Abstract
We have identified and characterized a potent new nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) of human immunodeficiency virus type 1 (HIV-1) that also is active against HIV-2 and which interferes with virus replication by two distinct mechanisms. 1-(3-Cyclopenten-1-yl)methyl-6-(3,5-dimethylbenzoyl)-5-ethyl-2,4-pyrimidinedione (SJ-3366) inhibits HIV-1 replication at concentrations of approximately 1 nM, with a therapeutic index of greater than 4 × 106. The efficacy and toxicity of SJ-3366 are consistent when evaluated with established or fresh human cells, and the compound is equipotent against all strains of HIV-1 evaluated, including syncytium-inducing, non-syncytium-inducing, monocyte/macrophage-tropic, and subtype virus strains. Distinct from other members of the pharmacologic class of NNRTIs, SJ-3366 inhibited laboratory and clinical strains of HIV-2 at a concentration of approximately 150 nM, yielding a therapeutic index of approximately 20,000. Like most NNRTIs, the compound was less active when challenged with HIV-1 strains possessing the Y181C, K103N, and Y188C amino acid changes in the RT and selected for a virus with a Y181C amino acid change in the RT after five tissue culture passages in the presence of the compound. In combination anti-HIV assays with nucleoside and nonnucleoside RT and protease inhibitors, additive interactions occurred with all compounds tested with the exception of dideoxyinosine, with which a synergistic interaction was found. Biochemically, SJ-3366 exhibited a Ki value of 3.2 nM, with a mixed mechanism of inhibition against HIV-1 RT, but it did not inhibit HIV-2 RT. SJ-3366 also inhibited the entry of both HIV-1 and HIV-2 into target cells. On the basis of its therapeutic index and multiple mechanisms of anti-HIV action, SJ-3366 represents an exciting new compound for use in HIV-infected individuals.
The structurally diverse class of nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) includes compounds which are among the most potent anti-human immunodeficiency virus (anti-HIV) agents identified (for reviews, see references 18 to 20). The therapeutic utility of these anti-HIV compounds, however, is severely compromised by the rapid appearance of drug-resistant virus isolates in patients (33) and dose-limiting toxic effects, such as macropapular rashes (26). Similarly, the growth of HIV in cell culture in the presence of the NNRTIs yields rapid selection of drug-resistant viruses (33). The high degree of specificity of the interaction of these compounds at the hydrophobic nonnucleoside binding site on the HIV type 1 (HIV-1) RT results in the ability of single amino acid changes in the NNRTI binding pocket to reduce or eliminate the inhibitory activity of the compound (14, 15, 24, 38). Amino acid changes in the RT which affect the efficacies of the NNRTIs include A98G, L100I, K101E, K103N, V106A, V108I, E138K, T139I, Y181C, Y188C, G190A, F227L, and P236L (33).
The effective use of NNRTIs in patients is dependent on defining appropriate combinations of agents which will prevent or retard the selection of drug-resistant viruses or which will result in the selection of drug-resistant virus isolates in which mutation of critical amino acid residues renders the RT less fit to support virus reproduction (22, 23, 30, 35). NNRTIs may also be useful as part of a combination anti-HIV strategy with a highly potent NNRTI and additional anti-HIV-1 agents in therapy-naive patients. The potential for the therapeutic use of the NNRTIs in patients has recently been reviewed (19, 20). Clinical results reported for nevirapine as a component of a three-drug regimen in patients has highlighted the possible benefits from the development of additional novel or more potent NNRTIs (13). Although the use of NNRTIs alone is not warranted, other possible strategies include the use of these compounds as topical microbicides to prevent the sexual transmission of HIV, for postexposure prophylaxis, or as a first-line therapeutic option for the treatment of patients without eliminating future therapy options. Recently, a new and potentially exciting role for the class of NNRTIs has been defined: the reported therapeutic potential of nevirapine to prevent the neonatal transmission of HIV (25).
A variety of structurally distinct NNRTIs have been identified (4, 5, 20, 21), and medicinal chemistry efforts have continued in an effort to identify the structural features of the NNRTIs responsible for anti-HIV activity in order to select for a new generation of compounds with improved pharmacologic and antiviral properties. Our investigations with the NNRTIs have also focused on means of retarding or inhibiting the selection of drug-resistant viruses by defining the sensitivities of resistant virus and purified RT to the compounds and selecting potentially effective NNRTI combinations (7, 9, 10, 16, 28, 29, 32, 41). The 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT)-type NNRTIs were among the first NNRTIs to be discovered and evaluated (1–3, 9, 17, 27, 31, 34, 39). SJ-3366 was identified through synthetic efforts based on modification of the N-1 homocyclic moiety of the HEPT analogs and has resulted in a series of compounds with therapeutic indices that reach 106 (H. S. Kwon, S. H. Lee, J. W. Lee, D. W. Kang, S. G. Chung, E. H. Cho, J. A. Turpin, T. L. Stup, and R. W. Buckheit, Jr., unpublished data). Distinct from all of the other reported NNRTIs, the SJ compounds have significant activity against HIV-2, with therapeutic indices in the range of 10,000 to 50,000. Data presented herein indicate that SJ-3366 interferes with viral replication by multiple mechanisms, and its significant potency alone and in combination with other active anti-HIV agents, especially against drug-resistant virus strains, suggests that this compound may have significant therapeutic potential in HIV-infected patients.
MATERIALS AND METHODS
Cells and viruses.
The established human cells, laboratory-derived virus isolates (including drug-resistant virus isolates), and low-passage clinical virus isolates used in these evaluations have previously been described in detail (10, 12). These cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Fresh human cells were obtained from the American Red Cross (Baltimore, Md.). The low-passage clinical strains of HIV-2 were obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases.
Materials.
1-(3-Cyclopenten-1-yl)methyl-6-(3,5-dimethylbenzoyl)-5-ethyl-2,4-pyrimidinedione (SJ-3366) was synthesized as described previously (Kwon et al., unpublished data), and the structure of the compound is provided in Fig. 1. Crystalline stock materials were stored at −70°C and solubilized in 100% dimethyl sulfoxide. All stocks were diluted at least 400-fold prior to performance of drug susceptibility assays. The compounds used in combination assays included zidovudine (AZT), dideoxyinosine (ddI), lamivudine (3TC), nevirapine, ritonavir, indinavir, and nelfinavir. Enzyme-linked immunosorbent assay (ELISA) plates were purchased from Coulter Immunotech (Hialeah, Fla.). Materials required for the performance of RT inhibition assays and anti-HIV assays and for the growth and maintenance of established and fresh human cells have been described previously (6, 10).
FIG. 1.
Structure of SJ-3366.
Antiviral and cross-resistance assays.
The HIV inhibitory activities of the compounds were evaluated as described previously (10) by microtiter anti-HIV assays with CEM-SS cells or fresh human peripheral blood mononuclear cells (PBMCs), which quantify the ability of a compound to inhibit HIV-induced cell killing or HIV replication. Quantification was performed with the tetrazolium dye 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2M-tetrazolium-5-carboxanilide (XTT), which is metabolized to a colored formazan product by viable cells, RT, and/or p24 ELISA. Antiviral and toxicity data are reported as the quantity of drug required to inhibit 50% of virus-induced cell killing or virus production (IC50) and the quantity of drug required to reduce cell viability by 50% (TC50).
Combination anti-HIV assays.
Analysis of drug combinations was performed with CEM-SS cells acutely infected with the IIIB strain of HIV-1 as described previously (10) by the anti-HIV assay methodology described above. Statistical evaluations were performed with MacSynergy II software (37). The results of the drug combination assays are presented three dimensionally for each combination concentration, yielding a surface of activity that extends above (synergy) or below (antagonism) the plane of additive interaction. The volume of the surface is calculated and expressed as a synergy volume (micromolar squared percent) calculated at the 95% confidence interval (37). For these studies, synergy is defined as drug combinations that yield synergy volumes greater than 50 μM2%. Slightly synergistic and highly synergistic anti-HIV activity have been defined as yielding synergy volumes of 50 to 100 and >100 μM2%, respectively. Synergy volumes between 0 and 50 μM2% are considered additive, and synergy volumes less than 0 μM2% are considered antagonistic.
Selection of drug-resistant strains.
Resistant virus isolates were selected in cell culture by serial passage of the IIIB strain of HIV-1 in CEM-SS cells in the presence of increasing concentrations of antiviral compound. The initial selection was performed with a drug concentration of two times the IC50 of the compound as determined by the microtiter anti-HIV assay. With successive passages the drug concentration was increased twofold to enhance the selective pressure on the virus. Upon selection of a drug-resistant virus isolate, cross-resistance testing was performed by the methods described for the performance of antiviral assays. Resistance has been defined in this study as a greater than fivefold increase in the IC50 compared to the activity of the compound against the wild-type (IIIB) isolate.
Analysis of RT mutations.
Resistance-engendering mutations were identified by the direct sequencing of PCR products amplified from the RT region of proviral DNA obtained from acutely infected CEM-SS cells. A PCR-amplified product was prepared from the first 750 bp of the RT gene with the A′-NE1′ primer set. Single-stranded, biotinylated DNA was purified from this product with avidin-conjugated supraparamagnetic beads (Dynabeads; M280; Dynal). Direct sequencing by dideoxynucleotide chain termination was performed with each of the appropriate G, A, C, and T dideoxy sequencing mixes by using the Sequenase T7 polymerase kit (U.S. Biochemicals) with 7-deaza-dGTP to resolve compression artifacts, α-33P, and five sets of overlapping primers obtained with a primer analysis software package (Oligo 4.04; National Bioscience, Inc.). Evaluation of the resulting sequencing gels was accomplished with Millipore's automated gel scanning system. RT sequences from drug-resistant isolates were aligned with the parental wild-type HIV-1 strain IIIB and strain HXB2 RTs by using Millipore/Bioimage software run on a Sun Microsystem's Sparc 10 Station microcomputer.
RT inhibition assays.
Analysis of the drug sensitivity of RT containing defined amino acid substitutions was performed as described previously (6). Evaluation of the activities of compounds by using homopolymer and heteropolymer templates was performed as described previously (8). For the Ki studies, HIV-1 RT activity was measured in 50-μl reaction mixtures containing 50 mM Tris (pH 8.0), 50 mM KCl, 10 mM MgCl2, 4 mM β-mercaptoethanol, 3% glycerol, 1 mg of bovine serum albumin per ml, 6.6 μg of primed 16S rRNA from Escherichia coli per ml, 10 μM dATP, 10 μM dCTP, 10 μM dGTP, and various concentrations of [3H]dTTP (36). Purified recombinant HIV-1BH10 RT was used for these experiments (40). The Km for dTTP was 1.67 μM, and that for the rRNA template was 0.66 μg/ml.
Binding and fusion inhibition assays.
HeLa–CD4–LTR–β-galactosidase cells, which use a Tat protein-induced transactivation of the β-galactosidase gene driven by the HIV-1 long terminal repeat (LTR) promoter, were used to quantify both the binding of infectious virus to cells and cell-cell fusion events. Both of these assays have been described previously (11). Infected cells form syncytia that can be easily counted microscopically after incubation with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The HIV binding inhibition assay involved plating of 104 HeLa–CD4–LTR–β-galactosidase cells in a 200-μl volume in flat-bottom, 96-well microtiter plates. The cells were incubated overnight, and the medium was removed and replaced with 100 μl of various concentrations of SJ-3366 or control compound. One hour later 100 μl of virus-containing medium was added to each well. Cells were incubated for an additional hour, and the monolayer was washed extensively to remove unbound virus and extracellular compound. At 48 h, the cells were fixed and stained with X-Gal. Blue multinuclear cells were then counted under an inverted microscope. The cell-cell fusion inhibition assay was also performed in flat-bottom, 96-well microtiter plates. HeLa–CD4–LTR–β-galactosidase cells (5 × 103) were added to each well, and the cells were incubated with test compound for 1 h prior to the addition of 5 × 103 HL2/3 cells. The cells were incubated for an additional 48 h and fixed and stained with X-Gal. Blue syncytia were counted microscopically. Staining of the cells was performed by fixing the cells with a solution of 1% formaldehyde–0.2% glutaraldehyde and staining the fixed cells with 4 μM potassium ferrocyanide, 4 μM potassium ferricyanide, 2 μM MgCl2, and 0.4% X-Gal in phosphate-buffered saline. Transactivation of β-galactosidase expression was also monitored by ELISA (5 Prime-3 Prime, Boulder, Colo.). Cell extracts were prepared by freezing-thawing and assayed for β-galactosidase activity according to the manufacturer's recommendations. The results of the ELISA were quantified spectrophotometrically at 405 nm with a Molecular Devices Vmax microtiter plate reader.
RESULTS
Efficacy and toxicity of SJ-3366 against HIV-1.
The activity of SJ-3366 was evaluated in established and fresh human cells infected with both laboratory-derived and clinical strains of HIV-1, HIV-2, and simian immunodeficiency virus (SIV). Nevirapine (an NNRTI) and AZT (a nucleoside RT inhibitor [NRTI]) were used as positive anti-HIV control compounds and were evaluated in parallel. These data are summarized in Table 1. SJ-3366 was determined to be highly active against HIV-1 in the T cells CEM-SS, H9, and MT2, the B-cell line AA5, the monocytic line U937, and the T-cell–B-cell hybrid line 174×CEM. The activities (IC50s) of the compounds against HIV-1 ranged from 0.0009 to 0.01 μM. SJ-3366 was nontoxic to all cells at concentrations up to 1,135 μM, at which point compound precipitation in cell culture eliminated the ability to measure toxicity. Under highly optimized assay conditions in CEM-SS cells infected with the IIIB strain of HIV-1, the efficacy and toxicity values obtained for SJ-3366 yield a therapeutic index of approximately 4 × 106. SJ-3366 initially precipitated in the test wells at approximately 125 to 250 μM, but cell viability was not affected until the higher test doses (1,135 μM). AZT and nevirapine exhibited the expected levels of activity in each of the cell lines.
TABLE 1.
Range of anti-HIV activity of SJ-3366 in established human cell lines
| Virus (isolate) | CELLb | IC50
(μM)a
|
||
|---|---|---|---|---|
| SJ-3366 | Nevirapine | AZT | ||
| HIV-1 (IIIB) | CEM-SS | 0.0009 | 0.1 | 0.003 |
| HIV-1 (RF) | CEM-SS | 0.001 | 0.2 | 0.004 |
| HIV-1 (SK1) | CEM-SS | 0.002 | 0.2 | 0.005 |
| HIV-1 (IIIB) | 174×CEM | 0.003 | 0.1 | 0.03 |
| HIV-1 (IIIB) | MT2 | 0.001 | 0.1 | 0.04 |
| HIV-1 (IIIB) | U937 | 0.01 | 0.3 | 0.003 |
| HIV-1 (IIIB) | AA5 | 0.009 | 0.7 | 0.01 |
| HIV-2 (ROD) | CEM-SS | 0.17 | >38 | 0.005 |
| SIV (DeltaB670) | 174×CEM | >100 | >38 | 0.02 |
The results presented were obtained from a single representative antiviral assay, with appropriate nevirapine and AZT control values selected from a minimum of two antiviral assays. We have demonstrated that the standard error between multiple antiviral assays averaged less than 10% of the respective mean IC50.
Cellular phenotypes: CEM-SS, T cell; 174×CEM, T-cell–B-cell fusion; U937, macrophage; AA5, Epstein-Barr virus-infected B-cell.
SJ-3366 was also evaluated in fresh human peripheral blood leukocytes and monocytes/macrophages infected with a variety of low-passage clinical virus isolates (Table 2). The compound was determined to be equally active against clinical virus strains, including viruses representative of the various HIV-1 subtypes (subtypes A through F) found worldwide, syncytium-inducing and non-syncytium-inducing viruses, and T-tropic and monocyte/macrophage-tropic viruses. The activity of SJ-3366 ranged from 0.0002 to 0.003 μM in fresh human PBMCs and was approximately 0.003 μM in fresh monocytes/macrophages infected with clinical strains of HIV-1. The toxicity of SJ-3366 in fresh human cells was also determined to be similar to that observed in the established cell lines (>1,000 μM). The average therapeutic index determined for SJ-3366 in the fresh human cell assays was approximately 106.
TABLE 2.
Range of anti-HIV activity of SJ-3366 in fresh human cells
| Virus | Isolate | Cellb | IC50
(μM)a
|
||
|---|---|---|---|---|---|
| SJ-3366 | Nevirapine | AZT | |||
| HIV-1 | WEJO | PBMC | 0.0002 | 0.06 | 0.02 |
| HIV-1 | ROJO | PBMC | 0.008 | 0.3 | 0.01 |
| HIV-1 | TEKI | PBMC | 0.001 | 0.1 | 0.05 |
| HIV-1 | SLKA | PBMC | 0.0008 | 0.2 | 0.02 |
| HIV-1 | Subtype A | PBMC | 0.0014 | 0.06 | 0.008 |
| HIV-1 | Subtype B | PBMC | 0.0006 | 0.07 | 0.01 |
| HIV-1 | Subtype C | PBMC | 0.002 | 0.05 | 0.007 |
| HIV-1 | Subtype D | PBMC | 0.0008 | 0.1 | 0.01 |
| HIV-1 | Subtype E | PBMC | 0.0008 | 0.007 | 0.01 |
| HIV-1 | Subtype F | PBMC | 0.003 | 0.2 | 0.05 |
| HIV-1 | BaL | MM | 0.0003 | 0.4 | 0.03 |
| HIV-1 | ADA | MM | 0.0003 | 0.2 | 0.01 |
| HIV-2 | CDC310342 | PBMC | 0.003 | NDc | 0.006 |
| HIV-2 | CDC310319 | PBMC | 0.04 | ND | 0.003 |
| HIV-2 | CBL20 | PBMC | 0.48 | ND | 0.002 |
The results presented were obtained from a single representative antiviral assay, with appropriate nevirapine and AZT control values selected from a minimum of two antiviral assays. We have demonstrated that the standard error between multiple antiviral assays averaged less than 10% of the respective mean IC50.
Cellular phenotypes: PBMC, fresh PBMCs; MM, fresh peripheral blood monocytes.
ND, not determined.
The effect of serum proteins was evaluated by adding human serum albumin (HSA), human type AB serum, bovine serum albumin, and/or alpha-acidic glycoprotein (AAGP) to our standard anti-HIV assays. As can be seen in Table 3, the addition of these proteins had little effect on the potency of SJ-3366 against HIV-1. A 10-fold loss of efficacy was observed in cultures which contained HSA and AAGP.
TABLE 3.
Effect of serum proteins on efficacy of SJ-3366 against HIV-1 and HIV-2
| Assaya | IC50
(μM)b
|
||
|---|---|---|---|
| SJ-3366 versus HIV-1 | SJ-3366 versus HIV-2 | AZT versus HIV-1/HIV-2 | |
| No additives | 0.002 | 0.0004 | 0.01/0.02 |
| SJ-3366 + human type AB serum | 0.001 | 0.001 | 0.06/0.05 |
| SJ-3366 + AAGP | 0.002 | 0.001 | 0.06/0.06 |
| SJ-3366 + BSA | 0.003 | 0.002 | 0.09/0.04 |
| SJ-3366 + HSA + AAGP | 0.020 | NDc | 0.02/ND |
Human type AB serum at 50%, AAGP at 1 mg/ml, bovine serum albumin (BSA) at 20 mg/ml, and HSA at 40 mg/ml.
The results presented were obtained from a single representative antiviral assay selected from a minimum of two antiviral assays. We have demonstrated that the standard error between multiple antiviral assays averaged less than 10% of the respective mean IC50.
ND, not determined.
Efficacy and toxicity of SJ-3366 against HIV-2 and SIV.
In addition to its wide therapeutic index against HIV-1 strains, SJ-3366 also was determined to be active in cell-based assays against the ROD strain of HIV-2 (Table 1). Although the activity against HIV-2 was approximately 200-fold less than that observed against HIV-1, SJ-3366 still exhibited a therapeutic index of >20,000 against HIV-2 (IC50, 0.17 μM; TC50, >1,135 μM). Additional assays were performed with low-passage clinical strains of HIV-2 (Table 2), yielding IC50s that ranged from 0.003 to 0.5 μM. The specific activity of SJ-3366 against HIV-2 is one of the features that distinguishes this compound from the remaining members of the NNRTI class of compounds. Similar to the results obtained against HIV-1, the addition of serum proteins had little effect on the activity of SJ-3366 against HIV-2 (Table 3). The addition of HSA and AAGP to in vitro assays resulted in a fivefold loss of activity of SJ-3366 against HIV-2 in CEM-SS cell culture. SJ-3366 was not active against SIV at concentrations up to 100 μM (Table 1).
Mechanism of antiviral activity.
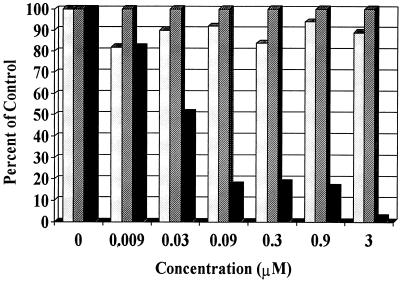
Mechanistic assays with SJ-3366 indicated that the compound inhibited HIV-1 RT when evaluated in a biochemical RT inhibition assay with either a homopolymeric poly(rC)-oligo(dG) or a heteropolymeric rRNA template-primer assay system. SJ-3366 exhibited Ki values of 2.7 and 3.8 nM in replicate assays with a mixed mechanism of RT inhibition (both the Km and the Vmax values were affected by the compound). In these assays, SJ-3366 specifically inhibited HIV-1 RT but was inactive against HIV-2 RT. These inhibitory values were similar to the effective concentration of SJ-3366 in cell culture against HIV-1. In addition to its ability to inhibit RT, SJ-3366 inhibited virus attachment in assays with HeLa–LTR–β-galactosidase cells when assayed with either HIV-1 or HIV-2 (Fig. 2). The effective concentrations for inhibition of virus attachment (IC50s, 100 to 200 nM) were similar to the inhibitory concentrations obtained for cell-based assays with HIV-2. SJ-3366 did not inhibit the enzymatic activity of HIV-1 integrase, protease, or RNase H (data not shown) and also did not inhibit cell-cell fusion at concentrations up to 100 μM (Fig. 2). On the basis of its inability to suppress virus production in chronically infected cells, SJ-3366 did not inhibit late-stage virus reproduction events (data not shown). Limited pretreatment assays, in which SJ-3366 was removed by extensive washing following an overnight culture with target cells, demonstrated that the compound had to be continuously present in order to be effective.
FIG. 2.
Inhibition of virus attachment and cell-cell fusion quantified as described in Materials and Methods. ░⃞, fusion; ▨, toxicity; █, attachment.
Interaction of SJ-3366 combined with other anti-HIV agents.
Anti-HIV assays were performed with SJ-3366 combined with a variety of anti-HIV agents including the NRTIs AZT, ddI, and 3TC, the NNRTI nevirapine, and the protease inhibitors ritonavir, indinavir, and nelfinavir. A summary of the combination anti-HIV data obtained from the MacSynergy II evaluations is presented in Table 4. Data obtained from the MacSynergy II evaluations are presented in synergy volume units (micromolar squared percent) at the 95% confidence interval, as described in Materials and Methods. Unlike the majority of the NNRTIs that we have evaluated, SJ-3366 primarily exhibited additive interactions with other active anti-HIV agents. Most particularly, little synergy was observed when SJ-3366 was tested in combination with other NRTIs and NNRTIs. Synergy was observed with the combination of SJ-3366 with the nucleoside analog ddI (mean synergy volume, 129 μM2%) but not with any of the other RT inhibitors tested. Similarly, additive interactions were also observed with three protease inhibitors, although slightly synergistic inhibition of HIV-1 was apparently achieved with SJ-3366 and nelfinavir (mean synergy volume, 63.5 μM2%). Antagonistic anti-HIV interactions or synergistic toxicity was not observed with any of the drug combinations evaluated.
TABLE 4.
Activity of SJ-3366 in combination with other anti-HIV agents
| Drug used in combination with SJ-3366 (mechanism of action) | Synergy vol (μM2%)a | Antiviral interaction |
|---|---|---|
| AZT (NRTI) | 18 ± 2 | Additive |
| ddI (NRTI) | 129 ± 19 | Synergistic |
| 3TC (NRTI) | 13.5 ± 5.5 | Additive |
| Nevirapine (NNRTI) | 28 ± 22 | Additive |
| Ritonavir (PIb) | 34.5 ± 4.5 | Additive |
| Indinavir (PI) | 43.5 ± 3.5 | Additive |
| Nelfinavir (PI) | 68.5 ± 27.5 | Slightly synergistic |
Mean synergy volume calculated from two replicate combination anti-HIV assays. Synergy volumes were calculated by the Prichard and Shipman (37) MacSynergy II program at the 95% confidence interval.
PI, protease inhibitor.
Sensitivities of NNRTI-resistant viruses to SJ-3366.
SJ-3366 and the positive control compounds nevirapine and AZT were evaluated for their activities against viruses which had been selected in cell culture for resistance to a variety of NNRTIs (Table 5). In this study, viruses with amino acid changes L100I, T139I, and M184V remained completely sensitive to SJ-3366. Slight reductions in efficacy were observed when SJ-3366 was tested against viruses with amino acid changes K101E (18-fold) or V108I (18-fold). Greater levels of resistance were observed when SJ-3366 was tested against viruses with the amino acid changes A98G plus V108I (70-fold), P236L (33-fold), Y188H (55-fold), K103N (>50-fold), and Y181C (10- to 375-fold). The various results obtained with several strains of virus that possess the Y181C amino acid change in the RT suggests that other phenotypic or genotypic properties of the resistant virus may play a role in the overall level of resistance achieved. In addition, the sensitivities of these viruses to SJ-3366 are distinct from the sensitivity patterns observed with nevirapine. These data also suggest that resistance to SJ-3366 results in only minor changes (10- to 400-fold) in therapeutic indices relative to the therapeutic index of the compound against wild-type virus (>4 × 106).
TABLE 5.
Sensitivity of viruses resistant to HIV-1-specific inhibitors to SJ-3366
| Resistant isolate (mutation)b | IC50 (μM
[fold resistance])a
|
||
|---|---|---|---|
| SJ-3366 | Nevirapine | AZT | |
| IIIB (control) | 0.002 | 0.01 | 0.05 |
| Oxathiin carboxanilide (L100I) | 0.008 (S) | 0.1 (10) | 0.04 |
| TSAO-costatolide (K101E) | 0.05 (18) | 0.4 (40) | 0.004 |
| UC10-costatolide (K103N) | >0.14 (>50) | >10 (>1000) | 0.003 |
| Thiazolobenzimidazole (V108I) | 0.05 (18) | 0.3 (30) | 0.04 |
| TIBO-R82150 (A98G/V108I) | 0.1 (70) | 0.6 (60) | 0.05 |
| Calanolide A (T139I) | 0.01 (S) | 0.01 (S) | 0.01 |
| Diphenylsulfone (Y181C) | >0.1 (>50) | 5.9 (590) | 0.01 |
| Nevirapine (Y181C) | 0.085 (100) | >38 (>3,800) | 0.03 |
| Pyridinone (Y181C/L103N) | 0.4 (180) | >38 (>3,800) | 0.01 |
| E-BPTU (Y181C) | 0.8 (375) | 1.9 (190) | 0.03 |
| UC38 (Y181C) | 0.02 (10) | 1.9 (190) | 0.01 |
| 3TC (M184V) | 0.0006 (S) | 0.01 (S) | 0.02 |
| Costatolide (Y188H) | 0.1 (55) | NDc | 0.004 |
| HEPT (P236L) | 0.028 (33) | 0.02 (S) | 0.01 |
The results presented were obtained from a single representative antiviral assay, with appropriate nevirapine and AZT control values selected from a minimum of two antiviral assays. We have demonstrated that the standard error between multiple antiviral assays averaged less than 10% of the respective mean IC50. Fold resistance values were calculated by determination of the ratio of the activity of each compound against the drug-resistant virus isolate to the activity against wild-type isolate IIIB (control) performed in parallel. Fold resistance values indicated as greater than indicate that the compound was inactive at the greatest nontoxic concentration tested; S, the virus remained sensitive to the agent.
TSAO, [2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2", 2"-dioxide)]-β-d-pentofuranosyl; TIBO, tetrahydro-imidazo-[4,5,1-jk][1,4]-benzodiazepin-2(M)-thione.
ND, not determined.
Confirmation of the results of these assays was obtained by evaluating the activity of each compound against viruses or purified RT with single amino acid changes introduced by site-directed mutagenesis (Table 6). In these assays, only the effect of the specific amino acid change in the RT is evaluated since these changes are placed into a common genetic background (isolate NL4-3). In these assays, SJ-3366 remained completely active against viruses or RTs with amino acid changes L74V, A98G, L100I, V106I, V108I, T139I, and V179D. Viruses with amino acid changes at position Y181 were determined to be slightly resistant, while changes at K101E, K103N, and Y188C resulted in significant levels of resistance to SJ-3366. In addition, viruses with amino acid changes that confer resistance to AZT and ddI exhibited enhanced sensitivity to SJ-3366. Virus with the L74V amino acid change was 10-fold more sensitive, while virus with the four AZT resistance-engendering amino acid changes (D67N, K70R, T215Y, and K219Q) was approximately 3-fold more sensitive. The addition of the NNRTI-specific amino acid changes L100I or Y181C to this background of four amino acid changes that engender resistance to AZT resulted in further enhancement of SJ-3366 activity so that it was 10- to 20-fold more sensitive than wild-type virus (isolate NL4-3). Once again, these results demonstrate significant differences in efficacy profiles between SJ-3366 and nevirapine.
TABLE 6.
Sensitivities to SJ-3366 of virus isolates with defined amino acid changes
| Virus isolate or mutation | IC50 (μM
[fold resistance])a
|
||
|---|---|---|---|
| SJ-3366 | Nevirapine | AZT | |
| NL4-3 | 0.002 | 0.02 | 0.001 |
| L74V | 0.0002 (S) | 0.02 (S) | 0.005 |
| A98G | 0.0006 (S) | 0.2 (10) | 0.01 |
| L100I | 0.0002 (S) | 0.1 (5) | 0.003 |
| K101E | >0.14 (>83) | 0.3 (15) | 0.002 |
| K103N | >0.14 (>83) | >2 (>100) | 0.004 |
| V106I | 0.0006 (S) | >2 (>100) | 0.003 |
| V108I | 0.003 (S) | 0.06 (S) | 0.008 |
| T139I | 0.002 (S) | 0.002 (S) | 0.003 |
| V179D | 0.001 (S) | 0.03 (S) | 0.003 |
| Y181C | 0.009 (5) | 1.3 (65) | 0.01 |
| Y188C | 0.06 (33) | 3.2 (160) | 0.006 |
| 4×AZTb | 0.0006 (S) | 0.02 (S) | 0.8 |
| 4×AZT + Y181C | 0.0003 (S) | 0.8 (40) | 0.04 |
| 4×AZT + L100I | 0.0001 (S) | 0.03 (S) | 0.02 |
The results presented were obtained from a single representative antiviral assay, with appropriate nevirapine and AZT control values selected from a minimum of two antiviral assays. We have demonstrated that the standard error between multiple antiviral assays averaged less than 10% of the respective mean IC50. Fold resistance values were calculated by determination of the ratio of the activity of each compound against the drug-resistant virus isolate to the activity against wild-type isolate NL4-3 (control) performed in parallel. Fold resistance values indicated as greater than indicate that the compound was inactive at the greatest nontoxic concentration tested; S, the virus remained sensitive to the agent.
4×AZT, virus with the four amino acid changes that engender resistance to AZT.
Evaluation of the activity of SJ-3366 against purified RT with mutations introduced by site-directed mutagenesis yielded results similar to those obtained with the mutagenized viruses (data not shown). Again, the fold resistance values obtained in these assays yield small changes in the therapeutic index for SJ-3366. As expected from the mechanistic assays, SJ-3366 was inactive at concentrations up to 100 μM against purified HIV-2 RT.
Selection and characterization of drug-resistant virus isolates.
Viruses resistant to SJ-3366 were selected in CEM-SS cells infected with the IIIB strain of virus after a short time of culture in the presence of increasing concentrations of each compound. SJ-3366 initially yielded a virus isolate with the Y181C amino acid change in the RT. This virus was approximately 100-fold resistant to SJ-3366. Replicate selections were performed to address the reproducibility of the resistance selection process. In each of 10 selection assays, the Y181C amino acid change appeared in the RT by passage 4 to 5.
DISCUSSION
A medicinal chemistry program centered upon the modification of the N-1 homocyclic moiety of the HEPT analogs resulted in the synthesis of a variety of highly potent anti-HIV compounds (Kwon et al., unpublished data). One analog, SJ-3366, was found to be a unique and highly potent NNRTI of HIV-1. SJ-3366 was active at subnanomolar to low nanomolar concentrations against all clinical and laboratory-derived strains of HIV-1, remained active when challenged at a high multiplicity of infection, and lacked inhibitory activity against SIV. Unlike the many NNRTIs reported to date, SJ-3366 also exhibited significant activity against HIV-2. Although SJ-3366 was 200-fold less active against HIV-2 compared with its activity against HIV-1, SJ-3366 possessed a therapeutic index of 20,000 when it was evaluated against HIV-2. The presence of serum proteins had no appreciable effect on the anti-HIV-1 or anti-HIV-2 activity of SJ-3366. At most, a 10-fold loss of activity was detected in assays with the addition of AAGP and HSA, which have been reported to have the most relevance to human drug treatment.
SJ-3366 inhibited HIV-1 RT in biochemical inhibition assays with purified RT with a Ki value of approximately 3 nM. Kinetic evaluations indicated that the mode of action of SJ-3366 against purified HIV-1 RT was of a complicated mixed mode of inhibition in which SJ-3366 affected both the Km and the Vmax of the reaction. SJ-3366 did not inhibit purified HIV-2 RT. Biochemical and cell-based mechanistic assays for determination of how SJ-3366 inhibited HIV-2 demonstrated that SJ-3366 effectively inhibited the attachment of both HIV-1 and HIV-2 to target cells but that it had no inhibitory activity against other viral enzymes such as protease, integrase, or RNase H. SJ-3366 inhibited the attachment of both HIV-1 and HIV-2 at IC50s identical to those observed in cell-based antiviral assays with HIV-2, yielding a therapeutic index of approximately 20,000, consistent with a specific antiattachment mechanism of action. SJ-3366 was inactive in assays that measured its ability to inhibit the fusion of infected and uninfected cells. Thus, SJ-3366 has two primary mechanisms of action. Inhibition of RT is effective for HIV-1, while inhibition of attachment occurs for both HIV-1 and HIV-2.
Unlike most of the NNRTIs that we have evaluated, when evaluated in anti-HIV assays with combinations of drugs, SJ-3366 and a variety of other anti-HIV agents primarily yielded additive interactions. In our hands, most NNRTIs yield significantly synergistic interactions with other RT inhibitors. The results with SJ-3366 are much more reminiscent of our experience with surface-active compounds that inhibit virus attachment or fusion (Julie L. Russel, and Robert W. Buckheit, Jr., unpublished observations). Synergistic inhibition of HIV-1 was achieved with the combination of SJ-3366 and the NRTI ddI, but all other assays with combinations of NRTIs, and NNRTIs, and protease inhibitors yielded additive antiviral interactions. Slightly synergistic interactions between SJ-3366 and nelfinavir were also detected. These results also suggest that the compound has antiviral properties that give it activity similar to the activities of surface-active agents in these assays. SJ-3366 exhibited properties similar to those observed with typical NNRTIs when it was evaluated against resistant strains of virus. Many of the amino acid changes in the RT that confer resistance to NNRTIs yield reductions in the antiviral activity of SJ-3366. Amino acid changes at positions K103, K101, and Y188 have the greatest detrimental effect on the activity of the compound. Unlike results normally achieved with a variety of NNRTIs, a significant discordance exists between data obtained with biological strains of resistant virus selected in cell culture and data obtained with viruses constructed by site-directed mutagenesis. Whereas high levels of resistance were detected when the activity of SJ-3366 was evaluated against biological strains selected in vitro, only three amino acid changes affected the inhibitory activity of SJ-3366 when it was tested against viruses with single amino acid changes introduced by site-directed mutagenesis. Furthermore, much lower quantitative levels of resistance were observed with these NL4-3-derived resistant strains. A more typical pattern of resistance was observed for the positive control NNRTI compound nevirapine, and the biologically derived and site-directed mutagenesis-derived mutant virus isolates had similar levels of resistance to nevirapine. The discordance in the results for these groups of viruses may result from an enhanced sensitivity of NL4-3 to the attachment inhibition mechanism of action of SJ-3366 or may be due to variations in the ability of the selected viruses to be inhibited at the step of virus entry. Certainly, the process of selection of drug-resistant viruses in cell culture may result in changes in viral fitness, cytopathogenicity, and replication rate which may render them more resistant to an attachment inhibitor by the accumulation of non-RT amino acid changes. The differences in fold resistance between the two panels of viruses are most notable with the various viruses possessing the Y181C amino acid change in the RT. With these Y181C-containing viruses, fold resistance values of 5-fold (site-directed mutagenesis-derived mutant), 10-fold (UC38 resistant), >50-fold (diphenylsulfone resistant), and 375-fold (E-BPTU [1-benzyloxymethyl-5-ethyl-6-(α-pyridylthis)uracil] resistant) were observed. Interestingly, the most resistant virus in the group was selected with the compound E-BPTU, which is a related HEPT-type compound (9). Sensitivity testing also suggests that SJ-3366 is slightly more active against viruses that are resistant to the NRTIs (AZT and ddI), including NRTI-resistant viruses with added NNRTI-specific mutations. These results would suggest that the changes in RT caused by resistance-engendering mutations result in an enzyme that is more sensitive to SJ-3366-mediated inhibition.
Resistance selection assays performed with SJ-3366 routinely yield a virus that possesses the Y181C amino acid change in the RT after approximately five to six passages in cell culture. Interestingly, the fold resistance values calculated suggest increasing levels of resistance of the serially passaged virus prior to the appearance of this initial change in the RT and fold resistance increases to nearly 100-fold prior to changes in RT. These data might suggest that the initial resistance selection process for SJ-3366 involves the ability of the virus to evade the attachment mechanism of inhibition. Most of our data with the panels of resistant strains of virus would also suggest that fold resistance values of 100- to 1,000-fold would not occur with only the Y181C amino acid change. Thus, we believe it is likely that amino acid changes in other HIV proteins, such as gp120 and gp41, will be found to explain the increase in fold resistance up until the first passage at which the Y181C change is detected. Interestingly, of the three amino acid changes that would be predicted to yield high-level resistance to SJ-3366, K101E, K103N, and Y188C do not appear during long-term passage of virus in the presence of SJ-3366. These data likely suggest that these amino acid changes cause replication disadvantages to the virus and, therefore, that the virus selects other pathways to achieve resistance and the ability to grow in the presence of high concentrations of the compound.
The HEPT compounds, of which SJ-3366 is a member, were among the first NNRTIs reported, and one member of this class (MKC-442) has progressed into human clinical trials. These compounds are among the most active inhibitors of HIV-1. None of the reported HEPT-type compounds, however, have demonstrated significant activity against HIV-2, suggesting that the specific modifications made at the N-1 homocyclic moiety of HEPT have resulted in this expanded range of activity. Structure-activity relationships among 80 analogs of SJ-3366 for efficacy in cytopathic effect assays for HIV-1 and HIV-2, as well as inhibition of RT and virus attachment, have been performed and are being reported elsewhere (R. W. Buckheit, Jr., unpublished data).
SJ-3366 represents a highly potent and unique member of the class of NNRTIs. In addition to joining thiocarboxanilide, the quinoxaline HBY097, and efavirenz as the most active NNRTIs described in the literature (20), SJ-3366 also possesses multiple mechanisms of anti-HIV action, exhibits distinct differences in activity against NNRTI-resistant strains compared to those of typical NNRTIs, remains active in the presence of high concentrations of protein, and exhibits positive interactions with NRTI, NNRTI, and protease inhibitor compounds. All of these antiviral features suggest that the compound may represent a potential clinical candidate for anti-HIV therapy.
ACKNOWLEDGMENTS
We gratefully acknowledge Barbara Toyer and Michelle Wenzel for drug preparation support and Diana Markle for assistance with the preparation of the manuscript.
REFERENCES
- 1.Baba M, Shigeta S, Tanaka H, Miyasaka T, Ubasawa M, Umezu K, Walker R T, Pauwels R, De Clercq E. Highly potent and selective inhibition of HIV-1 replication by 6-phenylthiouracil derivatives. Antivir Res. 1992;17:245–264. doi: 10.1016/0166-3542(92)90021-v. [DOI] [PubMed] [Google Scholar]
- 2.Baba M, Shigeta S, Yuasa S, Takashima H, Sekiya K, Ubasawa M, Tanaka H, Miyasaka T, Walker R T, De Clercq E. Preclinical evaluation of MKC-442, a highly potent and specific inhibitor of human immunodeficiency virus type 1 in vitro. Antimicrob Agents Chemother. 1994;38:688–692. doi: 10.1128/aac.38.4.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba M, Tanaka H, De Clercq E, Pauwels R, Balzarini J, Schols D, Nakashima H, Perno C F, Walker R T, Miyasaka T. Highly specific inhibition of human immunodeficiency virus type 1 by a novel 6-substituted acyclouridine derivative. Biochem Biophys Res Commun. 1989;165:1375–1381. doi: 10.1016/0006-291x(89)92756-3. [DOI] [PubMed] [Google Scholar]
- 4.Balzarini J, Karlsson A, Perez-Perez M J, Camarasa M J, Tarpley W G, De Clercq E. Treatment of human immunodeficiency virus type 1 (HIV-1)-infected cells with combinations of HIV-1-specific inhibitors results in a different resistance pattern than does treatment with single-drug therapy. J Virol. 1993;67:5353–5359. doi: 10.1128/jvi.67.9.5353-5359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini J, Karlsson A, Vandamme A M, Perez-Perez M J, Zhang H, Vrang L, Oberg B, Backbro K, Unge T, San-Felix A. Human immunodeficiency virus type 1 (HIV-1) strains selected for resistance against the HIV-1-specific [2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-spiro-5"-(4"-amino-1",2"-oxathiole-2",2"-dioxide)]-beta-d-pentofuranosyl (TSAO) nucleoside analogues retain sensitivity to HIV-1-specific nonnucleoside inhibitors. Proc Natl Acad Sci USA. 1993;90:6952–6956. doi: 10.1073/pnas.90.15.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer P L, Currens M J, McMahon J B, Boyd M R, Hughes S H. Analysis of nonnucleoside drug-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1993;67:2412–2420. doi: 10.1128/jvi.67.4.2412-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckheit R W, Jr, Snow M J, Fliakas-Boltz V, Kinjerski T L, Russell J D, Pallansch L A, Brouwer W G, Yang S S. Highly potent oxathiin carboxanilide derivatives with efficacy against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus isolates. Antimicrob Agents Chemother. 1997;41:831–837. doi: 10.1128/aac.41.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckheit R W, Jr, Fliakas-Boltz V, Decker W D, Roberson J L, Stup T L, Pyle C A, White E L, McMahon J B, Currens M J, Boyd M R, Bader J P. Comparative anti-HIV evaluation of diverse HIV-1-specific reverse transcriptase inhibitor-resistant virus isolates demonstrates the existence of distinct phenotypic subgroups. Antivir Res. 1995;26:117–132. doi: 10.1016/0166-3542(94)00069-k. [DOI] [PubMed] [Google Scholar]
- 9.Buckheit R W, Jr, Fliakas-Boltz V, Yeagy-Bargo S, Weislow O, Mayers D L, Boyer P L, Hughes S H, Pan B C, Chu S H, Bader J P. Resistance to 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives is generated by mutations at multiple sites in the HIV-1 reverse transcriptase. Virology. 1995;210:186–193. doi: 10.1006/viro.1995.1330. [DOI] [PubMed] [Google Scholar]
- 10.Buckheit R W, Jr, Kinjerski T L, Fliakas-Boltz V, Russell J D, Stup T L, Pallansch L A, Brouwer W G, Dao D C, Harrison W A, Schultz R J, Bader J P, Yang S S. Structure-activity and cross-resistance evaluations of a series of human immunodeficiency virus type 1-specific compounds related to oxathiin carboxanilide. Antimicrob Agents Chemother. 1995;39:2718–2727. doi: 10.1128/aac.39.12.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckheit R W, Jr, Roberson J L, Lackman-Smith C, Wyatt J R, Vickers T A, Ecker D J. Potent and specific inhibition of HIV envelope-mediated cell fusion and virus binding by G quartet-forming oligonucleotide (ISIS 5320) AIDS Res Hum Retrovir. 1994;10:1497–1506. doi: 10.1089/aid.1994.10.1497. [DOI] [PubMed] [Google Scholar]
- 12.Byrnes V W, Sardana V V, Schleif W A, Condra J H, Waterbury J A, Wolfgang J A, Long W J, Schneider C L, Schlabach A J, Wolanski B S. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob Agents Chemother. 1993;37:1576–1579. doi: 10.1128/aac.37.8.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carr A, Cooper D A. Current clinical experience with nevirapine for HIV infection. Adv Exp Med Biol. 1998;394:299–304. doi: 10.1007/978-1-4757-9209-6_27. [DOI] [PubMed] [Google Scholar]
- 14.Cohen K A, Hopkins J, Ingraham R H, Pargellis C, Wu J C, Palladino D E, Kinkade P, Warren T C, Rogers S, Adams J. Characterization of the binding site for nevirapine (BI-RG-587), a nonnucleoside inhibitor of human immunodeficiency virus type-1 reverse transcriptase. J Biol Chem. 1991;266:14670–14674. [PubMed] [Google Scholar]
- 15.Condra J H, Emini E A, Gotlib L, Graham D J, Schlabach A J, Wolfgang J A, Colonno R J, Sardana V V. Identification of the human immunodeficiency virus reverse transcriptase residues that contribute to the activity of diverse nonnucleoside inhibitors. Antimicrob Agents Chemother. 1992;36:1441–1446. doi: 10.1128/aac.36.7.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currens M J, Gulakowski R J, Mariner J M, Moran R A, Buckheit R W, Jr, Gustafson K R, McMahon J B, Boyd M R. Antiviral activity mechanism of action of calanolide A against the human immunodeficiency virus. J Pharmacol Exp Ther. 1996;279:645–651. [PubMed] [Google Scholar]
- 17.Danel K, Larsen E, Pedersen E B. Synthesis and potent anti-HIV-1 activity of novel 6-benzyluracil analogues of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine. J Med Chem. 1996;39:2427–2431. doi: 10.1021/jm9600499. [DOI] [PubMed] [Google Scholar]
- 18.De Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs) for the treatment of human immunodeficiency virus type 1 (HIV-1) infections: strategies to overcome drug resistance development. Med Res Rev. 1996;16:125–157. doi: 10.1002/(SICI)1098-1128(199603)16:2<125::AID-MED1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.De Clercq E. What can be expected from non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the treatment of human immunodeficiency virus type 1 (HIV-1) infections? Med Virol. 1996;6:97–117. doi: 10.1002/(SICI)1099-1654(199606)6:2<97::AID-RMV168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.De Clercq E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antivir Res. 1998;38:153–179. doi: 10.1016/s0166-3542(98)00025-4. [DOI] [PubMed] [Google Scholar]
- 21.Dueweke T J, Pushkarskaya T, Poppe S M, Swaney S M, Zhao J Q, Chen I S, Stevenson M, Tarpley W G. A mutation in reverse transcriptase of bis(heteroaryl)piperazine-resistant human immunodeficiency virus type 1 that confers increased sensitivity to other nonnucleoside inhibitors. Proc Natl Acad Sci USA. 1993;90:4713–4717. doi: 10.1073/pnas.90.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan N, Rank K B, Slade D E, Poppe S M, Evans D B, Kopta L A, Olmsted R A, Thomas R C, Tarpley W G, Sharma S K. A drug resistance mutation in the inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase impairs DNA synthesis and RNA degradation. Biochemistry. 1996;35:9737–9745. doi: 10.1021/bi9600308. [DOI] [PubMed] [Google Scholar]
- 23.Gerondelis P, Archer R H, Palaniappan C, Reichman R C, Fay P J, Bambara R A, Demeter L M. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J Virol. 1999;73:5803–5813. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grob P M, Wu J C, Cohen K A, Ingraham R H, Shih C K, Hargrave K D, McTague T L, Merluzzi V J. Nonnucleoside inhibitors of HIV-1 reverse transcriptase: nevirapine as a prototype drug. AIDS Res Hum Retrovir. 1992;8:145–152. doi: 10.1089/aid.1992.8.145. [DOI] [PubMed] [Google Scholar]
- 25.Guay L A, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler M G, Mofenson L, Miotti P, Dransfield K, Bray D, Mmiro F, Jackson J B. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 26.Havlir D, Cheeseman S H, McLaughlin M, Murphy R, Erice A, Spector S A, Greenough T C, Sullivan J L, Hall D, Myers M, Lamson M, Richman D D. High-dose nevirapine: safety, pharmacokinetics, and antiviral effect in patients with human immunodeficiency virus infection. J Infect Dis. 1995;171:537–545. doi: 10.1093/infdis/171.3.537. [DOI] [PubMed] [Google Scholar]
- 27.Kim D K, Gam J, Kim Y W. Synthesis and anti-HIV-1 activity of a series of 1-(alkoxymethyl)-5-alkyl-6-(arylselencnyl)uracils and -2-thiouracils. Heterocyclic Chem. 1999;14:1275–1282. [Google Scholar]
- 28.Kinjerski T L, Buckheit R W., Jr The role of genotypic heterogeneity in wild type virus populations on the selection of nonnucleoside reverse transcriptase inhibitor-resistant viruses. Antivir Res. 1997;33:109–115. doi: 10.1016/s0166-3542(96)01008-x. [DOI] [PubMed] [Google Scholar]
- 29.Kinjerski T L, Pallansch L A, Buckheit R W., Jr Isolation and characterization of HIV-1 isolates resistant to oxathiin carboxanilide analogs: evaluation of variables in the resistance selection process. Antivir Chem Chemother. 1996;7:261–269. [Google Scholar]
- 30.Kleim J P, Bender R, Kirsch R, Meichsner C, Paessens A, Riess G. Mutational analysis of residue 190 of human immunodeficiency virus type 1 reverse transcriptase. Virology. 1994;200:696–701. doi: 10.1006/viro.1994.1233. [DOI] [PubMed] [Google Scholar]
- 31.Mai A, Artico M, Massa S, Novellino E, Greco G, Loi A G, Tramontano E, Marongiu M E, La Colla P. 5-Alkyl-2-(alkylthio)-6-(2,6-dihalophenylmethyl)-3,4-dihydropyrimidin-4(3H)-ones: novel potent and selective dihydro-alkoxy-benzyl-oxopyrimidine derivatives. J Med Chem. 1999;42:619–627. doi: 10.1021/jm980260f. [DOI] [PubMed] [Google Scholar]
- 32.McMahon J B, Gulakowski R J, Weislow O S, Schultz R J, Narayanan V L, Clanton D J, Pedemonte R, Wassmundt F W, Buckheit R W, Jr, Decker W D. Diarylsulfones, a new chemical class of nonnucleoside antiviral inhibitors of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:754–760. doi: 10.1128/aac.37.4.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellors J W, Larder B A, Schinazi R F. Mutations in HIV-1 reverse transcriptase and protease associated with drug resistance. Int Antivir News. 1995;3:8–13. [Google Scholar]
- 34.Miyasaka T, Tanaka H, Baba M, Hayakawa H, Walker R T, Balzarini J, De Clercq E. A novel lead for specific anti-HIV-1 agents: 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine. J Med Chem. 1989;32:2507–2509. doi: 10.1021/jm00132a002. [DOI] [PubMed] [Google Scholar]
- 35.Olmsted R A, Slade D E, Kopta L A, Poppe S M, Poel T J, Newport S W, Rank K B, Biles C, Morge R A, Dueweke T J, Romero D L, Thomas R C, Sharma S K, Tarpley W G. (Alkylamino) piperidine bis(heteroaryl) piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIBwith reduced replication phenotypes. J Virol. 1996;70:3698–3705. doi: 10.1128/jvi.70.6.3698-3705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker W B, White E L, Shaddix S C, Ross L J, Buckheit R W, Jr, Germany J M, Secrist III J A, Vince R, Shannon W M. Mechanism of inhibition of human immunodeficiency virus type 1 reverse transcriptase and human DNA polymerases α, β, and gamma by the 5′-triphosphates of carbovir, 3′-azido-3′-deoxythymidine, 2′,3′-dideoxyguanosine and 3′-deoxythymidine. A novel RNA template for the evaluation of antiretroviral drugs. J Biol Chem. 1991;266:1754–1762. [PubMed] [Google Scholar]
- 37.Prichard M N, Shipman C., Jr A three-dimensional model to analyze drug-drug interactions. Antivir Res. 1990;14:181–206. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 38.Saag M S, Emini E A, Laskin O L, Douglas J, Lapidus W I, Schleif W A, Whitley R J, Hildebrand C, Byrnes V W, Kappes J C. A short-term clinical evaluation of L-697,661, a non-nucleoside inhibitor of HIV-1 reverse transcriptase. L-697,661 Working Group. N Engl J Med. 1993;329:1065–1072. doi: 10.1056/NEJM199310073291502. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H, Baba M, Ubasawa M, Takashima H, Sekiya K, Nitta I, Shigeta S, Walker R T, De Clercq E, Miyasaka T. Synthesis and anti-HIV activity of 2-, 3-, and 4-substituted analogues of 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) J Med Chem. 1991;34:1394–1399. doi: 10.1021/jm00108a023. [DOI] [PubMed] [Google Scholar]
- 40.White E L, Parker W B, Ross L J, Shannon W M. Lack of synergy in the inhibition of HIV-1 reverse transcriptase by combinations of the 5′-triphosphates of various anti-HIV nucleoside analogs. Antivir Res. 1993;22:295–308. doi: 10.1016/0166-3542(93)90039-l. [DOI] [PubMed] [Google Scholar]
- 41.Yang S S, Fliakas-Boltz V, Bader J P, Buckheit R W., Jr Characteristics of a group of nonnucleoside reverse transcriptase inhibitors with structural diversity and potent anti-human immunodeficiency virus activity. Leukemia. 1995;9:75–85. [PubMed] [Google Scholar]