Abstract
The association between lactic acid bacteria (LAB) and their immunostimulatory effects has attracted considerable attention; however, it remains unclear whether LAB can induce interferon-lambdas (IFN-λs) in human epithelial cells under conditions that do not mimic infection. In this study, we first employed a reporter assay to screen for a potential strain capable of inducing IFN-λ3 among 135 LAB strains derived from traditional Japanese pickles. Next, we assessed the strain’s ability to induce the expression of IFN-λ genes and interferon-stimulated genes (ISGs), and to produce IFN-λs. As a result, we screened and isolated Enterococcus casseliflavus KB1733 (KB1733) as a potential strain capable of inducing IFN-λ3 expression. Furthermore, we clarified that KB1733 induced the expression of IFN-λ genes and ISGs related to antiviral functions, and that KB1733 induced IFN-λ1 and -λ3 expression in a dose-dependent manner up to 10 μg/mL. In addition, KB1733 significantly increased IFN-λ1 production compared to Enterococcus casseliflavus JCM8723T, which belongs to the same genera and species as KB1733. In conclusion, we isolated a unique LAB strain from traditional Japanese pickles that is capable of stimulating IFN-λ production, although further study is needed to investigate how KB1733 protects against viruses in mice and humans.
Keywords: screening, pickles, Enterococcus casseliflavus KB1733, heat-killed bacteria, interferon-lambda
1. Introduction
Lactic acid bacteria (LAB) have historically been important mainly because of their utility in fermentation and food preservation as well as their health benefits. According to the definition suggested by the Food and Agriculture Organization (FAO)/World Health Organization (WHO), health-promoting LAB are called probiotics and are considered non-pathogenic live microorganisms that confer health benefits to the host when administered in adequate amounts [1]. Most LAB employed as probiotics belong to the genera Lactobacillus and Enterococcus [2]; however, their features are known to differ according to the strain [3]. Many studies have focused on the immunostimulatory effects of probiotics [4,5,6], especially on innate immune responses, and some of them have been found to have anti-infectious effects in humans and animals [7,8]. Furthermore, heat-killed LAB were also reported to show immune-modulatory properties similar to those of probiotics [9,10], suggesting that these could be used more effectively than live LAB because of their pharmaceutical advantages in terms of transport and storage [11]. Therefore, isolating unique heat-killed LAB strains that can strengthen the immune system may contribute to the development of functional foods and drinks that people could utilize to prevent infectious diseases in their daily lives [12].
The type III interferon (IFN) family, including IFN-λ1, -λ2, -λ3 (also known as interleukin (IL)29, IL28A, and IL28B, respectively) and -λ4 [13], has recently attracted a lot of attention as a crucial factor in the innate immune system. One subunit of the heterodimeric receptor of IFN-λ, IFN-λ receptor 1 (IFNLR1), is strongly expressed in the epithelial cells of mice and humans [14,15,16], and IFN-λ has been reported to provide antiviral protection to anatomic barriers such as the respiratory, hepatic, and gastrointestinal mucosa [17,18]. In addition, IFN-λ was found to readily induce the expression of IFN-stimulated genes (ISGs) such as myxovirus resistance 1 (Mx 1) and oligoadenylate synthetase 1 (OAS1) in epithelial cells of most body sites [19,20]. Furthermore, a previous in vitro study indicated that IFN-λ exhibited a dose-dependent inhibitory effect on SARS-CoV-2 in Vero cells [21]. Another in vivo study showed that IFN-λ treatment prevented and cured persistent enteric murine norovirus infection [22]. Recently, Murata et al. found that the nucleotide analogs, hepatitis B virus (HBV) polymerase inhibitors, induced IFN-λ3 in the gastrointestinal (GI) tract [23], which showed anti-HBV effects [24]. Serum IFN-λ3 levels were reported to be increased in patients with acute hepatitis E virus (HEV), while the HEV itself induced IFN-λ3 in colon cancer cell lines [25].
These results suggest that inducing IFN-λ production in epithelial cells before viral invasion could contribute to infection prevention. Previous studies have investigated the effects of LAB on IFN-λ induction in human dendritic cells [26] or porcine epithelial cells under conditions that mimic infection [27]. However, few studies have focused on the effects of LAB that can induce IFN-λs in human epithelial cells without mimicking infection. The aim of this study was to isolate potent and unique LAB strains with the ability to induce IFN-λs and exert antiviral effects on human intestinal epithelial cells. To our knowledge, this is the first study to investigate the effect of heat-killed LAB on IFN-λ production in human intestinal epithelial cells.
2. Materials and Methods
2.1. To Screen for the LAB Strain Capable of Inducing the Gene Expression of IFN-λ3
2.1.1. Strains, Growth Conditions, and Sample Preparation
A total of 135 LAB strains were evaluated during the screening (Supplementary Table S1). All LAB strains belonged to the Department of Nature and Wellness Research, Innovation Division, KAGOME CO., LTD. (Nasushiobara, Tochigi, Japan). Our strains were isolated from traditional Japanese pickles collected throughout Japan, stored at −80 °C, and freeze-dried until use [28]. The control strain, Lactococcus lactis subsp. lactis JCM5805T [26], was provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. Each bacterium was inoculated with 1% (v/v) of a thawing glycerol stock in 10 mL de Man, Rogosa, and Sharpe (MRS) broth (Difco, Detroit, MI, USA) and incubated for 20 h at 30 °C. Next, 4 mL of cell culture was inoculated in 36 mL MRS medium and incubated for 20 h at 30 °C. The cell cultures were washed with sterilized saline and water, followed by centrifugation at 4 °C and 2590× g for 10 min (himac22G, R10A2, Eppendorf Himac Technologies, Hitachinaka, Ibaraki, Japan). To prepare heat-killed bacteria, washed bacteria were autoclaved at 100 °C for 30 min (Iwaki, Tokyo, Japan) and freeze-dried. Each freeze-dried sample was diluted with Dulbecco’s modified Eagle’s medium (DMEM) (FUJIFILM Wako Pure Chemical Co., Ltd., Osaka, Japan) to a concentration of 10 μg/mL and used in the assay. Adefovir dipivoxil (ADV) (Funakoshi, Tokyo, Japan) was used as a positive control because it had previously been reported to induce IFN-λs in WiDr cells [23]. Dimethyl sulfoxide (DMSO, 0.1%) (Sigma-Aldrich, St. Louis, MO, USA) was used as a negative control.
2.1.2. Cells
We used a WiDr-Luc-IL28B cell line produced by Murata, K. The WiDr cells, which are human colon epithelial cell lines, were transfected with pGL4.28 (luc2CP/minP/Hygro) carrying the IL-28B promoter region [29]. The WiDr-Luc-IL28B cells were maintained in D-MEM supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY, USA) and 0.5% hygromycin B (FUJIFILM Wako Pure Chemical Co., Ltd., Osaka, Japan) at 37 °C in the 5% CO2 gassed incubator. WiDr cells were selected for the screening and the functional assays of IFN-λs since WiDr cells were the most potent cell lines to induce IFN-λs among the five human colon cancer cell lines we tested [23].
2.1.3. Cell Culture with Samples and Reporter Assay for IFN-λ3
WiDr-Luc-IL28B cells were seeded at 5 × 105 cells/well, with a total volume of 100 μL in a 96-well black plate, CulturPlate-96F (PerkinElmer, Inc., Waltham, MA, USA), and incubated at 37 °C in a 5% CO2 gassed incubator for 48 h. After removal of the culture supernatants, the prepared samples were added into each well (4 wells/sample) and co-cultured for another 48 h. Then, after removal of the culture supernatant, the DMEM medium was added to each well at a volume of 50 μL/well, followed by the Steady-Glo® Luciferase Assay System (Promega, Madison, WI, USA) at a volume of 50 μL/well; the 96-well black plate was then left for 5 min at 21 °C. After 5 min, the plate was placed in a CentroXS3 LB960 microplate luminometer (Berthold Japan, Tokyo, Japan), and the luciferase activity in each well was measured.
2.2. To Assess the Ability of Enterococcus casseliflavus KB1733 to Produce IFN-λs and Induce Expressions of IFN-λ Genes and ISGs
2.2.1. Strains, Growth Conditions, and Sample Preparation
KB1733 was selected for further analysis. The control strain, Enterococcus casseliflavus JCM8723T, was provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. The growth conditions, sample preparation, and positive control were the same as those described above. For the production of IFN-λ1, -λ2, and -λ3, each freeze-dried sample was diluted with D-MEM at concentrations of 0, 1, 3, 10, and 30 μg/mL and applied to the assay. For the expression assay of IFN-λ genes and ISGs, each freeze-dried sample was diluted with D-MEM at a concentration of 10 μg/mL.
2.2.2. Cells
WiDr cells [23] were maintained in D-MEM (FUJIFILM Wako Pure Chemical, Osaka, Japan) supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY, USA) and 0.5% penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C in an incubator with 5% CO2.
2.2.3. Cell Culture with Samples
The WiDr cells were seeded at 1 × 106 cells/well, with a total volume of 2 mL in a 6-well plate (AGC Techno Glass, Tokyo, Japan), and were incubated at 37 °C in a 5% CO2 gassed incubator for 48 h. After removal of the culture supernatants, the prepared samples were added into each well (1 well/sample) and co-cultured for another 48 h. The supernatant of the medium co-cultured with samples was used for the assay of IFN-λ production. Cells on a 6-well plate were used for the expression assay of IFN-λ genes and ISGs.
2.2.4. Assay for Production of IFN-λ1, -λ2, and -λ3
The levels of IFN-λ1 and -λ2 in the supernatant were quantified by enzyme-linked immunosorbent assay (ELISA) using commercial kits, according to the manufacturer’s instructions. The IL-29 and the IL-28A Human ELISA Kits (Thermo Fisher Scientific, Waltham, MA, USA) were used for the IFN-λ1 and IFN-λ2 measurements, respectively.
The level of IFN-λ3 in the supernatant was quantified using a chemiluminescence enzyme immunoassay (CLEIA). The assay was performed according to a previously described method [30].
2.2.5. Assay for Expressions of IFN-λ Genes and ISGs
Real-time PCR (RT-qPCR) was performed to quantify the selected mRNAs in the WiDr cells. Total RNA was extracted from all samples using TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. The cDNA template was synthesized from the extracted RNA using the PrimeScript® RT Reagent Kit (TaKaRa Bio, Kusatsu, Shiga, Japan) following the manufacturer’s instructions. The thermal cycling conditions for the reverse transcription reaction were 15 min at 37 °C, followed by 5 s at 85 °C.
mRNA expression of IFN-λs, ISGs, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping gene was measured using real-time PCR performed on a 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, CA, USA) with TB Green® Premix Ex Taq II (TaKaRa Bio, Shiga, Japan). The PCR thermal cycling conditions were 30 s at 95 °C, followed by 40 cycles of PCR reaction at 95 °C for 5 s and 60 °C for 30 s. Relative gene expression was calculated as fold induction compared to GAPDH expression. The primer sequences used are listed in Table 1.
Table 1.
Primers sequences 1.
| Genes | Primer Sequence (5′→3′) | |
|---|---|---|
| Human IL-29 (IFN-λ1) | F | GCCTCCTCACGCGAGACCTC |
| R | GGAGTAGGGCTCAGCGCATA | |
| Human IL-28A (IFN-λ2) | F | TCTGGAGGCCACCGCTGACA |
| R | TGGGCTGAGGCTGGATACAG | |
| Human IL-28B (IFN-λ3) | F | TGGCCCTGACGCTGAAGGTT |
| R | CGTGGGCTGAGGCTGGATAC | |
| Homo sapiens MX dynamin like GTPase 1 (MX1) | F | TACCAGACTCCGACACGAGTTCC |
| R | GATTTGCTGTTTCACGATTGTCTCA | |
| Homo sapiens 2′-5′-oligoadenylate synthetase 1 (OAS1) | F | AGAGCCTCATCCGCCTAGTCAA |
| R | GCTCCCAAGCATAGACCGTCA | |
| Interferon stimulated gene 15 (ISG15) | F | TGGACAAATGCGACGAACCTC |
| R | CTGCGGCCCTTGTTATTCCTC | |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | F | TGGTGAAGACGCCAGTGGA |
| R | GCACCGTCAAGGCTGAGAAC | |
1 F: Forward, R: Reverse.
2.3. Statistical Analysis
All statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan) [31]. EZR is a modified version (version 2.5–1) of the R Commander, designed to add statistical functions frequently used in biostatistics. A dose or time-course assessment of KB1733 (n = 6) was performed using the Steel test, and the treatment group data were compared with the control group data. Strain (KB1733 and JCM8723T) specificity for IFN-λ1 production (n = 4) was analyzed using the Tukey–Kramer method. All values are expressed as mean ± standard error (SE) (n = 4–6). In all analyses, p-values < 0.05 were considered statistically significant.
3. Results
3.1. To Screen for the LAB Strain Capable of Inducing the Gene Expression of IFN-λ3
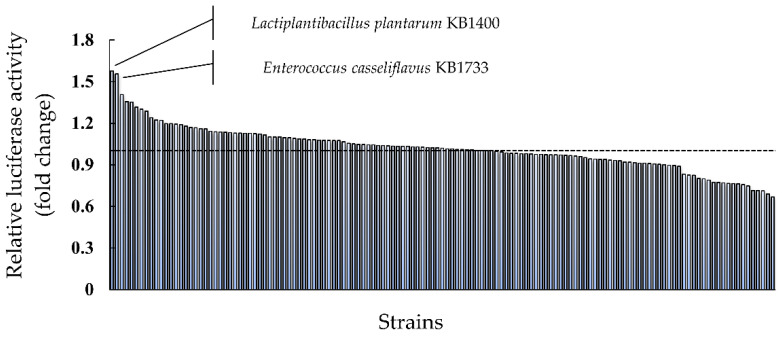
We screened for a potent LAB strain that could induce the expression of IFN-λ3 using a reporter assay. Figure 1 and Supplementary Table S2 show the relative luciferase activity of each LAB strain (vs. JCM5805T). Among the 135 LAB strains evaluated in this study, two strains—Enterococcus casseliflavus KB1733 (KB1733) and Lactiplantibacillus plantarum KB1400—showed an approximately 1.5-fold higher induction of the IFN-λ3 gene expression as compared to that of JCM5805T. We performed the same experiment focusing on these two strains and found that only KB1733 induced luciferase activity at the same level as the first screening result (Supplementary Table S2).
Figure 1.
Screening for the potent LAB strain with an ability to induce the expression of the IFN-λ3 gene among 135 strains. Dotted line indicates the value of relative luciferase activity of the control as 1.0. The value for each of the 135 strains is listed in Supplementary Table S2. Data from a single experiment are shown.
3.2. To Assess the Ability of Enterococcus casseliflavus KB1733 to Induce IFN-λ Production and Expressions of IFN-λ Genes and ISGs
3.2.1. Gene Expressions of IFN-λs
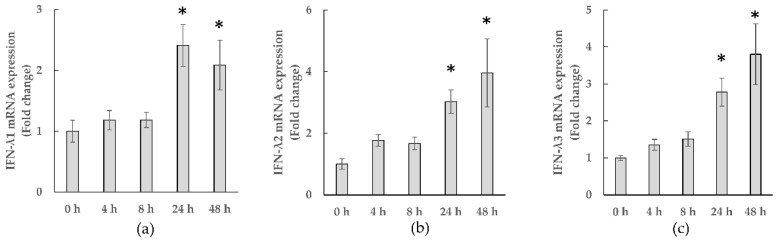
Figure 2 shows the time-course of IFN-λ gene expression in WiDr cells co-cultured with KB1733. KB1733 significantly increased the expression of all three genes (IFN-λ1, -λ2, and -λ3) at 24 h and 48 h compared to that at 0 h.
Figure 2.
Gene expressions of (a) IFN-λ1, (b) IFN-λ2, and (c) IFN-λ3. The values are shown as means ± SE. Asterisks indicate a significant difference when compared to 0 h (p-value < 0.05). Data from six independent experiments are shown.
3.2.2. Production of IFN-λ1, -λ2, and -λ3
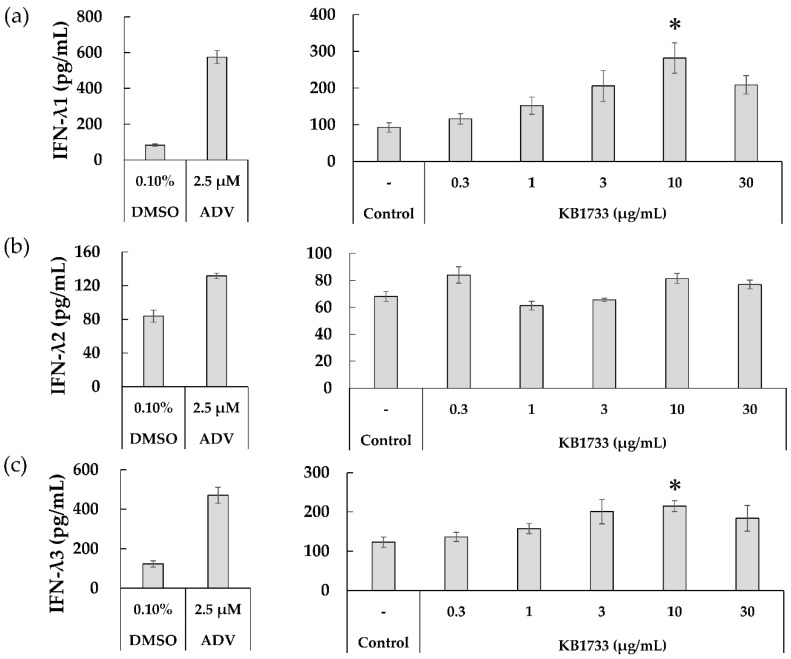
After checking the gene expression of IFN-λs in the WiDr cells co-cultured with KB1733, we assessed the dose-dependent effects of KB1733 on the production of IFN-λ1 and -λ2 using ELISA and of IFN-λ3 using CLEIA. Figure 3 shows the production of IFN-λ1, -λ2, and -λ3 in the cell culture supernatant after 48 h of KB1733 treatment. First, we confirmed that the assay worked by employing ADV and DMSO as positive and negative controls, respectively. KB1733 significantly induced both IFN-λ1 and λ3 at a concentration of 10 μg/mL (Figure 3a,c), although it did not significantly induce IFN-λ2 (Figure 3b).
Figure 3.
Production of (a) IFN-λ1, (b) IFN-λ2, and (c) IFN-λ3 after 48 h of KB1733 treatment. DMSO: Dimethyl sulfoxide employed as a negative control, ADV: Adefovir dipivoxil employed as a positive control. The values are shown as mean ± SE. Asterisks represent a significant difference when compared to control (DMEM) (p-value < 0.05). Data from six independent experiments are shown.
3.2.3. Expressions of ISGs
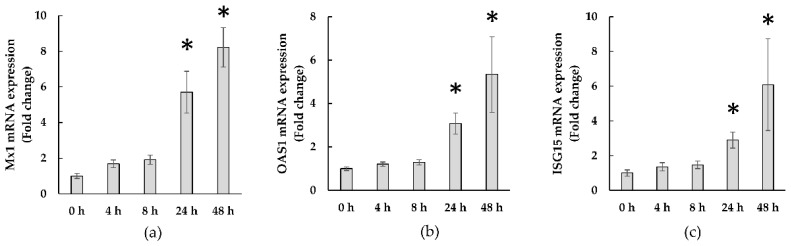
Figure 4 shows the time-course of ISG expression in WiDr cells co-cultured with KB1733. KB1733 significantly increased the expression of all three genes (Mx 1, OAS1, and ISG15) at 24 h and 48 h compared to that at 0 h.
Figure 4.
Gene expressions of (a) Mx1, (b) OAS1, and (c) ISG15. The values are shown as means ± SE. Asterisks mean a significant difference when compared to 0 h (p-value < 0.05). Data from six independent experiments are shown.
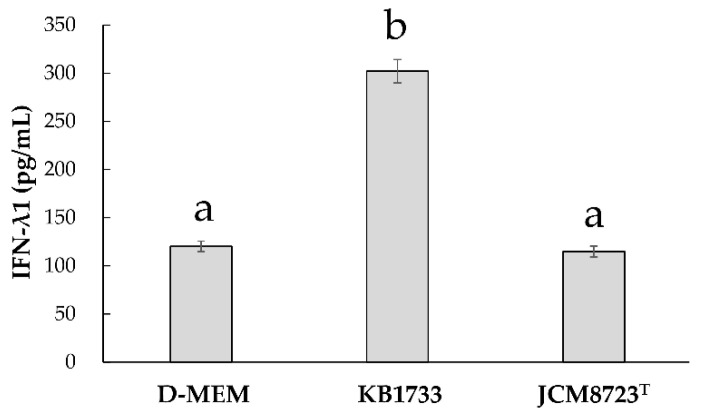
3.2.4. Strain Specificity for IFN-λ1 Production
To identify the strain specificity for IFN-λ production, we assessed the difference in IFN-λ1 production between KB1733 and JCM8723T.
Figure 5 shows IFN-λ1 production by the WiDr cell co-cultured with KB1733 or JCM8723T. IFN-λ1 production with D-MEM, KB1733, and JCM8723T were 120.3 ± 10.9, 302.0 ± 24.4, and 114.7 ± 11.4 pg/mL, respectively. KB1733 significantly increased IFN-λ1 production compared to D-MEM or JCM8723T. However, JCM8723T did not significantly increase IFN-λ1 production compared to D-MEM.
Figure 5.
Strain specificity for IFN-λ1 production after 48 h of KB1733 or JCM8723T addition (n = 4). The values are shown as mean ± SE. The treatments are significantly different if they do not share the same letter (a,b) (p-value < 0.05). Data from four independent experiments are shown.
4. Discussion
The association between LAB and its immunostimulatory effects has attracted considerable attention; however, there are few studies analyzing whether LAB can induce IFN-λ expression in human epithelial cells without mimicking infection. In the present study, we aimed to identify heat-killed LAB with the potential ability to induce IFN-λ and antiviral-related genes in WiDr cell lines without stimulation by mimicking viral RNAs. We selected KB1733, which exhibited the strongest ability to induce IFN-λ3 gene expression among a total of 135 strains isolated from traditional Japanese pickles. We further clarified that KB1733 could induce IFN-λ1 and -λ3 production in a dose-dependent manner up to 10 μg/mL. In addition, KB1733 induced the expression of genes such as IFN-λs and ISGs related to antiviral functions.
Fermented foods such as pickles and miso (fermented soy) are common in Japanese cuisine and have been recognized as safe for consumption. Pickles are foods fermented with LAB [32]. Fermented vegetables such as pickles are therefore potential sources of beneficial LAB. KB1733 was derived from a pickle, wasabina-duke, made from Brassica juncea. Although 10 other strains evaluated in this study were derived from the same source of isolation as KB1733, they did not enhance luciferase activity as high as this strain. Therefore, the IFN-λ-stimulating effects of LAB seem to depend on the strain itself rather than the source of isolation. In addition, we investigated the difference in IFN-λ1 production between KB1733 and JCM8723T and found that IFN-λ1 production induced by KB1733 was significantly higher than that induced by JCM8723T. This means that the IFN-λ1-inducing effect did not depend on the genera and species but on strain specificity. The results of this study prove that we were able to successfully isolate a unique LAB strain from traditional Japanese pickles to stimulate IFN-λ production.
This study showed that KB1733 induced the production of IFN-λ1 and -λ3 in a dose-dependent manner up to 10 μg/mL, but this dose-dependency was not observed at a higher dose. The underlying mechanisms remain unclear, but a plausible explanation could be that a higher dose of KB1733 could impair IFN-λ production. This phenomenon is consistent with a previous study’s findings that Lactiplantibacillus pentosus S-PT84 could induce interleukin-12 in a dose-dependent manner, but not at a higher dose [33]. This issue requires further exploration in the future.
Although KB1733 enhanced the expression of IFN-λs, it did not significantly induce IFN-λ2 protein. A previous study comparing the biological activity of different IFN-λ subtypes clearly showed that IFN-λ3 was equal to or more potent than IFN-λ1 with respect to antiviral activity [34]. This study also showed that the specific activity of IFN-λ3 was 16-fold higher than that of IFN-λ2 despite a 96% sequence similarity between IFN-λ2 and -λ3 [34]. We considered that in terms of antiviral activity, KB1733 was a potent immunostimulant since it could promote the secretion of IFN-λ1 and -λ3 rather than IFN-λ2.
Furthermore, the expression of IFN-λ genes and ISGs was upregulated by KB1733. The results showed that the expression of all three ISGs (Mx1, OAS1, and ISG15) evaluated in this study at 24 h at the latest was significantly enhanced by KB1733. Moreover, our findings are consistent with a previous review describing IFN-λ-induced ISG expression [35]. Namely, KB1733 seems to induce ISGs, at least via IFN-λ induction. ISGs interfere with viral replication and provide immune defense to the host [23,35]. Considering these factors, KB1733 might be a possible candidate for enhancing host immunity against viral infections. In fact, Indo et al. showed that pre-treatment of porcine epithelial cells with Legilactobacillus salivarius strains before a rotavirus challenge could enhance the gene expression of IFN-λ3, Mx 1, and OAS1 and significantly reduced both the rotavirus titer and infection rate [27]. In addition, Murata et al. investigated the inducing sites of IFN-λ3 using several cell lines that originated from different organs such as colon, lungs, skin, stomach, liver, and lymphocytes; they concluded that orally administered ADV induced IFN-λ3 in the GI tracts, which in turn induced ISGs in hepatocytes [23] although the precise mechanism remains unclear. When this phenomenon is extrapolated to KB1733, the putative mechanism of action for KB1733 application is presumably as follows: orally administered KB1733 reaches intestinal epithelial cells and induces IFN-λ production there. Subsequently, IFN-λs induced by KB1733 could, directly or indirectly through ISG, provide protection to the host against viral infections in the GI tract as well as other organs such as the liver or the lungs, to which IFN-λs are carried via blood flow. We intend to further explore how and whether KB1733 exerts protective effects against viral infection.
Nonetheless, it is unclear how KB1733 induces IFN-λ production and/or expression. In this study, we used the heat-killed and freeze-dried powder of KB1733 to investigate its IFN-λ-inducing effects. The powder likely includes DNA, RNA, and peptidoglycan derived from KB1733. Previous research has shown that nucleic acids derived from E. faecalis EC-12 induce IL-12 production in a murine macrophage cell line [36]. Another study showed that peptide glycans purified from Lactobacillus salivarius Ls33 induced IL-10 production in mice [37]. As described above, some studies have indicated a relationship between LAB components and their immunomodulatory properties. In this study, we did not clarify how KB1733 induced IFN-λ1 and -λ3 production. However, based on the fact that nucleotide analogs [23] or HEV [25] induce IFN-λ3 in the GI tract, it is intriguing to consider that host cells can recognize KB1733 as viral mimetic.
This study had several limitations. First, the application of KB1733 in the food industry requires several safety assessments. The genus Enterococci is assigned to risk group 2, which contains microorganisms that carry virulence factors [38]. Therefore, they are considered to be reservoirs for the dissemination of antibiotic resistance and pathogenic genes throughout the food chain [39]. In the future, we should perform a whole genome analysis of KB1733 to clarify whether this strain harbors antibiotic and virulence genes. Second, we only performed a screening for 133 strains once. Therefore, the possibility that we had lost certain hit strains inducing the gene expression of IFN-λ3 could not be eliminated; however, this screening test enabled us to quickly isolate Enterococcous casseliflavus KB1733 from our LAB libraries. Third, although we observed significant induction of IFN-λ genes by KB1733 in colon cancer cell lines as well as ISGs, which are the downstream genes of IFN-λs, we did not investigate whether these inductions of cytokines and ISGs are sufficient to elicit favorable responses against viral invasion. Therefore, further studies are required to translate the use of KB1733 to human applications aimed at improving human health.
5. Conclusions
Among the screened 135 isolates derived from traditional Japanese pickles, Enterococcus casseliflavus KB1733 exhibited the ability to induce IFN-λ and ISG gene expression. In this study, we isolated a unique LAB strain from traditional Japanese pickles capable of stimulating IFN-λ production, although further studies are needed to investigate how KB1733 can provide viral protection in mice and humans.
Acknowledgments
We thank Sachiyo Igari, Hiromi Kaneko, Miku Sasanuma, and Yuri Yasutake (Department of Nature and Wellness Research, Innovation Division, KAGOME CO., LTD., Nasushiobara, Tochigi, Japan) for their technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10040827/s1. Table S1: Sources of isolation, genera, species, and names of the 135 strains evaluated in this study. Table S2: Relative luciferase activities of the 135 LAB strains.
Author Contributions
Conceptualization, S.S. (Shohei Satomi) and K.M.; methodology, S.S. (Shohei Satomi), D.K., S.S. (Shigenori Suzuki) and K.M.; validation, S.S. (Shohei Satomi) and D.K.; formal analysis, S.S. (Shohei Satomi) and D.K.; investigation, S.S. (Shohei Satomi) and D.K.; resources, S.S. (Shohei Satomi), D.K., M.S., M.M. and K.M.; data curation, S.S.(Shohei Satomi) and D.K.; writing—original draft preparation, S.S. (Shohei Satomi); writing—review and editing, S.S. (Shohei Satomi), D.K., T.I., M.S., M.M., S.S. (Shigenori Suzuki) and K.M.; visualization, S.S. and D.K.; supervision, T.I., S.S. (Shigenori Suzuki) and K.M.; project administration, S.S. (Shigenori Suzuki); funding acquisition, S.S. (Shigenori Suzuki). All authors have read and agreed to the published version of the manuscript.
Funding
This study received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study as well as the bacterial strains used are available upon request from the corresponding author.
Conflicts of Interest
KAGOME CO., LTD. supported this work and provided support in the form of salaries for the authors S.S. (Shohei Satomi), D.K., T.I., and S.S. (Shigenori Suzuki).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Joint FAO/WHO Working Group . Guidelines for the Evaluation of Probiotics in Food: Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Food and Agriculture Organization of the United Nations; London, ON, Canada: 2002. [Google Scholar]
- 2.Mulaw G., Tessema T.S., Muleta D., Tesfaye A. In Vitro Evaluation of Probiotic Properties of Lactic Acid Bacteria Isolated from Some Traditionally Fermented Ethiopian Food Products. Int. J. Microbiol. 2019;2019:7179514. doi: 10.1155/2019/7179514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos C.L., Thorsen L., Schwan R.F., Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36:22–29. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki E., Suzuki S., Fukui Y., Yajima N. Cell-bound exopolysaccharides of Lactobacillus brevis KB290 enhance cytotoxic activity of mouse splenocytes. J. Appl. Microbiol. 2015;118:506–514. doi: 10.1111/jam.12686. [DOI] [PubMed] [Google Scholar]
- 5.Fukui Y., Sasaki E., Fuke N., Nakai Y., Ishijima T., Abe K., Yajima N. Effect of Lactobacillus brevis KB290 on the cell-mediated cytotoxic activity of mouse splenocytes: A DNA microarray analysis. Br. J. Nutr. 2013;110:1617–1627. doi: 10.1017/S0007114513000767. [DOI] [PubMed] [Google Scholar]
- 6.Villena L., Aso H., Rutten V.P.M.G., Takahashi H., van Eden W., Kitazawa H. Immunobiotics for the Bovine Host: Their Interaction with Intestinal Epithelial Cells and Their Effect on Antiviral Immunity. Front. Immunol. 2018;9:326. doi: 10.3389/fimmu.2018.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waki N., Yajima N., Suganuma H., Buddle B.M., Luo D., Heiser A., Zheng T. Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett. Appl. Microbiol. 2014;58:87–93. doi: 10.1111/lam.12160. [DOI] [PubMed] [Google Scholar]
- 8.Waki N., Matsumoto M., Fukui Y., Suganuma H. Effects of probiotic Lactobacillus brevis KB290 on incidence of influenza infection among schoolchildren: An open-label pilot study. Lett. Appl. Microbiol. 2014;59:565–571. doi: 10.1111/lam.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N., Russell W.M., Douglas-Escobar M., Hauser N., Lopez M., Neu J. Live and Heat-Killed Lactobacillus rhamnosus GG: Effects on Proinflammatory and Anti-Inflammatory Cytokines/Chemokines in Gastrostomy-Fed Infant Rats. Pediatr. Res. 2009;66:203–207. doi: 10.1203/PDR.0b013e3181aabd4f. [DOI] [PubMed] [Google Scholar]
- 10.Sang L.X., Chang B., Wang B.Y., Liu W.X., Jiang M. Live and heat-killed probiotic: Effects on chronic experimental colitis induced by dextran sulfate sodium (DSS) in rats. Int. J. Clin. Exp. Med. 2015;8:20072–20078. [PMC free article] [PubMed] [Google Scholar]
- 11.Piqué N., Berlanga M., Miñana-Galbis D. Health Benefits of Heat-Killed (Tyndallized) Probiotics: An Overview. Int. J. Mol. Sci. 2019;20:2534. doi: 10.3390/ijms20102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satomi S., Waki N., Arakawa C., Fujisawa K., Suzuki S., Suganuma H. Effects of Heat-Killed Levilactobacillus brevis KB290 in Combination with β-Carotene on Influenza Virus Infection in Healthy Adults: A Randomized Controlled Trial. Nutrients. 2021;13:3039. doi: 10.3390/nu13093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermant P., Michiels T. Interferon-λ in the context of viral infections: Production, response and therapeutic implications. J. Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L., Schnepf D., Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 16.Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e10000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 19.Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardoso N.P., Mansilla F.C., Benedetti E., Turco C.S., Barone L.J., Iserte J.A., Soria I., Baumeister E., Capozzo A.V. Bovine Interferon Lambda Is a Potent Antiviral Against SARS-CoV-2 Infection in vitro. Front. Vet. Sci. 2020;7:603622. doi: 10.3389/fvets.2020.603622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nice T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., Virgin H.W. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murata K., Asano M., Matsumoto A., Sugiyama M., Nishida N., Tanaka E., Inoue T., Sakamoto M., Enomoto N., Shirasaki T., et al. Induction of IFN-λ3 as an additional effect of nucleotide, not nucleoside, analogues: A new potential target for HBV infection. Gut. 2018;67:362–371. doi: 10.1136/gutjnl-2016-312653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada N., Murayama A., Shiina M., Aly H.H., Iwamoto M., Tsukuda S., Watashi K., Tanaka T., Moriishi K., Nishitsuji H., et al. Anti-viral effects of interferon-λ3 on hepatitis B virus infection in cell culture. Hepatol. Res. 2020;50:283–291. doi: 10.1111/hepr.13449. [DOI] [PubMed] [Google Scholar]
- 25.Murata K., Kang J.H., Nagashima S., Matsui T., Karino Y., Yamamoto Y., Atarashi T., Oohara M., Uebayashi M., Sakata H., et al. IFN-λ3 as a host immune response in acute hepatitis E virus infection. Cytokine. 2020;125:154816. doi: 10.1016/j.cyto.2019.154816. [DOI] [PubMed] [Google Scholar]
- 26.Jounai K., Ikado K., Sugimura T., Ano Y., Braun J., Fujiwara D. Spherical lactic acid bacteria activate plasmacytoid dendritic cells immunomodulatory function via TLR9-dependent crosstalk with myeloid dendritic cells. PLoS ONE. 2012;7:e32588. doi: 10.1371/journal.pone.0032588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Indo Y., Kitahara S., Tomokiyo M., Araki S., Islam M.A., Zhou B., Albarracin L., Miyazaki A., Ikeda-Ohtsubo W., Nochi T., et al. Ligilactobacillus salivarius Strains Isolated From the Porcine Gut Modulate Innate Immune Responses in Epithelial Cells and Improve Protection Against Intestinal Viral-Bacterial Superinfection. Front. Immunol. 2021;12:652923. doi: 10.3389/fimmu.2021.652923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki S., Kimoto-Nira H., Suganuma H., Suzuki C., Saito T., Yajima N. Cellular fatty acid composition and exopolysaccharide contribute to bile tolerance in Lactobacillus brevis strains isolated from fermented Japanese pickles. Can. J. Microbiol. 2014;60:183–191. doi: 10.1139/cjm-2014-0043. [DOI] [PubMed] [Google Scholar]
- 29.Sugiyama M., Tanaka Y., Wakita T., Nakanishi M., Mizokami M. Genetic Variation of the IL-28B Promoter Affecting Gene Expression. PLoS ONE. 2011;6:e26620. doi: 10.1371/journal.pone.0026620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiyama M., Kimura T., Naito S., Mukaide M., Shinauchi T., Ueno M., Ito K., Murata K., Mizokami M. Development of specific and quantitative real-time detection PCR and immunoassays for λ3-interferon. Hepatol. Res. 2012;42:1089–1099. doi: 10.1111/j.1872-034X.2012.01032.x. [DOI] [PubMed] [Google Scholar]
- 31.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rezac S., Kok C.R., Heermann M., Hutkins R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018;24:1785. doi: 10.3389/fmicb.2018.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonaka Y., Izumo T., Izumi F., Maekawa T., Shibata H., Nakano A., Kishi A., Akatani K., Kiso Y. Antiallergic Effects of Lactobacillus pentosus strain S-PT84 mediated by modulation of Th1/Th2 immunobalance and induction of IL-10 production. Int. Arch. Allergy Immunol. 2008;145:249–257. doi: 10.1159/000109294. [DOI] [PubMed] [Google Scholar]
- 34.Dellgren C., Gad H.H., Hamming O.J., Melchjorsen J., Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 35.Schoggins J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 36.Inoue R., Nagino T., Hoshino G., Ushida K. Nucleic acids of Enterococcus faecalis strain EC-12 are potent Toll-like receptor 7 and 9 ligands inducing interleukin-12 production from murine splenocytes and murine macrophage cell line J774. 1. FEMS Immunol. Med. Microbiol. 2011;61:94–102. doi: 10.1111/j.1574-695X.2010.00752.x. [DOI] [PubMed] [Google Scholar]
- 37.Macho Fernandez E., Valenti V., Rockel C., Hermann C., Pot B., Boneca I.G., Grangette C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–1059. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 38.EC Directive 2000/54/EC of the European Parliament and of the council of 18 September 2000 on the protection of workers from risks related to exposure to biological agents at work. Off. J. Eur. Union L. 2000;262:21–45. [Google Scholar]
- 39.Jahan M., Zhanel G.G., Sparling R., Holley R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015;199:78–85. doi: 10.1016/j.ijfoodmicro.2015.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study as well as the bacterial strains used are available upon request from the corresponding author.