Abstract
The mutant prevention concentration (MPC) represents a threshold above which the selective proliferation of resistant mutants is expected to occur only rarely. A provisional MPC (MPCpr) was defined and measured for five fluoroquinolones with clinical isolates of Streptococcus pneumoniae. Based on their potential for restricting the selection of resistant mutants, the five fluoroquinolones, in descending order, were found to be moxifloxacin > trovafloxacin > gatifloxacin > grepafloxacin > levofloxacin. For several compounds, 90% of about 90 clinical isolates that lacked a known resistance mutation had a value of MPCpr that was close to or below the serum levels that could be attained with a dosing regimen recommended by the manufacturers. Since MPCpr overestimates MPC, these data identify moxifloxacin and gatifloxacin as good candidates for determining whether MPCpr can be used as a guide for choosing and eventually administering fluoroquinolones to significantly reduce the development of resistance.
Antibiotic resistance among human pathogens now occurs in almost every bacterial species for which antibiotic therapies exist (15, 16, 26). In the case of Streptococcus pneumoniae, resistance to penicillin and erythromycin has become so widespread that clinicians have started to use the fluoroquinolones for therapy. As a result, drug resistance is now emerging against the quinolones (4). More active fluoroquinolones are becoming available, but new treatment strategies must accompany use of those agents to halt the selection of resistant mutants before the entire quinolone class of drugs becomes ineffective.
It has been suggested that if bacterial cells must attain two concurrent resistance mutations for growth in the presence of a quinolone, then few mutants would be selectively amplified because double mutations should rarely occur (19, 21, 28, 29). For example, bacterial populations may reach 1010 cells in human infections (1, 9, 15), but at a mutation frequency of 10−7, more than 1014 bacteria (107 × 107) would be required to detect two concurrent fluoroquinolone-resistant mutations. When we examined the effect of fluoroquinolone concentration on the selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus, we found a concentration with each organism at which no mutant was recovered when more than 1010 cells were applied to agar plates (5). This drug concentration, which we designated as the mutant prevention concentration (MPC), would require a bacterial cell to develop more than one resistance mutation for growth. Thus at concentrations above the MPC, a bacterial population size greater than that normally present during infection would be necessary to observe outgrowth of a resistant mutant. Since fluoroquinolone structure affects the value of the MPC (5), it appeared that the MPC might serve as a simple measure of antibiotic potency that incorporates the ability of a compound to restrict selection of resistant mutants (28).
In principle MPC also represents a dosing threshold above which mutants should rarely arise; use of MPC would add consideration of the development of resistance to the traditional goal of clearing infection. With a clinical isolate of Mycobacterium tuberculosis, we found that MPCs for two new C-8-methoxy fluoroquinolones are below the maximum attainable drug concentration in serum (6). Thus the possibility exists that fluoroquinolones might be administered such that serum drug concentrations in patients exceed the MPC. Whether this is true for other quinolone-pathogen combinations and for large numbers of clinical isolates is unknown.
Examination of large numbers of clinical isolates generally involves measurement of antibiotic potency in terms of the MIC. With the agar dilution method, approximately 105 CFU is applied to each of a series of agar plates containing various antibiotic concentrations (18). The concentration that allows no colony formation is taken as the MIC. Measurement of MPC is carried out using the same strategy except that more cells, on the order of 1010, are applied to agar plates. Consequently, it should be possible to perform MPC measurements for a large number of clinical isolates. Those measurements could then be compared with published values of pharmacokinetic parameters to determine whether and for how long the serum drug concentration would be above the MPC. The relationship between pharmacokinetics and MPC could then be used to identify compounds for further examination of the inability to restrict selection of resistance.
In the present work, we tested five fluoroquinolones (gatifloxacin, grepafloxacin, levofloxacin, moxifloxacin, and trovafloxacin) with clinical isolates of S. pneumoniae. A hierachy of potency was clear. For the most active compounds, moxifloxacin and gatifloxacin, a provisional MPC (MPCpr), which overestimates MPC by about twofold, was below the maximum serum drug concentrations attained with the doses recommended for streptococcal pneumonia. Thus these fluoroquinolones may be useful in clinical trials to determine the utility of MPC in reducing the development of resistance. To facilitate further testing of MPC, we describe an empirical relationship that will allow MPC to be calculated from standard MIC measurements made by the agar dilution method.
MATERIALS AND METHODS
Bacterial strains and bacteriological methods.
Isolates of S. pneumoniae were obtained from the Clinical Microbiology Division, Royal University Hospital, Saskatoon, Saskatchewan, Canada. No preselection criterion was used that would favor inclusion or exclusion of resistant isolates, and care was taken to avoid obtaining more than one specimen from a given patient. The MIC was determined by the standard twofold agar dilution method (18).
For MPC measurements, starter cultures were spread on blood agar plates (six plates per isolate) (PML, Richmond, Canada) and incubated overnight (18 to 24 h) at 35 to 37°C in 5% CO2. Bacterial cells were then transferred from the plates to 500 ml of Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.), followed by overnight incubation at 35 to 37°C in 5% CO2. After incubation, cultures were estimated to have concentrations of ≥3 × 108 CFU per ml by turbidity measurements. Cultures were concentrated by centrifugation at 5,000 × g for 30 min and resuspended in 3 ml of Todd-Hewitt broth. Aliquots of 200 μl, containing ≥1010 CFU, were applied to tryptic soy agar plates containing 5% sheep red blood cells (18). For each experiment, agar dilution plates were prepared by incorporating fluoroquinolones at seven concentrations into the tryptic soy agar-sheep red blood cell plates (plates were stored at 4°C and used within 7 days of preparation). Each experiment included the fully susceptible control strain S. pneumoniae ATCC 49619. Inoculated plates were incubated for 24 h at 35 to 37°C in 5% CO2 and then screened for growth. All plates were reincubated for an additional 24 h and reexamined. MPCpr was recorded as the lowest antibiotic concentration that allowed no growth. In a few cases a thin film was observed after incubation on plates with high fluoroquinolone concentrations. When these plates were washed with a growth medium that was reapplied to drug-free agar, no growth was observed. These plates were scored as negative for growth. All MPCpr determinations were made in duplicate, and the results were identical.
Fluoroquinolones.
Sources of the fluoroquinolones were Bayer AG (moxifloxacin), Bristol-Myers Squibb (gatifloxacin), Glaxo-Wellcome (grepafloxacin), Johnson-Ortho (levofloxacin), and Pfizer, Inc. (trovafloxacin). Powdered forms of each compound were dissolved according to manufacturers' instructions. Stock solutions were used as fresh preparations or from samples stored at −70°C.
DNA isolation, amplification and nucleotide sequence determination.
Selected isolates of S. pneumoniae were grown on brain heart infusion agar (Difco) containing 10% defibrinated sheep blood (Hemostat Laboratories, Dixon, Calif.) following high-density inoculation. Incubation was overnight at 37°C with 5% CO2. Bacteria grown as confluent lawns were recovered from agar plates by washing with 2 ml of Todd Hewitt broth per plate. Cells were concentrated by centrifugation, washed once with lysis buffer (50 mM Tris-HCl [pH 8.0] and 5 mM EDTA), and resuspended in 400 μl of lysis buffer per plate. Then 50 μl of 10% sodium dodecyl sulfate and 20 μl of proteinase K (10 mg/ml) were added, and the mixture was incubated, first at 55°C for 30 min and then at 37°C for 1.5 h. Cell lysates were extracted with phenol, then with phenol:chloroform (1:1), and finally with chloroform. DNA was precipitated with ethanol and recovered by centrifugation. DNA was then dissolved in Tris-EDTA buffer (10 mM Tris–HCl [pH 8.0] and 1 mM EDTA) and treated with a final concentration of 100 μg of RNase A per ml for 1 h at 37°C. DNA was reprecipitated with ethanol and dissolved in Tris-EDTA buffer. The nucleotide sequences of the quinolone-resistance-determining regions of parC and gyrA were determined with an automated DNA sequencer using primer SP-parC.seq (5′ TCA GCG CCG TAT TCT TTA TTC TAT G 3′) and primer SP-gyrA.seq (5′ TCG AGA TGG CTT AAA ACC TGT TCA C 3′) after PCR amplification of DNA fragments using primers SP-parCfwd (5′ GTC TAA CAT TCA AAA CAT GTC CCT G 3′), SP-parCrev (5′ TCT TTC TCC GTA TCG TCA AAG TTC 3′) for parC and SP-gyrAfwd (5′ TGT CAA TCT GAC AAA GGA GAT GAA G 3′), and SP-gyrArev (5′ CCA GTT GCT CCA TTA ACC AAA AG 3′) for gyrA.
RESULTS AND DISCUSSION
Estimation of MPC.
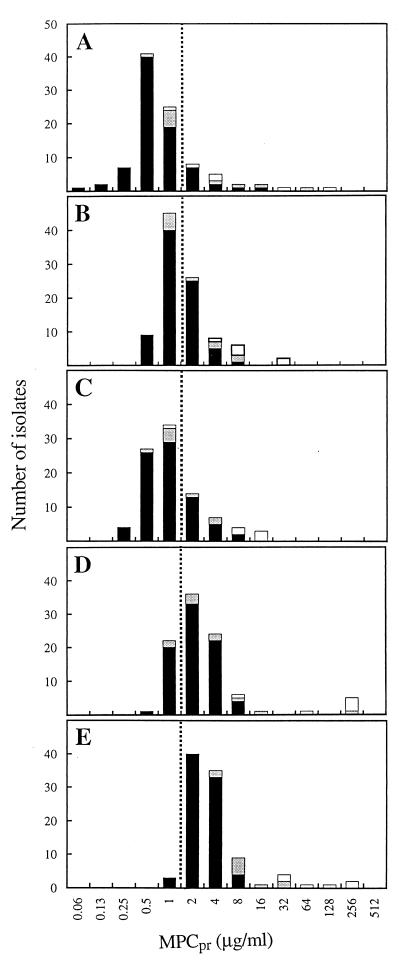
In preliminary work with S. pneumoniae strain ATCC 49619, we found that moxifloxacin-resistant mutants are recovered only within a narrow, twofold drug concentration range even when more than 1010 cells were examined. A similar finding was obtained previously with another strain (23) and with clinafloxacin, a C-8-chlorine fluoroquinolone (20). A narrow drug concentration range for mutant selection led us to expect that agar plates used in a standard twofold dilution analysis with clinical isolates would exhibit either confluent growth or no colonies, at least for the C-8-methoxy compounds moxifloxacin and gatifloxacin. When 1010 cells were applied per plate, a sharp drop in growth was seen over a range of one dilution. When the minimal concentration at which no colony was recovered (MPCpr) was plotted against the number of isolates, distinct peaks were seen in the distribution (Fig. 1). The peak for moxifloxacin appeared at the lowest drug concentration; therefore, moxifloxacin was the most potent fluoroquinolone by this assay.
FIG. 1.
Distribution of S. pneumoniae isolates with respect to MPCpr. MPCpr was determined for each isolate by the agar dilution method. White areas of bars represent isolates containing parC mutations known to confer resistance; shaded regions represent isolates containing parC mutations that have not been demonstrated by genetic tests to confer quinolone resistance; solid regions represent unsequenced isolates. Dashed line is for alignment of panels. Panels: A, moxifloxacin; B, gatifloxacin; C, trovafloxacin; D, grepafloxacin; and E, levofloxacin.
We consider the values of MPCpr shown in Fig. 1 to be provisional because the high inocula used could have raised the drug concentration required to prevent the isolation of mutants (e.g., crowding may have interfered with access of the fluoroquinolone to the cells). To test this idea, four isolates were examined by applying 108 cells to each of 100 agar plates at a concentration of moxifloxacin or levofloxacin that was one dilution lower than that of MPCpr. In each case no growth was observed, indicating that MPCpr overestimates MPC. These data also argue against complications due to autolysis occurring at high cell density, since that would have made MPCpr an underestimation of MPC. In a separate study using a laboratory isolate of S. pneumoniae (ATCC 49619), various fluoroquinolone concentrations were tested with dilute bacterial cultures and many agar plates so that mutants were recovered as single colonies. The MPC was 0.5 μg of moxifloxacin per ml and 2.6 μg of levofloxacin per ml, respectively (X. Li, X. Zhao, and K. Drlica, unpublished observation). With this S. pneumoniae strain, the MPCpr was 1 μg of moxifloxacin per ml and 4 μg of levofloxacin per ml, using the twofold agar dilution assay described in Materials and Methods (data not shown). Consequently, MPCpr overestimates MPC by about twofold for both the most active and the least active compounds in the present study. This small inoculum effect was deemed acceptable for the present analysis because the ability to load more cells on agar plates greatly reduced the number of plates necessary for examination of large numbers of isolates.
Since working with large numbers of S. pneumoniae is cumbersome, we determined an empirical relationship between MPCpr and the MIC of the drug so that the MPCpr of a drug can be calculated from the MIC of the drug. When the MIC of a drug was measured by the agar dilution method, most of the isolates exhibited a four- to eightfold difference between the MIC and MPCpr (two to three dilutions) of the five fluoroquinolones tested (Table 1). The ratio was slightly higher for trovafloxacin and grepafloxacin. When the ratio of the MPCpr to the MIC of a drug was calculated using values that exceeded those for 90% of the isolates (MPCpr90 and MIC90), the ratio was 8 for three of the compounds and 16 for trovafloxacin and grepafloxacin (Table 2). Similar ratios were obtained for the MPCpr and the MIC of these drugs with the laboratory strain ATCC 49619 (data not shown). When cells were distributed to more agar plates and when smaller quinolone concentration increments were used to obtain single resistant colonies. The MPC/MIC ratio for moxifloxacin and levofloxacin with strain ATCC 49619 was lower (three- to fourfold, using an MIC of the drugs that blocks growth of 99% of the CFUs; X. Li, X. Zhao, and K. Drlica, unpublished observation). We conclude that the ratios listed in Table 2 are overestimates of the ratio of the MPC to the MIC of the drugs. Therefore, using the numbers in Table 2 to calculate the MPCpr from the MIC of the drugs will provide a conservative estimate of MPC. Additional studies are required with other S. pneumoniae populations to determine whether the ratios we calculated are generally applicable to this bacterial species.
TABLE 1.
Relationship between MIC and MPCpr for clinical isolates of S. pneumoniae
| Fluoroquinolone | No. of isolatesa having ratios of MPCpr/MICb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | |
| Moxifloxacin | 2 | 10 | 35 | 29 | 12 | 3 | 3 | 3 | 2 |
| Gatifloxacin | 2 | 23 | 41 | 21 | 9 | 3 | 0 | 1 | 0 |
| Trovafloxacin | 1 | 1 | 27 | 31 | 22 | 9 | 4 | 2 | 0 |
| Grepafloxacin | 0 | 5 | 13 | 27 | 31 | 15 | 3 | 1 | 0 |
| Levofloxacin | 2 | 36 | 41 | 11 | 6 | 2 | 1 | 1 | 1 |
Ratio of MPCpr to MIC was determined for each isolate, and the number of isolates having the indicated values of the ratio was tabulated. This data set includes strains that have known resistance mutations, since the mutations have little effect on the ratio. Data for a few isolates were not obtained for every compound, causing the total number of isolates tabulated to vary among the compounds.
MPCpr and MIC were determined by plating either 1010 (MPCpr) or 105 (MIC) CFU on agar containing twofold dilutions of fluoroquinolone.
TABLE 2.
Fluoroquinolone potency based on MPCpr
| Fluoroquinolone | MPCpr50a (μg/ml) | MPCpr90a (μg/ml) | MIC90b (μg/ml) | MPCpr90/ MIC90 |
|---|---|---|---|---|
| Moxifloxacin | 0.5 | 2 | 0.25 | 8 |
| Trovafloxacin | 1 | 4 | 0.25 | 16 |
| Gatifloxacin | 1 | 4 | 0.5 | 8 |
| Grepafloxacin | 2 | 8 | 0.5 | 16 |
| Levofloxacin | 4 | 8 | 1 | 8 |
MPCpr in which 50% (MPCpr50) or 90% (MPCpr90) of the isolates had a value lower than the one given (determined from data in Fig. 1, omitting data from six resistant mutants, as indicated in the text).
MIC in which 90% of the isolates had a value lower than the one given (determined using the isolates described in Fig. 1, omitting data from six resistant mutants, as indicated in the text).
Some of the increased incidence of fluoroquinolone-resistant S. pneumoniae (4) has been associated with treatment of penicillin-resistant cases of pneumonia (discussed in 4). For the newer fluoroquinolones the value of MIC90 is unaffected by pneumococcal resistance to penicillin and/or other agents (3). Likewise, no increase in MPCpr was associated with the presence of penicillin resistance in the present data set (data not shown). Thus potential cross-resistance between the two classes of compounds does not appear to be a problem.
Fluoroquinolone-resistant mutants.
Some of the isolates examined at high inoculum concentrations required exceptionally high concentrations of fluoroquinolone to prevent colony formation (Fig. 1, open and shaded areas of bars). Since ciprofloxacin and levofloxacin have been used extensively in Canada as therapy for streptococcal pneumonia and since many fluoroquinolone-resistant isolates have emerged (4), we suspected that at least some of the isolates in the present study contained mutations in the target genes, parC (topoisomerase IV) and/or gyrA (gyrase). To test this idea, DNA was obtained from 22 isolates, and the nucleotide sequences of the quinolone-resistance-determining regions of the two genes were determined. As shown in Table 3, 17 isolates were parC mutants, 7 of which also contained a gyrA mutation. Six of the parC mutants (isolate numbers 10, 12, 13, 15, 16, and 18) contained alleles known from genetic studies (10, 11, 16, 21) to confer resistance (predicted amino acid changes of Ser-79 to Phe and Asp-83 to Asn). Of these, three also contained a GyrA alteration (Ser-81 changed to Phe or Tyr) known to confer resistance. Since MPC is based on the recovery of colonies grown from wild-type populations (5), the six resistant mutants (white portions of bars in Fig. 1) were excluded from determination of MPCpr90 (Table 2). For 11 other isolates the nucleotide sequence predicted changes of Ser-52 to Gly, Asn-91 to Asp, or Lys-137 to Asn in the ParC protein (shaded portion of bars in Fig. 1). To our knowledge, genetic studies have not been performed that attribute resistance to these alleles. Consequently, we did not exclude these strains or ones lacking a parC or gyrA mutation from the determination of MPCpr90. These isolates may contribute to the finding that MPCpr90 in Table 2 overestimates MPC.
TABLE 3.
Quinolone-resistance alleles associated with high values of MPC
| Isolate no. | MPC (μg/ml)a
|
Changes in QRDRb
|
|||||
|---|---|---|---|---|---|---|---|
| Gati | Grepa | Levo | Moxi | Trova | parC | gyrA | |
| Wtc | 2 | 4 | 4 | 1 | 1 | ||
| 10 | 4 | 256 | 32 | 4 | 16 | D83N (GAT to AAT) | S81F (TCC to TTC) |
| 12 | 8 | >256 | 32 | 4 | 16 | S79F (TCT to TTT) | S81Y (TCC to TAC) |
| 13 | 32 | 64 | 128 | 32 | 8 | S79F (TCT to TTT) | S114G (AGT to GGT) |
| S52G (AGC to GGA) | |||||||
| N91D (AAC to GAC) | |||||||
| 15 | 8 | 8 | 32 | 1 | 1 | S79F (TCT to TTT) | None |
| 16 | 32 | 256 | 32 | >128 | 16 | S79F (TCT to TTT) | S81F (TCC to TTC) |
| 18 | 8 | >256 | 64 | 2 | 8 | S79F (TCT to TTT) | None |
| 27 | 8 | >256 | 16 | >64 | 4 | S52G (AGC to GGC) | S114G (AGT to GGT) |
| N91D (AAC to GAC) | |||||||
| 33 | 2 | 1 | 4 | 1 | 2 | K137N (AAG to AAT) | None |
| 35 | 4 | 4 | 8 | 4 | 4 | K137N (AAG to AAT) | None |
| 36 | 1 | 1 | 4 | 8 | 1 | K137N (AAG to AAT) | None |
| 37 | 1 | 2 | >512 | 4 | 1 | K137N (AAG to AAT) | None |
| 42 | 1 | 4 | 32 | 1 | >16 | K137N (AAG to AAT) | None |
| 43 | 0.5 | 1 | 8 | 2 | 8 | None | None |
| 48 | 1 | 2 | 8 | 1 | 1 | K137N (AAG to AAT) | None |
| 51 | 1 | 2 | 8 | 0.5 | 1 | K137N (AAG to AAT) | None |
| 64 | 1 | 4 | 8 | 2 | 1 | None | None |
| 74 | 8 | 16 | 8 | 16 | >8 | S52G (AGC to GGC) | None |
| 78 | 0.5 | 1 | 8 | 0.5 | 1 | None | None |
| 87 | 1 | 1 | 8 | 0.5 | 0.5 | None | None |
| 89 | 4 | 8 | 8 | 1 | 1 | S52G (AGC to GGC) | S114G (AGT to GGT) |
| 91 | 1 | 2 | 32 | 1 | 0.5 | S52G (AGC to GGC) | S114G (AGT to GGT) |
| N91D (AAC to GAC) | |||||||
| 103 | 4 | 4 | 8 | 2 | 2 | None | None |
Gati, gatifloxacin; Grepa, grepafloxacin; Levo, levofloxacin; Moxi, moxifloxacin; and Trova, trovafloxacin.
QRDR, quinolone-resistance-determining region (17). Amino acid in wild-type protein is indicated before its number in the protein, followed by the amino acid change. D, aspartic acid; F, phenylalanine; G, glycine; K, lysine; N, asparagine; S, serine; Y, tyrosine.
Wild-type strain ATCC 49619.
When the average MIC for the resistant isolates was determined, the five fluoroquinolones could be ranked in terms of potency, in descending order, with moxifloxacin = trovafloxacin > gatifloxacin > grepafloxacin > levofloxacin (not shown). A similar order in potency was seen with a separate collection of resistant mutants (12), although in that case grepafloxacin was, on average, more potent than gatifloxacin. Variants with identical predicted amino acid changes occasionally exhibited different response patterns of susceptibility to the five fluoroquinolones (Table 3). These differences, which were reproducible, probably arose from unidentified mutations present in some isolates but absent from others.
Relationship of MPCpr to pharmacokinetics.
For MPC to be a therapeutically useful parameter, its value must be below the serum and tissue drug concentrations attained following administration of drug doses that are safe for patients. Recommended doses and pharmacokinetic parameters for the five fluoroquinolones are listed in Table 4. Since trovafloxacin and grepafloxacin have been withdrawn from the market, they are not considered further here. Moxifloxacin, the most potent of the compounds tested, has a maximum serum drug concentration of 4.5 μg/ml, about twice that of MPCpr for more than 90% of the isolates. Since the half-life of moxifloxacin is 12 h, daily dosing should keep concentrations of moxifloxacin in serum above the MPCpr for most of the treatment time. This should help restrict the selection of resistant mutants. Since MPCpr overestimates MPC by about twofold and since quinolone concentration in relevant tissue may be higher than in serum (J. Andrews, D. Honeybourne, G. Jevons, and R. Wise, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A29, 1998), gatifloxacin may also restrict the selection of resistant mutants, especially if administered twice daily. We speculate that levofloxacin may require higher doses, perhaps administered twice daily, to attain the same potency with respect to restricting selection of resistant mutants. Clinical trials are now required to test these ideas.
TABLE 4.
Relationship of pharmacokinetics and MPC
| Fluoroquinolone | MPCpr90 (μg/ml) | Dosea (mg) | Cmaxb (μg/ml) | t1/2b (h) | Source |
|---|---|---|---|---|---|
| Moxifloxacin | 2 | 400 | 4.5 | 12 | Ref. 25 |
| Gatifloxacin | 4 | 400 | 4.2 | 8 | WWWc |
| Trovafloxacin | 4 | 200 | 3.1 | 12 | WWWd |
| Grepafloxacin | 8 | 600 | <2.7e | 14 | Ref. 8 |
| Levofloxacin | 8 | 500 | 5.7 | 8 | WWWf |
Recommended dose for streptococcal pneumonia.
Maximum drug concentration, at steady state after multiple, once-daily doses at the level recommended for streptococcal pneumonia except for grepafloxacin (see footnote e).
U.S. prescribing information for Tequin (gatifloxacin), available at http://www.tequin.com/.
U.S. prescribing information for Trovan (trovafloxacin), available at http://www.trovanpi.html.
No steady state pharmacokinetic profile for grepafloxacin at a 600-mg multiple, once-daily dose was available; the value of the maximum drug concentration listed was obtained with a dose of 800 mg.
U.S. prescribing information for Levaquin (levofloxacin), available at http://www.levaquin.com/.
The serum drug concentrations listed in Table 4 represent total concentrations. Fluoroquinolones bind to serum proteins, so additional corrections may be necessary when particular compounds are compared. The extent of binding varies among the compounds, but in general less than half of the total drug that is present is bound (27); consequently, protein binding probably has little effect on the conclusions reached above. A future refinement may utilize bioassays rather than high-pressure liquid chromatography to determine relevant concentrations.
Conclusions.
The endpoint of antibiotic dosing strategies, such as those based on area under the inhibitory concentration curve (reviewed in 24), has often been clearance of infection, measured either as survival in animal models or loss of symptoms in humans. Widespread development of resistance suggests that consideration should also be given to restricting the selection of resistant mutants. One approach has utilized the ratio of maximum concentration of drug in serum to the MIC of the drugs; values have been found that restrict mutant outgrowth in vitro (2) and correlate with animal survival (7). However, this ratio does not take into account the time at which concentration is high, which may be important for preventing the development of resistance. Consequently, we have been examining the possibility of using a new potency parameter termed the MPC, the concentration that allows no growth of first-step mutants. Compounds and dosing protocols can readily be compared for the time that drug concentration in tissue exceeds the MPC.
The data described above show that with S. pneumoniae MPCpr can be below the serum drug concentration that can be safely achieved for moxifloxacin and gatifloxacin. This may also be true for levofloxacin if the compound is administered more frequently and at higher levels. Thus one of the criteria has been met for using the MPC to slow the development of quinolone resistance. Whether keeping relevant tissue concentrations of this drug above the MPC will actually restrict the selection of mutants in animal models or in human patients has not been determined.
For some antibiotic-pathogen combinations, the MPC may not fall below tissue drug concentrations that are achievable with safe doses. Although the antibiotic often may clear infection, for such cases enrichment of resistant mutants will gradually erode the utility of the agent, especially if administered to tens of millions of patients each year. Such antibiotics can be preserved by combination therapy if such therapy is instituted before resistance becomes too extensive (for discussion see 28).
Measurement of MPC with S. pneumoniae is cumbersome, so we determined an empirical relationship with the MIC of the fluoroquinolones examined, a more readily determined parameter. Use of this relationship should help determine whether use of a given compound as therapy against a particular population of S. pneumoniae will restrict the development of resistance, provided that no silent target mutations are present (see below). It may be possible to extend this conclusion to individual isolates when susceptibility measurements from clinical laboratories can be related to the MIC of different drugs as determined by agar dilution.
An important question is whether moxifloxacin- or gatifloxacin-based therapy for S. pneumoniae infection is influenced by the continued use of older compounds such as ciprofloxacin and levofloxacin. The latter agents have topoisomerase IV as their primary target, while the primary target for gatifloxacin and moxifloxacin is gyrase (10, 22, 23). This difference in target means that first-step mutants selected by ciprofloxacin and levofloxacin may remain susceptible to gatifloxacin and moxifloxacin (22). However, we have shown with Escherichia coli that a parC resistance allele, which has no effect on the MIC of the drugs, can increase by orders of magnitude the frequency at which resistant mutants are selected by C-8-methoxy fluoroquinolones (29). The effect of the parC mutation is to require only one resistance mutation rather than two for bacterial growth in the presence of high concentrations of the C-8-methoxy compound. This principle appears to hold for S. pneumoniae (X. Li, X. Zhao, and K. Drlica, unpublished observation and 23). Thus we expect continued use of ciprofloxacin and levofloxacin to seriously shorten the useful lifespan of moxifloxacin and gatifloxacin; although the latter two compounds may be potent enough to treat infection caused by parC mutants (selected by ciprofloxacin and levofloxacin), parC gyrA double mutants would be readily selected by moxifloxacin or gatifloxacin if the pathogens already contain a parC resistance allele. Thus, slowing the development of resistance may involve careful management of compounds within the same general class.
ACKNOWLEDGMENTS
We thank J. de Azavedo, M. Gennaro, and S. Kayman for critical comments on the manuscript, D. Leciuk for excellent clerical assistance, and S. Borsos for technical support.
This work was supported by grants to J.B. from Bayer AG and to K.D. from the National Institutes of Health (AI35257) and Bayer AG.
REFERENCES
- 1.Bingen E, Lambert-Zechovsky N, Mariani-Kurkdjian P, Doit C, Aujard Y, Fournerie F, Mathieu H. Bacterial counts in cerebrospinal fluid of children with meningitis. Eur J Clin Microbiol Infect Dis. 1990;9:278–281. doi: 10.1007/BF01968060. [DOI] [PubMed] [Google Scholar]
- 2.Blaser J, Stone B, Groner M, Zinner S. Comparative study with enoxacin and netilmicin in a pharmacodynamic model to determine importance of ratio of antibiotic peak concentration to MIC for bactericidal activity and emergence of resistance. Antimicrob Agents Chemother. 1987;31:1054–1060. doi: 10.1128/aac.31.7.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondeau J. A review of clinical trials with fluoroquinolones with an emphasis on new agents. Expert Opin Investig Drugs. 2000;9:383–413. doi: 10.1517/13543784.9.2.383. [DOI] [PubMed] [Google Scholar]
- 4.Chen D K, McGeer A, DeAzavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Y, Zhao X, Kreiswirth B, Drlica K. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000;44:2581–2584. doi: 10.1128/aac.44.9.2581-2584.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drusano G L, Johnson D, Rosen M, Standiford H. Pharmacodynamics of a fluoroquinolone antimicrobial agent in a neutropenic rat model of Pseudomonas sepsis. Antimicrob Agents Chemother. 1993;37:483–490. doi: 10.1128/aac.37.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efthymiopoulos C. Pharmacokinetics of grepafloxacin. J Antimicrob Chemother. 1997;40(Suppl A):35–43. doi: 10.1093/jac/40.suppl_1.35. [DOI] [PubMed] [Google Scholar]
- 9.Feldman W. Concentrations of bacteria in cerebrospinal fluid of patients with bacterial meningitis. J Pediatr. 1976;88:549–552. doi: 10.1016/s0022-3476(76)80003-0. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootz T, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Porter R. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones M E, Sahm D F, Martin N, Scheuring S, Heisig P, Thornsberry C, Köhrer K, Schmitz F-J. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from worldwide surveillance studies during the 1997-1998 respiratory season. Antimicrob Agents Chemother. 2000;44:462–466. doi: 10.1128/aac.44.2.462-466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy S B. Multidrug resistance—a sign of the times. N Engl J Med. 1998;338:1376–1378. doi: 10.1056/NEJM199805073381909. [DOI] [PubMed] [Google Scholar]
- 14.Milatovic D, Braveny I. Development of resistance during antibiotic therapy. Eur J Clin Microbiol. 1987;6:234–244. doi: 10.1007/BF02017607. [DOI] [PubMed] [Google Scholar]
- 15.Mitchison D A. Drug resistance in mycobacteria. Br Med Bull. 1984;40:84–90. doi: 10.1093/oxfordjournals.bmb.a071952. [DOI] [PubMed] [Google Scholar]
- 16.Munoz R, de la Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistant phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved Standard M7–A5. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 19.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestova E, Beyer R, Cianciotto N P, Noskin G A, Peterson L R. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob Agents Chemother. 1999;43:2000–2004. doi: 10.1128/aac.43.8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestova E, Millichap J, Noskin G, Peterson L. Intracellular targets of moxifloxacin: a comparison with other fluoroquinolones. J Antimicrob Chemother. 2000;45:583–590. doi: 10.1093/jac/45.5.583. [DOI] [PubMed] [Google Scholar]
- 24.Schentag J. Antimicrobial action and pharmacokinetics/pharmacodynamics: the use of AUIC to improve efficacy and avoid resistance. J Chemother. 1999;11:426–439. doi: 10.1179/joc.1999.11.6.426. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan J T, Woodruff M, Lettieri J, Agrarwal V, Krol G J, Leese P T, Watson S, Heller A H. Pharmacokinetics of a once-daily oral dose of moxifloxacin (Bay 12-8039), a new enantiomerically pure 8-methoxy quinolone. Antimicrob Agents Chemother. 1999;43:2793–2797. doi: 10.1128/aac.43.11.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F, McGowan J E. The epidemiology of bacterial resistance to antimicrobial agents. In: Evans A S, Brachman P S, editors. Bacterial infections of humans. New York, N.Y: Plenum Press; 1998. pp. 83–93. [Google Scholar]
- 27.Turnbridge J. Pharmacokinetics and pharmacodynamics of fluoroquinolones. Drugs. 1999;58(Suppl. 2):29–36. doi: 10.2165/00003495-199958002-00006. [DOI] [PubMed] [Google Scholar]
- 28.Zhao, X., and K. Drlica. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 29.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]