Abstract
Peptide deformylase (PDF) is an essential bacterial metalloenzyme which deformylates the N-formylmethionine of newly synthesized polypeptides and as such represents a novel target for antibacterial chemotherapy. To identify novel PDF inhibitors, we screened a metalloenzyme inhibitor library and identified an N-formyl-hydroxylamine derivative, BB-3497, and a related natural hydroxamic acid antibiotic, actinonin, as potent and selective inhibitors of PDF. To elucidate the interactions that contribute to the binding affinity of these inhibitors, we determined the crystal structures of BB-3497 and actinonin bound to Escherichia coli PDF at resolutions of 2.1 and 1.75 Å, respectively. In both complexes, the active-site metal atom was pentacoordinated by the side chains of Cys 90, His 132, and His 136 and the two oxygen atoms of N-formyl-hydroxylamine or hydroxamate. BB-3497 had activity against gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis, and activity against some gram-negative bacteria. Time-kill analysis showed that the mode of action of BB-3497 was primarily bacteriostatic. The mechanism of resistance was via mutations within the formyltransferase gene, as previously described for actinonin. While actinonin and its derivatives have not been used clinically because of their poor pharmacokinetic properties, BB-3497 was shown to be orally bioavailable. A single oral dose of BB-3497 given 1 h after intraperitoneal injection of S. aureus Smith or methicillin-resistant S. aureus protected mice from infection with median effective doses of 8 and 14 mg/kg of body weight, respectively. These data validate PDF as a novel target for the design of a new generation of antibacterial agents.
Ribosome-mediated synthesis of proteins starts with a methionine residue. In prokaryotes, the amino group of the methionyl moiety carried by the initiator tRNAfMet is N formylated by formyltransferase prior to its incorporation into a polypeptide. Consequently, N-formylmethionine is always present at the N terminus of a nascent bacterial polypeptide. However, most mature proteins do not retain the N-formyl group or the terminal methionine residue. Following translation, the formyl group is hydrolyzed by peptide deformylase (PDF), which is necessary for further processing at the N terminus by methionine aminopeptidase (32). Deformylation is therefore a crucial step in bacterial protein biosynthesis, and PDF is essential for bacterial growth (23). The gene encoding PDF (def) is present in all sequenced pathogenic bacterial genomes and has no mammalian counterpart, making it an attractive target for antibacterial chemotherapy. Although the enzyme has been known for 30 years, it has proved difficult to isolate and characterize due to its apparent instability. Recently, two X-ray crystal structures and a solution structure of PDF have been determined (5, 9, 12), identifying PDF as a new class of metalloenzyme related in structure to the metalloproteinase superfamily. PDF has been shown to utilize iron as the catalytic metal. With iron as the active-site metal ion, the enzyme is extremely unstable; iron, however, can be replaced by nickel or cobalt to yield a stable enzyme that retains full activity (4, 29, 30). In each case, the metal is coordinated by two histidines of an active-site HEXXH motif, a conserved cysteine, and a water molecule (4).
A number of PDF inhibitors have been reported, but most do not possess antibacterial activity (13, 18, 24). However, recently it has been shown that the natural antibiotic actinonin, a hydroxamic acid pseudopeptide, is a potent inhibitor of PDF (11). In addition, a series of β-sulfonyl and β-sulfinylhydroxamic acid derivatives have been shown to be potent PDF inhibitors with in vitro antibacterial activity (1). As a result of our previous experience with mammalian matrix metalloproteinases (6), we have accumulated an extensive library of potential metalloenzyme inhibitors. Using this library, we have identified a novel compound, BB-3497, which is a potent and selective inhibitor of PDF and inhibits the growth of several clinically relevant bacterial pathogens. BB-3497 is orally bioavailable and is active in systemic models of Staphylococeus aureus infection in the mouse. The X-ray crystal structures of both actinonin and BB-3497 have been determined, and these data should facilitate the design of novel inhibitors with improved pharmacokinetic and antibacterial properties.
MATERIALS AND METHODS
Antimicrobial agents.
BB-3497, 2R-[(formyl-hydroxy-amino)-methyl]-hexanoic acid (1S-dimethylcarbamoyl-2,2-dimethylpropyl)amide, was prepared at British Biotech Pharmaceuticals Ltd. as described in patent application WO 99/39704. The purity was determined to be >97% by high-pressure liquid chromatography. BB-3497 was freely soluble in water to at least 20 mg/ml. Actinonin, ampicillin, chloramphenicol, carbenicillin, ofloxacin, and vancomycin were obtained from Sigma (Poole, United Kingdom).
PDF purification.
Escherichia coli def was cloned into the expression vector pET24a(+) (Novagen, Inc., Madison, Wis.) by standard procedures (31) and was used to transform BL21(DE3) cells. Cultures of BL21(DE3) cells harboring pET24-PDF were induced with 1 mM isopropyl-β-d thiogalactopyranoside (IPTG) for 3 h at 37°C. All steps were carried out at 0 to 4°C unless otherwise indicated. Cells were disrupted by sonication in the presence of 50 mM HEPES (pH 7.5)–5 mM NiCl2 (buffer A), and the resulting suspension was cleared by centrifugation at 20,000 × g for 15 min. The lysate was dialyzed against buffer A, and the precipitate was removed by centrifugation at 20,000 × g for 15 min. PDF was then bound to Q-Sepharose and eluted with a 0 to 0.5 M KCl gradient. Active fractions were pooled and then dialyzed against buffer A. PDF was further purified by size exclusion chromatography using Superdex 75, and the active fractions were pooled. Under these conditions, the native metal exchanges for the stable Ni2+ form. Determination of the metal content by inductively coupled plasma mass spectrometry revealed 0.8 Ni2+ ion per polypeptide (data not shown).
Enzyme assays.
PDF in vitro assays were performed with a final volume of 100 μl containing 8 ng of PDF, 80 mM HEPES (pH 7.4), 0.7 M KCl, 0.035% Brij, 1 mM NiCl2, and 4 mM f-Met-Ala-Ser; incubation was at 37°C for 30 min. The free amino group of the product (Met-Ala-Ser) was detected using fluorescamine by the addition of 50 μl of 0.2 M sodium borate (pH 9.5) followed by 50 μl of fluorescamine (0.2 mg/ml in dry dioxane). Fluorescence was quantified with an SLT Fluostar plate reader using an excitation wavelength of 390 nm and an emission wavelength of 495 nm. Vehicle controls plus or minus enzyme provided the 0 and 100% inhibition values, respectively. The data were analyzed by conversion of the fluorescence units to percent inhibition, and the inhibitor concentration was plotted against percent inhibition. The concentration (nanomolar) of inhibitor required to decrease enzyme activity by 50% (IC50) was determined. Matrix metalloproteinases were prepared as described previously (10) and assayed using a coumarin-labeled peptide (19). Angiotensin I-converting enzyme and enkephalinase were assayed as described previously (8, 14).
In vitro microbiological analysis.
MICs were determined by a broth microdilution method (26) with a starting inoculum of 5 × 105 CFU/ml for all isolates. Mueller-Hinton broth (Oxoid) adjusted with divalent cations to final concentrations, per liter, of 20 mg of Ca2+ and 10 mg of Mg2+ (CSMHB) was used unless otherwise indicated. Organisms were incubated at 35°C for 20 h, and the MIC was defined as the lowest concentration of antimicrobial agent inhibiting visible growth. For Streptococcus pneumoniae and Haemophilus influenzae, brain heart infusion broth supplemented with 2% horse serum (Oxoid) and 20 mg of NAD (Sigma) per liter was used, and the organisms were incubated for 24 h. Bacterial strains were obtained from the American Type Culture Collection. Methicillin-resistant S. aureus and vancomycin-resistant Enterococcus faecalis were clinical isolates. E. coli DH5α [F′ Δ(lacZYA-argF)U169 deoR endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 (φ80dlacZΔM15)] was obtained from Life Technologies Ltd. BL21 (DE3) [B F− dcm ompT hsdS (rB− mB−) gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1)] was obtained from Novagen Inc., and TG1 [supE hsdΔ5 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] was obtained from New England Biolabs, Inc. E. coli TG1 ΔacrB was constructed using the pKO3-based system (21) obtained from George Church, Harvard Medical School. E. coli D22 [envA1 proA23 lac-28 tsk-81 trp-30 his-51 rpsL173 (strR) tufA1 ampCp-l] was obtained from the E. coli Genetic Stock Center. E. coli DH5α Δfmt was selected as a spontaneously occurring mutant and contains a four-base deletion within the fmt coding sequence.
Molecular techniques and sequence analysis.
Molecular techniques, including cloning, PCR, and DNA purification, were performed by standard protocols (31). DNA sequences of cloned or PCR-amplified fragments were determined on both strands using an ABI PRISM dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS according to the manufacturer's instructions. The products were analyzed using an ABI 377 PRISM sequence analysis system (Perkin-Elmer Applied Biosystems).
Time-kill analysis.
The test strains were grown overnight at 37°C in CSMHB and diluted with fresh broth prewarmed to 37°C to yield a staring inoculum of approximately 106 CFU/ml. BB-3497 and control antibiotics were added at final concentrations four- and eightfold above their MICs and cultures were incubated with agitation at 37°C. Parallel cultures containing no antibiotic served as controls. Colony counts were determined at intervals by serial dilution and plating techniques. Antibiotic carryover was eliminated by using a dilution factor of at least 100.
Spontaneous mutation frequencies.
Organisms were grown in CSMHB to the exponential phase. Strains were concentrated by centrifugation, and approximately 109 CFU was spread over the surface of Mueller-Hinton agar (Oxoid) containing BB-3497 at two or four times the appropriate agar dilution MIC. Colonies were counted after 48 h of incubation at 37°C. Spontaneous mutation frequencies were determined by dividing the number of colonies on antibiotic-containing plates by the number of CFU originally plated.
Pharmacokinetics.
BB-3497 and actinonin were formulated at 20 and 10 mg/ml, respectively, in water. Three rats in each group were dosed orally (p.o.) with BB-3497 at 100 mg/kg of body weight or actinonin at 50 mg/kg. Blood samples (0.5 ml) were taken at 0.25, 0.5, 1, 2, 4, 6, and 24 h postdose. Plasma was harvested and protein was precipitated before analysis by liquid chromatography and mass spectrophotometry.
In vivo systemic infection model.
Infection models were performed at MDS Panlabs Pharmacology Services (Bothell, Wash.). Groups of 10 ICR-derived male mice were inoculated intraperitoneally (i.p.) with S. aureus (Smith) ATCC 19636 at 1.6 × 106 CFU/0.5 ml/mouse or methicillin-resistant S. aureus ATCC 33591 at 5 × 107 CFU/0.5 ml/mouse; organisms were suspended in broth containing 5% mucin (type II from porcine stomach, M 2378, lot 48H0596; Sigma). BB-3497 (100, 60, 30, 10, 6, and 3 mg/kg), ofloxacin, and vehicle (sterile water or saline) control were administered p.o. or intravenously (i.v.) to test animals 1 h after bacterial challenge. Mortality was recorded after 7 days. In all experiments, at least 9 of 10 animals died in the control group. The amount of antibiotic, in milligrams per kilogram of body weight, required to cure 50% of the infected animals (ED50) was determined.
Crystallization and structural determination.
Crystals of the PDF-actinonin and PDF–BB-3497 complexes grew in a monoclinic form under the following conditions. Hanging drops were formed by mixing 5 μl of complex solution (10 mg of PDF per ml, 20 mM inhibitor, 50 mM HEPES [pH 7.5]) with 5 μl of reservoir solution (25 to 32% polyethylene glycol 4000, 0.1 M sodium citrate [pH 5.6], 0.2 M ammonium acetate) at room temperature. The crystals were similar to those reported previously (9) and belonged to space group C2, with three independent polypeptide chains in the asymmetric unit, and the following cell dimensions: for the PDF-actinonin complex—a = 138.6 Å, b = 63.1 Å, c = 85.6 Å, and β = 121.4; and for the PDF–BB-3497 complex—a = 143.2Å, b = 64.5Å, c = 85.2Å, and β = 123.1°. Data were collected to 1.75 and 2.1 Å from single crystals of the actinonin and BB-3497 complexes, respectively, at 100 K; samples were transferred, prior to freezing, to a solution containing 33% polythylene glycol 4000, 15% glycerol, 0.1 M sodium citrate (pH 5.6), and 0.2 M ammonium acetate using Ni-filtered, double-mirror-focused Cu Kα X-rays; and data were collected on a MAR345 image plate system. Data were processed using the HKL suite of programs (28) and then the CCP4 suite of programs (3). Both complex structures were solved by molecular replacement using the program AMORE with the E. coli PDF structure 1DFF (9) as the trial model. Three copies of the PDF polypeptide were placed at the positions indicated by the molecular replacement solutions and refined as separate rigid bodies using the TNT package (33). Subsequent positional and isotropic temperature factor refinement was done with the maximum-likelihood option of REFMAC (25); solvent molecules were added automatically using ARP (20) with an acceptance criterion of a B factor of <60. The electron density for residues 165 to 168 in both complexes was indistinct, and these residues were omitted from the refinement. See Tables 2 and 3 for the data collection and refinement statistics, respectively.
TABLE 2.
Data collection statisticsa for structural determination
| Drug | Resolution (Å) | Rsymm | I/ςI | Completeness (%) | No. of independent reflections | Multiplicity |
|---|---|---|---|---|---|---|
| Actinonin | 14.0–1.75 | 0.044 (0.19) | 13.6 (3.9) | 85.9 (86.8) | 54,729 (3,634) | 1.95 |
| BB-3497 | 20–2.1 | 0.04 (0.24) | 16.2 (3.9) | 94.7 (92.7) | 36,273 (2,368) | 1.95 |
Data in parentheses are for the highest-resolution shell (1.79 to 1.75 Å for the actinonin complex and 2.15 to 2.10 Å for the BB-3497 complex).
TABLE 3.
Refinement statistics for structural determination
| Drug | Rfactor (all reflections) | Rfree (5% of reflections) | rms for deviation bonds (Å) | rms for deviation angles (°) | No. of non-H atoms | No. of solvent atoms |
|---|---|---|---|---|---|---|
| Actinonin | 0.19 | 0.25 | 0.013 | 2.437 | 4,767 | 726 |
| BB-3497 | 0.21 | 0.27 | 0.014 | 1.043 | 4,097 | 79 |
Nucleotide sequence accession numbers.
The coordinates of the PDF-actinonin and PDF–BB-3497 structures have been deposited in the Protein Data Bank under accession numbers 1G2A and 1G27 respectively.
RESULTS
Identification of PDF inhibitors.
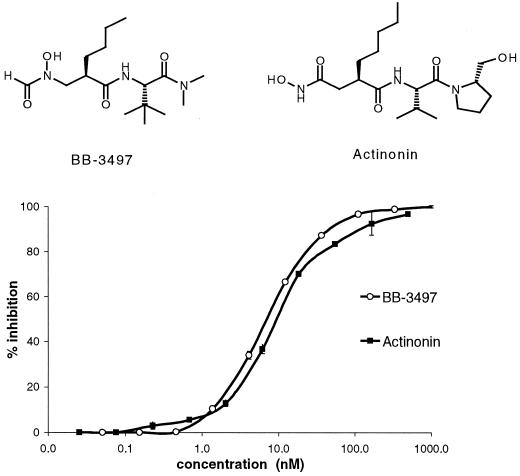
A library of compounds featuring metal-chelating groups was screened for agents that specifically inhibited PDF activity and had antibacterial activity. Several compounds were identified from this screen, including the natural hydroxamic acid antibiotic actinonin, a recently described PDF inhibitor (11). Also identified was a related N-formyl-hydroxylamine derivative, BB-3497, whose metal binding group closely mimics the N-formyl substrate of PDF and showed a strong structural resemblance to the known PDF substrate fMet-Ala-Ser. Actinonin and BB-3497 were both potent inhibitors of E. coli PDF.Ni in an in vitro assay, with IC50s of 10 and 7 nM, respectively, and were highly selective for PDF over other mammalian metalloenzymes (Table 1 and Fig. 1).
TABLE 1.
Metalloenzyme inhibition profiles for actinonin and BB-3497
| Compound | IC50 (nM)a for:
|
||||||
|---|---|---|---|---|---|---|---|
| PDF.Ni | MMP-1 | MMP-2 | MMP-3 | MMP-7 | Enkephalinase | ACE | |
| BB-3497 | 7 | 2,000 | 15,000 | >100,000 | >100,000 | 50,000 | >100,000 |
| Actinonin | 10 | 1,000 | 3,000 | 6,000 | 60%b | 7,000 | >100,000 |
MMP, matrix metalloproteinases; ACE, angiotensin I-converting enzyme.
Percent inhibition at 100 μM.
FIG. 1.
Structures of BB-3497 and actinonin and effects on the activity of E. coli PDF.Ni. Error bar, standard deviation.
Structural determination of actinonin and BB-347 bound to PDF.
To elucidate the interactions that contribute to the binding affinity of actinonin and BB-3497 for PDF, we purified the PDF.Ni enzyme, cocrystallized it with actinonin and BB-3497, and determined the resulting structures using molecular replacement (Tables 2 and 3 and Fig. 2). In both the PDF-actinonin and the PDF–BB-3497 structures, the three molecules in the asymmetric unit were very closely related, except for small deviations in regions 64 to 69 and the C-terminal helix at regions 147 to 164. The root mean square (rms) for deviations between the remaining 140 Cα atoms for both structures was approximately 0.25 Å. The inhibitors bound in a similar fashion in each complex, lying in a cleft on the enzyme surface and approximately within the active site, confirming the locations of the S1′, S2′, and S3′ binding pockets in PDF.
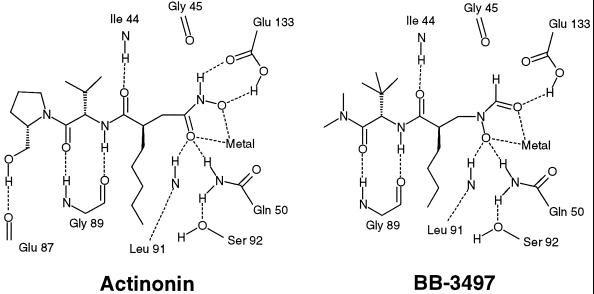
FIG. 2.
Actinonin and BB-3497 bound to the active site of E. coli PDF.
In both complexes, the Ni atom is pentacoordinated by the two O atoms of the hydroxamate group of actinonin or those of the N-formyl-hydroxylamine of BB-3497, the N-ɛ2 atoms of the side chains of His 132 and His 136, and the Sγ atom of Cys 90 (Fig. 2). The average oxygen nickel binding distances are 2.1 Å (to the carbonyl oxygen atom of the N-formyl-hydroxylamine or the nitrogen-bound oxygen atom of the hydroxamate) and 2.3 Å (to the nitrogen-bound oxygen atom of the N-formyl-hydroxylamine or the carbonyl oxygen atom of the hydroxamate), which are within the normal range of oxygen zinc binding distances observed for hydroxamate inhibitors bound to matrix metalloproteinases (2) and thermolysin (17). Hydrogen bonds are also made to the hydroxamate or the N-formyl-hydroxylamine by the side chains of Glu 133 and Gln 50 and the main-chain NH of Leu 91. In both inhibitor complexes, hydrogen bonds are made between the main-chain NH of Ile 44 and the P1′ carbonyl and also between the main-chain carbonyl oxygen and NH groups of Gly 89 and the P2′ NH and carbonyl groups, respectively. The hydrophobic S1′ pocket is delineated by the residues Ile 44, Ile 86, Glu 88, Leu 125, Ile 129, and His 132 and is occupied by the n-pentyl or n-butyl side chain of actinonin or BB-3497, respectively. In the P2′ position, the side chain (isopropyl in actinonin and tert-butyl in BB-3497) is mainly exposed to solvent but does make van der Waals interactions with the side chain of Arg 97, which adopts a slightly different conformation in each complex. Similarly, most of the inhibitor atoms at the P3′ position are solvent accessible, with one face of the pyrrolidine ring in actinonin or the tertiary amine in BB-3497 packing against the side chains of Ile 44 and Leu 125. In the PDF-actinonin complex, a final hydrogen bond is made between the terminal alcohol group and the main-chain carbonyl oxygen of Glu 87 (Fig. 2).
In vitro microbiological properties of BB-3497 and actinonin.
BB-3497 showed superior in vitro antibacterial activity relative to actinonin, particularly against gram-negative bacteria (Table 4). To determine if the antibacterial activity of these compounds was due to their inhibition of PDF, we measured the MICs of actinonin and BB-3497 for an E. coli strain with a null deletion mutation of the formyltransferase gene (fmt). Strains which lack transformylase activity use methionine-tRNAiMet to initiate protein synthesis, albeit inefficiently, and thus have no requirement for deformylase activity. Such mutations are known to severely inhibit the growth of E. coli (15). In accordance with the prediction for an antibiotic whose mode of action is inhibition of PDF, the E. coli fmt mutant was totally resistant to actinonin and BB-3497 (Table 5). In addition, when def was introduced into E. coli on a plasmid and expression was induced from a strong promoter, the MICs of actinonin and BB-3497 increased significantly compared to those of control antibiotics (Table 5). Taken together, these experiments confirm the mode of action of actinonin and BB-3497.
TABLE 4.
In vitro activities of actinonin and BB-3497
| Bacterial straina | MIC (μg/ml) of:
|
|
|---|---|---|
| Actinonin | BB-3497 | |
| Staphylococcus aureus ATCC 29213 (MSSA) | 32 | 16 |
| Staphylococcus aureus ATCC 6538 (MSSA) | 32 | 4 |
| Staphylococcus aureus (MRSA) | 32 | 16 |
| Staphylococcus epidermidis ATCC 27626 | 2 | 4 |
| Enterococcus faecalis ATCC 29212 | 64 | 32 |
| Enterococcus faecalis (Vanr) | 128 | 8 |
| Streptococcus pneumoniae ATCC 49619 | 32 | 8 |
| Haemophilus influenzae ATCC 49247 | 2 | 0.25 |
| Escherichia coli ATCC 25922 | 64 | 8 |
| Escherichia coli TG1 (parent) | 128 | 8 |
| Escherichia coli TG1 (ΔacrB) | 1 | 0.125 |
| Escherichia coli D21 (parent) | 128 | 8 |
| Escherichia coli D22 (envA1) | 8 | 0.25 |
| Klebsiella pneumoniae ATCC 13883 | 128 | 8 |
| Enterobacter cloacae ATCC 13047 | >128 | 8 |
| Pseudomonas aeruginosa ATCC 27853 | 128 | 128 |
MSSA, methicillin-sensitive S. aureus; MRSA, methicillin-resistant S. aureus.
TABLE 5.
MICs of actinonin and BB-3497 against an E. coli formyltransferase (fmt) mutant and E. coli BL21(DE3) strains harboring a control plasmid (pET24) or pET24-PDFa
| Strain | MIC (μg/ml)a of:
|
|||
|---|---|---|---|---|
| Actinonin | BB-3497 | Chloramphenicol | Carbenicillin | |
| DH5α (parent) | 64 | 4 | 8 | 8 |
| DH5α Δfmt | >512 | >128 | 4 | 4 |
| BL21(DE3)(pET24) | 100 | 8 | 2 | 1 |
| BL21(DES)(pET24-PDF) | 400 | 32 | 2 | 1 |
| BL21(DE3)(pET24-PDF) + IPTG | >800 | 128 | 1 | 1 |
MICs were determined using a standard microdilution method with Trypticase soy broth. For induction of PDF expression, 0.1 mM IPTG was added to the growth medium.
The bacteriostatic or bactericidal effects of BB-3497 against S. aureus ATTC 29213 and E. coli ATCC 25922 were assessed with a 24-h time-kill analysis. At four and eight times the MIC, both strains showed <1-log-unit decreases in viable counts. The control antibiotic ampicillin or vancomycin showed bactericidal activity, with a decrease of more than 3-log units in viable counts for E. coli or S. aureus, respectively. Thus, as reported previously for actinonin (11), BB-3497 has a bacteriostatic mode of action.
Mechanism of resistance to BB-3497.
The spontaneous mutation frequencies of S. aureus ATTC 29213 and E. coli 25922 were determined with BB-3497. Bacteria were plated on antibiotic-containing plates at two and four times the agar MIC for each organism. BB-3497-resistant mutants arose at frequencies of 1 × 10−7 for E. coli and 2 × 10−7 for S. aureus. For S. aureus, all the resistant strains were highly resistant to BB-3497 (Table 6). The DNA sequence of the defA-fmt operon and the defB gene of the S. aureus strains was determined. All the resistant strains had mutations within the fmt gene that would result in the expression of a truncated and presumably non-functional formyltransferase. These results were similar to those previously reported for actinonin-resistant mutants (22). For E. coli, two phenotypes were evident: first, strains with two- to fourfold increases in the MIC but with normal growth properties, and second, very resistant strains that grew slowly. The sequence of the def-fmt operon was determined for selected strains. For the highly resistant, slowly growing class of mutant, one strain had a missense mutation in the fmt gene, and the others had mutations that would result in the expression of a truncated formyltransferase (Table 6). DNA sequence analysis of a mutant with only a two- to fourfold increase in the MIC showed that the def-fmt operon was identical to that in the wild-type strain (Table 6).
TABLE 6.
Genotypic characterization and doubling times of BB-3497-resistant S. aureus and E. coli mutantsa
| Strain | fmt genotype | Doubling time (min) | MIC (μg/ml) |
|---|---|---|---|
| S. aureus | |||
| ATCC 29213 | Wild type | 27 | 32 |
| SA 2 | Frameshift at Ile 155 | 39 | >256 |
| SA 6 | Glu 197>Stop | 37 | >256 |
| SA 11 | Frameshift at Ile 183 | 35 | >256 |
| SA 13 | Frameshift at Ser 68 | 35 | >256 |
| E. coli | |||
| ATCC 25922 | Wild type | 30 | 8 |
| EC 23 | Wild type | 32 | 32 |
| EC 34 | Gln 65>Stop | 72 | >256 |
| EC 37 | Ala 10>Pro | 71 | >256 |
| EC 55 | Frameshift at Lys 45 | 60 | >256 |
All strains were wild type for def.
The doubling times of the resistant strains grown in rich media with agitation were determined (Table 6). The E. coli fmt mutants had doubling times in the exponential phase of growth that were at least twice that of the wild type but were not as disabled as previously reported for laboratory-adapted E. coli K-12 strains with fmt disruptions (15, 27). The S. aureus fmt mutants grew 30 to 40% more slowly than the parental strain, similar to previously reported results (22).
In vivo activity of BB-3497.
Actinonin and its derivatives were never developed for the treatment of infections due to their poor bioavailability and consequent lack of in vivo efficacy (7). This information was confirmed in our own studies with rats, for which we were unable to detect actinonin in the blood following a 50-mg/kg p.o. dose. In contrast, BB-3497 was rapidly and well absorbed following p.o. administration to rats at a dose of 100 mg/kg (maximum concentration of drug in serum, 24 mg/liter; area under the concentration-time curve from 0 to 24 h, 34 mg · h/liter). In light of its favorable pharmacokinetic properties, BB-3497 was tested in a murine systemic S. aureus infection model. A single i.v. or p.o. dose of BB-3497, given 1 h after an i.p. injection of S. aureus Smith, rescued mice from infection with an ED50 of 7 and 8 mg/kg, respectively (Table 7). When tested against a methicillin-resistant strain of S. aureus, BB-3497 administered p.o. had an ED50 of 14 mg/kg, which compared favorably to that of ofloxacin, which had an ED50 of 10 mg/kg. These results demonstrate the potential of this new class of antibiotic for the treatment of bacterial infections.
TABLE 7.
Protection of mice by the PDF inhibitor BB-3497 in a model of systemic S. aureus infection
| S. aureus | Compound | Route | ED50 (mg/kg) |
|---|---|---|---|
| Smitha | BB-3497 | i.v. | 7 |
| Smithb | Ofloxacin | i.v. | 2 |
| Smith | BB-3497 | p.o. | 8 |
| Smith | Ofloxacin | p.o. | 10 |
| Methicillin resistantc | BB-3497 | p.o. | 14 |
| Methicillin resistantd | Ofloxacin | p.o. | 10 |
BB-3497 MIC, 1 μg/ml.
Ofloxacin MIC, 0.3 μg/ml.
BB-3497 MIC, 3 μg/ml.
Ofloxacin MIC, 0.3 μg/ml.
DISCUSSION
Inhibition of deformylase activity is an attractive target for antibiotic therapy, as it is an essential function that is ubiquitous in pathogenic bacteria and as it has no mammalian counterpart. Recently, a number of inhibitors of PDF have been described to have antibacterial activity (1, 11); however, to date none has been shown to have activity in animal models of infection. In this study, by screening a library of metalloenzyme inhibitors for inhibitors of PDF with antibacterial activity, we have identified the previously described hydroxamic acid derivative actinonin and a novel N-formyl-hydroxylamine derivative, BB-3497. BB-3497 and actinonin are potent inhibitors of E. coli PDF and were highly selective for PDF over the other mammalian metalloenzymes tested. Both also show a strong structural resemblance to the known PDF substrate fMet-Ala-Ser. To understand how these inhibitors bind to PDF, X-ray crystal structures of BB-3497 and actinonin bound to PDF were determined.
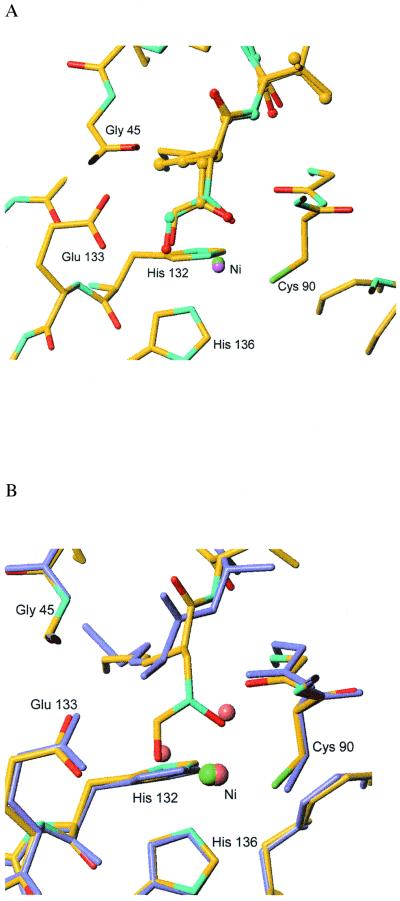
In both complexes, the active-site Ni atom is pentacoordinated by the two O atoms of the hydroxamate group of actinonin or those of the N-formyl-hydroxylamine of BB-3497 and by the side chains of Cys 90, His 132, and His 136. The alkyl chains which mimic the methonine side chain of the natural substrate lie along the hydrophobic S1′ pocket. The P2′ and P3′ side chains are largely exposed to solvent and are therefore attractive sites for modification to improve the properties of the molecules. From the structures it is apparent that the two oxygen atoms of the N-formyl-hydroxylamine of BB-3497 or those of the hydroxamate moiety of actinonin occupy approximately the same positions as the two Ni-bound water molecules seen in the structure of E. coli PDF. Ni complexed with Met-Ala-Ser (4) (Fig. 3). The position of the nitrogen-bound oxygen of the N-formyl-hydroxylamine of BB-3497 (or the carbonyl oxygen of the hydroxamate of actinonin) corresponds to that of the formyl group of the substrate, which is presumed to bind to an oxyanion hole created by interactions with the main-chain amide of Leu 91, the side-chain amide of Gln 50, and the Ni atom. The position of the carbonyl oxygen of the N-formyl-hydroxylamine (or the nitrogen-bound oxygen of the hydroxamate of actinonin) is close to that of the water molecule that is presumed to act as the attacking hydroxide nucleophile in the hydrolytic cycle of the enzyme. This information would suggest that the tight binding of BB-3497 or actinonin to PDF is derived in part by mimicking of the structure of critical elements of the active site chemistry. Since the structure that we observed is not that of the true transition state, it is difficult to be certain of the implications of these findings for the enzyme's mechanism. Nevertheless, the pentacoordinated nickel is consistent with the structure of the product complex of PDF and with a pentacoordinated transition state, as proposed by Becker et al. (4). However, analysis of the crystal structure of a substantially weaker H-phosphonate inhibitor bound to PDF revealed that the metal center is tetrahedrally coordinated in this transition state mimic and that the metal hydrogen bonding network is less complex than that in BB-3497 and actinonin (16). These factors probably contribute to the difference in activity between these inhibitors. Further studies are required to resolve these differences.
FIG. 3.
Overlay of X-ray crystal structures of actinonin, BB-3497, and Met-Ala-Ser bound to the active site of E. coli PDF. (A) Overlay of the nickel-inhibitor binding site in the PDF–BB-3497 complex (atom colors, sticks, Ni in green) with the actinonin structure (atom colors, ball and stick, Ni in purple), showing the similarity in nickel coordination of the hydroxamate in actinonin and the N-formyl-hydroxylamine in BB-3497; for clarity, only the protein atoms of the BB-3497 complex are shown. (B) Overlay of the PDF–BB-3497 structure (atom colors, Ni in green) with that of the product Met-Ala-Ser bound to the active site (slate blue, Ni and water molecules; pink, PDB access code 1bs6), showing the similarity in the positions of the water molecules in the Met-Ala-Ser–PDF structure and the oxygen atoms in the N-formyl-hydroxylamine in the PDF–BB-3497 structure.
BB-3497 and actinonin show activity predominantly against gram-positive pathogens, although BB-3497 does have activity against E. coli, Enterobacter cloacae, and Klebsiella pneumoniae. The potential to obtain significant potencies with the PDF inhibitor class of antibiotics against gram-negative bacteria, however, was evidenced by the potencies of actinonin and BB-3497 against an E. coli strain in which the gene encoding the AcrB multidrug efflux pump had been deleted or against a “leaky” E. coli envA1 strain, in which the outer membrane was compromised (Table 4). Similar improvements in potency have been obtained with BB-3497 for an oprM efflux pump mutant of Pseudomonas aeruginosa (S. Salama, personal communication). For gram-negative bacteria, it is well known that a combination of the negatively charged outer membrane and membrane-bound multidrug efflux pumps can act synergistically to reduce the activities of many antibiotics.
The mechanism of resistance of BB-3497 and other PDF inhibitors via mutations in the formyltranferase gene fmt was predicted from earlier genetic analysis of the formylation-deformylation cycle (23) and as previously reported for actinonin (22). For both E. coli and S. aureus, resistant mutants were readily isolated in vitro. The viability of the E. coli fmt mutants was clearly compromised, with low growth rates, as previously reported (15, 27). In contrast, the S. aureus strains were only modestly compromised in vitro; however, the growth of S. aureus fmt mutants has been shown to be significantly attenuated in a mouse abscess model (22). Similarly, BB-3497-resistant S. aureus strains require a 25- to 100-fold higher inoculum to establish infection in a mouse systemic infection model (British Biotech Pharmaceuticals Ltd., unpublished data). Thus, bypass of the normal methionine formylation-deformyation cycle places S. aureus at a considerable disadvantage in vivo. An important question for the PDF class of antibiotic will be to determine how quickly fmt mutants are selected during therapy.
Actinonin has been known as an antibiotic for nearly 40 years; however, early attempts to develop a series of hydroxamic acid analogues of actinonin never reached clinical development due to poor in vivo activity (7). By screening our metalloenzyme inhibitor library, which is enriched with more “drug-like” compounds, we isolated a compound with good pharmacokinetic properties. The reasons for these improved pharmacokinetic properties of BB-3497 relative to actinonin are unclear but may be related to stability within the gastrointestinal tract and transport across the gastrointestinal tract wall. We have previously hypothesized for matrix metalloproteinase inhibitors that feature a P2′ tert-leucine that the tert-butyl moiety shields the neighboring amide groups, thereby reducing hydrogen bonding to bulk solvent and improving absorption (6). The tert-butyl group may also prevent undesirable proteolytic attack on the amides. In contrast, the corresponding substituent in actinonin is isopropyl, which has a markedly weaker steric shielding effect.
To validate PDF as a target for antibacterial chemotherapy, we investigated BB-3497 in models of systemic S. aureus infection. BB-3497 showed significant antibacterial activity when given as a single dose 1 h after the start of infection. As predicted, this activity was independent of the bacterial resistance of the strain and was noted for both methicillin-sensitive and methicillin-resistant S. aureus. The activities of BB-3497 were similar via the i.v. or p.o. route, reflecting the good oral bioavailability of this compound. The activity also compared favorably with that of the control antibiotic ofloxacin (Table 7).
In conclusion, we have identified BB-3497 as a potent PDF inhibitor with good selectivity for mammalian metalloenzymes and activity against gram-negative and gram-positive pathogens, including multidrug-resistant strains. BB-3497 is well absorbed following p.o. administration and is effective in animal models of infection, validating the potential of PDF as an antibacterial target. Knowledge of the mode of binding of PDF inhibitors will greatly facilitate the design of further synthetic PDF inhibitors with improved antibacterial potency and in vivo efficacy. A key aspect of this design will be a detailed understanding of bacterial cell wall penetration and the processes that regulate the cytosolic concentration of the drug in bacteria. Details of the observed structure-activity relationship of BB-3497 and optimization of antibacterial activity will be described elsewhere.
ACKNOWLEDGMENTS
We thank Ian Johnson, British Biotech Pharmaceuticals Ltd., for construction of the E. coli acrB strain and Stephen Chandler, British Biotech Pharmaceuticals Ltd., for help and advice on the PDF assay.
REFERENCES
- 1.Apfel C, Banner D W, Bur D, Dietz M, Hirata T, Hubschwerlen C, Locher H, Page M G, Pirson W, Rosse G, Specklin J L. Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J Med Chem. 2000;43:2324–2331. doi: 10.1021/jm000018k. [DOI] [PubMed] [Google Scholar]
- 2.Babine E, Bender S L. Molecular recognition of protein-ligand complexes: applications to drug design. Chem Rev. 1997;97:1359–1472. doi: 10.1021/cr960370z. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, S. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. Sect. D 50:760–763. [DOI] [PubMed]
- 4.Becker A, Schlichting I, Kabsch W, Groche D, Schultz S, Wagner A F. Iron center, substrate recognition and mechanism of peptide deformylase. Nat Struct Biol. 1998;5:1053–1058. doi: 10.1038/4162. [DOI] [PubMed] [Google Scholar]
- 5.Becker A, Schlichting I, Kabsch W, Schultz S, Wagner A F. Structure of peptide deformylase and identification of substrate binding site. J Biol Chem. 1998;273:11413–11416. doi: 10.1074/jbc.273.19.11413. [DOI] [PubMed] [Google Scholar]
- 6.Beckett R P, Davidson A H, Drummond A H, Huxley P, Whittaker M. Recent advances in matrix metalloproteinase inhibitor research. Drug Discov Today. 1996;1:16–26. [Google Scholar]
- 7.Broughton B J, Chaplen P, Freeman W A, Warren P J, Wooldridge K R H, Wright D E. Studies concerning the antibiotic actinonin. Part VIII. Structure-activity relationships in the actinonin series. J Chem Soc Perkin Trans 1. 1975;1975:857–860. [PubMed] [Google Scholar]
- 8.Carmel A, Yaron A. An intramolecularly quenched fluorescent tripeptide as a fluorogenic substrate of angiotensin I-converting enzyme and bacterial dipeptidyl carboxypeptidase. Eur J Biochem. 1978;87:265–273. doi: 10.1111/j.1432-1033.1978.tb12375.x. [DOI] [PubMed] [Google Scholar]
- 9.Chan M K, Gong W, Rajagopalan P T, Hao B, Tsai C M, Pei D. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry. 1997;36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- 10.Chandler S, Coates R, Gearing A, Lury J, Wells G, Bone E. Matrix metalloproteinases degrade myelin basic protein. Neurosci Lett. 1995;201:223–226. doi: 10.1016/0304-3940(95)12173-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen D Z, Patel D V, Hackbarth C J, Wang W, Dreyer G, Young D C, Margolis P S, Wu C, Ni Z J, Trias J, White R J, Yuan Z Y. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 12.Dardel F, Ragusa S, Lazennec C, Blanquet S, Meinnel T. Solution structure of nickel-peptide deformylase. J Mol Biol. 1998;280:501–513. doi: 10.1006/jmbi.1998.1882. [DOI] [PubMed] [Google Scholar]
- 13.Durand D J, Gordon Green B, O'Connell J F, Grant S K. Peptide aldehyde inhibitors of bacterial peptide deformylases. Arch Biochem Biophys. 1999;367:297–302. doi: 10.1006/abbi.1999.1274. [DOI] [PubMed] [Google Scholar]
- 14.Florentin D, Sassi A, Roques B P. A highly sensitive fluorometric assay for “enkephalinase,” a neutral metalloendopeptidase that releases tyrosine-glycine-glycine from enkephalins. Anal Biochem. 1984;141:62–67. doi: 10.1016/0003-2697(84)90425-1. [DOI] [PubMed] [Google Scholar]
- 15.Guillon J M, Mechulam Y, Schmitter J M, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao B, Gong W, Rajagopalan P T, Zhou Y, Pei D, Chan M K. Structural basis for the design of antibiotics targeting peptide deformylase. Biochemistry. 1999;38:4712–4719. doi: 10.1021/bi982594c. [DOI] [PubMed] [Google Scholar]
- 17.Holmes M A, Matthews B W. Binding of hydroxamic acid inhibitors to crystalline thermolysin suggests a pentacoordinate zinc intermediate in catalysis. Biochemistry. 1981;20:6912–6920. doi: 10.1021/bi00527a026. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y J, Rajagopalan P T, Pei D. H-phosphonate derivatives as novel peptide deformylase inhibitors. Bioorg Med Chem Lett. 1998;8:2479–2482. doi: 10.1016/s0960-894x(98)00443-0. [DOI] [PubMed] [Google Scholar]
- 19.Knight C G, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 20.Lamzin V S, Wilson K S. Automated refinement of protein models. Acta Crystallogr Sect D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 21.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolis P S, Hackbarth C J, Young D C, Wang W, Chen D, Yuan Z, White R, Trias J. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinnel T, Patiny L, Ragusa S, Blanquet S. Design and synthesis of substrate analogue inhibitors of peptide deformylase. Biochemistry. 1999;38:4287–4295. doi: 10.1021/bi982622r. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov G N, Vagin A A, Dodson E J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D. 1996;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Newton D T, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Ragusa S, Blanquet S, Meinnel T. Control of peptide deformylase activity by metal cations. J Mol Biol. 1998;280:515–523. doi: 10.1006/jmbi.1998.1883. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan P T, Pei D. Oxygen-mediated inactivation of peptide deformylase. J Biol Chem. 1998;273:22305–22310. doi: 10.1074/jbc.273.35.22305. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Solbiati J, Chapman-Smith A, Miller J L, Miller C G, Cronan J E., Jr Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J Mol Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- 33.Tronrud D E, Ten Eyck L F, Matthews B W. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr Sect A. 1987;43:489–501. [Google Scholar]