Abstract
We describe a rapid and simple novel phenotypic assay for drug susceptibility of human immunodeficiency virus type-1 (HIV-1) using a CCR5-expressing HeLa/CD4+ cell clone 1-10 (MAGIC-5). MAGIC-5 cells produced large amounts of HIV-1 in culture supernatants, which enabled us to perform the phenotypic resistance assay. Determination of HIV-1 susceptibility to various protease inhibitors (PI) and nucleoside reverse transcriptase inhibitors was completed within 15 days in T-cell-tropic (X4) and macrophage-tropic (R5) viruses using fresh plasma samples containing at least 104 copies/ml. The nucleotide sequence of the envelope V3 region of HIV-1 in plasma was almost identical to that of the virus isolated by MAGIC-5 cells, suggesting a lack of selection bias in our assay. The assay variability was confined to within five-fold in all drugs examined. Accordingly, we used a 10-fold increase in the 50% inhibitory concentration as the cutoff value for viral resistance in the present assay. HIV-1 resistant to lamivudine, which was not detected by conventional genotypic assays, was isolated. In HIV-1 with PI-associated primary amino acid substitutions, our assay showed that drug resistance profiles correlated well with previously reported genotypic-assay data. Furthermore, our assay provided comprehensive results regarding PI resistance in the presence of multiple mutations. The novel assay successfully quantified the level of resistance of clinical HIV-1 isolates to a battery of anti-HIV drugs, indicating its clinical usefulness, particularly in patients who failed to respond to antiretroviral chemotherapy.
The morbidity and mortality associated with AIDS are declining with the use of highly active antiretroviral therapy (HAART) (26). The benefits of HAART are attributed to efficient virus suppression and the consequent increase in CD4 counts (28). However, the potency of certain HAART agents against human immunodeficiency virus type 1 (HIV-1) has diminished in some patients due to single or multiple viral mutations (5, 17, 20, 23, 25). Therefore, detection of HAART-resistant viruses is important for better management of the disease (1, 9, 13, 27).
A phenotypic drug susceptibility assay using peripheral blood mononuclear cells (PBMC) was standardized by the AIDS Clinical Trial Group for use in clinical trials (15). However, the original phenotypic assay using PBMC is time-consuming, labor-intensive, and expensive. Therefore, the AIDS Clinical Trial Group method was subsequently modified and automated to reduce time and labor. On the other hand, sequencing of HIV-1 to identify specific genetic changes that confer resistance to HAART (18), namely, the genotypic resistance assay, is widely used in many laboratories instead of the conventional phenotypic assay because it is easier to perform. A few genotypic methods are currently being developed commercially for use as simple kits (32). However, the results of these assays do not always reflect the clinical course of the disease because they are deduced only from the given mutations and/or obtained from the major population of quasispecies. Several groups have developed recombinant-virus assays in which chimeric viruses generated by homologous recombination of PCR-derived sequences and the molecular clone are assessed phenotypically (2, 11, 16, 30). This method could become one of the most reliable assays of viral resistance in well-equipped laboratories in the future.
HeLa/CD4-positive cells were first established by Chesebro and Wehrly (3) for quantitative measurement of HIV-1 and were subsequently used as a drug susceptibility test termed the quantitative plaque reduction assay (4, 19). This reduction assay is simple and reproducible but can only analyze a T-cell-line-tropic (T-tropic) phenotype, namely, CXCR4-dependent viruses (7), because HeLa/CD4 cells lack CCR5, which is the second receptor for the macrophage tropic (M-tropic) phenotype (6).
In order to determine the drug susceptibilities of both phenotypes, we introduced CCR5 into HeLa/CD4 cells with the expressing vector pEFBOSbsrHuCCR5. In the present study, we describe the establishment of a novel phenotypic drug resistance assay using CCR5-expressing HeLa/CD4 cells. In the process of testing several clones, we identified MAGIC-5A 1-10 (MAGIC-5), a clone that produced a large amount of HIV-1 RNA in the supernatant of the primary culture of clinical isolates. Subsequently, the clone was used to develop a rapid and simple phenotypic resistance assay.
MATERIALS AND METHODS
Clinical materials.
Blood samples were obtained from patients attending the outpatient clinic of the AIDS Clinical Center, International Medical Center of Japan. Plasma HIV-1 loads were measured with the Amplicor HIV-1 Monitor kit (Nippon-Roche, Tokyo, Japan), according to the method recommended by the manufacturer, using fresh samples obtained on the same day. Informed consent was obtained from all patients.
Cells.
MAGI-CCR-5 cells were kindly provided by H. Mitsuya, National Cancer Institute, National Institutes of Health, Bethesda, Md. MAGIC-5 cells were independently developed through the following procedure. The parental cell line, MAGI (HeLa-CD4-LTR-β-gal), was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Md., and from M. Emerman. MAGI cells were bioengineered to express the β-chemokine receptor CCR5 by transfecting the cells with the expression vector pEFBOSbsrHuCCR5. In this process, the expression of human CCR5 is enhanced by the elongation factor 1α promoter and selected with blasticidin. Several clones were selected and tested for functional CCR5 expression using HIV-1JR-FL.
Reagents.
Nucleoside reverse transcriptase (RT) inhibitors (NRTIs) zidovudine (AZT) and stavudine (d4T) were purchased from Sigma (Tokyo, Japan), and lamivudine (3TC) was kindly provided by Glaxo Wellcome (Tokyo, Japan). HIV-1 protease inhibitors (PIs) ritonavir (RTV), saquinavir (SQV), and nelfinavir (NFV) were generously provided by Abbott Laboratories (Chicago, Il.), Roche Research Center (Nutley, N.J.), and Agouron (San Diego, Calif.), respectively.
Primary cultures with plasma samples.
MAGIC-5 cells were maintained in complete medium (Dulbecco's modified Eagle's medium [GIBCO BRL, Tokyo, Japan] supplemented with 10% fetal calf serum, 100 U of penicillin G-sodium/ml, and 100 μg of streptomycin [GIBCO BRL]/ml; 100 μg of hygromycin B [GIBCO BRL]/ml and 200 μg of geneticin [GIBCO BRL]/ml; or 1 μg of blasticidin [Funakoshi, Tokyo, Japan]/ml for selection of β-Gal, CD4, and CCR5, respectively). The cells were detached with 0.25% trypsin and 1 mM EDTA (GIBCO BRL) and precultured at a density of 2 × 104/well in a 48-well tissue culture plate with complete medium the day before infection. The cells were 30 to 40% confluent on the day of infection. Preliminary tests showed that the sensitivity cutoff value ranged between 104 and 106 copies/ml.
Within 30 min of blood sample withdrawal, 1 ml of fresh plasma was centrifuged at 37,800 × g for 90 min at 4°C. The supernatant was removed, and the virus pellet was resuspended with 300 μl of infection medium (complete medium containing 20 μg of DEAE-dextran [5 prime →3 prime Inc., Boulder, Colo.] per ml). The complete medium filling the preculture plate was replaced with infection medium. MAGIC-5 cells were incubated overnight at 37°C in a 5% CO2 incubator. The infection medium was removed from each well, and complete medium was added. The plate was changed every 2 or 3 days when the cells reached 80 to 90% confluence. The culture supernatants were stored at −80°C as the virus stock for further studies.
Virus stock titration.
Cells were counted and dispensed into a 96-well tissue culture plate at 104/well with complete medium the day before infection. The virus stock was thawed quickly and diluted with the infection medium and then incubated in triplicate for 48 h at 37°C in a 5% CO2 incubator. The medium was removed, and the cells were fixed with phosphate-buffered saline containing 1% formaldehyde and 0.2% glutaraldehyde at room temperature. The cells were then stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (TaKaRa Shuzo Co., Shiga, Japan). The blue cells in each well were counted, and the diluted virus stocks that produced 100 to 300 blue focus units (BFU) were used for the drug susceptibility assays. Then, titration of the virus stock was measured based on single-round replication by 48 h of incubation.
Drug susceptibility assays using MAGIC-5 cells.
The susceptibilities of HIV-1 to AZT, 3TC, d4T, RTV, SQV, and NFV were determined by using triplicate samples of infected MAGIC-5 cells in each assay. Each drug was prepared in five serial 10-fold dilutions with infection medium. MAGIC-5 cells were seeded (104 /well) and cultured in a 96-well tissue culture plate for 24 h. The cells were exposed to the diluted virus stock for 2 h at 37°C in a 5% CO2 incubator. Media with and without various concentrations of the test drug were added to each well. β-Galactosidase is driven by the HIV-1 long terminal repeat in MAGIC-5 cells. To determine the susceptibilities to various NRTIs, the cells were fixed after 48 h and stained based on single-round replication. However, since PIs inhibit the maturation of progeny virus, even the PI-susceptible virus has a single-round replication and drives the transfected long terminal repeat. If the virus is sensitive to PI, the progeny virus must be immature and not infection competent. Then, to determine the susceptibility to PIs, the cells were cultured for 78 h for multiple-round replication, and the supernatant was transferred to another 96-well tissue culture plate in order to determine the virus load in the supernatant. In the second step, the virus was cultured to examine the capability for de novo infection in a drug-free medium. The cells were then fixed after 48 h of culture and stained. Infected cells were counted after incubation with X-Gal under an inverted microscope. A decrease in the number of blue cells per well, therefore, reflected the inhibitory activity of the test drug. The 50% inhibitory concentration (IC50) of each drug was estimated from plots of the percentage of BFU reduction versus drug concentration. Control cultures free of drugs were prepared in all assays, and the infectivity of the input virus was confirmed to be 100 to 300 BFU/well at the end of assays for both NRTIs and PIs. When the infectivity of the input virus was >300 BFU/well in the control culture, the assay for PIs was repeated with the diluted virus. Drug susceptibility assays with PBMC were performed using the method described by Japour et al. (15).
Sequence analysis of pol gene mutations.
Viral RNA was extracted from the plasma with a High Pure Viral RNA kit (Nippon-Roche) as instructed by the manufacturer. The HIV-1 pol gene was amplified by PCR using the One Step RNA PCR kit (TaKaRa) according to the instructions provided by the manufacturer. The sequences of primer sets for the first PCR (DRPO1 and DRRT4) and the second PCR (DRPO2 and DRRT15) were as follows (8); DRPO1, 5′-CCAACAGCCCCACCAGA (MN pol positions, 2155 to 2171); DRRT4, 5′-TTCTGTTAGTGCTTTGGTT (MN pol positions, 3419 to 3437); DRPO2, 5′-ATTTTCAGGCCCATTTTTTGA (MN pol positions, 2706 to 2726); and DRRT15, 5′-TCCCACTAACTTCTGTATGTC (MN pol positions, 3330 to 3350). The resultant PCR products were purified with SUPREC-01 (TaKaRa), according to the instructions provided by the manufacturer, after 1% agarose gel electrophoresis. Sequencing was performed with an automatic sequencer (Applied Biosystems, Chiba, Japan) using a Taq dideoxy terminator cycle sequence kit (Applied Biosystems). Amino acid sequences were deduced by the Genetyx-Win program version 3.1 (Software Development, Tokyo, Japan).
Analysis of HIV-1 V3 sequence.
Viral RNA from HIV-1 isolates from MAGIC-5 cultures or from plasma was extracted by the SMI Test (Nihon Genetex, Tokyo, Japan) using the protocol provided by the manufacturer. The V3 domain of HIV-1 gp120 was amplified by PCR with the One Step RNA PCR kit. The sequences of primer sets for the first (YT001 and MK650) and the second (SI01 and SI02) PCRs were as follows (14): YT001, 5′-ACAATTTCTGGGTCCCCTCCTGAGGA {North American and European Consensus [AMEU (C)] env positions, 1077 to 1092}; MK650, 5′-AATGTCAGCACAGTACAATGTACAC [AMEU (C) env positions, 708 to 732]; SI01, 5′-ATGGAATTAGGCCAGTAGTG [AMEU (C) env positions, 733 to 752]; and SI02, 5′-CTCCTAATTTTGTAACTAC [AMEU (C) env positions, 1013 to 1032]. Sequencing was performed using the method described above for the analysis of the pol gene.
Flow cytometric analysis.
MAGIC-5 cells were washed twice with phosphate-buffered saline (Ca, Mg −) containing 0.5% bovine serum albumin. The cells (105/tube) were incubated for 15 min at 4°C with 1 μg of anti-human immunoglobulin (Sigma)/tube. Then, the cells were incubated with Cy-chrome-labeled anti-human CD4 monoclonal antibody (Pharmingen, San Diego, Calif.), fluorescein isothiocyanate-labeled anti-CXCR4, or fluorescein isothiocyanate-labeled anti-CCR5 monoclonal antibodies (R&D Systems, Tokyo, Japan) for 40 min at 4°C. After being washed, the cells were fixed with 1% paraformaldehyde and analyzed by flow cytometry (Coulter, Hialeah, Fla.).
Nucleotide sequence accession numbers.
Accession numbers of nucleotide sequences were assigned at the DDBJ as follows: AB037015 to AB037079.
RESULTS
Expression of CD4, CXCR4, and CCRS and viral culture.
We analyzed the expression of CD4, CXCR4, and CCR5 on MAGIC-5 and MAGI-CCR5 (National Institutes of Health) cells by flow cytometry (Table 1). We also challenged MAGIC-5 and MAGI-CCR5 cells with HIV-1 strains NL432 and BAL and plasma samples. Higher expression levels of CD4 and CCR5 were noted on MAGIC-5 than on MAGI-CCR5 cells (Table 1), indicating that the efficiency of virus isolation by MAGIC-5 cells was superior to that by MAGI-CCR5 cells for both type strains and clinical isolates. Although the expression of CXCR4 on MAGI-CCR5 cells was slightly higher than that on MAGIC-5 cells, HeLa cells constitutively express CXCR4, and the difference could not influence the sensitivity. Therefore, we used MAGIC-5 cells in subsequent studies.
TABLE 1.
Comparison of expression of CD4, CXCR4, and CCR5 and of virus isolation in MAGIC 5 and MAGI-CCR5 cells
| Cell type | Expression (%)
|
No. of blue cellsa
|
||||||
|---|---|---|---|---|---|---|---|---|
| CD4 | CXCR4 | CCR5 | NL432 | BAL | Plasma 1 | Plasma 2 | Plasma 3 | |
| MAGIC 5 | 99 | 0.2 | 86.5 | >1,000 | >1,000 | >1,000 | >1,000 | >500 |
| MAGI-CCR5 | 78.5 | 1.8 | 1.1 | 128 ± 11 | 23 ± 3.5 | 0 | 0 | 0 |
The number of isolated viruses is expressed as mean ± standard deviation of triplicate determinants. Plasmas 1, 2, and 3 contained 104, 105, and 106 copies of HIV-1 RNA/ml, respectively.
Primary culture of clinical isolates using MAGIC-5 cells.
We examined HIV-1 isolated from 272 plasma samples using MAGIC-5 cells. Syncytia (blue cells) were observed with an inverted microscope in 150 samples. Isolation was considered positive when at least one blue cell was identified within 2 weeks. The positive result was further confirmed by detection of p24 antigen in the supernatant of culture medium. The mean latency to the peak titer of p24 antigen was 5.1 days. Since MAGIC-5 cells produced a large amount of infection-competent virus in the culture supernatant, we were able to use this cell line in our phenotypic drug resistance assay. As shown in Table 2, isolation of the virus from plasma samples was dependent on the plasma virus load; a minimum virus load of 104 copies/ml was necessary for the primary culture.
TABLE 2.
Viral cultures of plasma samples with MAGIC 5 cells
| Virus load (copies/ml) | No. positivea/tested | % Positive |
|---|---|---|
| >106 | 5/5 | 100 |
| ≦105 – <106 | 63/68 | 92 |
| ≦104 – <105 | 73/102 | 71 |
| ≦103 – <104 | 5/63 | 7 |
| <103 | 4/34 | 11 |
At least one blue cell was observed within 2 weeks in positive isolates. A large amount of infection-competent virus was present in the culture supernatant thereafter.
Selection through viral culture with MAGIC-5 cells.
HIV-1 exists as quasispecies in the host. In order to investigate any selection bias in the viral culture, we compared the sequences of the V3 region of ENV in viruses isolated by plasma culture with MAGIC-5 to those obtained from uncultured plasma by PCR followed by direct sequencing. In 8 out of 10 pairs examined, the sequences were identical (Table 3). All isolated viruses were genotypically deduced to be M tropic. Then, we applied the culture supernatant to MT-2 cells. We were able to obtain T-tropic viruses in cases 2 and 3. The V3 sequences of both viruses were compatible with the T-tropic genotype (data not shown). These results indicated that selection bias was not frequent and that MAGIC-5 cells could simultaneously isolate both CCR5-tropic (M-tropic) and CXCR4-tropic (T-tropic) viruses, even though T-tropic virus was a minor population in the plasma.
TABLE 3.
Comparison between PCR-derived and MAGIC 5-isolated V3 sequences of HIV-1 in plasma
| Case | V3 sequencesa | ||||
|---|---|---|---|---|---|
| 1 | P | CTRPNNNTRK | SVNIGPGRAF | YATGAIIGDI | RQAHC |
| M | –––––––––– | –––––––––– | –––––––––– | ––––– | |
| 2 | P | –––––––––– | –IH––––K–– | –––––––––– | ––––– |
| M | –––––––––– | –IH––––K–– | –––––––––– | ––––– | |
| 3 | P | –––––––––R | –IN––––––– | –––∗D––––– | ––––– |
| M | –––––––––R | –IN––––––– | –––∗D––––– | ––––– | |
| 4 | P | –––––––––– | –ISFA––Q–– | ––––Q––––– | ––––– |
| M | –––––––––– | –ISFA––Q–– | ––––Q––––– | ––––– | |
| 5 | P | –––––––––R | –IHM–––R–– | –G–∗D––––– | ––––– |
| M | –––––––––R | –IHM–––R–– | –G–∗D––––– | ––––– | |
| 6 | P | –––––––––R | ––SM–––K–– | FT––D––––– | ––––– |
| M | –––––––––R | ––SM–––K–– | FT––D––––– | ––––– | |
| 7 | P | –––––––––– | –IHM–L–––W | –T––G–––N– | ––––– |
| M | –––––––––– | –IHM–L–––W | –T––G–––N– | ––––– | |
| 8 | P | –––––––––– | –IHM–––G–– | ––––D––––– | ––––– |
| M | –––––––––– | –IHM–––G–– | ––––D––––– | ––––– | |
| 9 | P | ––––G––––– | GIH––––––– | ––––E–T––– | –––Y– |
| M | ––––G––––– | SIH––––––– | ––––E–T––– | –––Y– | |
| 10 | P | –––––––––– | GIH––––Q–– | ––––Q––––– | ––––– |
| M | –––––––P–– | GIH––––Q–– | ––––Q––––– | ––––– | |
P, PCR-derived V3 sequence; M, MAGIC 5-isolated V3 sequence. A dash indicates identity with case 1 P, an asterisk indicates a deletion, and an underline indicates an amino acid substitution within a pair.
IC50 of each drug for NL432.
We repeated our experiments five times to evaluate the reproducibility of the IC50 of each drug for NL432. In each experiment, the assay was performed in triplicate using the same virus stock. As shown in Table 4, the assay variability was confined to within five fold in all drugs examined. Accordingly, we used a 10-fold increase in IC50 as the cutoff value for viral resistance in the present assays. In each experiment, NL432 was used as a standard, and the data were expressed as fold increase, comparing the IC50s for NL432 with those for clinical isolates in each assay.
TABLE 4.
Reproducibility of MAGIC-5 assay with NL432
| Drug | Repetition no.a | IC50 (μM)
|
Overall mean ± SD | CVb(%) | ||||
|---|---|---|---|---|---|---|---|---|
| 1c | 2 | 3 | 4 | 5 | ||||
| AZT | 1 | 0.010 | 0.012 | 0.030 | 0.056 | 0.030 | ||
| 2 | 0.036 | 0.010 | 0.044 | 0.030 | 0.042 | |||
| 3 | 0.028 | 0.011 | 0.045 | 0.054 | 0.074 | |||
| Mean | 0.025 | 0.011 | 0.040 | 0.047 | 0.049 | 0.034 ± 0.02 | 46 | |
| 3TC | 1 | 1.20 | 1.20 | 0.52 | 0.26 | 1.20 | ||
| 2 | 0.40 | 0.42 | 1.00 | 0.58 | 0.42 | |||
| 3 | 1.80 | 1.20 | 0.86 | 0.70 | 0.85 | |||
| Mean | 1.13 | 0.94 | 0.79 | 0.51 | 0.82 | 0.84 ± 0.23 | 27 | |
| d4T | 1 | 2.1 | 1.4 | 2.6 | 2.2 | 2.6 | ||
| 2 | 2.3 | 2.2 | 1.8 | 2.0 | 1.3 | |||
| 3 | 1.9 | 2.3 | 1.4 | 1.2 | 2.1 | |||
| Mean | 2.10 | 1.97 | 1.93 | 1.80 | 2.00 | 1.96 ± 0.11 | 6 | |
| RTV | 1 | 0.028 | 0.027 | 0.010 | 0.090 | 0.010 | ||
| 2 | 0.021 | 0.013 | 0.100 | 0.060 | 0.013 | |||
| 3 | 0.030 | 0.024 | 0.022 | 0.042 | 0.046 | |||
| Mean | 0.026 | 0.021 | 0.044 | 0.064 | 0.023 | 0.03 ± 0.02 | 51 | |
| SQV | 1 | 0.0010 | 0.0010 | 0.0024 | 0.0010 | 0.0076 | ||
| 2 | 0.0044 | 0.0020 | 0.0042 | 0.0032 | 0.0086 | |||
| 3 | 0.0056 | 0.0062 | 0.0043 | 0.0040 | 0.0012 | |||
| Mean | 0.0037 | 0.0031 | 0.0036 | 0.0027 | 0.0058 | 0.0038 ± 0.001 | 31 | |
| NFV | 1 | 0.0028 | 0.0029 | 0.0038 | 0.0032 | 0.0021 | ||
| 2 | 0.0015 | 0.0025 | 0.0023 | 0.0025 | 0.0025 | |||
| 3 | 0.0022 | 0.0011 | 0.0012 | 0.0022 | 0.0020 | |||
| Mean | 0.0022 | 0.0022 | 0.0024 | 0.0024 | 0.0022 | 0.0023 ± 0.0001 | 5 | |
To examine reproducibility of the assay, experiments were repeated in triplicate five times.
CV, coefficient of variation.
Experiment number.
IC50s for viruses obtained from antiviral-naive patients.
As summarized in Table 5, the mean IC50 of each test drug was similar to the mean IC50 for NL432. However, when we analyzed the fold increase in the resistance of each clinical isolate to each drug relative to that of NL432, some viruses showed a 10-fold increase of IC50s (resistance) to SQV and NFV, even though they were isolated from antiviral-naive patients. The genotypes of all these viruses were similar to the AMEU (C) genotypes (no mutations) at the codons for the specific amino acids known as the source of drug resistance.
TABLE 5.
Variability of IC50s of clinical isolates obtained from antiviral-naive patientsa
| Drug | n | IC50 (μM)
|
Fold increaseb | |
|---|---|---|---|---|
| Mean ± SD | Range | |||
| AZT | 22 | 0.04 ± 0.03 | 0.003–0.15 | 0.1–5 |
| 3TC | 20 | 1.17 ± 0.90 | 0.1–3.2 | 0.12–3.86 |
| d4T | 21 | 2.36 ± 2.00 | 0.33–7.5 | 0.14–3.23 |
| RTV | 15 | 0.03 ± 0.02 | 0.01–0.08 | 0.33–2.67 |
| SQV | 16 | 0.02 ± 0.02 | 0.001–0.1 | 0.1–10 |
| NFV | 14 | 0.007 ± 0.009 | 0.001–0.03 | 0.3–10 |
All assays were performed in triplicate.
Fold increase was calculated by dividing the IC50 of each drug by the mean IC50 for NL432 in each assay.
Comparison of phenotypic and genotypic results.
In the following experiments, viruses obtained from antiviral-experienced patients were used. The RT- and protease (PR)-coding regions of uncultured PCR-derived viruses were sequenced. The genotypes were compared with the phenotypic data derived from MAGIC-5 cells. Phenotypic data of clinical isolates were expressed as fold increase relative to the IC50 for NL432 in each assay.
(i) AZT.
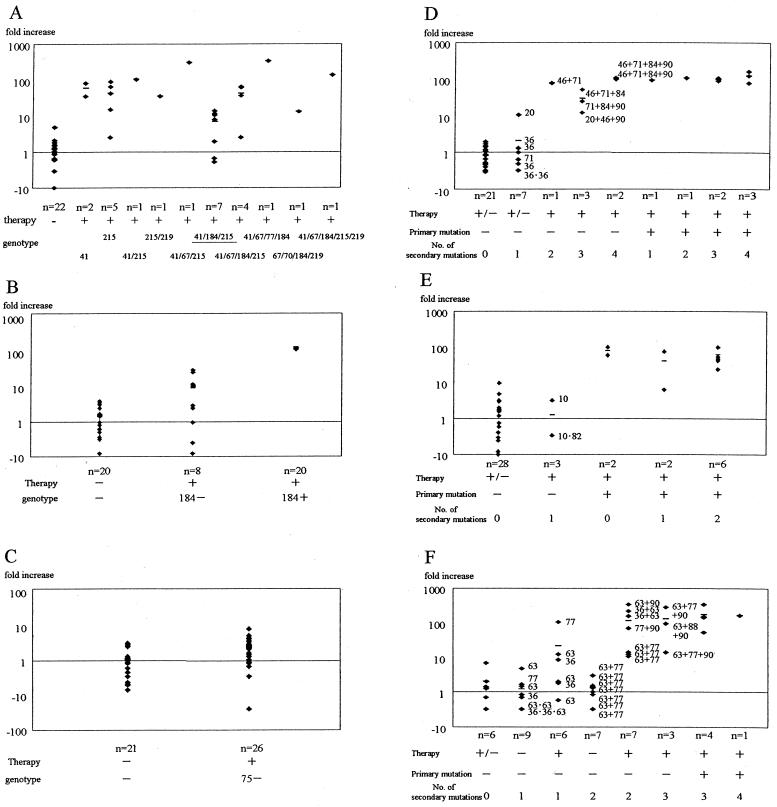
HIV-1 isolates with mutations at codons 41 and/or 215 of RT (primary mutations for resistance to AZT) were resistant to AZT (IC50, >10-fold increase) with the exception of one case (Fig. 1A). However, some but not all viruses with mutations at three sites (41, 184, and 215 [Fig. 1A]), were AZT susceptible (mean IC50, 8-fold; range, 0.53- to 14-fold), as reported previously (21). Further mutation at codon 67, 70, and/or 219 of RT, in addition to the above three sites, was associated with a further shift in IC50s up to 333-fold.
FIG. 1.
Relationship between resistance to AZT (A), 3TC (B), d4T (C), RTV (D), SQV (E), and NFV (F) and viral phenotype and genotype. The abscissas represent the genotypes of clinical isolates; n, number of isolates. The genotype numbers listed in panels A to C indicate the mutation sites of amino acids that confer drug resistance. Each numbers adjacent to a symbol in panels D to F indicates the mutation site of an amino acid that conferred drug resistance. Therapy, history of therapy using the respective drug. Phenotypic resistance is expressed as fold increase on the ordinate. The fold increase was calculated by dividing the IC50 of each drug by the mean IC50 for NL432 in each assay. The small horizontal bars represent the mean fold increase for each genotype. Some viruses with mutations at the sites underlined in panel A were AZT susceptible. +, present; −, absent; +/−, both treated and naive cases are included.
(ii) 3TC.
All isolates with one point mutation at codon 184 of RT (Fig. 1B) were resistant to 3TC (IC50, >120-fold). However, in cases where patients had been treated with NRTIs and the isolated virus showed no mutation at codon 184, the IC50s of some of these isolates demonstrated moderate resistance (up to 31-fold). These isolates had neither codon 44 and 118 mutations (12) nor other common polymorphism changes in RT (data not shown).
(iii) d4T.
In the case of d4T, none of the isolates showed a substitution at codon 75 of RT. In patients treated previously with NRTIs, the IC50S of some isolates were increased (Fig. 1C), albeit insignificantly. These isolates showed neither multinucleoside drug resistance mutations, such as at codon 151 (31), nor codon 69 insertion (22, 33).
(iv) RTV.
Viruses with only one secondary mutation at codon 36 or 71 were still sensitive to RTV, with the exception of one isolate, which had a mutation at codon 20 (11-fold) (Fig. 1D). Viruses with two or three secondary mutations but without a primary mutation exhibited moderate-to-high resistance (IC50, 12- to 106-fold). Mutation at codon 82 of the PR region, which is known as the primary mutation for resistance to RTV, was associated with a high resistance in all such isolates (IC50, >100-fold), regardless of the number of secondary mutations.
(v) SQV.
The presence of a single secondary mutation at codon 10 or codon 82 did not alter sensitivity to SQV (range, 0.34- to 3.2-fold) (Fig. 1E). In contrast, a mutation at codon 48 or 90 of the PR region, which are known as the primary mutations for resistance to SQV, was associated with acquired resistance in all but one case.
(vi) NFV.
A number of viruses isolated from antiretroviral-naive patients exhibited one or two secondary mutations at codons 36, 63, 77, and 63 plus 77 (Fig. 1F). These mutations were considered polymorphisms. However, in those patients who had previously received antiretroviral therapy, the IC50s for some of the isolates with one point mutation and all those with two mutations increased by more than 10-fold, even though the genotypes were similar (63 plus 77, for example). The primary mutation for resistance to NFV is at codon 30 of the PR region. Isolates with the primary mutation were resistance to NFV (IC50, >50-fold). The presence of three secondary mutations in the absence of a primary mutation was associated with an increase in IC50s of 11- to 333-fold. Most patients infected with these isolates had previously received PI-containing therapies other than NFV.
DISCUSSION
The available methods for the detection of drug resistance are usually classified into three types; genotypic (1, 23, 32), phenotypic (15), and recombinant (2, 11, 16, 30) virus assays. Among these, the genotypic assay is widely used in many laboratories because it is relatively rapid and easy. However, since this assay only detects specific known mutations, interpretation of the results is sometimes difficult in the presence of complex multiple mutations (24). Furthermore, genotypic changes cannot quantify the magnitude of resistance. Our data on PI resistance in the presence of one or two secondary mutations clearly demonstrated the superiority of our method to the genotypic resistance assay. A phenotypic assay using PBMC directly measures the growth of HIV in the presence of the test drug. However, the method is, in general, labor-intensive, time-consuming, expensive, and difficult to perform in clinical settings. Although the recombinant assay is promising, it is still difficult to perform in most clinical laboratories because transfection of the PCR-derived long fragment, for instance, is sometimes tricky.
In this report, we have described a novel phenotypic assay using MAGIC-5 cells and the application of the assay to test the susceptibilities of clinical isolates to PIs and NRTIs, with the exception of didanosine (ddI) and zalcitabine (ddC). In a series of preliminary experiments, we examined the susceptibilities of viruses to ddI or ddC using MAGIC-5 cells; however, the range of IC50S was very narrow, and the increase between NL432 and all clinical isolates, including those obtained from patients who failed to respond to ddI or ddC therapy, was limited to 10-fold (data not shown). Thus, we omitted further testing of ddI and ddC using this assay, although other investigators have reported that a high titer of input virus could deal with this problem (29). In phenotypic assays using PBMC, 2 to 4 weeks elapse before clinical isolates (primary cultures) become available, in addition to the time required for the preparation of phytohemagglutinin-stimulated PBMC. In contrast, the use of fresh plasma samples that contain >104 copies/ml allows the production by MAGIC-5 cells of large amounts of HIV-1 in the culture supernatant within 1 week on primary culture of clinical samples. Thus, our assay could be completed in about 2 weeks. In this study, we measured p24 antigen in the supernatant to confirm the positive results. However, in practice, positive results can be established by examination under the inverted microscope and detection of blue cells alone. Consequently, our phenotypic assay is rapid, simple, and cost-effective compared with PBMC assays. Since the variability of the present assay was limited to within 5-fold, we used a 10-fold increase in the IC50 as the cutoff value for viral resistance. Future clinical studies should further confirm the cutoff value. IC50S determined by phenotypic assays can be influenced by several factors. The introduction of automated procedures such as dilution of drugs, is likely to improve the reliability of the assay.
The susceptibility of clinical isolates to SQV and NFV varied in antiviral-naive patients, suggesting that some isolates have a natural resistance. However, specific mutations related to resistance to these drugs could not be detected by the genotypic assay. Alternatively, it is possible that the observed resistance was due to yet-unknown mutations that could not be identified in our genotypic assay. Harrigan et al. (10) recently indicated that the drug resistance profile could be used to predict the response to therapy in a community setting.
Our results using the novel assay correlated well with those of genotypic assays in most cases, except in the case of resistance to 3TC. We could not detect genotypic resistance to 3TC, and IC50S determined by our method indicated the resistance of some isolates, particularly in patients who had previously been treated with NRTIs, including 3TC. Resistance to AZT is known to be reversible when mutations accumulate at codons 41, 184, and 215 (21). The results of our assay were consistent with this phenomenon, at least in certain isolates. However, the IC50S of four out of seven isolates were more than 10-fold higher than those without such mutations. Furthermore, our results showed that this reversal effect was limited to this combination of mutations.
With regard to NRTIs, this is a single-round replication assay. Susceptibility to NRTIs in MAGI cells has been reported by other groups (19), which demonstrated similar IC50s determined by the standard PBMC culture method. For PIs, this is a two-step assay. The first step allows multiple-round replication with each drug, and the second step is a single-round replication assay without the drug. In a series of preliminary experiments, we examined the susceptibilities of clinical isolates, including viruses resistant to PIs, and compared the IC50s for them with the data obtained from the standard PBMC assay. The results of such analysis showed a close correlation between the results of two methods (data not shown), indicating that our method could be used for measuring susceptibility to PIs instead of the PBMC assay. The development of primary mutations in HIV-1 viruses was associated with a 100-fold increase in resistance to the drugs. These findings indicated that the results of the genotypic assay were consistent with those of the phenotypic assay.
Viruses acquire secondary mutations to accommodate changes in the local environment under drug pressure rather than to increase resistance to a particular drug (13). Clinical data demonstrated that only about 50% of patients previously treated with PIs but not NFV responded well to NFV-containing therapies (unpublished observation). Viruses isolated from such patients had no primary mutation for resistance to NFV but had acquired several secondary mutations that were identified by the genotypic assay. The clinical course in these patients lends support to our assay, since the IC50s determined by our method showed a shift to a highly resistant isolate. Interpretation of the results of the phenotypic assay is easier than that of results of the genotypic assay in these cases. These results also demonstrate the usefulness of our method when we considered the new PI-containing regimen after failure of the initial combination therapy.
In conclusion, we have developed a rapid and simple phenotypic assay for testing the susceptibility of HIV-1 to various anti-HIV agents, using MAGIC-5 cells. Our assay might be useful in clinical decision making with respect to the selection of PIs for antiviral-naive patients and patients who fail to respond to initial combination therapy. Further longitudinal clinical trials are necessary to confirm the clinical relevance of our assay.
ACKNOWLEDGMENTS
We thank Hiroaki Mitsuya of the National Institutes of Health for kindly providing MAGI-CCR5 cells and for critical reading of the manuscript. We also thank Gilbert Kaufmann and David A. Cooper of St. Vincent's Hospital for critical reading and H. Fraser and F. G. Issa for careful editing of the manuscript.
This study was supported by a grant-in-aid for AIDS research from the Ministry of Health and Welfare of Japan, by the Organization of Pharmaceutical Safety and Research (96-1), and by the Japanese Foundation for AIDS Prevention.
REFERENCES
- 1.Aizawa S, Gatanaga H, Ida S, Sakai A, Tanaka M, Takahashi Y, Hirabayashi Y, Oka S. Clinical benefits of resistance assay for HIV-specific protease inhibitors: when to check and in whom? AIDS. 1999;13:1278–1279. doi: 10.1097/00002030-199907090-00021. [DOI] [PubMed] [Google Scholar]
- 2.Boucher C A, Keulen W, van Bommel T, Nijhuis M, de Jong D, de Jong M D, Schipper P, Back N K T. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob Agents Chemother. 1996;40:2404–2409. doi: 10.1128/aac.40.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesebro B, Wehrly K, Metcalf J, Griffin D E. Use of a new CD4-positive HeLa cell clone for direct quantitation of infectious human immunodeficiency virus from blood cells of AIDS patients. J Infect Dis. 1991;163:64–70. doi: 10.1093/infdis/163.1.64. [DOI] [PubMed] [Google Scholar]
- 5.Craig C, Race E, Sheldon J, Whittaker L, Gilbert S, Moffatt A, Rose J, Dissanayeke S, Chirn G-W, Duncan I B, Cammack N. HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS. 1998;12:1611–1618. doi: 10.1097/00002030-199813000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Broder C C, Kennedy P, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.Gatanaga H, Oka S, Ida S, Wakabayashi T, Shioda T, Iwamoto A. Active HIV-1 redistribution and replication in the brain with HIV encephalitis. Arch Virol. 1999;144:29–43. doi: 10.1007/s007050050483. [DOI] [PubMed] [Google Scholar]
- 9.Gatanaga H, Aizawa S, Kikuchi Y, Tachikawa N, Genka I, Yoshizawa S, Yamamoto Y, Yasuoka A, Oka S. Anti-HIV effect of saquinavir combined with ritonavir is limited by previous long-term therapy with protease inhibitors. AIDS Res Hum Retrovir. 1999;15:1493–1498. doi: 10.1089/088922299309775. [DOI] [PubMed] [Google Scholar]
- 10.Harrigan P R, Hertogs K, Verbiest W, Pauwels R, Larder B A, Kemp S, Bloor S, Yip B, Hogg R, Alexander C, Montaner J S G. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS. 1999;13:1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 11.Hertogs K, de Bethune M-P, Miller V, Ivens T, Schel P, Cauwenberge A V, Eynde C V D, Gerwen V V, Azijin H, Houtte M V, Peters F, Staszewski S, Conant M, Bloor S, Kemp S D, Larder B A, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hertogs K, Bloor S, Vroey V D, den Eybde C V, Dehertogh P, Cauwenberge A V, Sturmer M, Alcorn T, Wegner S, Houtte M V, Miller V, Larder B A. A novel human immunodeficiency virus type 1 reverse transcriptase mutational pattern confers phenotypic lamivudine resistance in the absence of mutation 184V. Antimicrob Agents Chemother. 2000;44:568–573. doi: 10.1128/aac.44.3.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D for the International AIDS Society—USA Panel. Antiretroviral drug resistance testing in adults with HIV infection. Implication for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 14.Ida S, Gatanaga H, Shioda T, Nagai Y, Kobayashi N, Shimada K, Kimura S, Iwamoto A, Oka S. HIV type 1 V3 variation dynamics in vivo: long-term persistence of non-syncytium-inducing genotypes and transient presence of syncytium-inducing genotypes during the course of progressive AIDS. AIDS Res Hum Retrovir. 1997;13:1597–1609. doi: 10.1089/aid.1997.13.1597. [DOI] [PubMed] [Google Scholar]
- 15.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S the RV-43 study group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellam P, Larder B A. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 18.Larder B A, Kohli A, Kellam P, Kemp S D, Kronick M, Henfrey R D. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993;365:671–673. doi: 10.1038/365671a0. [DOI] [PubMed] [Google Scholar]
- 19.Larder B A, Chesebro B, Richman D D. Susceptibilities of zidovudine-susceptible and -resistant human immunodeficiency virus isolates to antiviral agents determined by using a quantitative plaque reduction assay. Antimicrob Agents Chemother. 1990;34:436–441. doi: 10.1128/aac.34.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 21.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 22.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codon 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzi P, Opravil M, Hirschel B, Chave L-P, Furrer H-J, Sax H, Perneger T V, Perrin L, Kaiser L, Yerly S the Swiss HIV Cohort Study. Impact of drug resistance mutations on virologic response to salvage therapy. AIDS. 1999;13:F17–F21. doi: 10.1097/00002030-199902040-00001. [DOI] [PubMed] [Google Scholar]
- 24.Miller V, Phillips A, Rottmann C, Staszewski S, Pauwels R, Hertogs K, de Bethune M-P, Kemp S D, Bloor S, Harrigan P R, Larder B A. Dual resistance to zidovudine and lamivudine in patients treated with zidovudine-lamivudine combination therapy: association with therapy failure. J Infect Dis. 1998;177:1521–1532. doi: 10.1086/515304. [DOI] [PubMed] [Google Scholar]
- 25.Molla A, Korneyeva M, Gao Q, Vasavanoda S, Schipper P J, Mo H-M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A B, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 26.Palella F J, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D the HIV Outpatient Study Investigators. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 27.Palmer S, Shafer R W, Merigan T C. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13:661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabound J M, Montaner J S G, Conway B, Rae S, Reiss P, Vella S, Cooper D, Lange J, Harris M, Wainberg M A, Robinson P, Myers M, Hall D. Suppression of plasma viral load below 20 copies/ml is required to achieve a long-term response to therapy. AIDS. 1998;12:1619–1624. doi: 10.1097/00002030-199813000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Richman D D, Johnson V A, Mayers D L, Shirasaka T, O'Brien M C, Mitsuya H. Measurement of susceptibility of HIV-1 to antiviral drugs, unit 12.9.1–12.9.21. In: Strober W, Shevach E, editors. Current protocols in immunology. New York, N.Y: Green Publishing Association and Wiley-Interscience; 1993. [Google Scholar]
- 30.Shi C, Mellors J W. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1997;41:2781–2785. doi: 10.1128/aac.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kaojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variations with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]