Abstract
A Staphylococcus aureus strain with low-level heteroresistance to vancomycin (designated MER) but susceptible to methicillin was isolated from an outpatient with conjunctivitis who did not receive any glycopeptide antibiotics. Incubation of the parent strain, MER, with increasing concentrations of vancomycin led to rapid selection of a stable progeny homogeneously resistant to vancomycin. Electron micrographs of strain MER showed enhanced cell wall thickness and abnormal septations typically seen with methicillin-resistant S. aureus having intermediate susceptibility to vancomycin.
Vancomycin-intermediate Staphylococcus aureus (VISA) strains are inhibited by vancomycin concentrations of 8 to 16 μg/ml as defined by the National Committee for Clinical Laboratory Standards (NCCLS) (8). The emergence of VISA has been described for clinical isolates of methicillin-resistant S. aureus in Japan (5), the United States (2), and Europe (10) and mainly from patients on vancomyin therapy. A second type of resistant strain is vancomycin-heterointermediate S. aureus, or hVISA. These strains are susceptible to vancomycin by NCCLS MIC criteria (≤4 μg/ml) but contain subpopulations (approximately 10−6) which can grow in the presence of ≥4 μg of vancomycin per ml (4). To our knowledge, no methicillin-susceptible S. aureus (MSSA) from a clinical sample with reduced susceptibility to vancomycin has been isolated or described so far. We report the first clinical case of an hVISA strain susceptible to methicillin from an outpatient who had not received any glycopeptide antibiotics or any other courses of antimicrobial agents in the preceding 3 months. Moreover, this patient had not been exposed to any person who had a potential risk factor for vancomycin resistance (e.g., a dialysis patient on vancomycin therapy) or been exposed to animals on avoparcin.
In December 1998, a 35-year-old woman wearing permanent contact lenses was admitted to the ophthalmology outpatient clinic at Edouard Herriot Hospital (Lyon, France) due to spontaneous conjunctivitis. She complained of severe eye pain which kept her awake the night before consulting the clinic. The patient had washed the infected eye with sterile physiologic salt solution but had never used any eye lotion containing antibiotics prior to bacteriologic sampling. An MSSA strain was isolated from the tears in the left eye and was designated parent strain MER. The patient was successfully treated with tobramycin eye lotion. The strain was identified as S. aureus by its ability to coagulate citrated rabbit plasma (bioMérieux, Marcy-l'Etoile, France) and to produce a clumping factor (Staphyslide Test; bioMérieux).
Strain MER was included in a prospective study on the prevalence of VISA strains at Edouard Herriot Hospital. It was thus screened on a Mueller-Hinton agar plate containing 2 μg of vancomycin per ml and read after 48 h of incubation at 35°C. MER was the only MSSA isolate that grew on the screen plate out of a total of 469 MSSA isolates tested. Broth microdilution MICs by the NCCLS method (8) and standard Etest method (inoculum of 0.5 McFarland) were 4 μg/ml for vancomycin and teicoplanin. MICs determined by an Etest macromethod (AB BIODISK, Solna, Sweden) with a heavy inoculum (2 McFarland), brain heart infusion agar, and 48-h incubation at 35°C (A. Bolmstrom, A. Karlsson, and P. Wong, 9th European Congress of Clinical Microbiology and Infectious Diseases, poster P104, 1999) were 6 μg/ml for vancomycin and 12 μg/ml for teicoplanin (Table 1). Reference VISA strains (S. aureus Mu50 [5] and LIM-2 [10]) and hVISA strains (S. aureus Mu3 [4]) and LIM-1 [10]) were also tested in parallel by the three methods (Table 1). When a 2-McFarland inoculum was used, the vancomycin and teicoplanin Etest MICs for VISA strains Mu50 and LIM-2 were 8 to 12 and 24 μg/ml, respectively, while Etest MICs for S. aureus ATCC 29213 (non-VISA quality control strain) remained in the susceptible range and were not affected by the higher inoculum.
TABLE 1.
MICs of vancomycin and teicoplanin for hVISA, VISA, and S. aureus ATCC 29213
| Strain | MIC (μg/ml) determined by:
|
|||||
|---|---|---|---|---|---|---|
| Etest witha:
|
||||||
| NCCLS broth microdilution
|
0.5-McFarland inoculum (MHA)
|
2-McFarland inoculum (BHI)
|
||||
| Vancomycin | Teicoplanin | Vancomycin | Teicoplanin | Vancomycin | Teicoplanin | |
| MER | 4 | 4 | 4 | 4 | 6 | 12 |
| MER-S6 | 8 | 8 | 4 | 24 | 12 | 48 |
| MER-S12 | 8 | 16 | 4 | 32 | 16 | 48 |
| MER-S20 | 8 | 16 | 8 | 32 | 24 | 64 |
| Mu3 | 2 | 8 | 2 | 8 | 4 | 16 |
| Mu50 | 4 | 8 | 4 | 8 | 12 | 24 |
| LIM-1 | 2 | 1 | 2 | 1.5 | 4 | 4 |
| LIM-2 | 4 | 8 | 2 | 1.5 | 8 | 24 |
| ATCC 29213 | 0.5 | 0.5 | 1 | 0.75 | 2 | 2 |
MHA, Mueller-Hinton agar plate; BHI, brain heart infusion plate.
Population analysis of strain MER performed according to the method of Hiramatsu et al. (4) revealed a heteroresistant subpopulation in the presence of 5 to 6 μg of vancomycin per ml (Fig. 1). Strain MER-S6, a progeny of parent strain MER growing on 6 μg of vancomycin per ml, was also subjected to population analysis. MER-S6 cells grew in the presence of 4 μg of vancomycin per ml, and 10−6 cells grew on 20 μg of vancomycin per ml. Strain MER-S12, a derivative of MER-S6 growing on 12 μg of vancomycin per ml, was more homogeneously resistant to vancomycin, and 10−3 cells grew on 20 μg of vancomycin per ml (Fig. 1). Strain MER-S20, a derivative of strain MER-S12 growing on 20 μg of vancomycin per ml, survived a concentration of 24 μg of vancomycin per ml (Fig. 1). S. aureus ATCC 29213, the susceptible quality control strain, was also subjected to four consecutive population analyses by selecting colonies that grew on the highest concentrations of vancomycin. The percentage of cells growing at 3 and 4 μg of vancomycin per ml increased after exposure to vancomycin, but none survived at more than 4 μg of vancomycin per ml (Fig. 1). Strain MER and its three derivatives exhibited strictly identical pulsed-field gel electrophoresis patterns (data not shown), confirming the clonality between all isolates.
FIG. 1.
Population analysis of parental hVISA strain MER, S. aureus ATCC 29213, and their derivatives. Bacteria grown overnight were serially diluted and plated on various concentrations of vancomycin-containing agar medium. MER-S6 was a MER-derived progeny selected at 6 μg of vancomycin per ml, MER-S12 was derived from MER-S6 selected at 12 μg of vancomycin per ml, and MER-S20 was derived from MER-S12 selected at 20 μg of vancomycin per ml. ATCC 29213-S3 was selected at 3 μg of vancomycin per ml, ATCC 29213-S4 was selected at 4 μg of vancomycin per ml, and ATCC 29213-S4′ was obtained by selecting ATCC 29213-S4 with 4 μg of vancomycin per ml. Horizontal arrows indicate the order in which the consecutive population analyses were performed.
MICs of vancomycin and teicoplanin for strains MER-S6, MER-S12, and MER-S20 were higher than those of the control VISA strains, Mu50 and LIM-2 (Table 1). Moreover, MER-S20 colonies appeared to be of two distinct colony types (large and small) but with identical glycopeptide MICs. Similar observations have been reported by Tenover (11).
Strain MER-S20 was subcultured daily on nonselective medium for 21 days. The population analyses for MER-S20 done 7, 14, and 21 days after such serial subcultures were similar and showed a reduction of 2 log10 of the countable subpopulations growing on medium containing ≥10 μg of vancomycin per ml (data not shown). Boyle-Vavra et al. reported that reversion of glycopeptide resistance occurred with all VISA isolates after 15 days of serial passages on nonselective medium (1). It is possible that 21 days of passaging was insufficient for observing reversion with strain MER-S20 or that reversion does not occur in all VISA strains.
MER was not resistant to oxacillin (MIC < 0.5 μg/ml) and did not harbor the mecA gene, as confirmed by PCR analysis using the procedure described by Murakami et al. (7). The MIC of oxacillin for MER-S20 was higher (8 μg/ml), although the mecA gene was still absent. The mechanism of VISA resistance may involve alterations in cell wall metabolism, thus affecting the activity of beta-lactam antibiotics. MER was susceptible to all antistaphylococcal antibiotics (gentamicin, tobramycin, kanamycin, chloramphenicol, tetracycline, erythromycin, lincomycin, pristinamycin, rifampin, fusidic acid, fosfomycin, ciprofloxacin, and trimethoprim-sulfamethoxazole) except benzylpenicillin due to inducible β-lactamase production; strain MER-S20 had an identical antibiotic susceptibility profile.
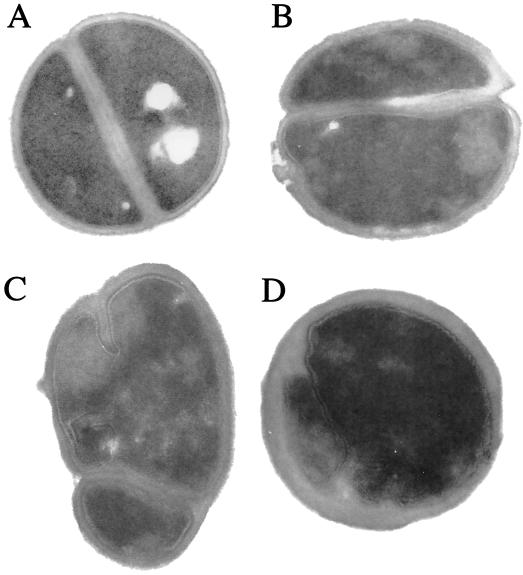
Cultures were grown to an optical density at 600 nm of 0.6 in brain heart infusion broth prior to processing for transmission electron microscopy, as described by Mani et al. (6). Electron micrographs of strain MER showed enhanced cell wall thickness, uneven surfaces, irregular shapes, and abnormal septations in contrast to the thin and regular cell wall morphology of the vancomycin-susceptible strain S. aureus ATCC 29213 (Fig. 2). When strains were grown in the presence of 2 μg of vancomycin per ml, the abnormalities observed with strain MER were slightly enhanced (Fig. 2) whereas ATCC 29213 remained unaffected (data not shown). The altered morphology described was more distinct for strain MER-S20 (Fig. 2).
FIG. 2.
Composite picture showing the morphology of S. aureus ATCC 29213 (A) susceptible to vancomycin and the morphological abnormalities of hVISA MER grown in the absence (B) or presence (C) of 2 μg of vancomycin per ml and of strain MER-S20 (D). All cultures were grown to an optical density at 600 nm of 0.6 in brain heart infusion broth prior to processing for transmission electron microscopic examination. Magnification, ×40,000 (original magnification, ×60,000).
S. aureus strains with increased potential for the development of glycopeptide resistance have shown cell wall abnormalities when grown in the presence of vancomycin (9). Our report indicates that even in a patient who had not received vancomycin, an S. aureus strain with altered cell wall structure associated with resistance to vancomycin could be detected after simple screening on vancomycin agar plates. Since the strain was methicillin susceptible, this report demonstrates that the genetic background for development of glycopeptide resistance is not strictly associated with the mecA gene. Parent strain MER is typically an hVISA strain that is susceptible to vancomycin by current NCCLS criteria, i.e., MIC ≤ 4 μg/ml, but contains subpopulations of cells at a frequency of 10−4 to 10−6 that can grow in the presence of 5 to 6 μg of vancomycin per ml. Stepwise passaging in increasing concentrations of vancomycin led to a rapid increase in the level of vancomycin resistance and the selection of a progeny homogeneously resistant to vancomycin. This observation reinforces the hypothesis that hVISA strains, such as MER, are precursors that can become homogeneously resistant to vancomycin (3, 4). The genetic basis for hVISA strains which appear to have a strain-specific capacity for transformation to the VISA phenotype remains to be characterized.
Acknowledgments
We thank Keichi Hiramatsu, François Denis, and Marie-Cécile Ploy for the gift of strains.
REFERENCES
- 1.Boyle-Vavra S, Berke S K, Lee J C, Daum R S. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother. 2000;44:272–277. doi: 10.1128/aac.44.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States 1997. Morb Mortal Wkly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 3.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum R S, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 4.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 6.Mani N, Tobin P, Jayaswal R K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J Bacteriol. 1993;175:1493–1499. doi: 10.1128/jb.175.5.1493-1499.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 9.Pfeltz R F, Singh V K, Schmidt J L, Batten M A, Baranyk C S, Nadakavukaren M J, Jayaswal R K, Wilkinson B J. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob Agents Chemother. 2000;44:294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ploy C M, Grelaud C, Martin C, de Lumley L, Denis F. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet. 1998;351:1212. doi: 10.1016/s0140-6736(05)79166-2. [DOI] [PubMed] [Google Scholar]
- 11.Tenover F C. VRSA, VISA, and GISA: the dilemma behind the name game. Clin Microbiol Newsl. 2000;22:49–53. [Google Scholar]