Abstract
Variation in the genome region coding for PLAG1 has well-documented associations with skeletal growth and age at puberty in cattle. However, the influence of PLAG1 on other economically important traits such as cow stayability has not yet been explored. Here we investigate the effect of PLAG1 variation on early and later in life female fertility, as well as size and growth, in a well-phenotyped Australian Brahman herd. Yearly pregnancy and productivity records were collected from 2,839 genotyped Brahman cows and used to generate fertility, growth, and weight phenotypes. A variant on chromosome 14 in PLAG1 (NC_037341.1:g.23338890G>T, rs109815800) was previously determined to be a putative causative mutation associated with variation in cattle stature. The imputed PLAG1 genotype at this variant was isolated for each animal and the effect of PLAG1 genotype on each trait was estimated using linear modeling. Regardless of how heifer fertility was measured, there was a significant (P < 0.05) and desirable relationship between the additive effects of PLAG1 genotype and successful heifer fertility. Heifers with two copies of the alternate allele (TT) conceived earlier and had higher pregnancy and calving rates. However, the effects of PLAG1 genotype on fertility began to diminish as cows aged and did not significantly influence stayability at later ages. While there was no effect of genotype on growth, PLAG1 had a negative effect on mature cow weight (P < 0.01), where females with two copies of the alternate allele (TT) were significantly smaller than those with either one or none. Selection emphasis on improved Brahman heifer fertility will likely increase the frequency of the T allele of rs109815800, which may also increase herd profitability and long-term sustainability through improved reproductive efficiency and reduced mature cow size.
Keywords: Brahman, heifer fertility, PLAG1, stayability
An allele of PLAG1 genotype had a significant and positive relationship with Brahman heifer fertility, although the effect on fertility diminished as cows aged. While there was no effect of PLAG1 genotype on growth, there was a significant and negative relationship of the allele improving heifer fertility with mature cow weight.
Introduction
Puberty is the first critical milestone in a beef cow’s reproductive life. Brahman cattle, and other Bos indicus breeds, tend to be older at the onset of puberty than their taurine counterparts. This is speculated to be an adaptive response allowing them to better withstand challenging environments (Rodrigues et al., 2002; Low et al., 2020). The average range that Brahmans reach puberty is reported to span anywhere between 582 d (Plasse et al., 1968) and 751 d (Johnston et al., 2009). In modern beef cattle production, this large range often results in management difficulties and the potential for economic losses. Despite its importance, the underlying genetic causes driving differences in age at puberty are poorly understood.
Age at puberty in Brahman and other Bos indicus-influenced beef females has previously been shown to be highly polygenic. Genome-wide association studies have identified many genes and gene networks that partially explain this variation (Fortes et al., 2010; Fortes et al., 2012; Hawken et al., 2012; Canovas et al., 2014). Variation in the pleomorphic adenoma gene 1 (PLAG1) has been consistently shown to explain a large proportion of variance associated with age at puberty in female Brahman cattle (Hawken et al., 2012; Fortes et al., 2013).
PLAG1 encodes a developmentally regulated zinc finger protein. The PLAG1 gene is a versatile transcription factor known to regulate many genes and pathways, among them, it has been shown to initiate transcription of IGF2 (Voz et al., 2000; Van Dyck et al., 2007) and IGF1R (Van Dyck et al., 2007). Within cattle, PLAG1 appears to be ubiquitously expressed (Nguyen et al., 2017; Li et al., 2020), generally at a very low level within adult animals, but with a significant increase in expression in fetal tissues from a variety of species (Hensen et al., 2004; Li et al., 2020).
PLAG1 variants have also been associated with height or skeletal size in a number of species, including humans, cattle, and mice (Hensen et al., 2004; Karim et al., 2011; Pryce et al., 2011; Fortes et al., 2013; Wood et al., 2014; Utsunomiya et al., 2017; Bouwman et al., 2018). For cattle, Karim et al. (2011) was the first to identify a quantitative trait locus on chromosome 14 with a major effect on bovine stature, identifying 8 possible candidate causal variants within PLAG1. Within that population of Holsteins, the candidate variants were in perfect linkage disequilibrium (LD). Using a multi-breed meta-analysis, Bouwman et al. (2018) was able to break down LD in this region of chromosome 14 and identified the single variant most highly associated with cattle stature, rs109815800 (G>T). Not only was this single-nucleotide polymorphism (SNP) the most highly associated sequence variant with cattle stature identified by Bouwman et al. (2018) but was also the most significant variant associated with cattle height reported by Utsunomiya et al. (2017) and was in perfect LD with the most significant SNP associated with growth and other pleiotropic effects, including age at puberty, reported by Fortes et al. (2013).
While the effect of the PLAG1 mutation has been previously reported for age at puberty and height, the effect on other economically important traits such as cow stayability has not yet been explored. Here we investigate the effect of the PLAG1 mutation on early and later in life female fertility, as well as size and growth, in a well-phenotyped Australian Brahman herd.
Materials and Methods
Animal ethics approval was obtained from the University of Queensland ethics board, animal ethics committee approval number QAAFI/270/17.
Animal and Herd Summary
For this study, a single herd of registered Brahman cattle in Central Queensland, Australia was used. Born between 1995 and 2018, females in this herd have been highly selected for improved fertility and early puberty, with all herd bulls ranking in the top 5% of the Australian Brahman breed for daughter fertility (BREEDPLAN, Agricultural Business Research Institute, Armidale, Australia). Heifers were first exposed to bulls at approximately 1 yr of age, with only a small proportion conceiving as yearlings (≤ 5% annually). All heifers were then exposed to bulls again at 2 yr of age. The breeding season, or joining period, begins on October 1 of each year and spans 4–5 mo, through the summer. To remain in the herd each cow must maintain a yearly calving schedule. All heifers were expected to have produced at least one calf by 3 yr of age, including those that may have first calved at 2 yr of age. Starting in 2013, pregnancy status of each female was determined via manual palpation by a trained technician approximately 5.5 mo after the start of the breeding season, and gestational maturity was recorded as fetal age in weeks.
Phenotypes
Calf performance and cow fertility records were recorded each year as part of normal management practice, with 2,839 individual cow’s records available for this study. These records were used to generate defined heifer, lifetime fertility, and weight phenotypes, along with potential covariates, described in Table 1.
Table 1.
Description of continuous versus binary fertility, growth, and size traits
| Continuous traits | |
|---|---|
| Weeks pregnant (actual) | Fetal age in weeks recorded via manual palpation at pregnancy diagnosis |
| Weeks pregnant (estimated) | A combination of actual fetal age in weeks, with estimated fetal age at the average date of pregnancy diagnosis in heifers born before 2011 |
| Heifer days to calving | Difference in days between the date of bull turn out at the beginning of the breeding season and first calving date |
| Age at first calving | Difference in days between first calving and date of cow’s birth |
| 200-d weight | Heifer weight recorded at approximately weaning |
| 400-d weight | Heifer weight recorded as approximately yearlings |
| 600-d weight | Heifer weight recorded at approximately the start of the breeding season, at ~2 yr of age |
| Mature cow weight (average ≥ 3 yr) | Average of all mature cow weight measurements collected at 3 yr of age or greater |
| Mature cow weight (average ≥ 5 yr) | Average of all mature cow weight measurements collected at 5 yr of age or greater |
| Average daily gain between 200- and 400-d weights |
The difference in weight measured at ~200- and 400-d of age, divided by number of days between measurements |
| Average daily gain between 200- and 600-d weights |
The difference in weight measured at ~200- and 600-d of age, divided by number of days between measurements |
| Average daily gain between 400- and 600-d weights |
The difference in weight measured at ~400- and 600-d of age, divided by number of days between measurements |
|
Binary traits
1 = successful, 0 = unsuccessful | |
|---|---|
| Heifer pregnancy | Ability to successfully conceive prior to 3 yr of age |
| Heifer calving | Ability to successfully calve by 3 yr of age |
| Heifer rebreed | Whether or not a heifer was able to successfully produce a calf the year following her first pregnancy |
| Stayability at 4 yr of age (Stay4) | Ability to produce two calves by 4 yr of age |
| Stayability at 5 yr of age (Stay5) | Ability to produce three calves by 5 yr of age |
| Stayability at 6 yr of age (Stay6) | Ability to produce four calves by 6 yr of age |
A range of fertility traits were measured and recorded in the herd. Heifer pregnancy was recorded as a binary trait based upon whether each heifer was successfully able to conceive prior to 3 yr of age. Pregnancy success was determined at time of yearly pregnancy diagnosis for all heifers born in 2011 and later. For any heifers born prior to 2011, calving records were used to determine pregnancy success. Among those heifers that had both a pregnancy diagnosis and calving record available, only 6% experienced pregnancy loss after pregnancy diagnosis, making calving success a good approximation in cases where pregnancy diagnosis records were unavailable. These records were used to define heifer calving as a separate binary trait based upon a heifer’s ability to give birth to a calf by approximately 3 yr of age. Heifer rebreed was scored as a binary trait based upon whether or not a heifer was able to successfully produce a calf the year following her first pregnancy. Heifers that first calved at 2 yr of age were not considered for this trait due to limited genotyped-phenotyped records (n = 1).
Heifer weeks pregnant, recorded as fetal age, was used as a proxy for heifer maturity, where it was assumed that heifers that were more advanced in gestation at a common time point would have likely been more mature at the start of the breeding season than heifers that were not as advanced in their gestation. Fetal age in weeks was recorded via manual palpation at pregnancy diagnosis for all heifers born in 2011 and later. For any heifers born before this, approximate fetal age was estimated as weeks gestation at the average pregnancy diagnosis date (April 15), given recorded date of calving, and assuming a 290-d gestation length.
Days to calving is a routinely recorded trait in Australian Brahmans and is defined as the number of days between the date of bull turn out at the beginning of the breeding season and calving date. Heifer days to calving was recorded as the number of days between first calving date and date of first bull exposure. Females that did not calve as a heifer, by 3 yr of age, were not considered. Age at first calving was only available for heifers with a recorded birth date and was calculated as the difference in days between first calving date and heifer birth date. Heifers that first calved at approximately 2 yr of age were not considered due to limited genotyped-phenotyped records (n = 38).
Stayability is traditionally defined as a cow’s probability of surviving to a specific age given the opportunity to first reach that age (Hudson and Van Vleck, 1981). Actual herd management practices requires that a cow must have first calved at least once by 3 yr of age and then maintain a yearly calving schedule thereafter. Binary stayability was defined as a cow’s ability to produce 2 calves by 4 years of age (Stay4), her ability to produce 3 calves by 5 years of age (Stay5), and her ability to produce 4 calves by 6 yr of age (Stay6). In this herd, stayability reflects the likelihood that a cow will not be culled for poor reproductive performance prior to a given age.
Heifer weight traits were recorded at industry standard time points: 200 d of age, 400 d of age, and 600 d of age. Birth weights were not recorded, as is common in extensive northern Australia beef cattle operations. Adjusted weights were adjusted to the specified age in days following the procedures recommended by Queensland DPI (2000) and the Beef Improvement Federation (BIF, 2018), using dam age adjustments recommended by the American Brahman Breeders Association (ABBA, 2015), and assuming a standard birthweight of 30 kg. Mature cow weight was, typically, recorded on a yearly basis. On average, Australian Brahman females are expected to have reached their full mature size between 4 and 5 yr of age (Ridley and Schatz, 2006). Due to a relatively low number of records, mature cow weight was averaged following 2 time points, 3 and 5 yr of age. Heifer average daily weight gain was calculated between 200- and 400-d weights, 200- and 600-d weights, and 400- and 600-d weights. Management-based covariates were also recorded, including management cohort (either year of birth or year + month of birth groups), age at joining, and age at trait recording (pregnancy test and weighing).
Genotyping
Genotypes of 2,839 females were generated using the Illumina Geneseek TropBeef V2 array (Neogen, Lincoln, NE). After quality control, with genotypes with QC score < 0.6 set to missing, monomorphic SNP excluded, and SNP with all heterozygous calls excluded, 50,045 SNPs were available. All genotypes were imputed to 709,000 SNPs from the Bovine HD array (following further quality control) using 4,506 cattle genotyped with the Bovine HD array (including a large number of Brahman, Droughtmaster and Santa Gertrudis cattle). Eagle (Loh et al., 2016) was used for phasing, and Minimac3 (Das et al., 2016) was used for imputation. The variant with the most significant effect on height for PLAG1 (NC_037341.1:g.23338890G>T, rs109815800) on chromosome 14 is among those genotyped on the Bovine HD array (Utsunomiya et al., 2017; Bouwman et al., 2018). The imputed PLAG1 genotype at this variant was isolated for each animal, with genotypes called as 0 = homozygous reference allele (GG), 1 = heterozygous (GT), and 2 = homozygous alternative allele (TT).
Model fitting and testing
To assess the relationship between each continuous trait (Table 1, Fig. 1) and the effect of PLAG1 genotype, a linear model was utilized using the lm function in R (version 3.6.2) (R Core Team, 2019). The gene action of PLAG1 was determined by comparing models fitting PLAG1 genotype call as either a continuous variable, assuming additive gene action (Karim et al., 2011), and as a categorical variable, to test for dominance effects (Littlejohn et al., 2012). Two phenotypes, one fertility and one weight trait, were used for this test: actual weeks pregnant (n = 773) and mature cow weight (averaged >3 yr of age) (n = 1047). The model for actual weeks pregnant included the fixed effects of PLAG1 genotype, age at pregnancy diagnosis, and adjusted 600-d weight. The model for mature cow weight included fixed effects of PLAG1 genotype and year of birth group. Least-squares means for each categorical genotype were calculated using the lsmeans package in R (Lenth, 2016), and a Tukey’s pairwise comparison was used to determine differences between allele combinations.
Figure 1.
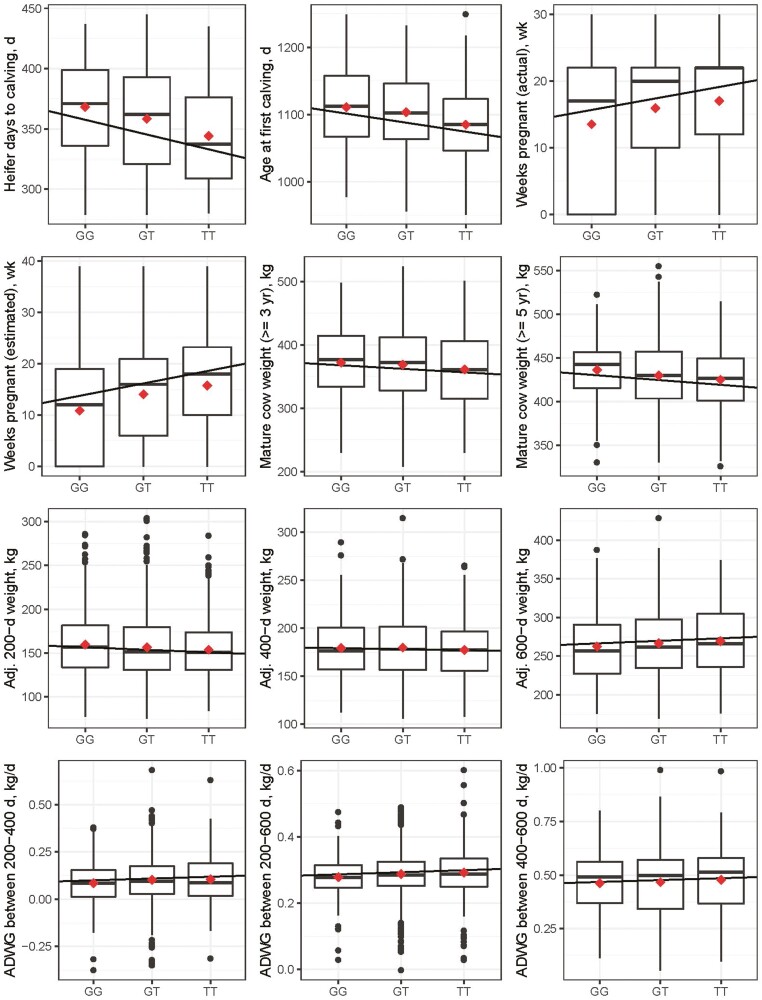
Continuous female fertility, growth, and size traits by PLAG1 rs109815800 genotype. The trait mean is represented by the red dot for each genotype category. To improve interpretation, 200-, 400-, and 600-d weights were adjusted (adj.) to the specified age in days following the procedures recommended by Queensland DPI (2000) using dam age adjustments recommended by ABBA (2015). ADWG = average daily weight gain.
The relationship between the additive effects of PLAG1 genotype call and the continuous traits (Table 1) were tested for significance. For each weight trait, fixed effects included the PLAG1 genotype call (fit as a continuous variable), management cohort (year of birth group), and age at weighing (for 200-, 400-, and 600-d weights only). Adjusted weights were not used as the dependent variable.
For each continuous fertility trait (Table 1), fixed effects included PLAG1 genotype call, fit as a continuous variable, year + month of birth group, either age at joining or age at trait measurement, and adjusted 600-d weight. Management cohort was considered as year + month of birth group, in order to account for the operation’s heifer management practices that grouped heifers by month of birth during the breeding season. Any contemporary groups with fewer than five animals were removed. Sum of squares for each fixed effect was calculated using anova in R (version 3.6.2) (R Core Team, 2019) and used to determine the proportion of phenotypic variance explained by PLAG1 genotype for each trait.
To assess the relationship between each binary fertility trait (Table 1) and the additive effect of PLAG1 genotype, a generalized linear model with a logit link was utilized, using the glm function in R (version 3.6.2) (R Core Team, 2019). Each binary model included the fixed effects of PLAG1 genotype call, fit as a continuous variable, and year + month of birth group, except for Stay6 that fit the effect of management cohort as year of birth group. This was done due to the low number of Stay6 records available for each year. Only contemporary groups containing at least 3 animals of each phenotype (0 and 1) were considered. Odds ratios were calculated from the log-odds estimate for each model, and then converted to a probability scale. Proportion of model deviance explained by each fixed effect was calculated using anova in R (version 3.6.2) (R Core Team, 2019).
Results
There were 2,839 animals with imputed genotypes at the putative causative loci. The PLAG1 rs109815800 SNP was not in Hardy–Weinberg equilibrium (P < 0.00001). Among genotyped females, the reference allele (G) to alternate allele (T) ratio was 0.475/0.525; ~18% were homozygous for the reference allele, 60% heterozygous, and 23% were homozygous for the alternate allele. This suggests that there may be selection preference for the alternate allele within this herd.
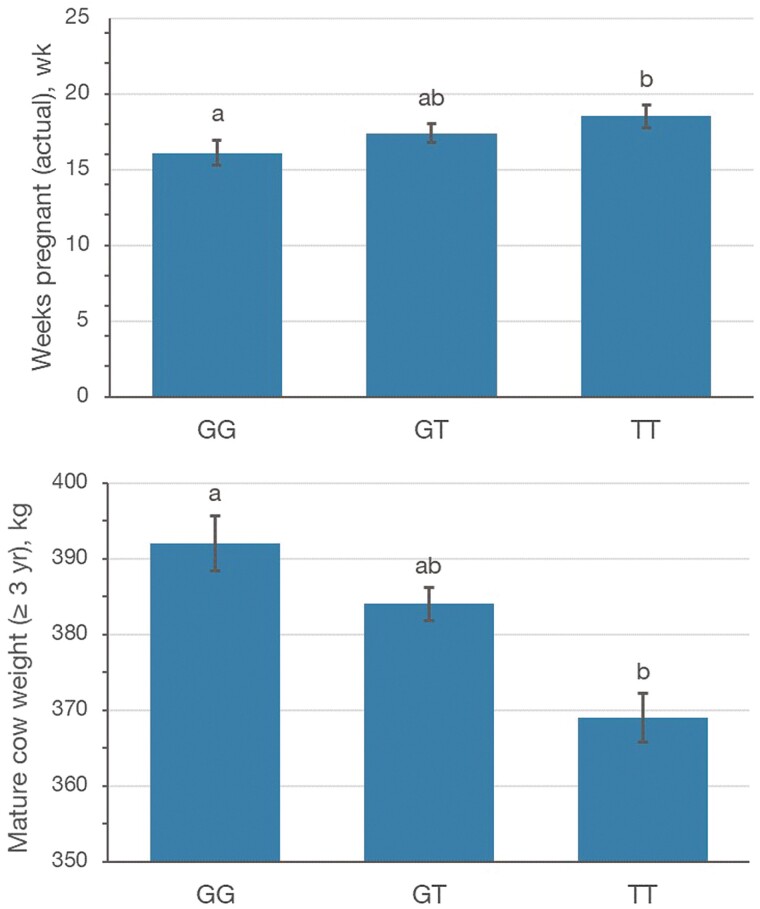
Regardless of how heifer fertility was measured, there was a significant and desirable relationship between the additive effects of PLAG1 genotype and successful heifer fertility (Tables 2 and 3). Heifers with 2 copies of the alternate allele (TT) conceived on average over a week sooner than contemporaries with only 1 copy (GT), and over 2 wk sooner than those with no copies of the alternate allele (GG) (Table 2). The additive effect of PLAG1 genotype significantly increased odds of successfully conceiving and successfully calving before 3 yr of age. Interestingly, among heifers that first calved at 2 yr of age (n = 38), 8% were homozygous for the reference allele, 39% heterozygous, and 53% were homozygous for the alternate allele. Heifers with this genotype also have an increased odds of successfully producing 2 calves by 4 yr of age (Table 3). As the number of alternate alleles in a heifer’s genotype increases, the likelihood of successful heifer pregnancy increased by 70%. There does not appear to be a significant dominance effect of PLAG1 genotype (Fig. 2).
Table 2.
Additive effect of PLAG1 genotype1 on continuous female fertility, growth, and size traits
| Trait | n | Additive effect of PLAG1 genotype 1 | SE | %Var 2 | Mean | SD |
|---|---|---|---|---|---|---|
| Weeks pregnant (actual), wk | 1498 | 1.06* | 0.44 | 0.61 | 15.74 | 9.44 |
| Weeks pregnant (estimated), wk | 2080 | 1.81** | 0.39 | 1.51 | 13.83 | 9.80 |
| Heifer days to calving, d | 1079 | −10.92*** | 1.78 | 1.76 | 356.89 | 42.12 |
| Age at first calving, d | 1110 | −8.90*** | 1.72 | 0.88 | 1100.51 | 57.85 |
| 200-d weight, kg | 2671 | −1.76* | 0.63 | 0.15 | 156.45 | 34.27 |
| 400-d weight, kg | 1781 | −1.28 | 0.86 | 0.08 | 179.21 | 30.97 |
| 600-d weight, kg | 1094 | 0.82 | 1.20 | 0.01 | 266.77 | 44.40 |
| Mature cow weight (≥3 yr), kg | 1201 | −10.88*** | 2.07 | 1.30 | 367.43 | 58.57 |
| Mature cow weight (≥5 yr), kg | 507 | −9.17* | 2.87 | 1.71 | 429.24 | 41.29 |
| ADWG3 between 200 and 400 d, kg/d | 1781 | 0.01 | <0.01 | 0.09 | 0.10 | 0.12 |
| ADWG3 between 200 and 600 d, kg/d | 1071 | <0.01 | <0.01 | 0.06 | 0.29 | 0.07 |
| ADWG3 between 400 and 600 d, kg/d | 982 | <−0.01 | <0.01 | <0.01 | 0.47 | 0.15 |
*P < 0.05; **P < 0.0001; ***P < 1 × 10−6.
PLAG1 rs109815800 genotypes: 0 = homozygous reference allele (GG), 1 = heterozygous (GT), and 2 = homozygous alternative allele (TT).
%Var = percent phenotypic variance explained by PLAG1 genotype.
ADWG = average daily weight gain.
Table 3.
| Trait | n | % Success | Log-odds estimate | SE | %Dev 3 | Odds ratio | 95% CI | Prob | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| Heifer pregnancy | 1388 | 0.71 | 0.53*** | 0.11 | 1.53 | 1.70 | 1.38–2.10 | 0.63 | 0.58–0.68 |
| Heifer calving | 933 | 0.66 | 0.47** | 0.13 | 1.16 | 1.61 | 1.25–2.07 | 0.62 | 0.56–0.67 |
| Heifer rebreed | 596 | 0.66 | 0.11 | 0.14 | — | — | — | — | — |
| Stayability: | |||||||||
| Four years | 734 | 0.46 | 0.39* | 0.13 | 0.95 | 1.48 | 1.15–1.90 | 0.60 | 0.54–0.66 |
| Five years | 379 | 0.38 | 0.32 | 0.19 | — | — | — | — | — |
| Six years | 259 | 0.59 | −0.22 | 0.23 | — | — | — | — | — |
*P < 0.01; **P < 0.001; ***P < 1 × 10−6.
PLAG1 rs109815800 genotypes: 0 = homozygous reference allele (GG), 1 = heterozygous (GT), and 2 = homozygous alternative allele (TT).
All binary traits; 1 = success, 0 = failure.
%Dev = percent model deviance explained by PLAG1 genotype.
Figure 2.
Least-squares means for continuous heifer fertility and weight traits by categorical PLAG1 rs109815800 genotype. (A, B) Significant pairwise differences of at least P < 0.05.
As females aged the impact of PLAG1 genotype on fertility decreased (Table 3). Once a heifer successfully produced her first calf, there was no relationship between her PLAG1 genotype and probability of successfully rebreeding during the subsequent breeding season. For this study, stayability was defined so that it accounted for first time heifer pregnancy success or failure. This is reflected in the significant and positive relationship between PLAG1 genotype and Stay4, but lack of relationship between PLAG1 and rebreed. However, as the influence of heifer pregnancy on a female’s potential to meet a stayability threshold diminished, in the cases of Stay5 and Stay6, so did the relationship between PLAG1 genotype and trait success. This suggests that the influence of PLAG1 is encountered early in a heifer’s life, likely on puberty, and does not directly affect her future reproductive performance.
For this study, mature cow size was assessed at 5 yr of age, and due to a relatively low number of records, also at 3 yr of age. The additive effect of PLAG1 genotype had a negative effect on mature cow weight (P < 0.01), where females with 2 copies of the alternate allele (TT) were significantly smaller than those with either one or none (Table 2). A relationship between PLAG1 genotype and growth rate was not observed, nor was there a relationship with either 400- or 600-d weight (Table 2). There was an additive effect (P < 0.05) of PLAG1 genotype on 200-d weight (Table 2).
Discussion
In this study, the effect of PLAG1 in Australian Brahmans was greatest on puberty and early life traits, with diminishing effects on reproduction as animals aged. Females with 2 copies of the alternate PLAG1 allele (rs109815800; T) were more moderate sized and experienced greater reproductive success as heifers. Females with 2 copies of reference PLAG1 allele (rs109815800; G) appeared to be larger, but significantly less productive as heifers. This pattern concurs with previous reports of this mutation and haplotypes carrying this mutation in Brahman (Fortes et al., 2013), Holstein (Karim et al., 2011; Littlejohn et al., 2012), Japanese Black (Nishimura et al., 2012), and Nellore (Utsunomiya et al., 2017).
PLAG1 was found to significantly influence size but did not have an effect on growth rate. In this Brahman herd the G allele of rs109815800 was associated with increased weaning weight, although this likely a result of the influence PLAG1 has on fetal size and birthweight (Littlejohn et al., 2012). In absence of available birthweight records for this study, this hypothesis cannot be tested. The G allele of rs109815800 was also associated with increased mature size. This is equivalent to previous reports of increased height and weight in Holstein (Karim et al., 2011; Fink et al., 2017). PLAG1 was previously found to not be associated with measures of growth, including feed intake, feed efficiency, residual feed intake, or Kleiber ratio in Holsteins (Fink et al., 2017), but has been associated with both greater feed intake and lower residual feed intake in other Australian Brahman animals (Fortes et al., 2013).
The allele associated with higher fertility (T) was more prevalent in this population of Australian Brahman than other Brahman populations (Fortes et al., 2013; Hayes and Daetwyler, 2019). This is consistent with this population having better fertility than other Australian Brahman herds. The higher proportion of favorable PLAG1 genotypes in this herd may be a causal driver of their increased fertility. Additionally, the PLAG1 allele may be increasing in this population in response to the strong selection pressure being applied, although this cannot be currently tested due to the relatively recent implementation of whole herd genotyping.
Not only is the PLAG1 rs109815800 SNP segregating within Australian Brahman, but it also has a large association with a number of economically important production traits. While the SNP is currently included on Illumina high density cattle SNP arrays, these results suggest it should also be included on lower density SNP panels. This is especially important when genotyping indicine-influenced cattle, and should improve the accuracy of genomic breeding values for female fertility. In particular, for genomic selection schemes that fit SNP effects individually, such as Bayesian models, that can account for SNP with large effect sizes.
Evidence from prior research strongly suggests that the G allele is of taurine origin and was likely introgressed into the Brahman breed through grading-up early in breed development (Fortes et al., 2013; Utsunomiya et al., 2017; Koufariotis et al., 2018). The proportion of taurine influence in the Australian Brahman genome has been estimated between 8.94% (Koufariotis et al., 2018) and 10% (Bolormaa et al., 2011). Koufariotis et al. (2018) identified that the region in the Brahman genome with the most extreme Bos taurus enrichment was on chromosome 14:22-42 Mb (UMD3.1), surrounding PLAG1. This very closely coincides with other reports of a 20 Mb region of depressed heterozygosity containing the taurine G allele of rs109815800 that was found in Brahmans (Fortes et al., 2013). Additionally, Utsunomiya et al. (2017) reported that the haplotype containing the taurine G allele coalesced within Brahmans ~121 yBP, which is consistent with a period of formation and grading-up of the breed (American Brahman Breeders Association).
Interestingly, Koufariotis et al. (2018) observed that prominent, historical Brahman bulls (born 1953-1989) were less likely to have this taurine introgression on chromosome 14 than younger bulls (born 1990–2005). This further suggests that the taurine introgression on chromosome 14 is not only recent but is also under ongoing selection within Australian Brahmans. This is likely driven by increased selection emphasis on size. Average liveweight at maturity and slaughter in Australian beef cattle has increased by approximately 30% from 1976–2018 (Fordyce et al., 2021), and there has been a 76% increase in 600-d weight breeding values from 1999 to 2018 within Australian Brahman (ABBA, 2018). Our results suggest that an unintended consequence of this introgression may have been reduced heifer fertility in Brahmans.
Large mature cow size and female fertility often have an antagonistic relationship, potentially leading to economic losses. Vargas et al. (1999) found that Brahman heifers of large frame size were significantly less reproductively efficient than small frame heifers. Large frame heifers were significantly older at puberty and had significantly reduced calving rates, weaning rates, calf survival rates, and kilogram of calf production per cow than small frame size heifers (Vargas, 1999). Larger cows are generally more expensive, as they consume more feed on an individual basis and in many situations, the marginal increase in calf weaning weights is not adequate to overcome the higher input costs of maintaining large cows (Lalman, 2018). This is particularly important in challenging production environments, such as the tropics and sub-tropics.
In order for cattle production to be sustainable, efficient resource use must balance economic profitability and environmental impact. Reduced inputs related to improved reproductive efficiency and reduced mature cow size associated with the T allele of rs109815800 may increase the sustainability of a herd. Both measures are highly correlated indicator traits for net methane production and will impact net greenhouse gas emissions. The lower the maintenance requirement and smaller the size of the cow, the less methane she will emit (Bell et al., 2012). Cows that have more calves over their lifetime dilute their own methane emissions over more kilograms of beef. As herd fertility improves, either fewer cows can be used to produce the same amount of product or the same number of cows can produce more product without an increase in emissions (Bell et al., 2012; Quinton et al., 2018). Furthermore, the longer the cow remains in the herd, and continues to produce calves, the more she dilutes the methane she emitted as a growing heifer (Quinton et al., 2018). Selection emphasis on improved Brahman heifer fertility is likely to increase the frequency of the T allele of rs109815800, which may also increase herd profitability and long-term sustainability through improved reproductive efficiency and reduced mature cow size.
Acknowledgment
The authors are very grateful to Mr Alf Collins Snr and his family for supplying records from the CBV Brahman herd for this study. They would also like to acknowledge Dr Michael D’Occhio for the helpful conversations and Dr Elizabeth Ross for assistance with editing this document. The authors are grateful for project funding support from the Australian Research Council, project LP160101626.
Glossary
Abbreviations:
- LD
linkage disequilibrium
- SNP
single-nucleotide polymorphism
Conflict of Interest
The authors declare no conflicts of interest.
Literature Cited
- ABBA. 2015. Brahman herd improvement records program manual. Houston, TX: American Brahman Breeders Association,. [Google Scholar]
- ABBA. 2018. Annual report. Rockhampton, Australia: Australian Brahman Breeder’s Association Ltd. [Google Scholar]
- Bell, M. J., Eckard R. J., and Pryce J. E.. . 2012. Breeding dairy cows to reduce greenhouse gas emissions. In: Livestock Production. London: InTechOpen; p. 47–58. doi: 10.5772/50395 [DOI] [Google Scholar]
- BIF. 2018. Guidelines for Uniform Beef Improvement Programs, 9th Rev. Ed. Beef Improvement Federation, Prairie, MS. https://beefimprovement.org/library-2/bif-guidelines [Google Scholar]
- Bolormaa, S., Hayes B. J., Hawken R. J., Zhang Y., Reverter A., and Goddard M. E.. . 2011. Detection of chromosome segments of zebu and taurine origin and their effect on beef production and growth. J. Anim. Sci. 89:2050–2060. doi: 10.2527/jas.2010-3363 [DOI] [PubMed] [Google Scholar]
- Bouwman, A. C., Daetwyler H. D., Chamberlain A. J., Ponce C. H., Sargolzaei M., Schenkel F. S., Sahana G., Govignon-Gion A., Boitard S., Dolezal M., . et al. 2018. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 50:362–367. doi: 10.1038/s41588-018-0056-5 [DOI] [PubMed] [Google Scholar]
- Canovas, A., Reverter A., DeAtley K. L., Ashley R. L., Colgrave M. L., Fortes M. R., Islas-Trejo A., Lehnert S., Porto-Neto L., Rincon G., . et al. 2014. Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS One 9:e102551. doi: 10.1371/journal.pone.0102551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., Forer L., Schönherr S., Sidore C., Locke A. E., Kwong A., Vrieze S. I., Chew E. Y., Levy S., McGue M., . et al. 2016. Next-generation genotype imputation service and methods. Nat. Genet. 48:1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, T., Tiplady K., Lopdell T., Johnson T., Snell R. G., Spelman R. J., Davis S. R., and Littlejohn M. D.. . 2017. Functional confirmation of PLAG1 as the candidate causative gene underlying major pleiotropic effects on body weight and milk characteristics. Sci. Rep. 7:44793. doi: 10.1038/srep44793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce, G., Shephard R., Moravek T., and McGowan M. R.. . 2021. Australian cattle herd: a new perspective on structure, performance and production. Anim Prod Sci. doi: 10.1071/AN20342 [DOI] [Google Scholar]
- Fortes, M. R. S., Kemper K., Sasazaki S., Reverter A., Pryce J. E., Barendse W., Bunch R., McCulloch R., Harrison B., Bolormaa S., . et al. 2013. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian beef cattle. Anim. Genet. 44:636–647. doi: 10.1111/age.12075 [DOI] [PubMed] [Google Scholar]
- Fortes, M. R. S., Lehnert S. A., Bolormaa S., Reich C., Fordyce G., Corbet N. J., Whan V., Hawken R. J., and Reverter A.. . 2012. Finding genes for economically important traits: brahman cattle puberty. Anim Prod Sci. 52:143–150. doi: 10.1071/AN11165 [DOI] [Google Scholar]
- Fortes, M. R. S., Reverter A., Zhang Y., Collis E., Nagaraj S. H., Jonsson N. N., Prayaga K. C., Barris W., and Hawken R. J.. . 2010. Association weight matrix for the genetic dissection of puberty in beef cattle. Proc Natl Acad Sci. USA 107:13642–13647. doi: 10.1073/pnas.1002044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken, R. J., Zhang Y. D., Fortes M. R. S., Collis E., Barris W. C., Corbet N. J., Williams P. J., Fordyce G., Holroyd R. G., Walkley J. R. W., . et al. 2012. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J. Anim. Sci. 90:1398–1410. doi: 10.2527/jas.2011-4410 [DOI] [PubMed] [Google Scholar]
- Hayes, B. J., and Daetwyler H. D.. . 2019. 1000 bull genomes project to map simple and complex genetic traits in cattle: applications and outcomes. Annu. Rev. Anim. Biosci. 7:89–102. doi: 10.1146/annurev-animal-020518-115024 [DOI] [PubMed] [Google Scholar]
- Hensen, K., Braem C., Declercq J., Van Dyck F., Dewerchin M., Fiette L., Denef C., and Van de Ven W. J. M.. . 2004. Targeted disruption of the murine Plag1 proto-oncogene causes growth retardation and reduced fertility. Dev. Growth Differ. 46:459–470. doi: 10.1111/j.1440-169x.2004.00762.x [DOI] [PubMed] [Google Scholar]
- Hudson, G. F. S., and Van Vleck L. D.. . 1981. Relationship between production and stayability in holstein cattle. J. Dairy Sci. 64:2246–2250. doi: 10.3168/jds.S0022-0302(81)82836-6 [DOI] [Google Scholar]
- Johnston, D. J., Barwick S. A., Corbet N. J., Fordyce G., Holroyd R. G., Williams P. J., and Burrow H. M.. . 2009. Genetics of heifer puberty in two tropical beef genotypes in northern australia and associations with heifer- and steer-production traits. Anim Prod Sci. 49:399–412. doi: 10.1071/EA08276 [DOI] [Google Scholar]
- Karim, L., Takeda H., Lin L., Druet T., Arias J. A. C., Baurain D., Cambisano N., Davis S. R., Farnir F., Grisart B., . et al. 2011. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43:405–413. doi: 10.1038/ng.814 [DOI] [PubMed] [Google Scholar]
- Koufariotis, L., Hayes B. J., Kelly M., Burns B. M., Lyons R., Stothard P., Chamberlain A. J., and Moore S.. . 2018. Sequencing the mosaic genome of brahman cattle identifies historic and recent introgression including polled. Sci. Rep. 8:17761. doi: 10.1038/s41598-018-35698-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalman, D. L., Wiseman A., and DeVuyst E.. . 2018. Implications of cow size change. In: Proc. of 67th Florida Beef Cattle Short Course, Gainsville, FL. p 45–50.
- Lenth, R. V. 2016. Least-squares means: the R package lsmeans. J Stat Softw. 69:33. doi: 10.18637/jss.v069.i01 [DOI] [Google Scholar]
- Li, Z., Wu M., Zhao H., Fan L., Zhang Y., Yuan T., He S., Wang P., Zhang Y., Sun X., . et al. 2020. The PLAG1 mRNA expression analysis among genetic variants and relevance to growth traits in chinese cattle. Anim. Biotechnol. 31:504–511. doi: 10.1080/10495398.2019.1632207 [DOI] [PubMed] [Google Scholar]
- Littlejohn, M., Grala T., Sanders K., Walker C., Waghorn G., Macdonald K., Coppieters W., Georges M., Spelman R., Hillerton E., . et al. 2012. Genetic variation in PLAG1 associates with early life body weight and peripubertal weight and growth in Bos taurus. Anim. Genet. 43:591–594. doi: 10.1111/j.1365-2052.2011.02293.x [DOI] [PubMed] [Google Scholar]
- Loh, P., Danecek P., Palamara P. F., Fuchsberger C., Reshef Y. A., Finucane H. K., Schoenherr S., Forer L., McCarthy S., Abecasis G. R., . et al. 2016. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 48:1443. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, W. Y., Tearle R., Liu R., Koren S., Rhie A., Bickhart D. M., Rosen B. D., Kronenberg Z. N., Kingan S. B., Tseng E., . et al. 2020. Haplotype-resolved genomes provide insights into structural variation and gene content in angus and brahman cattle. Nat. Commun. 11:2071. doi: 10.1038/s41467-020-15848-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L. T., Reverter A., Cánovas A., Venus B., Islas-Trejo A., Porto-Neto L. R., Lehnert S. A., Medrano J. F., Moore S. S., and Fortes M. R. S.. . 2017. Global differential gene expression in the pituitary gland and the ovaries of pre- and postpubertal brahman heifers. J. Anim. Sci. 95:599–615. doi: 10.2527/jas.2016.0921 [DOI] [PubMed] [Google Scholar]
- Nishimura, S., Watanabe T., Mizoshita K., Tatsuda K., Fujita T., Watanabe N., Sugimoto Y., and Takasuga A.. . 2012. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in japanese black cattle. BMC Genet. 13:40. doi: 10.1186/1471-2156-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasse, D., Warnick A. C., and Koger M.. . 1968. Reproductive behavior of Bos indicus females in a subtropical environment. I. Puberty and ovulation frequency in brahman and brahman X british heifers. J. Anim. Sci. 27:94–100. doi: 10.2527/jas1968.27194x [DOI] [PubMed] [Google Scholar]
- Pryce, J. E., Hayes B. J., Bolormaa S., and Goddard M. E.. . 2011. Polymorphic regions affecting human height also control stature in cattle. Genetics. 187:981–984. doi: 10.1534/genetics.110.123943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queensland DPI. 2000. Beef cattle recording and selection. Brisbane, Australia: . [Google Scholar]
- Quinton, C. D., Hely F. S., Amer P. R., Byrne T. J., and Cromie A. R.. . 2018. Prediction of effects of beef selection indexes on greenhouse gas emissions. Animal. 12:889–897. doi: 10.1017/S1751731117002373 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ridley, P. E. R., and Schatz T.. . 2006. Meeting post weaning market specifications for the live cattle export trade to South East Asia. Meat & Livestock Australia Ltd, Sydney, Australia. [Google Scholar]
- Rodrigues, H. D., Kinder J. E., and Fitzpatrick L. A.. . 2002. Estradiol regulation of luteinizing hormone secretion in heifers of two breed types that reach puberty at different ages. Biol. Reprod. 66:603–609. doi: 10.1095/biolreprod66.3.603 [DOI] [PubMed] [Google Scholar]
- Utsunomiya, Y. T., Milanesi M., Utsunomiya A. T. H., Torrecilha R. B. P., Kim E. -S., Costa M. S., Aguiar T. S., Schroeder S., do Carmo A. S., Carvalheiro R., . et al. 2017. A PLAG1 mutation contributed to stature recovery in modern cattle. Sci. Rep. 7:17140. doi: 10.1038/s41598-017-17127-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck, F., Declercq J., Braem C. V., and Van de Ven W. J. M.. . 2007. PLAG1, The prototype of the PLAG gene family: versatility in tumour development (Review). Int. J. Oncol. 30:765–774. doi: 10.3892/ijo.30.4.765 [DOI] [PubMed] [Google Scholar]
- Vargas, C. A., Olson T. A., C. C.Chase, Jr, Hammond A. C., and Elzo M. A.. . 1999. Influence of frame size and body condition score on performance of brahman cattle. J. Anim. Sci. 77:3140–3149. doi: 10.2527/1999.77123140x [DOI] [PubMed] [Google Scholar]
- Voz, M. L., Agten N. S., Van de Ven W. J. M., and Kas K.. . 2000. PLAG1, The main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 60:106–113. [PubMed] [Google Scholar]
- Wood, A. R., Esko T., Yang J., Vedantam S., Pers T. H., Gustafsson S., Chu A. Y., Estrada K., Luan J. A., Kutalik Z., . et al. 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46:1173–1186. doi: 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]