Campylobacter spp. have become the most important bacteria causing diarrhea in humans in many countries of the world (4). The most important species is Campylobacter jejuni, accounting for over 90% of the infections (4), with Campylobacter coli accounting for most of the remaining infections (6, 7). Macrolides are the drugs of choice for treatment of human infected with Campylobacter spp. Only limited prevalence of macrolide resistance has been detected for C. jejuni (1), especially that of human origin, while a higher prevalence has been found for C. coli (6, 7, 9, 10). Campylobacter spp. are considered zoonotic pathogens (1), and resistance among isolates in the animal reservoir could have implications for the treatment of infections in humans (8). The genetic background for macrolide resistance in Campylobacter spp. has not previously been identified. Some have reported indications of horizontal transfer of macrolide resistance between Campylobacter spp. (3), while others have not been able to transfer macrolide resistance (11).
Previously, a point mutation mediating macrolide resistance has been found in several other bacterial species (13). In this study we have defined the genetic background for macrolide resistance in 54 C. coli isolates from the Danish Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP) (2). All isolates were from 1997 and 1998. Among the isolates, 28 isolates were defined as resistant (MIC ≥ 8 μg/ml) and 26 isolates were defined as sensitive to erythromycin by MIC determinations performed as previously described (1). One isolate was from cattle, 4 were from broilers, and 49 were from pigs.
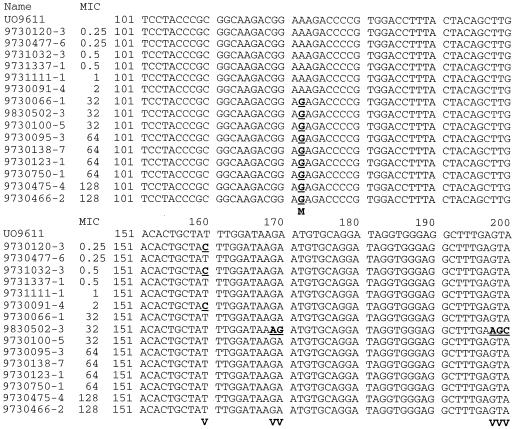
An internal part of the domain V of the 23S ribosomal DNA (rDNA) was amplified by PCR using the universal 23S primers (5′-GTAAACGGCGGCCGTAACTA-3′ and 5′-GACCGAACTGTCTCACGACG-3′) published by Leser et al. (5). An amplicon of 699 bp was obtained from each isolate. To verify the specificity of the primers and the presence of the previously published point mutation for macrolide resistance, amplicons from 15 of the 54 isolates (1 from cattle, 4 from broilers and 10 from pigs) were sequenced. Six of the isolates were sensitive to macrolide and 9 had a MIC of ≥32 μg/ml. From each of the 15 isolates a sequence of 532 bp, corresponding to positions 2109 to 2640 in the published sequence for 23S rDNA of C. coli (GenBank accession no. UO9611) and covering the domain V of the 23 S rDNA, was obtained. Several sequence variations were found in the amplified area (Fig. 1). In all the macrolide-resistant (MIC > 8 μg/ml) C. coli isolates a base substitution, G for A, was detected at position 2230 (corresponding to position 2058 in the nomenclature for Escherichia coli [GenBank accession no. J01695]). This indicates that the genetic background for macrolide resistance in the tested C. coli is the specific point mutation previously described for other bacterial species.
FIG. 1.
Sequences of a 699-bp PCR fragment of an internal area of the 23S rDNA from 15 selected C. coli isolates of animal origin from Denmark. The isolate names and the MIC for each isolate are indicated. The numbers indicate positions in the 532-bp sequenced area. UO9611 is the accession number for the sequence of 23S rDNA of C. coli found in GenBank. Underlined letters indicate sequences variations. Bold letters indicate either the position of the defined point mutation for macrolide resistance (M) or sequence variations (V).
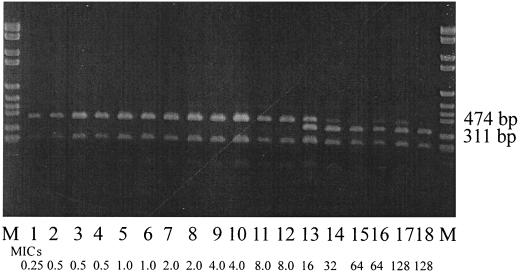
The mutation at position 2230 led to the appearance of an additional target for the nonpalindromic restriction enzyme BsmAI [↑(N)5 GAGACG]. Using this restriction enzyme the presence of this specific point mutation could be visualized by the appearance of an additional fragment in the digest of the 699-bp PCR amplicon using BsmAI. A wild-type strain should give rise to three fragments (of 474, 227, and 13 bp, respectively), while a mutated strain would have four fragments (of 311, 227, 163, and 13 bp, respectively). The presence of the observed point mutation was verified in the remaining 39 isolates, and the results for 18 strains are presented here (Fig. 2). Only in isolates with an erythromycin MIC of >8 μg/ml was the presence of the 311-bp fragment detected. In some of these isolates bands of both 474 and 311 bp appeared. This result was reconfirmed as not being partial digestion of the DNA (data not shown). Since Campylobacter spp. contain three copies of the rRNA genes (the rrnB operon) (12), this result indicates that not all copies are mutated in resistant isolates. It was, however, not possible to detect any association between the MIC and the number of operons with the mutation.
FIG. 2.
Restriction fragment from digestion with BsmAI of a 699-bp PCR fragment of an internal area of the 23S rDNA of C. coli. A 1% agarose gel was used. From left to right: Lanes M, DNA molecular-weight marker (Boehringer VI: 2,176, 1,766, 1,230, 1,033, 653 bp, 517, 453, 394, 298, 234, 220, and 154 bp); lane 1, isolate 9730120-3; lane 2, isolate 9730477-6; lane 3, isolate 9730091-4; lane 4, isolate 9730098-1; lane 5, isolate 9730100-3; lane 6, isolate 9730973-4; lane 7, isolate 9730138-3; lane 8, isolate 9730500-4; lane 9, isolate 9730462-1; lane 10, isolate 9730477-1; lane 11, isolate 9730489-2; lane 12, isolate 9730994-5; lane 13, isolate 9730478-1; lane 14, isolate 9730100-5; lane 15, isolate 9730138-6; lane 16, isolate 9730993-1; lane 17, isolate 9730466-2; lane 18, isolate 9730475-3. MICs (in micrograms per microliter) for the isolates are indicated.
REFERENCES
- 1.Aarestrup F M, Nielsen E M, Madsen M, Engberg J. Antimicrobial susceptibility patterns of thermophilic Campylobacter spp. from humans, cattle, and broilers in Denmark. Antimicrob Agents Chemother. 1997;41:2244–2250. doi: 10.1128/aac.41.10.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. DANMAP 1999. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Report. Copenhagen, Denmark: Danish Zoonosis Centre; 2000. [Google Scholar]
- 3.Ansary A, Radu S. Conjugal transfer of antibiotic resistances and plasmids from Campylobacter jejuni clinical isolates. FEMS Microbiol Lett. 1992;91:125–128. doi: 10.1016/0378-1097(92)90671-a. [DOI] [PubMed] [Google Scholar]
- 4.Lastovica A J, Skirrow M B. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 89–120. [Google Scholar]
- 5.Leser T D, Møller K, Jensen T K, Jorsal S E. Specific detection of Serpulina hyodysenteriae and potentially pathogenic weakly beta-haemolytic porcine intestinal pirochetes by polymerase chain reaction targeting 23S rDNA. Mol Cell Probes. 1997;11:363–372. doi: 10.1006/mcpr.1997.0129. [DOI] [PubMed] [Google Scholar]
- 6.McNulti C A M. The treatment of campylobacter infections in man. J Antimicrob Chemother. 1987;19:281–284. doi: 10.1093/jac/19.3.281. [DOI] [PubMed] [Google Scholar]
- 7.Mirelis B, Llovet T, Muñoz C, Navarro F, Prats G. Resistance in Salmonella and Campylobacter species to antimicrobial agents. Eur J Clin Microbiol Infect Dis. 1999;18:312. doi: 10.1007/s100960050286. [DOI] [PubMed] [Google Scholar]
- 8.Nachamkin I, Engberg J, Aarestrup F M. Diagnosis and antimicrobial susceptibility of Campylobacter spp. In: Nachamkin I, Blaser M J, editors. Campylobacter. 2nd ed. Washington, D.C.: American Society for Microbiology; 2000. pp. 45–66. [Google Scholar]
- 9.Secker D A. Erythromycin resistance only found in Campylobacter coli. J Antimicrob Chemother. 1983;12:414–415. doi: 10.1093/jac/12.4.414. [DOI] [PubMed] [Google Scholar]
- 10.Tajada P, Gomez-Garces J-L, Alós J-I, Balas D, Cogollos R. Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli to 12 β-lactam agents and combinations with β-lactamase inhibitors. Antimicrob Agents Chemother. 1996;40:1924–1925. doi: 10.1128/aac.40.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenover F C, Williams S, Goron K P, Nolan C, Plorde J J. Survey of plasmids and resistance factors in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 1985;27:37–41. doi: 10.1128/aac.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Vliet H M, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 13.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]