Abstract
Holomycin, a member of the pyrrothine class of antibiotics, displayed broad-spectrum antibacterial activity, inhibiting a variety of gram-positive and gram-negative bacteria, with the exception of Enterobacter cloacae, Morganella morganii, and Pseudomonas aeruginosa. The antibiotic lacked activity against the eukaryotic microorganisms Saccharomyces cerevisiae and Candida kefyr. Holomycin exhibited a bacteriostatic response against Escherichia coli that was associated with rapid inhibition of RNA synthesis in whole cells. Inhibition of RNA synthesis could have been a secondary consequence of inhibiting tRNA aminoacylation, thereby inducing the stringent response. However, the levels of inhibition of RNA synthesis by holomycin were similar in a stringent and relaxed pair of E. coli strains that were isogenic except for the deletion of the relA gene. This suggests that inhibition of RNA synthesis by holomycin could reflect direct inhibition of DNA-dependent RNA polymerase. Examination of the effects of holomycin on the kinetics of the appearance of β-galactosidase in induced E. coli cells was also consistent with inhibition of RNA polymerase at the level of RNA chain elongation. However, holomycin only weakly inhibited E. coli RNA polymerase in assays using synthetic poly(dA-dT) and plasmid templates. Furthermore, inhibition of RNA polymerase was observed only at holomycin concentrations in excess of those required to inhibit the growth of E. coli. It is possible that holomycin is a prodrug, requiring conversion in the cell to an active species that inhibits RNA polymerase.
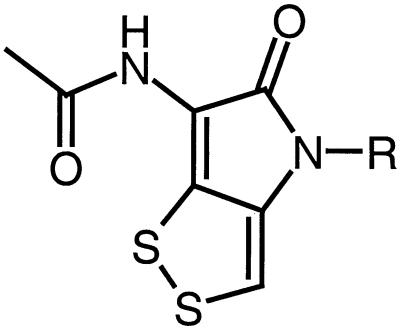
Thiolutin and holomycin (see Fig. 1) are members of the pyrrothine class of naturally occurring antibiotics that are characterized by the possession of a unique pyrrolinonodithiole nucleus (2). Although these antimicrobial agents were originally discovered more than 40 years ago (6, 13, 23, 26), relatively little is known about their mode of action. Limited studies with thiolutin suggest that the pyrrothines may act as inhibitors of DNA-dependent RNA polymerase. Thus, thiolutin preferentially inhibits RNA synthesis in both Saccharomyces cerevisiae (10) and Escherichia coli (14) and is reported to be a potent inhibitor of partially purified RNA polymerases from S. cerevisiae (10, 25). Furthermore, by monitoring the effects of thiolutin on the induction of β-galactosidase in E. coli, Khachatourians and Tipper (14) concluded that the antibiotic interfered with RNA chain elongation rather than with initiation of transcription.
FIG. 1.
Structure of thiolutin (where R is Me) and holomycin (where R is H).
However, there are several observations that cast doubt upon the hypothesis that thiolutin, and therefore the pyrrothines as a class, prevents microbial growth by interfering with the elongation of RNA transcripts catalyzed by RNA polymerase. For instance, Sivasubramanian and Jayaraman (24) were unable to reproduce the observations made by Khachatourians and Tipper (14) concerning the effects of thiolutin on β-galactosidase induction in E. coli. Thus, these authors (24) concluded that thiolutin inhibits initiation of RNA transcription rather than RNA chain elongation as proposed by Khachatourians and Tipper (14). Furthermore, in contrast to the observations for yeast (10, 25), thiolutin does not appear to inhibit E. coli RNA polymerase in vitro (24).
In view of the various discrepancies concerning the mode of action of thiolutin, we have reinvestigated the effects of the pyrrothine class on E. coli RNA polymerase, macromolecular synthesis, and induction of β-galactosidase in E. coli. For this purpose we chose to study holomycin, which is structurally closely related to thiolutin (Fig. 1) and is also a member of the pyrrothine antibiotic class. In addition to the effects of holomycin on biochemical processes in E. coli, we also report, for the first time, data on the antimicrobial spectrum and antibacterial properties of this member of the pyrrothine class.
MATERIALS AND METHODS
Organisms.
E. coli and Staphylococcus aureus laboratory strains used for studies on growth, survival, macromolecular synthesis, and β-galactosidase production in the presence of holomycin are listed in Table 1. E. coli K-12 strain CSH141 (16) was cultured on medium A, containing 0.4% (wt/vol) glycerol as a carbon source, to maintain the episome carrying the lac operon (16).
TABLE 1.
Descriptions of laboratory strains of E. coli K-12 and S. aureus and their susceptibilities to antimicrobial agents
| Strain | Description | Reference | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|---|---|
| Holomycin | Ciprofloxacin | Rifampin | Chloramphenicol | Cerulenin | |||
| E. coli | |||||||
| MG1655 | Prototroph | 29 | 0.2 | 0.62 | 1 | 6.25 | 62.5 |
| CF1652 | MG1655Δ relA251::kan | 29 | 0.2 | NDa | ND | ND | ND |
| CSH141 | ara Δ(gpt-lac) 5 supE rpsE [F′ lac+proA+proB+] | 16 | 2 | ND | 16.0 | ND | ND |
| S. aureus 8325-4 | NCTC 8325 cured of phages 11, 12, and 13 | 18 | 8 | 2.0 | 0.008 | 8 | 100 |
ND, not determined.
Additional bacteria used for more extensive susceptibility testing (Table 2) were maintained in the SmithKline Beecham culture collection. Candida kefyr NCPF3234, which is a standard control yeast for antifungal susceptibility testing, was obtained from the United Kingdom National Culture Collection for Pathogenic Fungi. S. cerevisiae 464 is a wild-type, clinical isolate obtained from a peritonitis patient at the University of Leeds and Leeds General Infirmary.
TABLE 2.
Antimicrobial activity of holomycin
| Organism | MIC (μg/ml)a |
|---|---|
| Staphylococcus aureus Oxford | 4 |
| Staphylococcus aureus Carter37 | 2 |
| Staphylococcus aureus F89 | 4 |
| Staphylococcus epidermidis PHLN20 | 1 |
| Streptococcus agalactiae Hester | 2 |
| Streptococcus pneumoniae ERY2 | 0.1 |
| Streptococcus pneumoniae PU7 | 0.3 |
| Streptococcus pyogenes CN10 | 16 |
| Enterococcus faecalis 1 | 4 |
| Enterobacter cloacae N1 | 64 |
| Escherichia coli DC0 | 2 |
| Escherichia coli DC2 | 0.5 |
| Klebsiella pneumoniae E70 | 8 |
| Proteus mirabilis C889 | 4 |
| Pseudomonas aeruginosa Dalgleish | 64 |
| Morganella morganii 1 | 64 |
| Moraxella catarrhalis 1502 | 0.3 |
| Moraxella catarrhalis Ravasio | 0.1 |
| Haemophilus influenzae Q1 | 0.3 |
| Haemophilus influenzae WM493 | 0.3 |
Determined by agar dilution.
Antibiotics and chemical and biochemical reagents.
Holomycin was obtained from Streptomyces clavuligerus IT1 (13) by SmithKline Beecham Pharmaceuticals at Brockham Park Research Centre, Surrey, United Kingdom. Cerulenin, chloramphenicol, ciprofloxacin, and rifampin were purchased from Sigma-Aldrich Co. Ltd. (Poole, United Kingdom). The following radiolabeled chemicals were purchased from Amersham Pharmacia Biotech (Amersham, United Kingdom): [methyl-3H]thymidine (70 to 85 Ci/mmol), [5-3H]uridine (25 to 30 Ci/mmol), [3,4-3H]glutamine (30 to 50 Ci/mmol), [3H]acetic acid, sodium salt (87 mCi/mmol), and [5-3H]UTP (14 Ci/mmol).
Poly(dA-dT) was purchased from Amersham Pharmacia Biotech, and plasmid pGEM β-gal was obtained from Promega Ltd., Southampton, United Kingdom. The enzyme β-galactosidase (EC 3.2.1.2.3, grade VI) was purchased from Sigma-Aldrich Co. Two types of E. coli RNA polymerase (EC 2.7.7.6) were purchased, one from Sigma (product no. R7394) and the other from Amersham Pharmacia Biotech (product no. E78022Y). The latter is a guaranteed sigma factor-saturated enzyme. All other chemical and biochemical reagents were obtained from standard commercial sources.
In vitro susceptibility testing.
The activity of holomycin against a range of bacterial clinical isolates was determined by standard agar dilution procedures. Blood agar base (Oxoid, Basingstoke, United Kingdom) was used for nonfastidious organisms. This medium was supplemented with 5% chocolated defibrinated horse blood (TCS Microbiology Ltd., Buckingham, United Kingdom) for growth of fastidious organisms. The inocula were 1-μl spots of an overnight broth culture containing 105 to 106 CFU. The MIC was defined as the lowest concentration of compound completely inhibiting visible growth after 18 to 24 h of incubation at 37°C. MIC determinations were also carried out, using a standard microdilution procedure, for E. coli K-12 strains MG1655 and CF1652 in morpholinepropanesulfonic acid (MOPS) minimal medium (17, 29) supplemented with 2 mg of all amino acids per liter, 10 g of glucose per liter, and 10% (wt/vol) brain heart infusion. Inocula contained 104 CFU. In the case of E. coli K-12 strain CSH141 and S. aureus 8325-4 (18), MICs were determined by broth microdilution in Iso-Sensitest medium (Oxoid) following incubation at 37°C for 24 h.
For direct comparison of the antibacterial and antifungal activities of holomycin and rifampin, sterile filter paper disks (diameter, 4 mm) containing 20 μg of holomycin or rifampin were prepared. These were then applied to plates seeded with suspensions of S. aureus 8325-4, C. kefyr NCPF3234, or S. cerevisiae 464. In the case of S. aureus the suspensions (109 CFU/ml) were prepared in Iso-Sensitest broth and then plated onto Iso-Sensitest agar. For the yeast strains, light suspensions were made in sterile distilled water, and each was spread over half the surface of a nutrient agar plate supplemented with yeast nitrogen base (Difco Ltd.), 2% (wt/vol) glucose, and 0.15% (wt/vol) asparagine. Cultures were incubated at 37°C for 24 h.
Effects of holomycin on bacterial viability and culture turbidity.
Studies to determine bactericidal activity were carried out on exponential-phase cultures of E. coli K-12 strain MG1655 grown in supplemented MOPS liquid medium. Samples (1 ml) were serially diluted in phosphate-buffered saline and were plated onto supplemented MOPS medium to which agar (1.5%, wt/vol) had been added. Colonies were counted after incubation at 37°C for 18 to 24 h. The turbidity of cultures of E. coli MG1655 growing in supplemented MOPS liquid medium was determined by measuring the absorbance at 675 nm in a Philips PU 8620 spectrophotometer.
Labeling of macromolecules in E. coli.
Bacteria were cultured in supplemented MOPS liquid medium. Radioactive precursors (1 μCi/ml for 3H-labeled and 0.1 μCi/ml for 14C-labeled compounds) were added during the early logarithmic phase and 3 min before the addition of inhibitors at four times their MICs, which were determined by the microdilution method. Incorporation of radioactivity into macromolecules was determined by following previously published procedures from this laboratory (3, 4, 19, 28). For DNA, RNA, and protein synthesis, this involved treatment of cells with trichloroacetic acid (5%, wt/vol) followed by collection of filtrates on glass fiber filters (Whatman GF/C) and quantification of radioactivity by liquid scintillation counting using OptiPhase Safe scintillation fluid (Wallac Ltd.) (3, 19, 28). For determination of lipid synthesis, cells were treated with chloroform and methanol, and radioactivity in the lipid fraction was determined as described previously (4). For each set of experiments an antibiotic known to specifically inhibit the pathway of interest was included as a positive control.
In vitro effects of rifampin and holomycin on purified E. coli β-galactosidase.
Reactions were performed in 1-ml volumes containing 5 μl of β-galactosidase (3,000 U/ml in 0.1 M sodium phosphate buffer, pH 7.5), 700 μl of Z buffer (16), 200 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) (4 mg/ml), and 95 μl of either rifampin or holomycin diluted in 0.1 M sodium phosphate buffer to yield final drug concentrations of 64 and 8 μg/ml, respectively (four times the MIC of each drug for strain CSH141). Controls contained 95 μl of sodium phosphate buffer. Reactions were initiated by the addition of ONPG, and the formation of o-nitrophenol was determined by measuring the absorbance of samples at 420 nm following incubation for 5 min at 28°C.
Effects of rifampin and holomycin on the production of β-galactosidase by E. coli CSH141.
The protocol for determining the effects of rifampin and holomycin on β-galactosidase production was based on previously published methods (14, 16, 22). An overnight culture of CSH141 grown in medium A containing glycerol (0.4%, vol/vol) was diluted in a fresh batch of the same medium and grown at 37°C with aeration to a culture turbidity of 0.2 optical density units at 600 nm. Cultures were induced with 10 mM isopropyl-β-d-thiogalactopyronoside (IPTG) (time zero) and 100-μl samples were taken at 30-s intervals for 10 to 15 min. Z buffer (0.9 ml) containing 20 μl of chloroform and 10 μl of sodium dodecyl sulfate (0.1%, wt/vol) was added to each sample and the mixtures were vortexed (10 s). Antibiotics were added 2 min after IPTG induction. Chloroform was removed by evaporation (37°C, 20 min) and β-galactosidase assays were initiated as described above by the addition of ONPG. Reactions were stopped after 35 min by the addition of 0.5 ml of 1 M sodium carbonate. Absorbance of the samples was measured at 420 and 550 nm to determine enzyme activities, which were expressed as Miller units.
Effect of holomycin on RNA polymerase in vitro.
RNA polymerase assays were performed essentially as described previously (7). This involved determining (i) the synthesis of poly(U · A) catalyzed by Sigma E. coli RNA polymerase in vitro using a synthetic poly(dA-dT) template and (ii) the in vitro synthesis of mRNA from wild-type promoters catalyzed by Amersham Pharmacia Biotech sigma-factor-saturated E. coli RNA polymerase using the superhelical plasmid template pGEM β-gal from Promega. Reaction mixtures (100 μl) containing 0.5 U of RNA polymerase were incubated with antibiotics at 37°C for 5 min prior to initiation of the assay by the addition of nucleoside triphosphates. Activity was monitored by the incorporation of [5-3H]UTP into trichloroacetic acid (5%, wt/vol)-precipitable material. Samples were filtered (Whatman GF/C) and dried, and radioactivity on the filters was determined by liquid scintillation counting using OptiPhase Safe scintillation fluid (Wallac Ltd.).
RESULTS
In vitro antimicrobial activity.
The activity of holomycin against a range of pathogenic bacteria was determined by agar dilution. Bacteria (Table 2) could be grouped as those that were highly susceptible to the antibiotic (MICs of 0.1 to 1 μg/ml), moderately susceptible (MICs of 2 to 8 μg/ml), and relatively unsusceptible (MICs of 16 to 64 μg/ml).
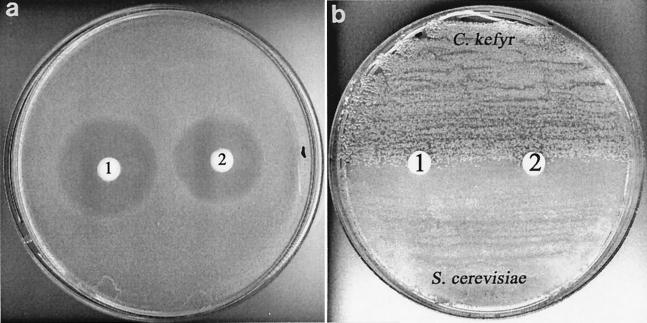
Thiolutin, which is an analog of holomycin (Fig. 1), has been reported to possess antifungal activity (10, 23). However, the antifungal activity of holomycin has not been determined. Comparative antibacterial and antifungal activities of holomycin were determined by placing paper disks impregnated with antibiotic (20 μg) onto plates seeded with S. aureus 8325-4 (Fig. 2a), C. kefyr (Fig. 2b), and S. cerevisiae (Fig. 2b). Disks containing rifampin (20 μg) were used as controls. Both holomycin and rifampin produced large zones of inhibition of staphylococcal growth consistent with the susceptibility of S. aureus 8325-4 to these antibiotics as determined by conventional susceptibility testing (Table 1). However, neither antibiotic demonstrated activity against the two yeast strains studied here, since confluent growth up to the antibiotic-containing disks occurred (Fig. 2b).
FIG. 2.
Effects of holomycin (20 μg, disk 1) and rifampin (20 μg, disk 2) on the growth of S. aureus 8325-4 (a) and C. kefyr NCPF3234 and S. cerevisiae (b). Note the zones of inhibition around both disks for S. aureus (a) and the absence of inhibition for both yeast strains (b).
Effects of holomycin on growth and survival of E. coli MG1655.
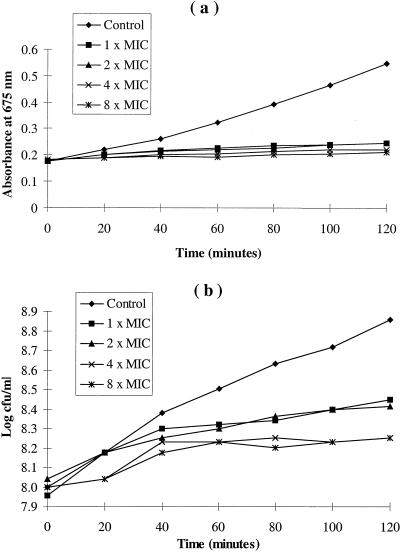
The effects of holomycin on the growth and survival of E. coli over a 2-h period were determined for liquid cultures of strain MG1655 grown in supplemented MOPS medium. The addition of 0.2 μg of holomycin/ml (equivalent to the MIC determined in liquid culture by the microdilution method) to early-logarithmic-phase cultures led to partial suppression of growth (Fig. 3). This was demonstrated by small increases in both culture turbidity and viable cell numbers compared to those of antibiotic-free controls. Addition of higher concentrations of holomycin (two to eight times the MIC equivalents) caused complete cessation of growth (Fig. 3a). This resulted from a bacteriostatic effect, since the number of viable cells did not decline upon exposure to the antibiotic, even at concentrations eight-fold higher than the MIC (Fig. 3b).
FIG. 3.
Effects of holomycin on the growth (a) and survival (b) of E. coli K-12 strain MG1655 during exposure to antibiotic for 2 h.
Effect of holomycin on macromolecular synthesis in E. coli MG1655.
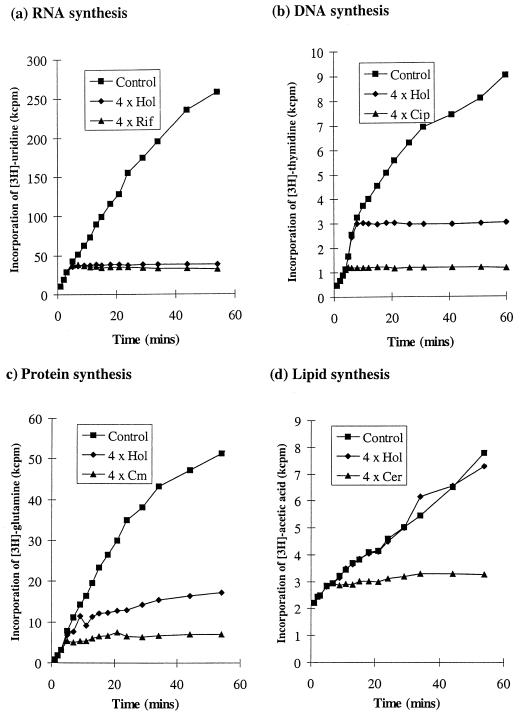
The effect of holomycin on the incorporation of radiolabeled precursors into RNA, DNA, protein, and lipid was determined (Fig. 4). In each case a specific inhibitor with a known mechanism of action was included as a positive control. These agents and holomycin were added to early-logarithmic-phase cultures of E. coli MG1655 at concentrations equivalent to four times their respective MICs determined by the microdilution method.
FIG. 4.
Effects of holomycin (Hol), rifampin (Rif), ciprofloxacin (Cip), chloramphenicol (Cm), and cerulenin (Cer) at four times their MICs on macromolecular synthesis in E. coli MG1655. (a) RNA synthesis determined by incorporation of [3H]uridine; (b) DNA synthesis determined by incorporation of [3H]thymidine; (c) protein synthesis determined by incorporation of [3H]glutamine; (d) lipid synthesis determined by incorporation of [3H]acetate. Antimicrobial agents were added at 3 min.
Within 5 min of addition of holomycin, incorporation of [3H]uridine into RNA ceased (Fig. 4a). Inhibition of RNA synthesis by rifampin demonstrated comparable kinetics (Fig. 4a). Addition of holomycin also resulted in complete inhibition of incorporation of [3H]thymidine into DNA, but this effect occurred at around 10 to 12 min after addition of the antibiotic (Fig. 4b), i.e., some 5 to 7 min after RNA synthesis had ceased. In contrast to holomycin, ciprofloxacin (a specific inhibitor of DNA synthesis) caused complete inhibition of [3H]thymidine incorporation within 5 min of addition (Fig. 4b). Holomycin also affected incorporation of [3H]glutamine into protein (Fig. 4c). However, inhibition was not evident until about 10 min after addition of the antibiotic and was incomplete, since incorporation of precursor continued throughout the experiment. In contrast, chloramphenicol blocked precursor incorporation within 3 min and achieved complete inhibition of incorporation by 20 min (Fig. 4c). Holomycin had no effect on the incorporation of [3H]acetic acid into lipid (Fig. 4d), whereas cerulenin (a specific inhibitor of fatty acid biosynthesis) caused complete inhibition of incorporation within 8 min of addition (Fig. 4d).
Effect of holomycin on RNA synthesis in E. coli CF1652, a relA null mutant.
The experiments described above indicate that holomycin preferentially inhibits RNA synthesis in E. coli. To determine whether the effect on RNA synthesis was the result of direct inhibition by the antibiotic or a secondary consequence of inducing stringency, the effect of holomycin on RNA synthesis in E. coli CF1652, a relA null mutant of E. coli MG1655 (29), was determined. Inhibition of RNA synthesis by holomycin was unaffected by the nature of the host strain. In both cases the addition of holomycin at four times the MIC resulted in 85 to 90% inhibition of RNA synthesis over a 30-min period of incubation with the antibiotic (data not shown).
Effects of rifampin and holomycin on β-galactosidase production by E. coli CSH141.
The effects of rifampin and thiolutin on the kinetics of β-galactosidase production in E. coli following induction of the lac operon have been used previously to demonstrate that these antibiotics inhibit the initiation and elongation phases, respectively, of mRNA transcription (15).
However, the effects of holomycin in such a system have not been determined. In order to approach this problem it was essential to demonstrate that neither rifampin nor holomycin had direct effects upon the activity of E. coli β-galactosidase. If the antibiotics caused enzyme inhibition, this would complicate interpretation of data obtained following induction of β-galactosidase in whole cells. We established that the purified E. coli enzyme was not affected by either holomycin or rifampin at concentrations equivalent to four times the MICs of these drugs for strain CSH141 (data not shown).
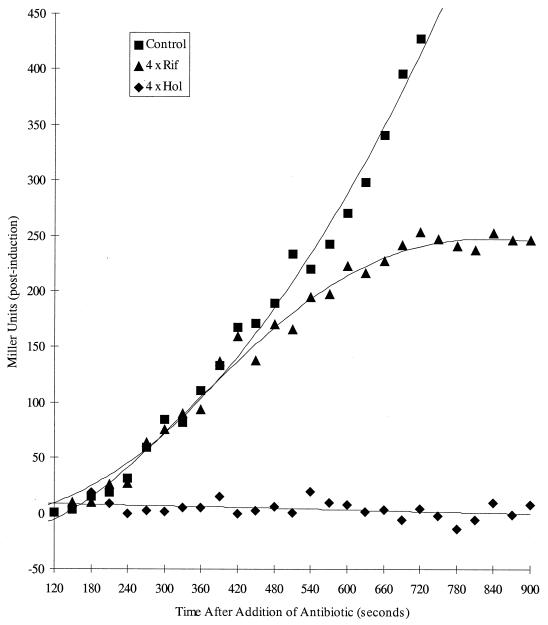
Consequently, β-galactosidase induction studies in the presence of the drugs were feasible. Cultures of strain CSH141 were induced with IPTG at time zero, and 2 min later portions of the induced bacteria were treated with rifampin or holomycin at four times the MIC of each drug. Control and drug-containing samples were assayed for β-galactosidase activity, and representative data are shown in Fig. 5. Synthesis of β-galactosidase ceased immediately in the holomycin-treated culture but continued for several minutes in the rifampin-inhibited culture before a plateau was reached. Addition of holomycin at later stages after induction with IPTG (e.g., 2 to 10 min postinduction) also led to rapid inhibition of enzyme production (data not shown).
FIG. 5.
Comparison of β-galactosidase synthesizing capacity of E. coli CSH141 treated with rifampin (Rif) and holomycin (Hol) at four times their MICs. Bacteria were induced with IPTG at time zero and drugs were added 120 s later. Samples were removed and β-galactosidase activities were determined as described in Materials and Methods.
Activity of holomycin against E. coli RNA polymerase in vitro.
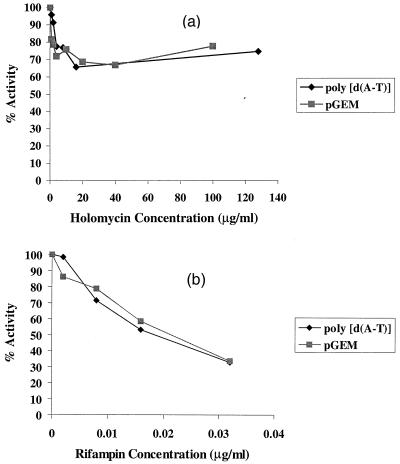
The activity of holomycin as an inhibitor of E. coli RNA polymerase in cell-free transcription assays was compared with that of rifampin in separate experiments using both a synthetic poly(dA-dT) template and natural promoters in the superhelical plasmid pGEM β-gal. Holomycin was a poor inhibitor of E. coli RNA polymerase when a synthetic template was used, achieving at most only a 35% reduction in the incorporation of radioactivity into poly(U · A) (Fig. 6a). However, since this system may not be indicative of transcription from normal bacterial promoters, experiments were also performed using sigma factor-saturated E. coli RNA polymerase and naturally occurring promoters in the plasmid vector pGEM β-gal. Holomycin was also a poor inhibitor of transcription in this system (Fig. 6a). Furthermore, virtually no inhibition of RNA synthesis was observed in either system in the presence of 0.2 to 2 μg of holomycin/ml. These concentrations correspond to the MIC of the antibiotic for E. coli strains (Tables 1 and 2). Consequently, inhibition of RNA synthesis in vitro by holomycin was observed only at antibiotic concentrations greatly in excess of the holomycin MIC for E. coli. In contrast to the weak enzyme inhibition displayed by holomycin, rifampin was a potent inhibitor of E. coli RNA polymerase in vitro (Fig. 6b). Rifampin displayed 50% inhibitory concentrations of approximately 0.02 μg/ml in both transcription assays. These data are in agreement with previous values reported in the literature for rifampin-sensitive RNA polymerase from E. coli (5, 8, 11, 21).
FIG. 6.
Effects of holomycin and rifampin on the activity of in vitro transcription systems using E. coli RNA polymerase with poly(dA-dT) and plasmid pGEM β-gal (pGEM) templates. Assays were performed as described in Materials and Methods. The data are means of duplicate determinations.
DISCUSSION
Thiolutin is the most extensively studied member of the pyrrothine class of antibiotics. It is reported to have a broad spectrum of antimicrobial activity, encompassing protozoa, yeast, pathogenic fungi, and bacteria, including both gram-positive and gram-negative species (10, 23). However, comparable studies on the antimicrobial spectrum of holomycin have not been reported. We observed that the antibacterial profile of holomycin was similar to that reported for thiolutin (23). Thus, thiolutin and holomycin are both effective against a number of gram-positive cocci and E. coli but less effective against Pseudomonas aeruginosa (Table 2). However, in contrast to previous observations on thiolutin (10, 23), we were unable to demonstrate that holomycin possessed antifungal activity. Although we have not reevaluated the antimicrobial activity of thiolutin, there is only a small structural difference between this antibiotic and holomycin (Fig. 1), which makes it difficult to explain why thiolutin apparently possesses antifungal activity whereas holomycin does not.
Although the effects on bacterial survival of antibiotics from the pyrrothine class have not been extensively studied, an early report (23) suggests that thiolutin exhibits a bactericidal response against a range of gram-positive and -negative bacterial species. However, a more recent report demonstrates that the antibiotic has bacteriostatic properties against E. coli (15). Our observations with holomycin are consistent with the more recent report on thiolutin. Thus, holomycin exhibited a bacteriostatic response against E. coli at concentrations up to eightfold higher than the MIC (Fig. 3b).
Studies on the mechanism of action of the pyrrothines have been limited to thiolutin. Experiments utilizing intact S. cerevisiae demonstrated that RNA synthesis stops immediately when cells are exposed to concentrations of thiolutin sufficient to prevent growth (10). However, under these conditions, inhibition of protein synthesis was more gradual. Since in vitro protein synthesis using yeast cell extracts was not inhibited by thiolutin, it was deduced that the antibiotic had a primary effect on RNA synthesis, probably by inhibiting DNA-dependent RNA polymerase (10). Indeed, subsequent studies utilizing purified yeast RNA polymerases confirmed that these enzymes were the likely targets of thiolutin and that initiation of RNA synthesis was disrupted (25). The effects of thiolutin on the kinetics of the appearance of β-galactosidase in E. coli following induction of the enzyme are also consistent with inhibition of RNA synthesis as the primary target of the antibiotic (14). However, since conflicting β-galactosidase induction profiles were obtained (14, 24), there is some doubt as to whether this reflects inhibition of the initiation or elongation steps of RNA synthesis.
We have now examined the effects of holomycin on β-galactosidase production in E. coli and compared the induction profile with that occurring in rifampin-inhibited cells. The results we obtained with rifampin agree with previous observations and are consistent with the inhibition of RNA synthesis being mediated through binding of the drug to RNA polymerase (5, 8, 11, 14, 21, 27). The data for holomycin suggest rapid inhibition of RNA chain elongation by the antibiotic and are thus in agreement with the findings of Khachatourians and Tipper (14), who concluded that thiolutin is also an RNA elongation inhibitor. If thiolutin and holomycin inhibit RNA elongation by interaction with RNA polymerase, their binding sites within the enzyme may well differ from that of rifampin, which is an inhibitor of transcriptional initiation (5, 11, 15, 27). This suggestion is consistent with the finding that thiolutin and holomycin are both active against S. aureus strains containing mutations in rpoB that confer resistance to rifampin (20).
The whole-cell RNA labeling studies utilizing [3H]uridine reported here for E. coli do not distinguish between the initiation and elongation processes of RNA synthesis. However, we noted that compared to the synthesis of other macromolecules, the synthesis of RNA was most susceptible to inhibition following exposure of E. coli K12 MG1655 (wild type) to holomycin (Fig. 4). These observations are consistent with RNA polymerase being the primary target of holomycin action in E. coli.
Although inhibition of RNA synthesis and hence transcription is sufficient to explain why we also observed substantial inhibition of protein synthesis in holomycin-treated cells, an alternative explanation is possible. If holomycin and other pyrrothines inhibit the aminoacylation of one or more tRNA species, then induction of the stringent response would rapidly lead to a shutdown of both RNA and protein synthesis in the cell (1, 9, 28). Stringency is invoked by an increase in the ratio of uncharged to charged tRNA (1, 9, 28), which causes a ribosome-bound enzyme, ppGpp synthase I, encoded by relA, to synthesize the regulatory nucleotide ppGpp (guanosine 5′, 3′-bispyrophosphate). Accumulation of this nutritional stress alarmone confers altered promoter specificity on RNA polymerase, resulting not only in suppression of RNA synthesis but also in suppression of the synthesis of proteins and other macromolecules (1, 9, 28).
If the antibacterial activity of holomycin and other pyrrothines results from inhibition of tRNA charging, then inhibition of RNA synthesis would not be a consequence of a direct effect on RNA polymerase. This hypothesis for the mode of action of pyrrothines could therefore be consistent with the apparent failure to demonstrate inhibition of E. coli RNA polymerase in vitro by thiolutin (24) or holomycin (reported here). However, data obtained in our study with a stringent and relaxed pair of E. coli strains (isogenic, except for deletion of the relA gene) demonstrate that synthesis of RNA is extensively inhibited by holomycin in both strains. This rules out an indirect effect of the antibiotic on RNA synthesis and favors the hypothesis that holomycin and other pyrrothines directly inhibit bacterial RNA synthesis. Furthermore, the finding that lipid synthesis continues in wild-type holomycin-inhibited E. coli (Fig. 4d) is not consistent with the hypothesis that holomycin induces stringency, since lipid synthesis is down regulated upon induction of the stringent response (1).
Several indirect lines of evidence point towards inhibition of the elongating activity of bacterial RNA polymerase by the pyrrothines. Nevertheless, thiolutin apparently does not directly inhibit the E. coli enzyme (24), and our results with holomycin using two types of transcription assay demonstrated only weak in vitro inhibition of RNA polymerase. It therefore appears that pyrrothines are not direct inhibitors of RNA polymerase. Our data are consistent with the possibility that pyrrothines may be prodrugs that are converted upon uptake into the bacterial cell to produce active RNA polymerase inhibitors. Indeed, an earlier report by Juhl and Clark (12) suggests that the pyrrothine antibiotic thiolutin may be a prodrug whose activation in E. coli requires sulfone oxidase (thd) activity. By inference, holomycin, which is a structural analog of thiolutin (Fig. 1), may be similarly activated by bacterial sulfone oxidases.
Although the target for pyrrothines or their drug-like metabolites has yet to be identified, the bacteriostatic nature of thiolutin and holomycin implies that the drug-target interaction is reversible. The possibility that the pyrrothines might represent leads for development as antibiotics has recently been discussed (20). However, it is clear that the prospects for development of these compounds will depend upon elucidating their precise mode of action in bacteria and determining whether they are truly specific for prokaryotic organisms.
ACKNOWLEDGMENTS
We thank Richard Barton for assistance with antifungal evaluation of holomycin, Nico Caggiano and Keith Miller for technical assistance, and Michael Cashel for the provision of strains MG1655 and CF1652.
REFERENCES
- 1.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 2.Celmer W D, Solomons I A. The structures of thiolutin and aureothricin, antibiotics containing a unique pyrrolinonodithiole nucleus. J Am Chem Soc. 1955;77:2861–2865. [Google Scholar]
- 3.Cherrington C A, Hinton M, Chopra I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J Appl Bacteriol. 1990;68:69–74. doi: 10.1111/j.1365-2672.1990.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 4.Chopra I. Induction of tetracycline resistance in Staphylococcus aureus in the absence of lipid synthesis. J Gen Microbiol. 1975;91:433–436. doi: 10.1099/00221287-91-2-433. [DOI] [PubMed] [Google Scholar]
- 5.Das A, DeVito J, Sparkowski J, Warren F. RNA synthesis in bacteria: mechanisms and regulation of discrete biochemical events at initiation and termination. In: Sutcliffe J, Georgopapadakou N H, editors. Emerging targets in antibacterial and antifungal chemotherapy. New York, N.Y: Chapman and Hall; 1992. pp. 68–116. [Google Scholar]
- 6.Ettlinger L, Gaumann E, Hutter R, Keller-Schierlein W, Kradlofer F, Neipp L, Prelog V, Zahner H. Stoffwechselprodukte von actinomyceten, holomycin. Helv Chim Acta. 1959;42:563–569. [Google Scholar]
- 7.Gross C, Engbaek F, Flammang T, Burgess R. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J Bacteriol. 1976;128:382–389. doi: 10.1128/jb.128.1.382-389.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann G, Honikel K O, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 9.Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiminez A, Tipper D J, Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin D J, Zhou Y N. Mutational analysis of structure-function relationship of RNA polymerase in Escherichia coli. Methods Enzymol. 1996;273:300–319. doi: 10.1016/s0076-6879(96)73027-6. [DOI] [PubMed] [Google Scholar]
- 12.Juhl M J, Clark D P. Thiophene-degrading Escherichia coli mutants possess sulfone oxidase activity and show altered resistance to sulfur-containing antibiotics. Appl Environ Microbiol. 1990;56:3179–3185. doi: 10.1128/aem.56.10.3179-3185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenig M, Reading C. Holomycin and an antibiotic (MM19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J Antibiot. 1979;32:549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 14.Khachatourians G G, Tipper D J. Inhibition of messenger ribonucleic acid synthesis in Escherichia coli by thiolutin. J Bacteriol. 1974;119:795–804. doi: 10.1128/jb.119.3.795-804.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khachatourians G G, Tipper D J. In vivo effect of thiolutin on cell growth and macromolecular synthesis in Escherichia coli. Antimicrob Agents Chemother. 1974;6:304–310. doi: 10.1128/aac.6.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 17.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 19.Oliva B, Maiese W M, Greenstein M, Borders D B, Chopra I. Mode of action of the cyclic depsipeptide antibiotic LL-AO341β1 and partial characterization of a Staphylococcus aureus mutant resistant to the antibiotic. J Antimicrob Chemother. 1993;32:817–830. doi: 10.1093/jac/32.6.817. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill A, Oliva B, Storey C, Hoyle A, Fishwick C, Chopra I. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:3163–3166. doi: 10.1128/aac.44.11.3163-3166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabussay D, Zillig W. A rifampicin resistant RNA-polymerase from E. coli altered in the β-subunit. FEBS Lett. 1969;5:104–106. doi: 10.1016/0014-5793(69)80305-4. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning; a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Seneca H, Kane J H, Rockenbach J. Bactericidal, protozoicidal and fungicidal properties of thiolutin. Antibiot Chemother. 1952;2:357–360. [PubMed] [Google Scholar]
- 24.Sivasubramanian N, Jayaraman R. Thiolutin resistant mutants of Escherichia coli. Are they RNA chain initiation mutants? Mol Gen Genet. 1976;145:89–96. doi: 10.1007/BF00331562. [DOI] [PubMed] [Google Scholar]
- 25.Tipper D J. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol. 1973;116:245–256. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Daehne W, Godtfredsen W O, Tybring L, Schaumburg K. New antibiotics containing the 1, 2-dithiolo[4, 3-b] pyrrole ring system. J Antibiot. 1969;22:233–236. doi: 10.7164/antibiotics.22.233. [DOI] [PubMed] [Google Scholar]
- 27.Wehrli W. Rifampin: mechanisms of action and resistance. Rev Infect Dis. 1983;5:S407–S411. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]
- 28.Wilson J M, Oliva B, Cassels R, O'Hanlon P J, Chopra I. SB205952, a novel semisynthetic monic acid analog with at least two modes of action. Antimicrob Agents Chemother. 1995;39:1925–1933. doi: 10.1128/aac.39.9.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]