the N-terminal IDPR of p53 protein, which changes from disordered to an -helical form (40–60 AAs) by interacting with Mdm2 protein (Xue et al. 1834). Membrane-interacting MoRFs are discussed in detail in “Coupled folding and binding mechanism” section.
Intrinsically Disordered Protein Domains (IDPDs)
Most protein domains are structured, but some can be entirely or largely unstructured on membranes, excluding the formation of -helices upon membrane binding. Such disordered domains are usually referred to as IDPDs. The IDPDs often comprise bulky, yet flexible parts of the IDPs/IDPRs (Fig. 2). The unstructured state of IDPDs is what makes them functional (van der Lee et al. 2014). Intriguingly, IDPDs are observed in many proteins (see Table 1) involved in membrane trafficking, suggesting that IDPD-containing proteins play a role in remodeling of cellular membranes that is still poorly understood. The action of IDPDs (i.e., bulky IDPRs) with the membrane is further discussed in “Entropic effects” section. Henceforth, we will use the term IDPDs in place of bulky IDPRs.
Table 1.
The potential involvement of intrinsically disordered protein domains (IDPDs) in membrane remodeling
| Protein | % IDPD (location) | Membrane interaction | Binding proteins | Protein function | References |
|---|---|---|---|---|---|
| Epsin1 | 78 (CT) | ENTH domain | Cargo, AP2, clathrin | CCP | Schmid and McMahon (2007) and Pietrosemoli et al. (2013) |
| AP180 | 28 (CT) | ANTH domain | Cargo, AP2, clathrin | CCP | Schmid and McMahon (2007) and Pietrosemoli et al. (2013) |
| Amphiphysin1 | 60 (middle) | N-BAR domain | AP2, clathrin, dynamin | CCP | Schmid and McMahon (2007) and Pietrosemoli et al. (2013) |
| SNX9 | 27 (middle) | PX-BAR domain | Dynamin, AP2, clathrin | CCP | Schmid and McMahon (2007) and Pietrosemoli et al. (2013) |
| Endophilin A1 | 33 (middle) | N-BAR domain | Dynamin, synaptojanin | CCP | Pietrosemoli et al. (2013) and Ambroso et al. (2014) |
| FCHo1 | 48 (middle) | F-BAR domain | Eps15, intersectin | CCP | Henne et al. (2010) |
| FBP17 | 11 (middle) | F-BAR domain | SNX2 | EGFR endocytosis | Tsujita et al. (2006) and Su et al. (2020) |
| Intersectin1 | 28 (middle) | PH, C2 domains | AP2, clathrin, Eps15, FCHo1/2 | CCP | Schmid and McMahon (2007) and McMahon and Boucrot (2011) |
| Auxilin | 45 (middle) | – | Hsc70, dynamin, clathrin | Clathrin uncoating | Schmid and McMahon (2007) and Pietrosemoli et al. (2013) |
| SEC16A | 71 (NT/CT) | – | SEC13/31, SEC23/24, Sar1 | COPII route | Whittle and Schwartz (2010) and Pietrosemoli et al. (2013) |
| SEC31A | 34 (middle) | – | SEC13, SEC23/24, SEC16 | COPII route | Fath et al. (2007) and Pietrosemoli et al. (2013) |
CT C-terminal, NT N-terminal, CCP Clathrin-coated pit, EGFR Epidermal growth factor receptor
Interestingly, a single IDPR-containing protein can contain all the motifs as mentioned above multiple times (see the example of Epsin1 protein in Fig. 2). SLiMs/MoRFs can both be located within IDPDs. Furthermore, SLiMs themselves can be seen in MoRFs (O’Shea et al. 2017). The existence of multiple motifs can function synergistically to enhance the binding affinity of IDPRs for their target partners. While short IDPRs containing individual SLiMs/MoRFs may act as linkers, longer IDPs/IDPRs can have multiple SLiMs/MoRFs, which may serve as a scaffold for generating large membrane curvature. For example, the accessory protein Epsin1 dynamically interacts with clathrin and adopter protein AP2 through multiple SLiMs that are located in the IDPD during clathrin-mediated endocytosis (CME) (Kalthoff et al. 2002a). The ordered ENTH domain of this protein contains an -MoRF motif that binds to the membrane.
IDPs/IDPRs-Membrane Interactions: A Biophysical Study
The interplay between proteins and lipid membranes is complex while determining the biological membrane properties. The studies reveal that proteins precisely generate the large membrane curvature, whereas they also can sense the curvature and bind to the specific geometrical cues (Has and Das 2021). In principle, there should be more curvature sensors than inducers because some sensors that are too flexible to bend the membrane may not be able to drive the curvature (Zimmerberg and Kozlov 2006). In reality, there are far fewer proteins that have been demonstrated to sense membrane curvature than those that deform membranes (Antonny 2011). It has been proposed that the same protein can induce and sense curvature under varying concentration regimes (Suetsugu and Gautreau 2012; Simunovic et al. 2018). Nonetheless, it remains unclear whether all proteins that induce curvature are, by default, curvature sensors (Madsen et al. 2010). This complex relationship between lipid membrane and proteins leads to the organization of proteins by the membrane and vice-versa. It is well known that protein–membrane interaction is primarily driven by the partitioning of hydrophobic residues into the membrane and also by electrostatic interactions between charged protein residues and polar headgroups of the lipids (Seelig 2004; White and Wimley 1998, 1999). It is not surprising that electrostatic interactions can be quite effective in guiding IDPs/IDPRs to their target membrane surfaces. In addition, environmental factors such as pH, salts, electrolytes, temperature, and other critical chemicals can influence the interaction between membrane and proteins. The presence of protein functional motifs/domains (MoRFs and/or IDPDs), protein intrinsic shape and surface density, lipid composition, packing, and membrane curvature all play a role in how IDPs/IDPRs interact with the membranes.
Coupled Folding and Binding Mechanism
A characteristic hallmark of IDPs/IDPRs function is the transition from an unfolded state in the solution to a more ordered form inside the membranes. The interaction of a protein with its binding partner results in a conformational change in the protein, known as the induced fit (IF) model. In contrast, several systems follow the conformational selection (CS) paradigm in which the binding partner chooses the suitable conformation (prefolded state) from an ensemble as a protein binds to its target (Berlow et al. 2015; Cornish et al. 2020). In general, conformational selection is expected in ordered proteins (Kim et al. 2007; Pozzi et al. 2012; Vogt et al. 2014) and appears to be comparatively rare for IDPs/IDPRs. Perhaps because the preferred model is determined by inherent secondary structure propensity (Arai et al. 2015), and IDPs/IDPRs in their free state often lack a very stable secondary structure.
The membrane protein folding event is investigated by molecular interactions between protein and lipid moieties. The energetics involved with the partitioning of AA residues from an aqueous solution into membrane interfacial region [thickness around 1.5 nm, this region comprises lipid headgroups and considerable bound water (Bowie 2005)] is a major driving force behind the IDPs/IDPRs folding. An essential component of this energetics is the reduction in free energy per AA residue at the membrane interface, which causes secondary structures to form. Here, the required energy is also termed partitioning energy, and it is provided by hydrophobic side chains of AAs (Wimley and White 1996). Often folding occurs when AAs are partitioned into membranes, resulting in the development of the secondary structures (mostly alpha helices) oriented parallel to membrane plane [Fig. 3A(a, b)]. This is known as the partitioning-folding coupling process (White et al. 2001). A question arises, why do AAs fold and remain in the membrane? Soluble IDPs/IDPRs form a continuum structure with various shapes, from random coil-like chains to premolten to molten globules (Uversky et al. 2012). In the case of coil-like conformation, it will have a few hydrogen-bonded peptide bonds that are thermodynamically unstable in the hydrophobic region of the membrane. The membrane-embedded IDPs/IDPRs will have to be minimally organized into backbone hydrogen bonds internally, most likely in an -helical secondary structure. The helical structure in the bilayer region resembles soluble PMG or MG (Kjaergaard 2015).
Fig. 3.
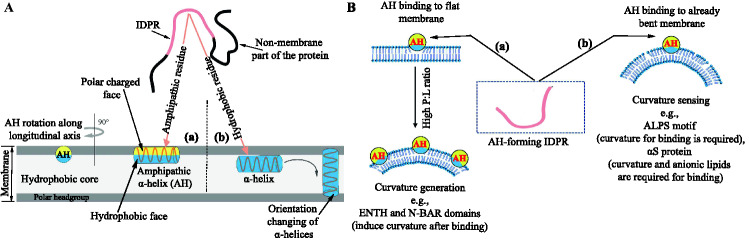
IDPR folding into alpha helices. A By folding-upon-binding transition, membrane-interacting IDPRs adopt amphipathic -helical (a), or only -helical structures (b). To find a better amphipathic orientation, AH is rotated along its longitudinal axis relative to its original conformation (leftmost). Reproduced from Cornish et al. (2020). Alpha helices are formed if helices are made of non-polar hydrophobic residues (b). Once helices are formed, they change the orientation and insert into the hydrophobic core of the membrane, referred to as TM helices. Next, TM helices are assembled into functional structures. The non-membrane part of the protein attached to the helix has not been shown in the schematic. Reproduced from White et al. (2001). B Lipid-binding mechanisms for AHs that sense and induce membrane curvature. (a) It is believed that proteins involved in curvature generation can bind to flat membranes by using hydrophobic and electrostatic interactions (specifically lipid for Epsin1). In high protein:lipid (P:L) ratios, the wedge effect, and the bilayer-couple mechanism cause curvature. (b) Some motifs, such as ALPS, require an already bent membrane for binding, as membrane insertion is driven only by hydrophobic interactions. In another example, the binding of S with its small and poorly hydrophobic residues and zwitterionic polar face is dependent on curvature and anionic lipids. Reproduced from Drin and Antonny (2010)
The helical structure can span the entire bilayer once or multiple times (i.e., TM proteins). Alternatively, it can only be embedded transiently into one of the membrane monolayers (i.e., peripheral membrane proteins). The -helical motifs composed primarily of non-polar and hydrophobic residues have generally traversed the membrane [Fig. 3A(b)], whereas helical motifs containing both hydrophobic and hydrophilic residues [known as amphipathic -helix (AH)] penetrate the bilayer region shallowly (Campelo et al. 2008; Mahata and Das 2017; Li 2018) [Fig. 3A(a)]. Various parameters, including the size of those residues’ side chains, their placement relative to the interface, and their helical periodicity, all influence the finer control of binding specificity (Antonny 2011). In the case of AHs insertion, hydrophobic and polar/charged residues are partitioned across the two faces of the helix. Membrane-inserting AHs lie parallel to the membrane, where their hydrophobic and polar/charged faces are respectively hidden and exposed to the cytosol and lipid headgroups (Giménez-Andrés et al. 2018), as shown in Fig. 3A(a). Here, electrostatic and hydrophobic interactions may play a role in protein binding to the membrane. Hydrophobic segments can penetrate the lipid bilayer, whereas polar residues can force lipid headgroups apart. Membrane bending can occur when anionic lipids interact with positively charged side chains on the polar face of the AH. On the other hand, the AHs consisting mainly of negatively charged residues cannot effectively interact with a flat membrane. AHs with negatively charged or even without charged residues are unable to induce the curvature, and such helices require a membrane to be already curved to insert. In this case, lipid headgroups are stretched apart, and thus the interaction is primarily driven by the hydrophobic effect (Drin et al. 2007; Drin and Antonny 2010; Gallop et al. 2006; Mesmin et al. 2007). From here, we can conclude that proteins can induce as well as sense the membrane curvature through their AH motifs [Fig. 3B(a,b)].
The amount of curvature increase is determined by two factors, i.e., the size of inserted AH and surface density of IDPs/IDPRs. A typical example of a disorder-to-order transition that senses and generates the curvature occurs in 100-residues N-terminal of S [an IDP in solution (Jensen et al. 2011), associated with Parkinson’s disease] (Davidson et al. 1998). This protein can integrate two separate membrane-binding AHs connected through a non-helical linker, making it potentially able to bridge two membranes in close proximity (Dikiy and Eliezer 2012). The binding of S with negatively charged membrane results in the formation of an N-terminal AH, which induces the curvature by changing the membrane surface area of the outer monolayer when an AH is inserted into the outer membrane monolayer (Braun et al. 2012). Note that an N-terminal AH is believed to be inserted into the membrane like a wedge, thereby inducing membrane bending (Peter et al. 2004). In-vitro investigations with model lipid vesicles have proven the transition to the alpha-helical state and the resultant generation of curvature (Ferreon et al. 2009; Varkey et al. 2010; Braun et al. 2012, 2014). However, the curvature sensitivity of S (Jensen et al. 2011) is more pronounced compared to its curvature-inducing potency [Fig. 3B(b)]. The binding of S on a flat membrane with sparse and shallow packing defects is hampered by a poorly developed hydrophobic face having multiple polar threonine residues, resulting in curvature sensitivity and preferred binding to negatively charged membranes (Pranke et al. 2011). If the membrane has no net charge, S interacts weakly with the membrane. Due to the lack of a requirement for lipids binding specificity or protein structural features, the authors (Jiang et al. 2013) speculate that S may induce curvature by protein–protein crowding mechanism (discussed in “Molecular crowding” section).
The curvature-sensing/inducing ability of many proteins and protein domains, such as BAR domains of Amph, Endo, PICK1, and MIM/ABBA (Peter et al. 2004; Lee et al. 2007; Hatzakis et al. 2009; Bhatia et al. 2009; Sorre et al. 2012; Herlo et al. 2018), epsin N-terminal homology (ENTH) domain of Epsin (Ford et al. 2002), amphipathic lipid packing sensor (ALPS) motifs of ArfGAP1 (Bigay et al. 2005) and Golgin GMAP-210 (Drin et al. 2008), ANTH (AP180 N-terminal homology) domain of CALM (Miller et al. 2015), small GTPases Arf1 and Sar1 (Lee et al. 2005; Lundmark et al. 2008; Beck et al. 2008), and Annexin B12 (AnxB12) (Fischer et al. 2007; Jensen et al. 2011) have been identified by their N-terminal AHs. In the case of BARs, these domains are isolated from the full-length proteins and shown to fold their membrane inserting IDPRs (i.e., -MoRFs) into AH motifs in the presence of lipids. Although BAR domains are usually well structured even when isolated in aqueous solutions, the presence of an AH coordinates to amplify the curvature sensing and induction (Peter et al. 2004; Gallop et al. 2006). However, the recent study shows that the presence of N-terminal AH of Endo has no significant contribution to its molecular curvature driving potency (Chen et al. 2016). Similarly, Epsin1 (clathrin pathway) capacity to induce large curvature through the ENTH domain by inserting its N-terminal AH (called H0) is doubted, and it can drive the curvature when bound to high surface coverage (discussed in “Surface density of IDPs/IDPRs on membrane surface” section) (Stachowiak et al. 2012; Snead et al. 2017; Busch et al. 2015). Furthermore, membrane curvature-sensing properties of Epsin1 and ArfGAP1/GMAP-210 arise, respectively, from an AH-containing structured ENTH domain and disordered ALPS motifs. It has been shown that the ALPS senses curvature by forming a stable AH upon binding to the membrane (Bigay et al. 2005; Drin et al. 2007). Note that membrane insertion in the case of ALPS is driven by only the hydrophobic effect [Fig. 3B(b)], as ALPS does not have charged residues in the hydrophilic region (Drin and Antonny 2010). In another example, Complexin (CPX) protein senses the curvature of synaptic vesicle (SV) membranes by tandem motifs found in C-terminal domain (CTD), i.e., a C-terminal (CT) and an adjacent AH motifs (Snead et al. 2014). Although CPX is thought to bind SV membranes rather than other membranes, its molecular mechanism remains largely unclear.
Besides AHs, proteins with disordered stretches of hydrophobic motifs are also capable of forming short wedged-shaped hydrophobic loops (HLs) in the membrane, which can cause the membrane to bend [see e.g., Has and Das (2021)]. The HLs are also expected to sense defects between lipid headgroups, and are more prevalent on highly curved membrane surfaces. Proteins with HLs such as PACSIN2 (Wang et al. 2009; Plomann et al. 2010), EHD2 (Daumke et al. 2007), Dynamin (Ramachandran and Schmid 2008; Ramachandran et al. 2009), FAPP (Cao et al. 2009; Lenoir et al. 2010), and Synaptotagmin1 (Syt1) (Martens et al. 2007) have shown their ability to sense/derive the membrane curvature. Many TM proteins are believed to sense/induce the membrane curvature by inserting hydrophobic helical segments, e.g., potassium channel KvAP (Aimon et al. 2014). Another example is M2 TM protein of influenza causes membrane scission by deep insertion of its AH motif (Martyna et al. 2017). Furthermore, some TM proteins such as reticulons, DP1/Yop1p, and Caveolins sense/induce the curvature by the insertion of short wedged-shaped -helical hairpins (Shibata et al. 2009) and AH motif (Brady et al. 2015; Breeze et al. 2016; Wang et al. 2021).
The capability of IDPs/IDPRs to adopt the -helical structure upon binding membranes has also been linked with disease. The hIAPP (Pannuzzo et al. 2013b) and A (Pannuzzo et al. 2013a), respectively, are involved in developing type 2 diabetes and Alzheimer’s disease. It has been claimed that the interaction of these proteins with the membrane leads to the formation and aggregation of membrane-embedded -helical peptides, which is suggested to be hazardous.
Many IDPRs also sense the curvature by binding to the membranes while remaining unfolded and interacting with charged lipid headgroups, covalently linked membrane lipids, or through ionic bridges. Examples include Annexin family proteins that bind to phosphatidylserine (PS)-rich membrane bridged by (Moreno-Pescador et al. 2019), Rhodopsin-like G protein-coupled receptors (GPCRs) (Escribá et al. 2007), Ras proteins (Larsen et al. 2015; Liang et al. 2019), and MARCKS-ED (Morton et al. 2013) bind to the membrane through lipid anchors. GPCRs also contain 7 TM domains and an AH (Huber et al. 2004). Membrane curvature sensing/generation due to the insertion of hydrophobic/amphipathic helical motifs for a plethora of TM and peripheral membrane proteins have been extensively reviewed recently (Has and Das 2021).
Membrane Curvature Modulation by Scaffolding
Many proteins, through the scaffolding mechanism, generate the large curvature either by their intrinsically curved shape or oligomerization (Has and Das 2021). They are also thought to sense the curvature through a scaffolding pathway in which a membrane having large curvature attracts such protein domains with matching curvature (Peter et al. 2004). These proteins induce forces capable of deforming a flat membrane, resulting in membrane fission or fusion in extreme cases (Snead et al. 2017; Snead and Eliezer 2019). To locally induce membrane curvature, the protein–membrane binding energy must be greater than the membrane bending energy (Zimmerberg and Kozlov 2006). The well-known examples of curvature-inducing proteins can be seen in the process of CME, where N-BAR domain-containing proteins Amph and Endo, with a large curved surface and strong affinity towards membrane surface (due to electrostatic interactions), facilitate the scaffold for curvature generation (Chernomordik and Kozlov 2003; Zimmerberg and Kozlov 2006). The bulky C-terminal domains remain unfolded (C-terminal IDPD) in physiological conditions for these proteins. Many scaffold-forming proteins oligomerize to create massive rigid structures and thus can enhance curvature generation, for instance, BAR domains (Peter et al. 2004; McDonald and Gould 2016), clathrin proteins (Takei et al. 1999), Dynamins (Roux et al. 2010), ESCRT machinery (Hurley and Hanson 2010), and the bacterial tubulin FtsZ (Sundararajan and Goley 2017). Note that clathrin induces the membrane curvature by indirect scaffolding, where membrane and clathrin are bridged by accessory/adopter proteins such as Epsin and AP180/CALM. Here, the structured ANTH domain of AP180 interacts with negatively charged membrane, whereas disordered CTD interacts with the N-terminal domain of clathrin heavy chain (Zhuo et al. 2010). As a result, AP180 appears to be a fuzzy complex, a disordered protein even when linked to a partner (Tompa and Fuxreiter 2008).
Membrane curvature modulation can occur due to the cooperated actions of structured and unstructured regions of membrane proteins. Accordingly, the curvature is driven by a combination of asymmetric binding of a partly structured scaffold protein or by inserting a partially structured protein into one of the bilayer monolayers and introducing lateral pressure through the unstructured, solvent-exposed segment (Fakhree et al. 2019a). For example, the cooperative action between structured and unstructured regions can be seen in curved BAR domain scaffold protein and S that is partially inserted into the membrane through the formation of an AH (Zeno et al. 2018; Fakhree et al. 2019b; Snead and Eliezer 2019).
Surface Density of IDPs/IDPRs on Membrane Surface
Molecular Crowding
Proteins not only generate curvature by the insertion of hydrophobic residues or scaffolding mechanisms but also can modulate the curvature at large or small membrane surface coverage. The fractional coverage can be computed by the product of the projected area of proteins on the membrane surface and the number of interacting proteins per unit membrane area (Zeno et al. 2019a). In the case of large surface coverage (or surface density), collision among the crowded adsorbed or anchored proteins leads to the generation of steric pressure (analogous to compressed gas) (Carnahan and Starling 1969; Carignano and Szleifer 1995), which provides an efficient force for expanding the membrane surface (Montesano et al. 2001; Scheve et al. 2013). Unless equally crowded molecules counter this pressure on the opposite membrane surface, the membrane will bend towards the more crowded surface, and this is commonly known as a molecular crowding (MC) mechanism (Fig. 4A), which happens at very large surface density (Stachowiak et al. 2012; Snead et al. 2017). For asymmetric protein distribution, increasing the curvature of the membrane expands the area of the outer leaflet of the bilayer, thus lowering the pressure. The crowding mechanism has been studied for ordered and disordered domains (IDPDs). This mechanism for disordered domains has been discussed in “Entropic effects” section.
Fig. 4.
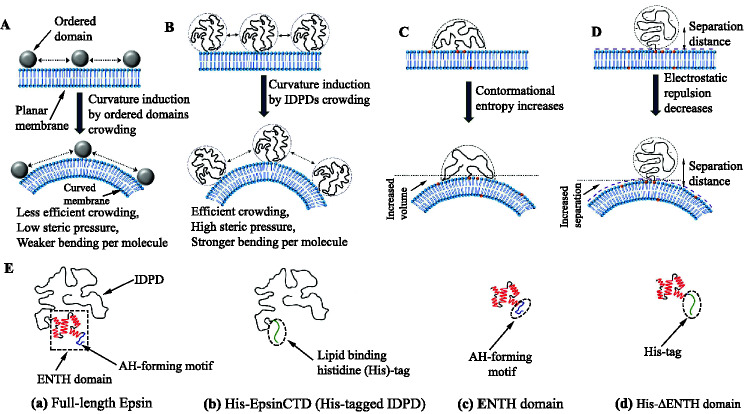
Schematic illustrations of crowding and entropic/electrostatic mechanisms by which proteins can induce/sense the membrane curvature. A Membrane bending by ordered protein domains. B Long IDPRs, i.e., IDPDs, drive membrane curvature. IDPDs crowding on membrane surfaces is more efficient than ordered domains of similar molecular weight. Reproduced from Snead and Stachowiak (2018). C Illustration of entropically driven curvature sensing by IDPDs. D Representing the electrostatically driven curvature sensing by IDPDs. Reproduced from Zeno et al. (2019b). E Schematics of Epsin1 and its domains employed for studying membrane curvature sensing (reproduced from Zeno et al. (2018)). Full-length Epsin1 comprises an ordered domain ENTH and an IDPD at the C-terminus (a). Curvature sensitivity of IDPD (b) and ENTH (c) are studied in isolation. In the case of IDPD, a lipid-binding engineered His-tag is attached near the N-terminal, represented by His-EpsinCTD. His-tag replaces the AH to investigate the ability of mutant ENTH (His-ENTH) in curvature sensing and induction (d). The curvature-sensing ability is found in the order of FL-Epsin > His-EpsinCTD > ENTH > His-ENTH (insensitive) (Zeno et al. 2018)
The curvature induction by a few ordered domains, including Epsin ENTH and AP180 ANTH, was investigated almost a decade ago (Stachowiak et al. 2012). The experimental approach confirmed that the ENTH domain with and without AH had no significant role in membrane curvature generation. However, curvature sensitivity was attributed to AH insertion. Moreover, no evidence of ENTH domain oligomerization was observed (Stachowiak et al. 2012), contrary to past reported data (Yoon et al. 2010). As a result, it may be argued that the ENTH domain is responsible for membrane curvature via the MC mechanism. At high surface coverage, the membrane deformation activity was also described by crowding of helix lacking ANTH domain (Stachowiak et al. 2012).
Entropic Effects
As discussed, through their intrinsic curved shape or AHs/HLs motifs, most of the proteins directly interact/bind to the membrane and sense/deform the curvature. Both of these mechanisms are dependent on the structural features of proteins. These structured domains often account for only a small portion of the mass of protein molecules that contain them (Busch et al. 2015). A substantial region of the same protein molecules that do not directly interact with the membrane remains disordered in solution (Dyson and Wright 2005). Thus, the folding transition has not been proven to occur in these disordered domains. There are several examples of accessory/adopter proteins in the CME pathway. The membrane-interacting regions of adopters are composed of structural motifs/domains (e.g., AH motifs and BAR domains). The other part of these proteins is membrane non-interacting domains that interact with clathrin, and another component of the coat, typically CTDs, are often IDPDs [see “Intrinsically disordered protein domains (IDPDs)” section and also Table 1] (Kalthoff et al. 2002b; Zhuo et al. 2010). Specifically, Epsin1 protein consists of an ENTH domain followed by an IDPD () (Kalthoff et al. 2002b). Similarly, AP180/CALM consists of an ANTH domain plus a substantial disordered domain () (Kalthoff et al. 2002b; Zhuo et al. 2010). Moreover, BAR domains are also typically associated with extensive disordered regions. For instance, Amph and FCHo1 have a crescent-shaped BAR domain and an IDPD () (Pietrosemoli et al. 2013; Zeno et al. 2018). Similarly, Sorting Nexin 9 (SNX9) contains a BAR domain and an IDPD of more than 100 AAs (Lundmark and Carlsson 2009). Finally, clathrin-case disassembly Auxilin protein contains more than 260 disordered AAs (Scheele et al. 2003). Generally speaking, IDPDs are exceptionally high in the CME pathway, where it has been recently reported that 30% of proteins contain IDPDs of . It includes the adaptors discussed above as well as other well-studied proteins such as Dynamin and Eps15 (Pietrosemoli et al. 2013). IDPDs are also seen in intracellular trafficking proteins. SEC16, for example, contains an IDPD of (Fath et al. 2007), which is assumed to be critical for capturing COPII components across a long distance. In addition, COPII coat component SEC31 has an IDPD of over 400 AAs that may serve as a binding site for SEC23/24 complex (Fath et al. 2007).
Curvature-sensing/induction properties of structured domains have been extensively studied previously (Peter et al. 2004; Bhatia et al. 2009). However, the unstructured domains have been disregarded in these studies, owing to the widespread belief that curvature sensing/induction necessitates specific structural motifs. As a result of new findings of biophysical roles for disordered domains (Busch et al. 2015; Snead et al. 2017; Zeno et al. 2018, 2019b), it has become even more critical to reconsider the mechanisms behind membrane remodeling in trafficking routes. Recently, it has been experimentally demonstrated that the IDPDs can potentially drive/sense the membrane curvature through a combination of entropic and electrostatic mechanisms (Busch et al. 2015; Zeno et al. 2018). The polymer-like behavior of IDPD facilitates a wide range of conformational diversity. When an IDPD is tethered onto the membrane surface, its chain entropy is considerably diminished, as the surface restricts the number of IDPD conformations (Lipowsky 1997; Nowicki et al. 2009). However, the geometric restriction is alleviated, and the IDPDs conformational entropy increases when the surface bends away from the protein and takes on a convex shape. Increasing chain entropy thus leads to IDPDs preferentially binding to curved surfaces (Fig. 4C) rather than planar surfaces (Zeno et al. 2018, 2019b).
At the large density, proteins become crowded on the membrane surface, and lateral steric pressure is generated between neighboring IDPDs. The membrane needs to be bent away from densely crowded proteins to alleviate this steric effect, which produces a larger average distance between them (Stachowiak et al. 2010, 2012), thus generating the curvature, as shown in Fig. 4B. A significant advantage of IDPDs is that their expanded conformations make them effective curvature inducers (Busch et al. 2015; Snead et al. 2017). IDPDs, which have much greater hydrodynamic radii than structured domains of identical molecular weight, are thought to contribute the most to membrane crowding because they occupy the most area on the membrane surface (Fig. 4A, B). The study reveals that the density of the protein in solution required to induce substantial membrane curvature is inversely proportional to IDPD’s projected area (Zeno et al. 2019a). The structured Epsin1 ENTH domain occupies a membrane surface area of 16 nm2 (Boucrot et al. 2012), while the disordered CTDs of Epsin and AP180 are projected to occupy 70 and 90 nm2, respectively, based on analytical modeling (Hofmann et al. 2012) as well as experiments (Kalthoff et al. 2002b). The projected area of the disordered domain of Amph is estimated as 75 nm2, while this area for N-BAR is approximately 24 nm2 per monomer, based on its crystal structure (Campelo et al. 2008; Zeno et al. 2018). We can summarize that IDPDs cause higher steric pressure, which causes a stronger membrane bending per molecule than ordered protein domains (see Fig. 4B).
In addition to entropic effects, a second mechanism that can sense/drive membrane curvature may also arise from electrostatic effects (Fig. 4D), especially when there is a substantial net negative charge in the disordered regions (Zeno et al. 2019b). Such IDPDs lead to a strong repulsive electrostatic interaction with a membrane containing anionic lipids (Sun and Drubin 2012). Average separation between IDPDs and the membrane surface increases when membrane curvature is large enough (convex shape), thus decreasing in repulsive interaction. It is believed that both entropic and electrostatic mechanisms work together to enhance the overall curvature sensitivity. For instance, adding anionic lipids to the membrane increases the overall curvature sensitivity of AP180CTD (Zeno et al. 2019b).
To investigate the curvature sensitivity of disordered CTDs (i.e., IDPDs) in isolation, the membrane-binding structured regions are truncated. Accordingly, it is concluded that the CTDs contain no significant motif for membrane binding. An N-terminal hexahistidine-tag (His-tag, for brevity) is added to protein CTDs for binding to the membrane containing nickel-chelating lipid DGS-Ni-NTA (1,2-dioleoyl-sn-glycero-3[N-(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl(nickel salt)) (Busch et al. 2015). Nickel nitrilotriacetic acid (Ni-NTA) headgroups of chelating lipid form a strong complex with His-tag to facilitate efficient membrane recruitment (Nye and Groves 2008). Importantly, His-tagged IDPDs do not interact to membranes lacking DGS-NTA lipids, suggesting that additional motifs or residues within the disordered proteins do not significantly interact with membranes (Zeno et al. 2018). Recently, a few His-tagged IDPDs (e.g., AP180CTD, EpsinCTD, and AmphCTD) have been shown to sense membrane curvature through entropic mechanism at comparable levels as ordered curvature-sensing domains. They can synergistically augment the sensitivity of these domains (Busch et al. 2015; Zeno et al. 2018; Zeno et al. 2019a; Zeno et al. 2019b; Zeno et al. 2021). For example, full-length Amphiphysin1 (FL-Amph) and Epsin1 (FL-Epsin) sense membrane curvature more effectively than their structured domains, N-BAR and ENTH, respectively (Zeno et al. 2018). Even CTD of Epsin1 has more sensitivity than the ordered ENTH domain. Figure 4E exhibits the cartoons of FL-Epsin and its domains that have been used to investigate the curvature sensitivity. Table 2 summarizes the curvature-sensing/induction mechanism by some ordered/disordered protein domains.
Table 2.
Mechanisms of membrane curvature sensing/generation by some ordered and disordered protein domains. Adapted from Zeno et al. (2018, 2019a, 2019b, 2021)
| Protein/domain | Domain types | Membrane-binding domain | Preferred lipid | Sensing | Induction |
|---|---|---|---|---|---|
| FL-Espin | OPD, IDPD | ENTH | AH, Entropic | MCd | |
| His-Epsina | OPD, IDPD | His-tag | DGS-NTA | Entropic | MC |
| wt-ENTHb | OPD | ENTH | AH | MC | |
| His-ENTH | OPD | His-tag | DGS-NTA | Insensitive | MC |
| His-EpsinCTDc | IDPD | His-tag | DGS-NTA | Entropic | MC |
| His-AP180CTDc | IDPD | His-tag | DGS-NTA | Entropic | MC |
| FL-Amph | OPD, IDPD | N-BAR | AH, IC, Entropic | AH, IC, OLG, MC | |
| Amph N-BAR | OPD | N-BAR | AH, IC | AH, IC, OLG | |
| His-AmphCTDc | IDPD | His-tag | DGS-NTA | Entropic | MC |
OPD Ordered protein domain, IDPD Intrinsically disordered protein domain, FL Full-length, MC Molecular crowding, IC Intrinsic curvature, OLG Oligomerization, His-tag Hexahistidine-tag, wt Wild-type, CTD C-terminal domain, Amph Amphiphysin, Phosphatidylinositol 4,5-bisphosphate, DGS-Ni-NTA 1,2-dioleoyl-sn-glycero-3[N-(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl(nickel salt)
aCurvature-sensing AH is replaced with His-tag
bWild-type Epsin is known to sense membrane curvature through inserting its AH
cStructured domains are truncated, and an N-terminal His-tag is added to protein CTDs for binding to the membrane
dThe molecular crowding of IDPDs on the membrane surface is more efficient than the OPDs
From the above discussion, we can conclude that the extent of each mechanism’s contribution to driving curvature is determined by the sizes of structured and unstructured domains, and protein surface density. The latter is determined by the bulk concentration and membrane affinity.
IDPs/IDPRs and Membrane Lipid Composition
It is thought that the lipid composition in a membrane can affect its physiological functions owing to the possibility of accommodating many different types of phospholipids in a bilayer. The major lipids found in eukaryotic membranes are phosphatidylcholine (PC), phosphatidylethanolamine (PE), sphingomyelin (SM), and PS. Additionally, two species are less abundant, phosphatidylinositol (PI) and phosphatidic acid (PA). Also, PIs are not only a source of phosphatidylinositols but also of phosphoinositides, a phosphorylated-phosphatidylinositol derivative which is known as phosphatidylinositol phosphate (PIP). It is important to note that lipid composition is different between cellular organelles and between their outer and inner monolayers. The PC and SM are mainly found on the cell’s outer membrane, whereas the PE is primarily found on the inner leaflet. Meanwhile, PS and PI are located at the inner PM monolayer (Alberts et al. 2002; Suetsugu et al. 2014).
These differences are introduced and maintained by membrane proteins at two different levels. A primary mechanism is the asymmetric incorporation of newly synthesized lipids in the bilayer of lipids. Flippases are notable proteins that contribute to maintaining asymmetry of the lipid composition (Devaux et al. 2008; Hankins et al. 2015). They are the enzymes that cause an asymmetric distribution of lipids between inner and outer membrane monolayers by transporting the lipid from extracellular to cytosolic leaflet. IDPRs are found in flippases and serve to connect the structured regions. Targeting the unstructured segment of a flippase (i.e., PglK) has been found to diminish its flipflop action (Perez et al. 2017). This suggests that the unstructured portions of these proteins are required for their function by potentially allowing the structured components of the enzyme to move around freely. The second mechanism involves the modification of lipid headgroups on the membrane surface. For example, Phosphatidylinositol phosphate kinases (PIPKs) are kinase proteins that phosphorylate phosphatidylinositol lipids on the membrane surface. A specificity loop in PIPKs consists of an unstructured region that dictates the PIPK substrate specificity (Muftuoglu et al. 2016). The PIP can be either , , or , depending on the PIPK class and type (Muftuoglu et al. 2016). Each PIP serves as a trigger for a specific protein–membrane interaction. For instance, is a specialized cue for early endosomes, whereas is a cue for PM inner monolayer (Muftuoglu et al. 2016). Among all PIPs, or simply is the most abundant phosphoinositide, accounting for 0.5–1% of total lipids, while the levels of other phosphoinositides are significantly lower (Suetsugu et al. 2014). is thought to play a key role in many cellular processes such as cytoskeleton remodeling, endocytosis, and membrane deformation, all of which are triggered by the interaction of PM with particular cytosolic IDPRs (Di Paolo and De Camilli 2006; Vicinanza et al. 2008; Rusinova et al. 2013).
The electrostatic characteristics of the membrane very much depend on the distribution of PS and PIP. PS can operate as a partial alternative for PIPs in membranes due to its electrostatic nature, especially when interacting with cationic motifs of IDPs and IDPR-containing proteins, for instance, AnxB12 binding to PS (Lizarbe et al. 2013). Membrane-interacting domains, such as N-BAR, F-BAR, I-BAR, E/ANTH, and some additional lipid-binding domains in BAR proteins like PH (pleckstrin homology) and PX (phox homology), as well as PH domain of Dynamin, are an example of cationic motifs that use electrostatic interactions (Suetsugu et al. 2014). However, appears to be preferred by these domains over PS (McLaughlin and Murray 2005; McMahon and Gallop 2005). PIPs are the preferred phospholipids for many proteins rather than PS because of their higher charge. It has been proposed that binding plays an important role in the correct targeting of these proteins to the PM (Ford et al. 2002). Several studies involving mutagenesis support this hypothesis, demonstrating that proteins with mutations in their binding sites do not localize appropriately (Ford et al. 2002). Several PIP-binding domains have been shown to deform liposomes into tubules and small vesicles. The PH domains of dynamin, for example, play an essential role in their electrostatic interactions with -containing membrane (Roux et al. 2010).
Assays to Measure IDPs/IDPRs–Membrane Interactions
Research has grown steadily into IDPs/IDPRs properties and behavior in recent years as researchers understand its importance in cellular physiology. These proteins interact with biological membranes and function as hubs in signaling and trafficking pathways (Cornish et al. 2020). In addition to being a valuable player in many biological processes, the study of IDPs/IDPRs–membrane interactions may also contribute to constituting novel drug targets (Ambadipudi and Zweckstetter 2016; Neira et al. 2017; Hosoya and Ohkanda 2021). Several recent methodological advances have enhanced our understanding of how and why different protein domains/motifs interact with specific curvatures. We briefly review a few tools that can characterize such interactions in the following sections.
In-Vitro IDPs/IDPRs–Membrane Interactions
In-vitro assays are often utilized in studies on protein potency to recognize or induce the membrane curvature. For this, artificial model membrane structures with fixed lipid composition, such as size-regulated liposomes (Has and Sunthar 2020) and supported lipid bilayers (SLBs), have been employed to construct bilayers free from protein constituents. A quantitative and reproducible analysis of protein–membrane interactions can be achieved through such in-vitro approaches (Carvalho et al. 2008; Salzer et al. 2017; Chand et al. 2019). Generally, proteins and membranes are made visible by being tagged with different fluorescent dyes.
Microscopy-based assays One of the basic approaches to dictate the mechanism of membrane deformation by any protein is using the plethora of microscopy-based techniques. To this aim, giant unilamellar vesicles (GUVs) are found to be a suitable choice since their usual diameters (a few microns to tens of microns) closely resemble cell membranes and may be examined using an optical microscope. Several BARs (Saarikangas et al. 2009; Tanaka-Takiguchi et al. 2013) and other proteins (Has and Das 2021) have been employed to study the membrane deformation. Nonetheless, merely visuals cannot be used to extract quantitative measurements of membrane shape induced by proteins.
Sedimentation, flotation, and PLiMAP assays Sedimentation assay (SA) (Peter et al. 2004; Carlton et al. 2004; Pylypenko et al. 2007), flotation assay (FA) (Bigay et al. 2005; Mesmin et al. 2007), or Proximity-based Labeling of Membrane-Associated Proteins (PLiMAP) assay (Jose et al. 2020) may be employed for measuring the protein affinity to the membranes of different mean curvatures. Liposomes of different curvatures are synthesized in these assays by extruding them through filters with predefined pore sizes, followed by incubation with proteins. A mixture of protein and extruded liposomes is either spun directly down (in sedimentation) or spun up in a sucrose density gradient (in flotation). Both assays work at a high-speed centrifugation spin to isolate the fraction of membrane-bound proteins. Further, a recent report (Jose et al. 2020) has obviated the requirement of ultracentrifugation spin using PLiMAP assay. In PLiMAP assay, a bi-functional reactive fluorescent lipid (RFL) is incorporated into the liposomes and used as an indicator for membrane–protein interactions. Upon photoactivation (exposed with ultraviolet light), the membrane-bound protein is crosslinked with RFL. The sample is then centrifuged at low speed. Eventually, the fraction of any protein (not just IDPs/IDPRs) bound to the liposomal membrane (in SA, FA, or PLiMAP) is quantified by protein analysis techniques (e.g., SDS-PAGE).
Even though SA is faster, more versatile, and requires fewer proteins and lipids, it has reduced sensitivity and cannot distinguish membrane-bound proteins from protein aggregates. Instead FA, which is less susceptible to protein aggregation, is recommended. Furthermore, PLiMAP assay can be integrated with existing liposome-based assays to improve the sensitivity. Unfortunately, these assays cannot investigate dynamic protein recruitment to curved membranes. Moreover, because the liposome sizes vary when they are extruded, it is difficult to assess the range of curvature-sensing ability of individual protein domains (Kunding et al. 2008; Simunovic et al. 2015).
Single liposome curvature or tethered vesicle assays It has been proven that single liposome curvature (SLiC) assays overcome the limitations of ensemble experiments (Bhatia et al. 2009; Madsen et al. 2010). In SLiC, liposomes (size 50–800 nm) doped with biotinylated BSA are immobilized through streptavidin–biotin linkages to a BSA-coated glass substrate (Stamou et al. 2003). The nanosized vesicles are isolated by tethering them onto passivated surfaces at low densities. Next, the solution of labeled protein is added in solution to the surface-tethered vesicles, and imaging is done using fluorescent/confocal microscopy after 20–30 min of incubation to achieve a faithful reconstruction of the liposome population (Zeno et al. 2019b; Larsen et al. 2020). In this study, vesicle diameters (d) are estimated from the intensities (I) of lipid fluorescent channel by the relation . The curvature-dependent protein binding to the membrane is estimated using integrated intensity ratios between protein and liposome (Bhatia et al. 2009). In the last decade, SLiC assay has been widely utilized to investigate the curvature-sensing/driving ability of many ordered/disordered proteins and protein domains, such as BAR domains (Bhatia et al. 2009), AHs containing proteins such as S and AnxB12 (Jensen et al. 2011), lipidated protein N-Ras (Larsen et al. 2015), C2AB domain of Syt1 (Larsen et al. 2020), AP180CTD (Zeno et al. 2019b), Epsin1CTD (Zeno et al. 2018), and Amph1CTD (Zeno et al. 2019a), as well as FL-Epsin, FL-Amph (Zeno et al. 2018), and clathrin (Zeno et al. 2021).
In SLiC, a large number of isolated/distinct membrane curvatures may be quickly recorded in a high-throughput manner (Bhatia et al. 2010; Madsen et al. 2010). Even though this method has opened up new avenues for systematically studying curvature-dependent protein dynamics, tiny vesicles have shown considerable heterogeneity in lipid composition (Larsen et al. 2011), which could contribute to measurement error as curvature rises. Finally, considering that SLiC often requires elaborate experimental procedures and technical expertise, they are a bit tedious to implement.
Supported lipid bilayers In addition to the studies as mentioned above based on vesicles, supported lipid bilayers (SLBs) constructed with predetermined topographies by lipid deposition on a solid hydrophilic substrate (such as glass, silica, and gold) have been routinely used to simulate biological membranes under more straightforward experimental conditions (Tanaka et al. 2020). To drive membrane curvature, SLBs use either static (Parthasarathy and Groves 2006) or switchable (Sanii et al. 2008) topographic patterns. These assays provide a means for probing membrane geometries that may otherwise not be possible using liposome-based assays (e.g., saddles, cylinders) (Ebrahimkutty and Galic 2019). The thin hydration layer between the lipid bilayer and the solid support might not accurately simulate the properties of the cytosolic fluid. As a result, the precision and usability of SLBs are limited. Using a nanobar-assisted SLB system has recently shown that the protein FBP17 senses the curvature. Still, its curvature sensitivity is mostly derived from its IDPD, not the structured F-BAR domain itself, contrary to popular perception (Su et al. 2020).
Tether-pulling assays In order to overcome the limitations associated with the previous approaches and to enhance the detection accuracy, tether-pulling assays (TPAs) have gained immense popularity in studying the dynamic recruitment of curvature-sensitive proteins (Ambroggio et al. 2010; Sorre et al. 2012; Ramesh et al. 2013; Prévost et al. 2015) and/or curvature generation by proteins (Sorre et al. 2012; Ramesh et al. 2013; Prévost et al. 2015; Knorr et al. 2014). In a TPA, a transient membrane nanotube or tether from a GUV is pulled with the help of an optical tweezer (OT). As a result, we may investigate transient protein enrichment as soon as the membrane is deformed. All BAR proteins that have been investigated experimentally thus far, such as N-BARs (Sorre et al. 2012; Zhu et al. 2012; Wu et al. 2014), F-BAR (Ramesh et al. 2013), and I-BAR (Prévost et al. 2015), are enriched on the tether. Furthermore, curvature-dependent enrichment of KvAP (Aimon et al. 2014), ArfGAP1, and Arf1 (Ambroggio et al. 2010) has also been reported. Although TPA has been mostly used for ordered proteins and protein domains, it can be easily implemented separately for disorder domains or proteins containing both ordered and disordered domains.
In-Vivo IDPs/IDPRs–Membrane Interactions
In-vitro approaches carried out on artificial lipid membranes have proved to be powerful tools for quantifying the function of proteins as curvature sensors/inducers. Still, they are restricted by a requirement to use model membranes of well-defined lipid composition, a constraint that fails to allow for asymmetry in lipid composition in biological membranes. To address such shortcomings, living cell assays (LCAs) can be used to explore how membrane curvature impacts cytoplasmic proteins and cellular processes under pristine physiological conditions. A variety of fluorescence- and electron-based techniques (Galic et al. 2012, 2014; Begemann and Galic 2016) can be used to investigate the protein sensitivity to brief- and/or long-lived membrane deformations in living and fixed cells. Alternatively, experiments that artificially generate membrane deformations in living cells have proven to be effective. For example, tethers or nanotubes can be extruded either directly from the cell PM (Breuer et al. 2019) or cell-derived giant PM vesicles (GPMVs) (Moreno-Pescador et al. 2019). Here, a brief membrane deformation is induced at a predefined location, and the subsequent curvature-dependent protein recruitment is investigated. In another approach, cells may be cultured on patterned nanostructure platforms, and fluorescence microscopy can be used to examine the protein dynamics on live cells (Galic et al. 2012; Li et al. 2019). Nanostructure- and tether-assisted LCAs, although being effective techniques, employ static membrane curvature, making real-time monitoring of a cell’s reaction to membrane deformation difficult. We note that the curvature sensing and induction by IDPs/IDPRs are yet to be explored using live cells.
Computational Approaches for IDPs/IDPRs–Membrane Interactions
When taken as a whole, each experimental strategy has its own set of benefits. Unfortunately, no single technique allows for the unbiased analysis of membrane curvature sensing/generation by proteins. So, what is the next step? Because experiments to quantify curvature-dependent IDPs/IDPRs–membrane interactions are still tricky, molecular dynamics (MD) modeling can help us learn more about these interactions. All-atom MD simulations can simulate how BAR-containing proteins drive membrane curvature in-silico, thanks to advanced force fields (Yu and Schulten 2013). Because all-atom MD simulation is computationally expensive, coarse-grained (CG) approximation models are frequently used for such complex systems (Das and Eliezer 2019). Using CG MD simulations, it has been demonstrated an aggregation behavior of N-BAR proteins on membrane curvature (Simunovic et al. 2013). MD studies have further revealed that the positive residues on BAR domains and membrane lipid components are found to be essential features in curvature-dependent protein distribution/sorting (Takemura et al. 2017; Stanishneva-Konovalova and Sokolova 2019). Despite their potency, MD simulations are limited to short spatiotemporal intervals. Continuum models (CM) can be an excellent option for expanding these limits and gaining more understanding of biological assumptions.
In contrast to MD simulations, which include discrete molecules, CM takes the atomistic data and averages it to get a continuous mass. A wealth of models, such as Dynamic Triangulated Monte Carlo (DTMC), have been developed in recent years that facilitate to bridge the molecular, mesoscopic, and cellular scales (Ebrahimkutty and Galic 2019). Using DTMC, it has been recently reported that both curvature sensing and induction can be found in the same system as a function of protein binding affinity on the membrane (Krishnan et al. 2019). The researchers can accurately link the model (Tourdot et al. 2014) to the experimental data (Shi and Baumgart 2015) using these approaches. For learning more about this rapidly evolving field, an interested reader is referred to reviews by Ramakrishnan et al. (2018).
Conclusions and Future Perspectives
In this review, we have focused on the interplay between IDPs/IDPRs and membrane and how this dynamic cross-talk is crucial for signaling and trafficking pathways (Cornish et al. 2020). Cellular membranes are lipid bilayers that are fluid-like in appearance and in which proteins are embedded or to which proteins are anchored that assist the membranes in performing their functions. It has been discussed how membrane lipids can potentially affect the binding and organization of proteins. Compared to structured domains, the IDPRs of identical molecular weight possess a larger projected area, more flexibility, and undergo disorder-to-order transition upon binding to the membranes. Cells use these characteristics for functional reasons.
We have discussed how IDPs/IDPRs are employed to regulate the membrane properties and thus influence membrane trafficking. Specifically, proteins and membranes interact in a two-way fashion. First, membrane curvature is generated by constituent proteins. Second, proteins sense the membrane curvature, or in other words, membrane curvature provides a cue to recruit distinct proteins selectively. It has become clear that curvature sensing and induction are two biophysical processes that are related but not the same. From various assays discussed in this article, we can state that curvature sensing is most effective when proteins are sparsely bound to the membranes (Sorre et al. 2012). On the other hand, proteins begin to induce curvature as their coverage of the membrane surface grows (Stachowiak et al. 2010, 2012). We have seen that full-length proteins have a larger capacity to sense/generate the membrane curvature than the structured and/or unstructured domains studied in isolation.
The current review underscores the need to research the synergistic connection between ordered and disordered protein domains in a larger context. The IDPRs are common in the protein machinery that controls membrane trafficking, and they are frequently paired with ordered domains in the same protein molecule (Pietrosemoli et al. 2013). In biophysical investigations of interactions of proteins with membrane, it has become more apparent that IDPRs cannot be overlooked. To date, only a few IDPRs have been identified as curvature sensors and drivers. Because IDPRs make up around one-third of the proteome, they are a diverse group of proteins, implying that there are still many more potential sensors and drivers of membrane curvature to be characterized. As a result, understanding the biophysical roles of IDPRs in membrane remodeling is an essential priority for future research. Using hybrid strategies that combine cellular and biophysical experiments with techniques that can evaluate the IDPRs contribution in the membrane environment like NMR and MD simulations will help researchers to understand the proteins involved in signaling and trafficking pathways in which they are found.
Acknowledgements
We gladly acknowledge IIT Palakkad’s faculty seed grant and the Ministry of Human Resource Development’s (MHRD) grant Scheme for Transformational and Advanced Research in Sciences (Grant No. STARS/APR2019/293).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chandra Has, Email: chandrahashbti@gmail.com.
P. Sivadas, Email: 222004006@smail.iitpkd.ac.in
Sovan Lal Das, Email: sovan@iitpkd.ac.in.
References
- Aimon S, Callan-Jones A, Berthaud A, et al. Membrane shape modulates transmembrane protein distribution. Dev Cell. 2014;28(2):212–218. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4. Garland Science; 2002. [Google Scholar]
- Ambadipudi S, Zweckstetter M. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin Drug Discov. 2016;11(1):65–77. doi: 10.1517/17460441.2016.1107041. [DOI] [PubMed] [Google Scholar]
- Ambroggio E, Sorre B, Bassereau P, et al. ArfGAP1 generates an Arf1 gradient on continuous lipid membranes displaying flat and curved regions. EMBO J. 2010;29(2):292–303. doi: 10.1038/emboj.2009.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroso MR, Hegde BG, Langen R. Endophilin A1 induces different membrane shapes using a conformational switch that is regulated by phosphorylation. Proc Natl Acad Sci USA. 2014;111(19):6982–6987. doi: 10.1073/pnas.1402233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chothia C, et al. SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Res. 2014;42(D1):D310–D314. doi: 10.1093/nar/gkt1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Arai M, Sugase K, Dyson HJ, et al. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA. 2015;112(31):9614–9619. doi: 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awile O, Krisko A, Sbalzarini IF, et al. Intrinsically disordered regions may lower the hydration free energy in proteins: a case study of nudix hydrolase in the bacterium deinococcus radiodurans. PLoS Comput Biol. 2010;6(7):e1000,854. doi: 10.1371/journal.pcbi.1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44(5):1185–1200. doi: 10.1042/Bst20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T, Capraro BR, Zhu C, et al. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu Rev Phys Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, et al. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc Natl Acad Sci USA. 2008;105(33):11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann I, Galic M. Correlative light electron microscopy: connecting synaptic structure and function. Front Synaptic Neurosci. 2016;8:28. doi: 10.3389/fnsyn.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlow RB, Dyson HJ, Wright PE. Functional advantages of dynamic protein disorder. FEBS Lett. 2015;589(19):2433–2440. doi: 10.1016/j.febslet.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia VK, Madsen KL, Bolinger PY, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009;28(21):3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia VK, Hatzakis NS, Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin Cell Dev Biol. 2010;21(4):381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, et al. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24(13):2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, et al. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149(1):124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438(7068):581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- Brady JP, Claridge JK, Smith PG, et al. A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proc Natl Acad Sci USA. 2015;112(7):E639–E648. doi: 10.1073/pnas.1415882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun AR, Sevcsik E, Chin P et al (2012) -synuclein induces both positive mean curvature and negative gaussian curvature in membranes. J Am Chem Soc 134(5):2613–2620. 10.1021/ja208316h [DOI] [PMC free article] [PubMed]
- Braun AR, Lacy MM, Ducas VC et al (2014) -synuclein-induced membrane remodeling is driven by binding affinity, partition depth, and interleaflet order asymmetry. J Am Chem Soc 136(28):9962–9972. 10.1021/ja5016958 [DOI] [PMC free article] [PubMed]
- Breeze E, Dzimitrowicz N, Kriechbaumer V, et al. A C-terminal amphipathic helix is necessary for the in vivo tubule-shaping function of a plant reticulon. Proc Natl Acad Sci USA. 2016;113(39):10902–10907. doi: 10.1073/pnas.1605434113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer A, Lauritsen L, Bertseva E, et al. Quantitative investigation of negative membrane curvature sensing and generation by I-BARs in filopodia of living cells. Soft Matter. 2019;15(48):9829–9839. doi: 10.1039/C9SM01185D. [DOI] [PubMed] [Google Scholar]
- Busch DJ, Houser JR, Hayden CC, et al. Intrinsically disordered proteins drive membrane curvature. Nat Commun. 2015;6(1):1–11. doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95(5):2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Coskun Ü, Rössle M, et al. Golgi protein FAPP2 tubulates membranes. Proc Natl Acad Sci USA. 2009;106(50):21121–21125. doi: 10.1073/pnas.0911789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignano M, Szleifer I. On the structure and pressure of tethered polymer layers in good solvent. Macromolecules. 1995;28(9):3197–3204. doi: 10.1021/ma00113a023. [DOI] [Google Scholar]
- Carlton J, Bujny M, Peter BJ, et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14(20):1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Carnahan NF, Starling KE. Equation of state for nonattracting rigid spheres. J Chem Phys. 1969;51(2):635–636. doi: 10.1063/1.1672048. [DOI] [Google Scholar]
- Carvalho K, Ramos L, Roy C, et al. Giant unilamellar vesicles containing phosphatidylinositol (4, 5) bisphosphate: characterization and functionality. Biophys J. 2008;95(9):4348–4360. doi: 10.1529/biophysj.107.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand S, Beales P, Claeyssens F, et al. Topography design in model membranes: where biology meets physics. Exp Biol Med. 2019;244(4):294–303. doi: 10.1177/1535370218809369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhu C, Kuo CJ, et al. The N-terminal amphipathic helix of endophilin does not contribute to its molecular curvature generation capacity. J Am Chem Soc. 2016;138(44):14616–14622. doi: 10.1021/jacs.6b06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72(1):175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Cornish J, Chamberlain SG, Owen D, et al. Intrinsically disordered proteins and membranes: a marriage of convenience for cell signalling? Biochem Soc Trans. 2020;48(6):2669–2689. doi: 10.1042/BST20200467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Zhang L, Wu Z, et al. Generation and sensing of membrane curvature: where materials science and biophysics meet. Curr Opin Solid State Mater Sci. 2013;17(4):164–174. doi: 10.1016/j.cossms.2013.06.002. [DOI] [Google Scholar]
- Dafforn TR, Smith CJ. Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep. 2004;5(11):1046–1052. doi: 10.1038/sj.embor.7400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Eliezer D. Membrane interactions of intrinsically disordered proteins: The example of alpha-synuclein. Biochim Biophys Acta Proteins Proteom. 2019;1867(10):879–889. doi: 10.1016/j.bbapap.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449(7164):923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, et al. Attributes of short linear motifs. Mol Biosyst. 2012;8(1):268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- Davidson WS, Jonas A, Clayton DF et al (1998) Stabilization of -synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 273(16):9443–9449. 10.1074/jbc.273.16.9443 [DOI] [PubMed]
- Devaux PF, Herrmann A, Ohlwein N, et al. How lipid flippases can modulate membrane structure. Biochim Biophys Acta Biomembr. 2008;1778(7–8):1591–1600. doi: 10.1016/j.bbamem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Diella F, Haslam N, Chica C, et al. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13(6580):603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- Dikiy I, Eliezer D. Folding and misfolding of alpha-synuclein on membranes. Biochim Biophys Acta - Biomembr. 2012;1818(4):1013–1018. doi: 10.1016/j.bbamem.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584(9):1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R et al (2007) A general amphipathic -helical motif for sensing membrane curvature. Nat Struct Mol Biol 14(2):138–146. 10.1038/nsmb1194 [DOI] [PubMed]
- Drin G, Morello V, Casella JF, et al. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320(5876):670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Ewright P. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–340. doi: 10.1371/journal.pcbi.1000854. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ebrahimkutty MP, Galic M. Receptor-free signaling at curved cellular membranes. BioEssays. 2019;41(10):1900068. doi: 10.1002/bies.201900068. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26(10):597–604. doi: 10.1016/S0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Escribá PV, Wedegaertner PB, Goñi FM, et al. Lipid-protein interactions in GPCR-associated signaling. Biochim Biophys Acta Biomembr. 2007;1768(4):836–852. doi: 10.1016/j.bbamem.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Jonasson K, von Heijne G, et al. Prediction of the human membrane proteome. Proteomics. 2010;10(6):1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- Fakhree MA, Blum C, Claessens MM. Shaping membranes with disordered proteins. Arch Biochem Biophys. 2019;677(108):163. doi: 10.1016/j.abb.2019.108163. [DOI] [PubMed] [Google Scholar]
- Fakhree MA, Engelbertink SA, van Leijenhorst-Groener KA, et al. Cooperation of helix insertion and lateral pressure to remodel membranes. Biomacromolecules. 2019;20(3):1217–1223. doi: 10.1021/acs.biomac.8b01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, et al. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129(7):1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Ferreon ACM, Gambin Y, Lemke EA et al (2009) Interplay of -synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci USA 106(14):5645–5650. 10.1073/pnas.0809232106 [DOI] [PMC free article] [PubMed]
- Fischer T, Lu L, Haigler HT, et al. Annexin B12 is a sensor of membrane curvature and undergoes major curvature-dependent structural changes. J Biol Chem. 2007;282(13):9996–10004. doi: 10.1074/jbc.M611180200. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419(6905):361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Fukuchi S, Homma K, Minezaki Y, et al. Intrinsically disordered loops inserted into the structural domains of human proteins. J Mol Biol. 2006;355(4):845–857. doi: 10.1016/j.jmb.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23(8):950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Galic M, Jeong S, Tsai FC, et al. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat Cell Biol. 2012;14(8):874–881. doi: 10.1038/ncb2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic M, Tsai FC, Collins SR, et al. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. Elife. 2014;3(e03):116. doi: 10.7554/eLife.03116.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25(12):2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Andrés M, Čopič A, Antonny B. The many faces of amphipathic helices. Biomolecules. 2018;8(3):45. doi: 10.3390/biom8030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grecco HE, Schmick M, Bastiaens PI. Signaling from the living plasma membrane. Cell. 2011;144(6):897–909. doi: 10.1016/j.cell.2011.01.029. [DOI] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S, et al. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- Hankins HM, Baldridge RD, Xu P, et al. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 2015;16(1):35–47. doi: 10.1111/tra.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Has C, Das SL. Recent developments in membrane curvature sensing and induction by proteins. Biochim Biophys Acta Gen Subj. 2021;1865(10):129971. doi: 10.1016/j.bbagen.2021.129971. [DOI] [PubMed] [Google Scholar]
- Has C, Sunthar P. A comprehensive review on recent preparation techniques of liposomes. J Liposome Res. 2020;30(4):336–365. doi: 10.1080/08982104.2019.1668010. [DOI] [PubMed] [Google Scholar]
- Hatzakis NS, Bhatia VK, Larsen J, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5(11):835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C. 1973;28(11–12):693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328(5983):1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlo R, Lund VK, Lycas MD, et al. An amphipathic helix directs cellular membrane curvature sensing and function of the BAR domain protein PICK1. Cell Rep. 2018;23(7):2056–2069. doi: 10.1016/j.celrep.2018.04.074. [DOI] [PubMed] [Google Scholar]
- Hofmann H, Soranno A, Borgia A, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(40):16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya Y, Ohkanda J. Intrinsically disordered proteins as regulators of transient biological processes and as untapped drug targets. Molecules. 2021;26(8):2118. doi: 10.3390/molecules26082118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Botelho AV, Beyer K, et al. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys J. 2004;86(4):2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Song F, Aung Z, et al. SLiM on Diet: finding short linear motifs on domain interaction interfaces in Protein Data Bank. Bioinformatics. 2010;26(8):1036–1042. doi: 10.1093/bioinformatics/btq065. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, et al. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Israelachvili JN. Intermolecular and surface forces. San Diego: Academic Press; 2015. [Google Scholar]
- Jensen MB, Bhatia VK, Jao CC et al (2011) Membrane curvature sensing by amphipathic helices: a single liposome study using -synuclein and annexin B12. J Biol Chem 286(49):42603–42614. 10.1074/jbc.M111.271130 [DOI] [PMC free article] [PubMed]
- Jiang Z, de Messieres M, Lee JC (2013) Membrane remodeling by -synuclein and effects on amyloid formation. J Am Chem Soc 135(43):15970–15973. 10.1021/ja405993r [DOI] [PMC free article] [PubMed]
- Jose GP, Gopan S, Bhattacharyya S, et al. A facile, sensitive and quantitative membrane-binding assay for proteins. Traffic. 2020;21(3):297–305. doi: 10.1111/tra.12719. [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, et al. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277(10):8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Groos S, Kohl R, et al. Clint: a novel clathrin-binding ENTH-domain protein at the Golgi. Mol Biol Cell. 2002;13(11):4060–4073. doi: 10.1091/mbc.e02-03-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Kalinowski SS, Marcinkeviciene J. A pre-steady state analysis of ligand binding to human glucokinase: evidence for a preexisting equilibrium. Biochemistry (Mosc ) 2007;46(5):1423–1431. doi: 10.1021/bi0617308. [DOI] [PubMed] [Google Scholar]
- Kjaergaard M. Can proteins be intrinsically disordered inside a membrane? Intrinsically Disord Proteins. 2015;3(1):e984,570. doi: 10.4161/21690707.2014.984570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliche J, Kuss H, Ali M et al (2021) Cytoplasmic short linear motifs in ACE2 and integrin 3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Science Signaling 14(665):eabf1117. 10.1126/scisignal.abf1117 [DOI] [PMC free article] [PubMed]
- Knorr RL, Nakatogawa H, Ohsumi Y, et al. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PloS one. 2014;9(12):e115,357. doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotta-Loizou I, Tsaousis GN, Hamodrakas SJ. Analysis of molecular recognition features (MoRFs) in membrane proteins. Biochim Biophys Acta Proteins Proteom. 2013;1834(4):798–807. doi: 10.1016/j.bbapap.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Kozlov MM, Campelo F, Liska N, et al. Mechanisms shaping cell membranes. Curr Opin Cell Biol. 2014;29:53–60. doi: 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan TS, Das SL, Kumar PS. Transition from curvature sensing to generation in a vesicle driven by protein binding strength and membrane tension. Soft Matter. 2019;15(9):2071–2080. doi: 10.1039/C8SM02623H. [DOI] [PubMed] [Google Scholar]
- Kunding AH, Mortensen MW, Christensen SM, et al. A fluorescence-based technique to construct size distributions from single-object measurements: application to the extrusion of lipid vesicles. Biophys J. 2008;95(3):1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Hatzakis NS, Stamou D. Observation of inhomogeneity in the lipid composition of individual nanoscale liposomes. J Am Chem Soc. 2011;133(28):10685–10687. doi: 10.1021/ja203984j. [DOI] [PubMed] [Google Scholar]
- Larsen JB, Jensen MB, Bhatia VK, et al. Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases. Nat Chem Biol. 2015;11(3):192–194. doi: 10.1038/nchembio.1733. [DOI] [PubMed] [Google Scholar]
- Larsen JB, Rosholm KR, Kennard C, et al. How membrane geometry regulates protein sorting independently of mean curvature. ACS Cent Sci. 2020;6(7):1159–1168. doi: 10.1021/acscentsci.0c00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122(4):605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kerff F, Chereau D, et al. Structural basis for the actin-binding function of missing-in-metastasis. Structure. 2007;15(2):145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Coskun Ü, Grzybek M, et al. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010;11(4):279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZL. Molecular dynamics simulations of membrane deformation induced by amphiphilic helices of Epsin, Sar1p, and Arf1. Chinese Phys B. 2018;27(3):038703. doi: 10.1088/1674-1056/27/3/038703. [DOI] [Google Scholar]
- Li X, Matino L, Zhang W, et al. A nanostructure platform for live-cell manipulation of membrane curvature. Nat Protoc. 2019;14(6):1772–1802. doi: 10.1038/s41596-019-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Mu H, Jean-Francois F, et al. Membrane curvature sensing of the lipid-anchored K-Ras small GTPase. Life Sci Alliance. 2019;2(4):e201900,343. doi: 10.26508/lsa.201900343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Currie SL, Rosen MK. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J Biol Chem. 2017;292(46):19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky R. Flexible membranes with anchored polymers. Colloids Surf A Physicochem Eng. 1997;128(1–3):255–264. doi: 10.1016/S0927-7757(96)03906-4. [DOI] [Google Scholar]
- Lizarbe MA, Barrasa JI, Olmo N, et al. Annexin-phospholipid interactions. functional implications. Int J Mol Sci. 2013;14(2):2652–2683. doi: 10.3390/ijms14022652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR. SNX9-a prelude to vesicle release. J Cell Sci. 2009;122(1):5–11. doi: 10.1242/jcs.037135. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Vallis Y, et al. Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J. 2008;414(2):189–194. doi: 10.1042/BJ20081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen KL, Bhatia VK, Gether U, et al. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584(9):1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- Mahata P, Das SL. Generation of wavy structure on lipid membrane by peripheral proteins: a linear elastic analysis. FEBS Lett. 2017;591(10):1333–1348. doi: 10.1002/1873-3468.12661. [DOI] [PubMed] [Google Scholar]
- Marinko JT, Huang H, Penn WD, et al. Folding and misfolding of human membrane proteins in health and disease: from single molecules to cellular proteostasis. Chem Rev. 2019;119(9):5537–5606. doi: 10.1021/acs.chemrev.8b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316(5828):1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- Martyna A, Bahsoun B, Badham MD, et al. Membrane remodeling by the M2 amphipathic helix drives influenza virus membrane scission. Sci Rep. 2017;7(1):1–12. doi: 10.1038/srep44695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Gould KL. Linking up at the BAR: oligomerization and F-BAR protein function. Cell Cycle. 2016;15(15):1977–1985. doi: 10.1080/15384101.2016.1190893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438(7068):605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Meier S, Blackledge M, Grzesiek S. Conformational distributions of unfolded polypeptides from novel NMR techniques. J Chem Phys. 2008;128(5):02B604. doi: 10.1063/1.2838167. [DOI] [PubMed] [Google Scholar]
- Mesmin B, Drin G, Levi S, et al. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry (Mosc) 2007;46(7):1779–1790. doi: 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Sámano-Sánchez H, Alvarado-Valverde J, et al. Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Science Signaling. 2021;14(665):eabd0334. doi: 10.1126/scisignal.abd0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Mathiasen S, Bright NA, et al. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell. 2015;33(2):163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Nishikawa K. Intrinsically disordered regions of human plasma membrane proteins preferentially occur in the cytoplasmic segment. J Mol Biol. 2007;368(3):902–913. doi: 10.1016/j.jmb.2007.02.033. [DOI] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23(2):105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, et al. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362(5):1043–1059. doi: 10.1016/j.bbapap.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Montesano G, Bartucci R, Belsito S, et al. Lipid membrane expansion and micelle formation by polymer-grafted lipids: scaling with polymer length studied by spin-label electron spin resonance. Biophys J. 2001;80(3):1372–1383. doi: 10.1016/S0006-3495(01)76110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Pescador G, Florentsen CD, Østbye H, et al. Curvature-and phase-induced protein sorting quantified in transfected cell-derived giant vesicles. ACS Nano. 2019;13(6):6689–6701. doi: 10.1021/acsnano.9b01052. [DOI] [PubMed] [Google Scholar]
- Morton LA, Yang H, Saludes JP, et al. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem Biol. 2013;8(1):218–225. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu Y, Xue Y, Gao X, et al. Mechanism of substrate specificity of phosphatidylinositol phosphate kinases. Proc Natl Acad Sci USA. 2016;113(31):8711–8716. doi: 10.1073/pnas.1522112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Wu N, Palczewski K. Vertebrate membrane proteins: structure, function, and insights from biophysical approaches. Pharmacol Rev. 2008;60(1):43–78. doi: 10.1124/pr.107.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579(15):3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Neira JL, Bintz J, Arruebo M, et al. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Sci Rep. 2017;7(1):1–15. doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki W, Nowicka G, Narkiewicz-Michałek J. Influence of confinement on conformational entropy of a polymer chain and structure of polymer-nanoparticles complexes. Polymer. 2009;50(9):2161–2171. doi: 10.1016/j.polymer.2009.02.044. [DOI] [Google Scholar]
- Nye JA, Groves JT. Kinetic control of histidine-tagged protein surface density on supported lipid bilayers. Langmuir. 2008;24(8):4145–4149. doi: 10.1021/la703788h. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, et al. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(S6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- Orosz F, Ovádi J. Proteins without 3D structure: definition, detection and beyond. Bioinformatics. 2011;27(11):1449–1454. doi: 10.1093/bioinformatics/btr175. [DOI] [PubMed] [Google Scholar]
- O’Shea C, Staby L, Bendsen SK, et al. Structures and short linear motif of disordered transcription factor regions provide clues to the interactome of the cellular hub protein radical-induced cell death1. J Biol Chem. 2017;292(2):512–527. doi: 10.1074/jbc.M116.753426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pannuzzo M, Milardi D, Raudino A et al (2013) Analytical model and multiscale simulations of a peptide aggregation in lipid membranes: towards a unifying description of conformational transitions, oligomerization and membrane damage. Phys Chem Chem Phys 15(23):8940–8951. 10.1039/c3cp44539a [DOI] [PubMed]
- Pannuzzo M, Raudino A, Milardi D et al (2013) -helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes. Sci Rep 3(1):1–10. 10.1038/srep02781 [DOI] [PMC free article] [PubMed]
- Parthasarathy R, Groves JT. Curvature and spatial organization in biological membranes. Soft Matter. 2006;3(1):24–33. doi: 10.1039/B608631D. [DOI] [PubMed] [Google Scholar]
- Pentony MM, Jones DT. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78(1):212–221. doi: 10.1002/prot.22504. [DOI] [PubMed] [Google Scholar]
- Perez C, Köhler M, Janser D, et al. Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nanobody. Sci Rep. 2017;7(1):1–9. doi: 10.1038/srep46641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303(5657):495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Phillips R, Ursell T, Wiggins P, et al. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459(7245):379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrosemoli N, Pancsa R, Tompa P. Structural disorder provides increased adaptability for vesicle trafficking pathways. PLoS Comput Biol. 2013;9(7):e1003,144. doi: 10.1371/journal.pcbi.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomann M, Wittmann JG, Rudolph MG. A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing. J Mol Biol. 2010;400(2):129–136. doi: 10.1016/j.jmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Pozzi N, Vogt AD, Gohara DW, et al. Conformational selection in trypsin-like proteases. Curr Opin Struct Biol. 2012;22(4):421–431. doi: 10.1016/j.sbi.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranke IM, Morello V, Bigay J et al (2011) -synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J Cell Biol 194(1):89–103. 10.1083/jcb.201011118 [DOI] [PMC free article] [PubMed]
- Prévost C, Zhao H, Manzi J, et al. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun. 2015;6(1):1–11. doi: 10.1038/ncomms9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, et al. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26(22):4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin’s GTP-dependent conformational changes to the membrane. EMBO J. 2008;27(1):27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Pucadyil TJ, Liu YW, et al. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20(22):4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan N, Bradley RP, Tourdot RW, et al. Biophysics of membrane curvature remodeling at molecular and mesoscopic lengthscales. J Phys : Condens Matter. 2018;30(27):273001. doi: 10.1088/1361-648X/aac702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh P, Baroji YF, Reihani SNS, et al. FBAR syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner. Sci Rep. 2013;3(1):1–6. doi: 10.1038/srep01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robustelli P, Piana S, Shaw DE. Developing a molecular dynamics force field for both folded and disordered protein states. Proceedings of the National Academy of Sciences. 2018;115(21):E4758–E4766. doi: 10.1073/pnas.1800690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Koster G, Lenz M, et al. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107(9):4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova R, Hobart EA, Koeppe RE, et al. Phosphoinositides alter lipid bilayer properties. J Gen Physiol. 2013;141(6):673–690. doi: 10.1085/jgp.201310960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykäläinen A, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19(2):95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Salzer U, Kostan J, Djinović-Carugo K. Deciphering the BAR code of membrane modulators. Cell Mol Life Sci. 2017;74(13):2413–2438. doi: 10.1007/s00018-017-2478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanii B, Smith AM, Butti R, et al. Bending membranes on demand: Fluid phospholipid bilayers on topographically deformable substrates. Nano Lett. 2008;8(3):866–871. doi: 10.1021/nl073085b. [DOI] [PubMed] [Google Scholar]
- Scheele U, Alves J, Frank R, et al. Molecular and functional characterization of clathrin-and AP-2-binding determinants within a disordered domain of auxilin. J Biol Chem. 2003;278(28):25357–25368. doi: 10.1074/jbc.M303738200. [DOI] [PubMed] [Google Scholar]
- Scheve CS, Gonzales PA, Momin N, et al. Steric pressure between membrane-bound proteins opposes lipid phase separation. J Am Chem Soc. 2013;135(4):1185–1188. doi: 10.1021/ja3099867. [DOI] [PubMed] [Google Scholar]
- Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448(7156):883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- Seelig J. Thermodynamics of lipid-peptide interactions. Biochim Biophys Acta - Biomembr. 2004;1666(1–2):40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Sharma R, Raduly Z, Miskei M, et al. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015;589(19):2533–2542. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Shi Z, Baumgart T. Membrane tension and peripheral protein density mediate membrane shape transitions. Nat Commun. 2015;6(1):1–8. doi: 10.1038/ncomms6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, et al. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- Simunovic M, Srivastava A, Voth GA. Linear aggregation of proteins on the membrane as a prelude to membrane remodeling. Proc Natl Acad Sci USA. 2013;110(51):20396–20401. doi: 10.1073/pnas.1309819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Voth GA, Callan-Jones A, et al. When physics takes over: BAR proteins and membrane curvature. Trends Cell Biol. 2015;25(12):780–792. doi: 10.1016/j.tcb.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Bassereau P, Voth GA. Organizing membrane-curving proteins: the emerging dynamical picture. Curr Opin Struct Biol. 2018;51:99–105. doi: 10.1016/j.sbi.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M, Evergren E, Callan-Jones A, et al. Curving cells inside and out: roles of BAR domain proteins in membrane shaping and its cellular implications. Annu Rev Cell Dev Biol. 2019;35:111–129. doi: 10.1146/annurev-cellbio-100617-060558. [DOI] [PubMed] [Google Scholar]
- Snead D, Eliezer D. Intrinsically disordered proteins in synaptic vesicle trafficking and release. J Biol Chem. 2019;294(10):3325–3342. doi: 10.1074/jbc.REV118.006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead WT, Stachowiak JC. Structure versus stochasticity-the role of molecular crowding and intrinsic disorder in membrane fission. Journal of Molecular Biology. 2018;430(16):2293–2308. doi: 10.1016/j.jmb.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead D, Wragg RT, Dittman JS, et al. Membrane curvature sensing by the C-terminal domain of complexin. Nat Commun. 2014;5(1):1–10. doi: 10.1038/ncomms5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead WT, Hayden CC, Gadok AK, et al. Membrane fission by protein crowding. Proc Natl Acad Sci USA. 2017;114(16):E3258–E3267. doi: 10.1083/jcb.201807119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B, Callan-Jones A, Manzi J, et al. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc Natl Acad Sci USA. 2012;109(1):173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Hayden CC, Sasaki DY. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proc Natl Acad Sci USA. 2010;107(17):7781–7786. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, et al. Membrane bending by protein-protein crowding. Nat Cell Biol. 2012;14(9):944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Stachowiak JC, Brodsky FM, Miller EA. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol. 2013;15(9):1019–1027. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamou D, Duschl C, Delamarche E, et al. Self-assembled microarrays of attoliter molecular vessels. Angew Chem. 2003;115(45):5738–5741. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- Stanishneva-Konovalova TB, Sokolova OS (2019) Effects of concentration on the F-BAR domain membrane binding as revealed by coarse-grained simulations. Proteins 87(7):561–568. 10.1002/prot.25678 [DOI] [PubMed]
- Stornaiuolo M, Lotti LV, Borgese N, et al. KDEL and KKXX retrieval signals appended to the same reporter protein determine different trafficking between endoplasmic reticulum, intermediate compartment, and Golgi complex. Mol Biol Cell. 2003;14(3):889–902. doi: 10.1091/mbc.e02-08-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome B, Hsu IS, Man MLC, et al. Short linear motifs in intrinsically disordered regions modulate HOG signaling capacity. BMC Syst Biol. 2018;12(1):1–13. doi: 10.1186/s12918-018-0597-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Zhuang Y, Miao X, et al. Comparative study of curvature sensing mediated by F-BAR and an intrinsically disordered region of FBP17. Iscience. 2020;23(11):101712. doi: 10.1016/j.isci.2020.101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012;22(3):141–150. doi: 10.1016/j.tcb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Takenawa T. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev. 2014;94(4):1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- Sun Y, Drubin DG. The functions of anionic phospholipids during clathrin-mediated endocytosis site initiation and vesicle formation. J Cell Sci. 2012;125(24):6157–6165. doi: 10.1242/jcs.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K, Goley ED. The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro. J Biol Chem. 2017;292(50):20509–20527. doi: 10.1074/jbc.M117.809939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, et al. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1(1):33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Takemura K, Hanawa-Suetsugu K, Suetsugu S, et al. Salt bridge formation between the I-BAR domain and lipids increases lipid density and membrane curvature. Sci Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-06334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Komikawa T, Yanai K, et al. Proteomic exploration of membrane curvature sensors using a series of spherical supported lipid bilayers. Anal Chem. 2020;92(24):16197–16203. doi: 10.1021/acs.analchem.0c04039. [DOI] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y, Itoh T, Tsujita K, et al. Physicochemical analysis from real-time imaging of liposome tubulation reveals the characteristics of individual F-BAR domain proteins. Langmuir. 2013;29(1):328–336. doi: 10.1021/la303902q. [DOI] [PubMed] [Google Scholar]
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Tompa P, Kalmar L. Power law distribution defines structural disorder as a structural element directly linked with function. J Mol Biol. 2010;403(3):346–350. doi: 10.1016/j.jmb.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Tourdot RW, Ramakrishnan N, Radhakrishnan R. Defining the free-energy landscape of curvature-inducing proteins on membrane bilayers. Phys Rev E. 2014;90(2):022717. doi: 10.1103/PhysRevE.90.022717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FC, Simunovic M, Sorre B, et al. Comparing physical mechanisms for membrane curvature-driven sorting of BAR-domain proteins. Soft Matter. 2021;17:4254–4265. doi: 10.1039/D0SM01573C. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172(2):269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN (2003) A protein-chameleon: conformational plasticity of -synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn 21(2):211–234. 10.1080/07391102.2003.10506918 [DOI] [PubMed]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta Proteins Proteom. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Finkelstein AV. Life in phases: intra-and inter-molecular phase transitions in protein solutions. Biomolecules. 2019;9(12):842. doi: 10.3390/biom9120842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Santambrogio C, Brocca S, et al. Length-dependent compaction of intrinsically disordered proteins. FEBS Lett. 2012;586(1):70–73. doi: 10.1016/j.febslet.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Vacic V, Oldfield CJ, Mohan A, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Kosol S, Lebrun P, et al. pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res. 2014;42(D1):D326–D335. doi: 10.1093/nar/gkt960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey J, Isas JM, Mizuno N, et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J Biol Chem. 2010;285(42):32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M, D’angelo G, Di Campli A, et al. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci. 2008;65(18):2833–2841. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt AD, Pozzi N, Chen Z, et al. Essential role of conformational selection in ligand binding. Biophys Chem. 2014;186:13–21. doi: 10.1016/j.bpc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, et al. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein pacsin/syndapin. Proc Natl Acad Sci USA. 2009;106(31):12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Clark LD, Gao Y, et al. Mechanism of membrane-curvature generation by ER-tubule shaping proteins. Nat Commun. 2021;12(1):1–15. doi: 10.1038/s41467-020-20625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta. 1998;1376(3):339–352. doi: 10.1016/S0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28(1):319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- White SH, Ladokhin AS, Jayasinghe S, et al. How membranes shape protein structure. J Biol Chem. 2001;276(35):32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU. Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol. 2010;190(3):347–361. doi: 10.1083/jcb.201003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- Woodsmith J, Kamburov A, Stelzl U. Dual coordination of post translational modifications in human protein networks. PLoS Comput Biol. 2013;9(3):e1002,933. doi: 10.1371/journal.pcbi.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Shi Z, Baumgart T. Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization. PLoS One. 2014;9(4):e93,060. doi: 10.1371/journal.pone.0093060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Brown CJ, Dunker AK, et al. (2013) Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim Biophys Acta Proteins Proteom. 1834;4:725–738. doi: 10.1016/j.bbapap.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Tong J, Lee PJ, et al. Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J Biol Chem. 2010;285(1):531–540. doi: 10.1074/jbc.M109.068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput Biol. 2013;9(1):e1002,892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeno WF, Baul U, Snead WT, et al. Synergy between intrinsically disordered domains and structured proteins amplifies membrane curvature sensing. Nat Commun. 2018;9(1):1–14. doi: 10.1038/s41467-018-06532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeno WF, Snead WT, Thatte AS, et al. Structured and intrinsically disordered domains within amphiphysin1 work together to sense and drive membrane curvature. Soft Matter. 2019;15(43):8706–8717. doi: 10.1039/C9SM01495K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeno WF, Thatte AS, Wang L, et al. Molecular mechanisms of membrane curvature sensing by a disordered protein. J Am Chem Soc. 2019;141(26):10361–10371. doi: 10.1021/jacs.9b03927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeno WF, Hochfelder JB, Thatte AS, et al. Clathrin senses membrane curvature. Biophys J. 2021;120(5):818–828. doi: 10.1016/j.bpj.2020.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Das SL, Baumgart T. Nonlinear sorting, curvature generation, and crowding of endophilin N-BAR on tubular membranes. Biophys J. 2012;102(8):1837–1845. doi: 10.1016/j.bpj.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo Y, Ilangovan U, Schirf V, et al. Dynamic interactions between clathrin and locally structured elements in a disordered protein mediate clathrin lattice assembly. J Mol Biol. 2010;404(2):274–290. doi: 10.1016/j.jmb.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7(1):9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]