Abstract
Infections with the human pathogenic basidiomycetous yeast Cryptococcus neoformans are often treated with fluconazole. Resistance to this antifungal agent has been reported. This study investigated the patterns of mutation to fluconazole resistance in C. neoformans in vitro. The MIC of fluconazole was measured for 21 strains of C. neoformans. The MICs for these 21 strains differed (0.25 to 4.0 μg/ml), but the strains were selected for this study because they exhibited no growth on plates of yeast morphology agar (YMA) containing 8 μg of fluconazole per ml. To determine their mutation rates, six independent cultures from a single original colony were established for each of the 21 strains. Each culture was then spread densely on a YMA plate with 8 μg of fluconazole per ml. A random set of putative mutants was subcultured, and the MIC of fluconazole was determined for each mutant. The 21 strains evinced significant heterogeneity in their mutation rates. The MICs of the putative mutants ranged widely, from their original MIC to 64 μg of fluconazole per ml. However, for this set of 21 strains, there was no significant correlation between the original MIC for a strain and the mutation rate of that strain; the MIC for the mutant could not be predicted from the original MIC. These results suggest that dynamic and heterogeneous mutational processes are involved in generating fluconazole resistance in C. neoformans.
Cryptococcus neoformans is an encapsulated basidiomycetous yeast capable of causing fatal infection in both immunocompetent and immunocompromised patients, including a prevalence of infection of up to 15% of patients with AIDS (9, 21). The most common clinical manifestation of C. neoformans infection, cryptococcal meningoencephalitis, is usually incurable in immunocompromised patients despite antifungal therapy (9, 21).
Fluconazole is currently the most widely used antifungal drug for maintenance therapy because it can be given orally, lacks major side effects, penetrates the central nervous system, and has broad efficacy against most pathogenic yeasts, including C. neoformans (5, 9). It perturbs the biosynthesis of ergosterol by blocking an alpha-14-demethylation step in the biosynthetic pathway (5, 29). However, fluconazole-resistant fungal pathogens are becoming increasingly common (12, 13, 17, 24, 29, 30, 32).
The relationship between in vitro resistance to fluconazole as measured by the MIC and clinical resistance as defined by treatment failure is not clearly understood. In many studies describing treatment failures, relapse isolates showed no increase in the MIC of fluconazole despite long-term treatment (7, 22, 26; M. E. Brandt, M. A. Pfaller, R. A. Hajjeh, et al., for the CDC Cryptococcal Disease Active Surveillance Group, Abstr. 37th Annu. Meet. Infect. Dis. Soc. Am., abstr. 395, 1999). In studies where molecular subtyping was performed, substantial differences in MIC and genotypes were observed between some serial isolates obtained before and during treatment with fluconazole, suggesting the possibility that susceptible strains were replaced by resistant, possibly exogenous strains (18, 28). In contrast, other studies reported the recovery from single patients of serial isolates with no detectable genotypic differences, and antifungal susceptibility testing revealed stepwise (two- to fourfold) increases in MIC over time (1, 3, 4, 13, 18, 22, 24, 26).
The objective of this study was to examine in vitro the patterns of mutation to fluconazole resistance in C. neoformans, to help clarify the observed clinical findings and to evaluate the use of the MIC as a possible indicator for the development of resistance. We defined fluconazole resistance in C. neoformans as an MIC of ≥32 μg/ml by the NCCLS standard in vitro protocol (23). Specifically, we were interested in the following questions. First, do different strains vary in their rates of mutation to fluconazole resistance? The answer to this question could explain why some strains are and others are not associated with an increase in fluconazole MIC during similar courses of antifungal treatment. Second, in single-mutation experiments, will only small increases (two- to fourfold) in MIC occur or will large increases in MIC develop (over eightfold)? Third, will plating strains on medium containing 8 μg of fluconazole per ml, a concentration that approximates the achievable level in cerebrospinal fluid (CSF) (5, 32), lead to the recovery of mutants for which the MICs are significantly greater than 8 μg/ml (e.g., ≥64 μg/ml)? Lastly, are strains with higher initial fluconazole MICs (e.g., 2 to 4 μg/ml) more likely to develop resistance than strains with low fluconazole MICs (e.g., 0.25 to 0.5 μg/ml)? Our expectation is that if a gradual, stepwise increase in MIC is the main process involved in generating fluconazole resistance in C. neoformans, we should observe a significant correlation between the original MIC and the mutation rate and between the original MIC and the MIC for the mutant. If the expectation is confirmed, measuring the MIC for an initial isolate might predict its likelihood of becoming resistant to fluconazole after the initiation of fluconazole treatment.
To investigate these issues, we selected a set of 21 strains, representing three serotypes (A, B, and D), from various geographic regions (9, 16). Eighteen strains were clinical isolates, two were isolated from the environment, and the origin of the other one is unknown. These strains were chosen from an initial pool of 101 strains representing all five serotypes, A, B, C, D, and AD. These 21 strains did not grow at all on yeast morphology agar (YMA) plates with 8 μg of fluconazole per ml. The no-growth phenotype at this concentration was necessary to estimate accurately the mutation rate to resistance on plates with 8 μg of fluconazole per ml. Mutation rates and the MICs for the mutants were measured and analyzed for all strains. Furthermore, random amplified polymorphic DNA (RAPD), obtained using two primers, was used to compare genotypes between mutants and the original strains.
MATERIALS AND METHODS
C. neoformans strains.
Strains used in this study were obtained from two resources: the culture collection of the Mycotic Diseases Branch at the Centers for Disease Control and Prevention in Atlanta, Ga., and the Medical Mycology Research Laboratory at the Duke University Medical Center in Durham, N.C. Initially, 101 strains representing at least 20 multilocus enzyme electrophoresis (MLEE) genotypes were screened (6–8). The MLEE genotype codes and the number of strains from each genotype tested in this study are presented in Table 1 (see below). Serotypes were determined for each strain by a commercial kit (Iatron) (16). For each strain, a single colony was suspended in 200 μl of sterile water by vortexing and 1 μl of liquid suspension was streaked onto YMA (Difco, Detroit, Mich.) plates containing 8 μg of fluconazole per ml. The plates were incubated for 48 h at 37°C and examined for evidence of growth under a microscope (magnification, ×100). Twenty-one strains showed no growth on plates with 8 μg of fluconazole per ml. This no-growth phenotype was necessary for accurately estimating the mutation rate to fluconazole resistance. For the other 80 strains, the fluconazole MICs for most strains were lower than 8 μg/ml but the strains underwent growth with at least one mitotic division per cell after plating. In addition, many produced sporadic visible colonies, with frequencies in the range of 1/100 to 1/1,000. Because this extra growth can inflate the number of cells plated, biasing our mutation rate estimates upward, the other 80 strains were not used in subsequent experiments. Table 2 (see below) lists the geographic origins, isolation sites on hosts' bodies, and serotypes of the 21 strains selected for this study. Strains CDC-MAS92-0368 and CDC-MAS92-0804 were serial isolates from CSF of the same patient in Georgia. These two isolates have been described previously (reference 7, patient RC20).
TABLE 1.
Number of strains initially screened for evidence of growth on 8 μg of fluconazole per mla
| Serotype | MLEE genotype | Prevalence (%)b | Total no. of strains | No. of strains that did not growg | No. of strains that grewg |
|---|---|---|---|---|---|
| A | ET-1 | 69 | 31 | 4 | 27 |
| ET-2 | 10 | 10 | 3 | 7 | |
| ET-3 | 5 | 5 | 5 | 0 | |
| ET-4 | 4 | 2 | 0 | 2 | |
| Unknown | 4 | 0 | 4 | ||
| AD | ET-5 | 6c | 3 | 0 | 3 |
| ET-6 | NAd | 1 | 0 | 1 | |
| ET-7 | 6c | 4 | 0 | 4 | |
| ET-21 | 6c | 5 | 0 | 5 | |
| D | ET-8 | 3e | 7 | 2 | 5 |
| ET-9 | 3e | 3 | 1 | 2 | |
| ET-10 | NA | 1 | 0 | 1 | |
| ET-11 | NA | 1 | 1 | 0 | |
| ET-12 | 3e | 3 | 3 | 0 | |
| Unknown | 4 | 0 | 4 | ||
| C | ET-13 | NA | 1 | 0 | 1 |
| ET-16 | NA | 2 | 0 | 2 | |
| B | ET-14 | NA | 1 | 0 | 1 |
| ET-15 | NA | 2 | 0 | 2 | |
| ET-17 | NA | 1 | 0 | 1 | |
| ET-18 | 1f | 2 | 0 | 2 | |
| ET-19 | NA | 1 | 1 | 0 | |
| Unknown | 5 | 1 | 4 | ||
| Unclear | Unknown | 2 | 0 | 2 | |
| Total | 101 | 21 | 80 |
Prevalence of each ET (enzyme type) in four regions of the United States as determined by active surveillance (8).
The prevalence of ET-5, ET-7, and ET-21 was determined as a group.
NA, not available.
The prevalence of ET-8, ET-9, and ET-12 was determined as a group.
The prevalence of ET-18, ET-29, and ET-30 was determined as a group.
Growth medium is YMA with 8 μg of fluconazole per ml.
TABLE 2.
Strains of C. neoformans used to estimate mutations to fluconazole resistance
| Strain | MLEE genotypea | Geographic origin | Sample | Serotype | Fluconazole MIC (μg/ml) in:
|
|
|---|---|---|---|---|---|---|
| ref. 6 | Present study | |||||
| CDC-B4964 | ET-1 | Zaire | CSF | A | 2 | |
| CDC-MAS92-0248 | ET-1 | United States | Blood | A | 16 | 2 |
| CDC-Y195-90 | ET-1 | Brazil | Unknown | A | 4 | |
| CDC-Y289-90 | ET-1 | Canada | CSF | A | 1 | |
| CDC-MAS92-0316 | ET-2 | United States | Blood | A | 1 | 1 |
| CDC-MAS92-0368 | ET-2 | United States | CSF | A | 0.5 | 1 |
| CDC-MAS92-0804 | ET-2 | United States | CSF | A | 0.5 | 2 |
| CDC-MAS92-0037 | ET-3 | United States | CSF | A | 4 | 0.25 |
| CDC-MAS92-0064 | ET-3 | United States | Blood | A | 2 | 1 |
| CDC-MAS92-0109 | ET-3 | United States | Blood | A | 2 | 0.5 |
| CDC-MAS92-0245 | ET-3 | United States | CSF | A | 4 | 1 |
| CDC-Y288-90 | ET-3 | Canada | CSF | A | 1 | |
| CDC-MAS92-0232 | ET-8 | United States | Blood | D | 2 | 0.5 |
| CDC-Y286-90 | ET-8 | Canada | CSF | D | 2 | |
| CDC-Y290-90 | ET-9 | Canada | CSF | D | 2 | |
| ATCC34875 | ET-11 | Denmark | Pigeon nest | D | 1 | |
| CDC-MAS92-0088 | ET-12 | United States | Blood | D | 1 | 2 |
| CDC-MAS92-0403 | ET-12 | United States | Blood | D | 1 | 1 |
| CDC-Y494-91 | ET-12 | United States | CSF | D | 1 | |
| CDC-B4496 | ET-19 | Australia | CSF | B | 4 | |
| E275 | Unknown | Australia | Eucalyptus | B | 2 | |
Susceptibility to fluconazole.
The MICs of fluconazole for all strains were determined by a standard broth microdilution method, M-27A, recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (23). The MICs for all strains were also determined by a method based on the colony size (36). For all strains, the MICs obtained from these two methods differed by no more than a twofold dilution, confirming that the rapid and statistically analyzable colony size method is comparable to the NCCLS method for C. neoformans. Indeed, in this and a previous study, we observed less variation among replicates in MICs determined by the colony size method than in MICs determined by the standard NCCLS protocol (36). However, to be consistent with most publications, only the MICs determined by the NCCLS microdilution method are presented (Table 2).
Rate of mutation to fluconazole resistance.
For each of the 21 strains, one 3-day-old colony was suspended in 500 μl of sterile water. Cell density was determined using a hemocytometer, and the cell suspension was aliquoted into six independent culture tubes, each of which contained 10 ml of yeast extract-peptone-dextrose (YEPD) broth. The typical inoculum size in each culture tube ranged from 1 × 104 to 2 × 104 cells. These cultures were incubated at 37°C on a shaker at 250 rpm for 72 h. Cells were then collected by centrifugation at 5,000 × g for 5 min. After the supernatant was discarded, the cells were washed twice with 5 ml of physiological saline (0.9% [wt/vol] sterile NaCl) by vortexing and recentrifugation. After the final wash, the cells were resuspended in 1.0 ml of physiological saline. One microliter of the final suspension was diluted, and the cells were counted in a hemocytometer to determine the concentration of cells in each of the six replicates for each strain. Appropriate dilutions were also plated on YEPD agar to determine the concentrations of viable cells for each culture. Both methods yielded the same results for each culture.
For each replicate, 950 μl of the suspension was evenly spread on a YMA plate containing 8 μg of fluconazole per ml. The plates were incubated at 37°C for 1 week, and visible mutant colonies were counted and recorded for each plate. The mutation rate, defined as the number of mutations per cell per generation, was calculated according to the following equation (2):
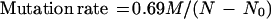 |
where M is the number of mutants produced when N0 cells increase in number to N cells.
MIC and the stability of putative mutants.
To ensure that resistant phenotypes were genetically stable in the absence of fluconazole, resistant colonies from each plate were streaked on YEPD plates without fluconazole. These putative resistant mutants were incubated at 37°C for 24 to 48 h, and single colonies were picked to determine the MIC as described above (23, 36). When available, up to three randomly collected colonies were examined from each plate. For 10 random mutants, three tests up to 1 month apart from each other were performed to examine the overall stability of the MIC within and among mutants.
Comparison of DNA fingerprints of the original and mutant isolates by RAPD.
Genomic DNA was extracted from all original isolates and selected mutants by a method described previously (35). Two oligonucleotides were used as single primers for PCR fingerprinting: (i) the OPA-03 oligonucleotide 5′-AGT CAG CCA C-3′, and (ii) the OPA-17 oligonucleotide 5′-GAC CGC TTG T-3′. Both primers were obtained from Operon Technologies. Amplifications were performed in volumes of 25 μl containing 10 ng of genomic DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl, 0.2 mM each dATP, dCTP, dTTP, and dGTP, 3 mM magnesium acetate, 10 ng of primer, and 1.5 U of AmpliTaq DNA polymerase. All PCRs were performed in a Perkin-Elmer thermal cycler (model 9700) with an initial denaturation of 97°C for 3 min followed by 45 cycles of 60 s at 93°C, 60 s at 36°C, and 120 s at 72°C and then a final cycle of 5 min at 72°C. Amplification products were separated by electrophoresis in 1.5% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer for 13 h at 2 V/cm. PCR products were detected by staining with ethidium bromide (0.5 μg/ml) and visualized under UV light.
RESULTS
Selection of strains.
Of the 101 strains initially screened, 21 showed no growth on YMA plates with 8 μg of fluconazole per ml (Table 1). There were several interesting features in Table 1. The five serotypes differed in their ability to grow in the presence of 8 μg of fluconazole per ml. For example, all 13 serotype AD strains and 3 serotype C strains had some growth. In the other three serotypes (A, B, and D), a variable proportion of strains within each group displayed some growth. Within a serotype, different MLEE genotypes had significantly different patterns of growth. For example, in the presence of 8 μg of fluconazole per ml, 27 of the 31 ET-1 strains grew while none of the 5 ET-3 strains grew (Table 1).
Initial fluconazole susceptibilities.
The initial MICs of fluconazole for all 21 strains are presented in Table 2. The MICs ranged from 0.25 to 4 μg/ml. Two strains, MAS92-0037 and MAS92-0248, showed 16- and 8-fold differences, respectively between these tests and those of Brandt et al. (Abstr. Annu. Meet. IDSA). The MICs for other strains were identical, or there was a two- to fourfold difference between the two tests (Table 2). The two- to fourfold difference between independent tests of the same strain by different laboratories is considered acceptable by the NCCLS microbroth dilution protocol (20, 23). Two factors favored the use of MIC results from the present study for subsequent analyses. First, in this study we also used the colony size method to determine the MICs for all strains. Results from the colony size method are highly reliable and do not suffer the disadvantages of the NCCLS method, such as high sensitivity to inoculum age and size (36). Second, 10 of the 21 strains were not previously tested for the fluconazole MIC.
This wide distribution of MIC among the 21 strains (0.25 to 4.0 μg/ml) offered a good opportunity to examine the relationship between initial MIC and mutation rates, as described below. Within the 21 strains selected for mutation rate study, there was no significant correlation of MIC and serotype, geographic origin, or isolation site (Table 1).
Mutation rate.
The mutation rates of the 21 strains were examined. After 7 days of growth at 37°C on 8 μg of fluconazole per ml, all distinctively visible colonies were scored as mutants. Table 3 presents the total number of cells plated, the total number of mutants obtained, and the rate of mutation to fluconazole resistance for each strain. The mean mutation rate of all 21 strains was 301.82 × 10−9. However, the standard deviation (SD) of the mutation rate was 762.55 × 10−9, over twice the value of the mean. When t tests (27) were applied, many pairwise comparisons between strains showed significantly different mutation rates (Table 3, pairwise statistical test results not shown).
TABLE 3.
Mutation rate and range of MIC to fluconazole in 21 strains of C. neoformans
| Strain | Total no. of cells (109) | Total no. of mutants | Mutation rate (10−9 [mean ± SD]) | Mutant MIC (μg/ml), median (range) |
|---|---|---|---|---|
| CDC-B4964 | 6.45 | 4 | 0.41 ± 0.21 | 32 (16–64) |
| CDC-MAS92-0248 | 5.93 | 0 | 0 | |
| CDC-Y195-90 | 7.28 | 73 | 6.92 ± 3.97 | 16 (16) |
| CDC-Y289-90 | 8.05 | 336 | 28.80 ± 29.24 | 8 (8–16) |
| CDC-MAS92-0316 | 0.48 | 21 | 30.17 ± 21.02 | 16 (8–16) |
| CDC-MAS92-0368 | 3.1 | 82 | 18.25 ± 8.39 | 8 (1–32) |
| CDC-MAS92-0804 | 1.5 | 21 | 9.66 ± 5.65 | 8 (4–16) |
| CDC-MAS92-0037 | 0.77 | 23 | 20.60 ± 11.12 | 8 (0.5–16) |
| CDC-MAS92-0064 | 1.91 | 180 | 80.18 ± 94.90 | 16 (4–32) |
| CDC-MAS92-0109 | 0.75 | 19 | 17.46 ± 11.18 | 4 (4–8) |
| CDC-MAS92-0245 | 0.4 | 562 | 969.44 ± 224.53 | 16 (8–16) |
| CDC-Y288-90 | 2.3 | 41 | 12.30 ± 2.37 | 16 (8–32) |
| CDC-MAS92-0232 | 0.4 | 36 | 62.10 ± 16.73 | 4 (4–8) |
| CDC-Y286-90 | 7.78 | 32 | 2.83 ± 1.01 | 8 (4–32) |
| CDC-Y290-90 | 5.3 | 122 | 15.87 ± 20.92 | 16 (8–16) |
| ATCC34875 | 21.21 | 192 | 6.24 ± 3.31 | 32 (8–64) |
| CDC-MAS92-0088 | 0.25 | 1,136 | 3,135.36 ± 98.36 | 32 (16–64) |
| CDC-MAS92-0403 | 4.93 | 0 | 0 | |
| CDC-Y494-91 | 0.28 | 664 | 1,636.28 ± 166.6 | 16 (8–16) |
| CDC-B4496 | 5.15 | 0 | 0 | |
| E275 | 0.3 | 124 | 285.19 ± 68.21 | 16 (8–32) |
The significant differences in mutation rate among strains were not correlated with serotype, geographic origin, or isolation site (Table 3, statistical tests not shown). For example, strain CDC-MAS92-0088 (serotype D) had the highest mutation rate, 3,135.36 × 10−9. In contrast, despite the large inoculum sizes, three strains produced no mutants in any of the six independent cultures (Table 3). These three strains were CDC-MAS92-0248 (serotype A), CDC-B4496 (serotype B), and CDC-MAS92-0403 (serotype D). Two serial strains, CDC-MAS92-0368 and CDC-MAS92-0804, from the same patient showed no significant difference in mutation rate (Table 3).
There was also significant heterogeneity in mutation rate among the six replicates of the same strains, as shown by the large SDs for many strains in Table 3. For example, among the six replicates of strain CDC-MAS92-0064, the mutation rates were 33.66 × 10−9, 52.44 × 10−9, 23.52 × 10−9, 48.30 × 10−9, 50.60 × 10−9, and 272.55 × 10−9. The SD among these replicates was 94.90 × 10−9, which exceeded the mean mutation rate of 80.18 × 10−9. Two other strains, Y289-90 and Y290-90, also showed similar patterns with SDs greater than their means. For the other 15 strains that produced mutants, the SDs in mutation rates ranged from about 3 to 70% of the mean.
Fluconazole MICs and stability of putative mutants.
Among the 3,668 mutants recorded (Table 3), a total of 252 random mutants were tested for the MIC of fluconazole. The 252 putative mutants included up to 3 from each of the six replicates of each original strain. All mutants grew on 8 μg of fluconazole per ml at 37°C when examined under a microscope (×100). The range and median of mutant MICs from each original strain are presented in Table 3.
Except for two mutants from the same replicate of strain CDC-MAS92-0368, the MICs for all mutants were elevated compared to those for their original strain. The increases in MIC were smaller for some mutants than for others. For example, one mutant from strain CDC-MAS92-0037 had a fluconazole MIC of 0.5 μg/ml, slightly higher than the MIC (0.25 μg/ml) for the original strain. Each of the other three mutants from CDC-MAS92-0037 had fluconazole MICs greater than 8 μg/ml.
The MICs for multiple mutants from the same culture were usually within a twofold range. However, in some strains, the MICs for random mutants from the same culture were very different. For example, the MICs for three mutants from the same plate of strain Y286-90 were 4, 8, and 32 μg/ml.
For most strains, mutants obtained from different replicates had different fluconazole MICs. However, there were exceptions. For instance, for all nine mutants from strain CDC-Y195-90, the MIC was the same, at 16 μg/ml. The median mutant MICs were not associated with serotype, geographic origin, or the original MIC (Tables 2 and 3).
All 10 putative random mutants tested multiple times during a 2-month period exhibited the same fluconazole MICs by both the NCCLS and colony size methods. Therefore, these mutant phenotypes are genetically stable.
Lack of correlation between the original MIC and the mutation rate.
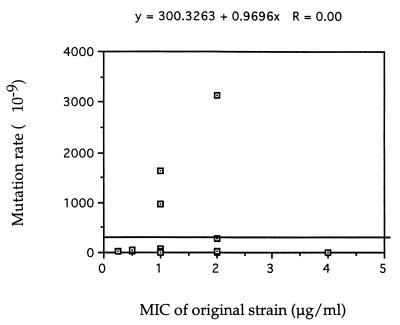
The extensive variation among the 21 strains in both the fluconazole MIC and the mutation rate allowed us to determine whether there was any correlation between the original MIC and the mutation rate to fluconazole resistance (27). As shown in Fig. 1, this analysis showed no correlation between the two variables among the 21 strains. When the data were analyzed separately by serotype, no positive correlation was found among the serotype A or D isolates (data not shown). Since only two serotype B strains were represented in this study, no correlation analysis was conducted for serotype B.
FIG. 1.
Scatter plot of fluconazole MIC and mutation rate to fluconazole resistance for 21 strains of C. neoformans.
Lack of significant correlation in fluconazole MICs between original strains and their derived mutants.
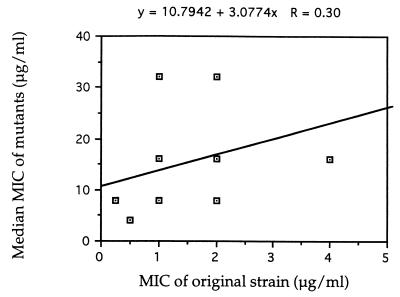
Because fluconazole MICs ranged widely among mutants from the same original strain, the correlation analysis was done only between the median MIC for mutants from each strain and the MIC for the original strain. The result of this analysis is presented in Fig. 2. This analysis demonstrated some correlation between these two MICs in this set of 21 strains. However, this correlation is not statistically significant (P > 0.10). Separate analyses of serotype A and serotype D strains did not yield any significant correlation in either group.
FIG. 2.
Scatter plot of fluconazole MIC for the original strains and median MIC for mutants of 18 C. neoformans strains.
DNA fingerprinting.
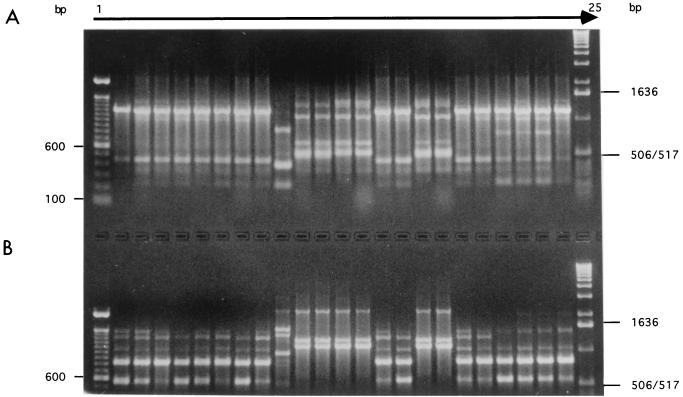
All 21 original strains and 252 mutants were genotyped using RAPDs obtained with primers OPA-03 and OPA-17. A representative picture of the PCR products is shown in Fig. 3. While there were some differences in band intensity between certain pairs of strains, no unambiguous change was observed when mutants were compared with their progenitors. This analysis also indicated no cross-contamination between strains during the multistep experimentation process.
FIG. 3.
Example of electrophoretic separation of RAPD fingerprints obtained by amplifying genomic DNA from 10 original strains of C. neoformans and 13 mutants using OPA-03 (5′ AGTCAGCCAC 3′) (A) and OPA-17 (5′-GACCGCTTGT-3′) (B) as single primers. Lanes 1 and 25 contain 100-bp and 1-kbp DNA ladders from GIBCO-BRL, respectively. Lanes 2 to 24 show RAPD profiles of strains CDC-MAS92-0064, CDC-MAS92-0064-mut1, CDC-MAS92-0064-mut2, CDC-MAS92-0064-mut3, CDC-MAS92-0064-mut4, CDC-MAS92-0064-mut5, CDC-Y288-90, CDC-Y288-90-mut1, CDC-B4496, CDC-Y494-91, CDC-Y494-91-mut1, CDC-MAS92-0088, CDC-MAS92-0088-mut1, CDC-MAS92-0109, CDC-MAS92-0109-mut1, CDC-MAS92-0232, CDC-MAS92-0232-mut1, CDC-MAS92-0245, CDC-MAS92-0245-mut1, CDC-MAS92-0316, CDC-MAS92-0316-mut1, CDC-MAS92-0804, and CDC-MAS92-0804-mut1 respectively.
DISCUSSION
This study investigated the patterns of mutation to fluconazole resistance in 21 strains of C. neoformans representing three serotypes, A, B, and D. Significant heterogeneity in mutation rates was observed among strains as well as among replicates within a single strain. For this set of strains, no statistically significant correlation was found between the original MIC and the mutation rate to fluconazole resistance. Randomly isolated mutants from a single strain often exhibited a wide range of fluconazole MICs. The results suggest dynamic and heterogeneous processes of mutation to fluconazole resistance in C. neoformans.
Growth on 8 μg of fluconazole per ml.
Significantly different patterns of growth on YMA with 8 μg of fluconazole per ml were observed among serotypes and among MLEE types within a serotype (Table 1). There might be two reasons for the different growth patterns. The first was due to the randomness of sampling. Aside from covering as many serotypes and genotypes as possible, the 101 strains initially screened were randomly picked from a total of over 400 strains. Whether the observed patterns of growth are representative for the whole collection or the global population of C. neoformans is unknown at present. The second possibility is that the different patterns of growth among serotypes and genotypes might reflect the underlying genetic differences among strains. For example, ET-1 is by far the most common MLEE type isolated from four regions in the United States (8). Compared to other MLEE types (e.g., ET-2 and ET-3), far more strains from ET-1 grew on 8 μg of fluconazole per ml (Table 1). Because the achievable fluconazole concentration in the CSF is estimated to be around 8 μg/ml, strains capable of growing on 8 μg of fluconazole per ml could have an advantage in the CSF compared with strains not able to grow, regardless of the MIC of fluconazole.
Discrepancies in MIC.
Two strains (CDC-MAS92-0248 and CDC-MAS92-0037) showed very different fluconazole MICs in two independent tests by two laboratories. There might be two reasons for this discrepancy. First, it is well known that the NCCLS protocol is sensitive to many factors, including the pH of the medium, the size and age of the inoculum, and the incubation time (12, 23). Differences in any one of these factors might influence the MIC reading (12, 23). Second, during clonal propagation and transfers between laboratories, random mutations occur. Some of these mutations might influence the strain sensitivity to fluconazole. The second possibility is rarely discussed but could be potentially important. For example, assuming a random mutation rate of 10−9 per base pair per cell division (2), with a genome size of about 23 × 106 bp in C. neoformans (31), a colony with 107 cells could potentially have 230,000 different genotypes, some of which might differ in their sensitivity to fluconazole. Therefore, there is a potential for generating new genotypes during every strain transfer event. Indeed, significant genetic and phenotypic differences were observed among clonal derivatives of a standard laboratory strain of C. neoformans (14). These clonal derivatives had been asexually transferred and maintained by different laboratories for unknown numbers of generations (14). In our case, given that we observed no growth on 8 μg of fluconazole per ml, the original MIC of 16 μg/ml for strain CDC-MAS92-0248 reported by Brandt et al. (Abstr. Annu. Meet. IDSA) might represent a genetically different colony from the one we used in this study. Similar possibilities could exist for strain CDC-MAS92-0037. Interestingly, no mutant was recovered from strain CDC-MAS92-0248 after plating about 6 × 109 cells on 8 μg of fluconazole per ml (Table 3).
Rates of mutation to fluconazole resistance.
The results in Table 3 indicated that some strains had very high mutation rates while others produced no resistant mutants. The significant heterogeneity among strains in mutation rates to drug resistance is similar to that found in bacteria (19). In most bacteria, the baseline mutation rate to antibiotic resistance is about 1 in 108 cells (19). However, mutator genotypes with high mutation rates (10−5 and 10−6) were found in clinical and natural samples of Escherichia coli and Salmonella pathogens. Strains of C. neoformans with high mutation rates found in this study might be similar to the mutator genotypes found in bacteria.
As far as we know, mutation rates to fluconazole resistance have not been accurately measured for multiple strains in other pathogenic yeast species. Typically, the mutation rate in eukaryotes is in the range of 10−5 to 10−6 per gene per generation (11). Mondon et al. (22) observed seven strains of C. neoformans with potentially very high mutation rates (from 0.7 to 4.6%) to fluconazole resistance, one strain from Israel and six serial isolates from an Italian patient with AIDS. In that study, single colonies in each of the seven strains were shown to contain mixed populations of C. neoformans for which the MICs were different, and some of the MICs exceeded 64 μg/ml (22). The six serial isolates from the Italian patient showed an increasing percentage of cells resistant to fluconazole at 64 μg/ml, with a strain from the sixth isolation having 4.6% resistant cells compared to 0.7% for a strain from the first isolation (22). However, the majority of resistant clonal populations in their study reverted to susceptible after being subcultured on drug-free media. Unlike their results, the MICs for the mutants recovered in the present study were stable and the two serial isolates (CDC-MAS92-0368 and CDC-MAS92-0804) from the same patient did not have significantly different mutation rates (Table 3). However, similar to the results of Mondon et al. (22), we observed no obvious association between the mutation rate to fluconazole resistance and serotype, geographic origin, or site of isolation.
These results are the first to demonstrate in vitro that resistance to fluconazole (MIC, ≥32 μg/ml) can emerge in strains of C. neoformans selected on 8 μg of fluconazole per ml. Although we are unable to accurately assess mutation rates to fluconazole resistance or range of fluconazole MICs in vivo, the results are consistent with the hypothesis that resistant strains for which the MICs are ≥32 μg/ml could arise in the CSF, possibly through a single mutation. When such an in vivo method becomes available, correlation between the in vitro mutation rate and the in vivo rate could be examined. If strong correlation exists, it might be possible to use the in vitro mutation rate measurements to predict the development of resistance in vivo and therefore improve clinical treatment with fluconazole.
The lack of a significant positive correlation between MIC and mutation rate is surprising. In addition, the MICs for the mutants were not significantly correlated with the MICs for the original strains (Tables 1 and 2; Fig. 2). Neither result is completely consistent with the hypothesis that fluconazole resistance develops through a process of stepwise or gradually increasing the MIC. Alternatively, the lack of correlation is consistent with the hypothesis that (i) there may be many mutational processes, some of which can increase the MIC dramatically, and (ii) mutation to fluconazole resistance is heterogeneous and dynamic in C. neoformans.
MIC and stability of mutants.
In this study, mutants obtained from a single strain often exhibited a wide range of fluconazole MICs. Many mutants derived from eight strains (Tables 2 and 3; CDC-MAS-92-0037, CDC-MAS-92-0064, CDC-MAS-92-0368, CDC-MAS-92-0804, CDC-Y195-90, E275, CDC-Y286-90, and CDC-Y290-90) demonstrated small increases (two- to fourfold) in fluconazole MIC above those for the original isolates. This finding is consistent with the hypothesis of small increases of MIC in single mutation experiments, as observed among serial isolates following treatment with fluconazole (1, 4, 13, 24, 29; Brandt et al., Abstr. Annu. Meet. IDSA). However, large increases (8- to 64-fold) in MIC over those for the original strains were also observed for many mutants, including some mutants from the eight strains mentioned above (Tables 2 and 3). It is likely that these different patterns of increased fluconazole MIC reflect different underlying mutations. Some mutations may cause major effects on fluconazole resistance, and others may cause small effects. The genetic basis of these differences is currently under investigation.
The results in Table 3 also suggest that some strains are capable of producing transient physiological adaptations or tolerance to fluconazole. Transient fluconazole resistance has been reported in Candida albicans by Marr et al. (20). In our study, strains CDC-MAS92-0037 and CDC-MAS92-0368 produced putative mutant colonies, but the MICs for these mutants were only 0.5 and 1 μg/ml, respectively. Despite the low MICs, these two mutants were capable of growing on YMA plates with 8 μg of fluconazole per ml, albeit very slowly. However, when more than 104 cells of these transient mutants were plated, no visible colony could be recovered after 72 h. Therefore, these transient mutations are probably different from the heteroresistant mutations found by Mondon et. al. (22).
Alternatively, phenotypic switching might have contributed to the transient physiological adaptations. Different colony morphology types resulting from phenotypic switching in three strains of C. neoformans were found to differ in their susceptibility to fluconazole (15). However, unlike the three discrete colony morphology types found by Goldman et al. (15), colony morphology among some mutants in this study was continuous and difficult to quantify. Further experiments are required to investigate whether these transient adaptations were the results of phenotypic switching.
The ability to undergo transient physiological adaptation and limited growth on 8 μg of fluconazole per ml could be important in allowing pathogens to survive and reproduce in CSF during fluconazole therapy. These adaptations might serve as intermediate stages or transition states that ultimately lead to the generation of stable resistant mutants.
Clinical relevance.
Successful fluconazole treatment of C. neoformans infections is influenced by both host factors (e.g., CD4 counts) and pathogen characteristics (9, 21). Under the same host conditions, infections with resistant strains will be harder to treat than those with susceptible strains. The heterogeneity in mutation rate and the wide range of MICs for mutants within and among strains found in this study are therefore relevant to empirical observations of various clinical outcomes in the treatment of cryptococcosis with fluconazole (9). Quantitative cultures of CSF are not routinely performed in clinical practice, but when they are performed, the yeast concentration is generally around 104 to 105 CFU/ml of CSF (25). Because the total volume of CSF in a host is between 130 and 150 ml (34), the total CFU in the CSF would be about 107, assuming that the yeasts are evenly distributed. Assuming a random mutational process and the rate of mutation as observed in this study (Table 3), mutation to fluconazole resistance might not be common or observable in the majority of clinical cases, as has been found in many studies (7, 18, 26, 33; Brandt et al., Abstr. Annu. Meet. IDSA).
In the absence of stable resistant mutations, transient physiological adaptations might occur. These adaptations could allow the infecting populations to survive and reproduce in vivo during fluconazole therapy. Even without transient physiological adaptations, the majority (27 of 31 [87%] in Table 1) of members of the most commonly recovered MLEE genotype (ET-1) could reproduce without mutation on plates containing 8 μg of fluconazole per ml. Therefore, mutation to resistance might not be necessary for these strains to survive and reproduce in hosts during fluconazole treatment. Furthermore, in the presence of mutations to fluconazole resistance, if the resistant mutants were not sufficiently competitive with the susceptible progenitors for in vivo survival and reproduction, they could be selectively purged by various host factors. Our results are therefore consistent with cases of treatment failures and relapses where the MIC for the relapse strain(s) was not elevated compared to the MIC for the pretreatment strain(s) (7, 10, 18, 26, 33).
In cases where genetically different strains were isolated before and after treatment with fluconazole (18, 28), the original strains might have had very low mutation rates and been unable to survive during fluconazole treatment. Subsequent reinfection by a more resistant or tolerant strain or by a strain with a higher mutation rate would explain changes in both the fluconazole MIC and the genotype of C. neoformans. Of the 21 strains analyzed here, 3 never developed resistant colonies, despite plating over 4.93 × 109 cells per strain (Table 3). Alternatively, the original infecting population might be heterogeneous, containing both susceptible and resistant subpopulations. Before fluconazole therapy, the fluconazole-susceptible subpopulation might dominate. The administration of fluconazole could select for the resistant subpopulation. Our in vitro results support the concept that subpopulations with different fluconazole MICs may exist in a clonal population derived from a single colony (see Results) (Table 3).
The results presented here clearly demonstrated that resistance to fluconazole in C. neoformans (MIC, ≥32 μg/ml) could develop after exposure to 8 μg of fluconazole per ml in vitro. The acquisition of this level of resistance (MIC, ≥32 μg/ml) did not require a stepwise (two- to fourfold) increase of MIC for 8 of the 21 strains. It should be emphasized that while the concentration of 8 μg/ml is the level of fluconazole achievable in the CSF (5, 32), the in vitro concentration of 8 μg/ml on solid and in liquid medium might not correspond to the in vivo situation. Unfortunately, there is currently no satisfactory method to accurately measure mutation rates to antibiotic resistance in vivo for any human pathogen.
Conclusions.
This study demonstrated that mutation to fluconazole resistance in C. neoformans in vitro is a dynamic and heterogeneous process. Significant heterogeneity in the mutation rate and the fluconazole MIC for mutants exist among strains as well as among replicates of the same strain. The results indicate that strains with higher fluconazole MICs (e.g., 2.0 to 4.0 μg/ml) are not necessary more likely to develop fluconazole resistance on 8 μg of fluconazole per ml than are strains with lower fluconazole MICs (e.g., 0.25 to 0.5 μg/ml). The results here are only partially consistent with the hypothesis that the development of fluconazole resistance in human pathogenic fungi is a gradual, multistep process. Multiple mechanisms of mutation to drug resistance in C. neoformans are likely. Whether the observed significant differences in mutation rates among strains in vitro are reproducible in vivo awaits investigation.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grants AI 25783 and AI 44975 from the National Institutes of Health.
We thank Tarek Hanna for technical assistance and Mary Brandt for providing strains and for her critical comments on an earlier version of the manuscript. Some of the CDC strains were collected through the CDC Cryptococcal Active Surveillance, a population-based active surveillance for cryptococcal disease. This laboratory is a component of the Duke University Mycology Research Unit.
REFERENCES
- 1.Armengou A, Porcar C, Mascaro J, Garcia-Bragaro F. Possible development of resistance to fluconazole during suppressive therapy for AIDS-associated cryptococcal meningitis. Clin Infect Dis. 1996;23:1337–1338. doi: 10.1093/clinids/23.6.1337-a. [DOI] [PubMed] [Google Scholar]
- 2.Atlas R M. Principles of microbiology. W. C. Dubuque, Iowa: Brown Publishers; 1997. [Google Scholar]
- 3.Berg J, Clancy C J, Nguyen M H. The hidden danger of primary fluconazole prophylaxis for patients with AIDS. Clin Infect Dis. 1998;26:186–187. doi: 10.1086/517056. [DOI] [PubMed] [Google Scholar]
- 4.Birley H D L, Johnson E M, McDonald P, Parry C, Caret P B, Warnock D W. Azole drug resistance as a cause of clinical relapse in AIDS patients with cryptococcal meningitis. Int J STD AIDS. 1995;6:353–355. doi: 10.1177/095646249500600510. [DOI] [PubMed] [Google Scholar]
- 5.Brammer K W, Farrow P R, Faulkner J K. Pharmacokinetics and tissue penetration of fluconazole in human. Rev Infect Dis. 1990;12:S318–S326. doi: 10.1093/clinids/12.supplement_3.s318. [DOI] [PubMed] [Google Scholar]
- 6.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt M E, Pfaller M A, Hajjeh R A, Graviss E A, Rees J, Spitzer E D, Pinner R W, Mayer L W Cryptococcal Disease Active Surveillance Group. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated cryptococcosis. J Infect Dis. 1996;174:812–820. doi: 10.1093/infdis/174.4.812. [DOI] [PubMed] [Google Scholar]
- 8.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W the CDC Cryptococcal Disease Active Surveillance Group. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 10.Casadevall A, Spitzer E D, Webb D, Rinaldi M G. Susceptibilities of serial Cryptococcus neoformans isolates from patients with recurrent cryptococcal meningitis to amphotericin B and fluconazole. Antimicrob Agents Chemother. 1993;37:1383–1386. doi: 10.1128/aac.37.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Rates of spontaneous mutations. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinel-Ingroff A, White T, Pfaller M A. Antifungal agents and susceptibility tests. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 1640–1652. [Google Scholar]
- 13.Franz R, Kelly S L, Lamb D C, Kelly D E, Ruhnke M, Morschhauser J. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franzot S P, Mukherjee J, Cherniak R, Chen L, Hamdan J S, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman D L, Fries B C, Franzot S P, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J Clin Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klepser M E, Ernst E J, Pfaller M A. Update on antifungal resistance. Trends Microbiol. 1997;5:372–375. doi: 10.1016/S0966-842X(97)01108-6. [DOI] [PubMed] [Google Scholar]
- 18.Klepser M E, Pfaller M A. Variation in electrophoretic karyotype and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987 to 1994. J Clin Microbiol. 1998;36:3653–3656. doi: 10.1128/jcm.36.12.3653-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 20.Marr K A, Lyons C N, Rustad T, Bowden R A, White T C. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob Agents Chemother. 1998;42:2584–2589. doi: 10.1128/aac.42.10.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mondon P, Petter R, Amalfitano G, Luzzati R, Concia E, Polacheck I, Kwon-Chung K J. Heteroresistance to fluconazole and voriconazole in Cryptococcus neoformans. Antimicrob Agents Chemother. 1999;43:1856–1861. doi: 10.1128/aac.43.8.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard. NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.Paugam A, Dupouy-Camet J, Blanche P, Gangneaux J P, Tourte-Schaefer C, Sicard D. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin Infect Dis. 1993;17:431–436. doi: 10.1093/clinids/19.5.975-a. [DOI] [PubMed] [Google Scholar]
- 25.Perfect J R, Durack D T, Gallis H A. Cryptococcemia. Medicine. 1983;62:98–109. doi: 10.1097/00005792-198303000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller M, Zhang J, Messer S, Tumberland M, Mbidde E, Jessup C, Ghannoum M. Molecular epidemiology and antifungal susceptibility of Cryptococcus neoformans isolates from Ugandan AIDS patients. Diagn Microbiol Infect Dis. 1998;32:191–199. doi: 10.1016/s0732-8893(98)00095-9. [DOI] [PubMed] [Google Scholar]
- 27.Sokal R R, Rohlf F J. Biometry: the principles and practices of statistics in biological research. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1981. [Google Scholar]
- 28.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistence, replacement and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanden Bossche H, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 30.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickes B L, Moore T D E, Kwon-Chung K J. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140:543–550. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 32.Wildfeuer A, Laufen H, Schmalreck A F, Yeates R A, Zimmermann T. Fluconazole: comparison of pharmacokinetics, therapy and in vitro susceptibility. Mycoses. 1997;40:259–265. doi: 10.1111/j.1439-0507.1997.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 33.Witt M D, Lewis R J, Larsen R A, Milefchik E N, Leal M A, Haubrich R H, Richie J A, Edwards Jr J E, Ghannoum M A. Identification of patients with acute AIDS-associated cryptococcal meningitis who can be effectively treated with fluconazole: the role of antifungal susceptibility testing. Clin Infect Dis. 1996;22:322–328. doi: 10.1093/clinids/22.2.322. [DOI] [PubMed] [Google Scholar]
- 34.Woodburne R T, Burkel W E. Essentials of human anatomy. 9th ed. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 35.Xu J, Ramos A R, Vilgalys R J, Mitchell T G. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol. 2000;38:1214–1220. doi: 10.1128/jcm.38.3.1214-1220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Vilgalys R J, Mitchell T G. Colony size can be used to determine the MIC of fluconazole for pathogenic yeasts. J Clin Microbiol. 1998;36:2383–2385. doi: 10.1128/jcm.36.8.2383-2385.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]