Abstract
Nowadays, the biosynthesis of metal nanoparticles, particularly from plants, has been gaining interest. In the present work, the methanolic extracts of leaves, fruits, and the pollen grains of Sabal blackburniana were used for the green synthesis of ZnO nanoparticles, which were early detected by the formation of precipitate and further confirmed by UV-vis spectroscopy, transmission electron microscopy (TEM), X-ray diffraction (XRD), Fourier transform infra-red (FT-IR) spectroscopy and zeta potential (ZP) studies. TEM analysis has shown different shapes, predominantly irregular small spherical narrow particles included in hexagonal structures with size ranging from 2.23 to 49.56 nm. The XRD pattern confirmed that all synthesized ZnO nanoparticles have wurtzite hexagonal structure with crystalline nature. The average particle crystallite sizes were 47.21, 47.67 and 47.8 nm. The UV-visible spectra showed λmax in the range of 354–368 nm, which indicated the presence of ZnO nanoparticles. The FT-IR analysis identifies the characteristic functional groups present on the surface of ZnO nanoparticles. The ZP determination demonstrated that all representative selected synthesized ZnONPs exhibited acceptable ZP values of −30.8 to −45.9 mV, which indicated their good stability. In addition, the anti-Alzheimer potential of the selected extracts and ZnONPs was evaluated by assessing acetylcholinesterase inhibitory activity in vitro according to the improved Ellman method. The results indicated that the selected extracts have acetylcholinesterase inhibitory activity, and highlighted the promising inhibitory potential of green-synthesized ZnONPs using pollen grains, fruits and leaves extracts; they exhibited a potent inhibitory effect with IC50 values 63.78 ± 1.04651, 81.985 ± 3.075 and 117.95 ± 6.858 ng ml−1 respectively in comparison to donepezil as standard (IC50 = 50.7 ± 5.769 ng ml−1). Dereplication analysis of the selected extracts was performed using LC-MS; metabolic profiling revealed the presence of 41 compounds belonging to various chemical classes: flavonoids, steroidal saponins, terpenoids, alkaloids, lignans, sterols and fatty acids. Docking these dereplicated metabolites against the human AChE showed that the non-glycosylated flavonoid class of compounds was able to achieve interesting binding modes inside the AChE active site; they are suggested to be associated with the observed anti-AChE activity of Sabal extracts. This study is the first report to shed light on the acetylcholinesterase inhibitory activity of green-synthesized ZnO nanoparticles of S. blackburniana metabolites.
Schematic diagram representing the current study.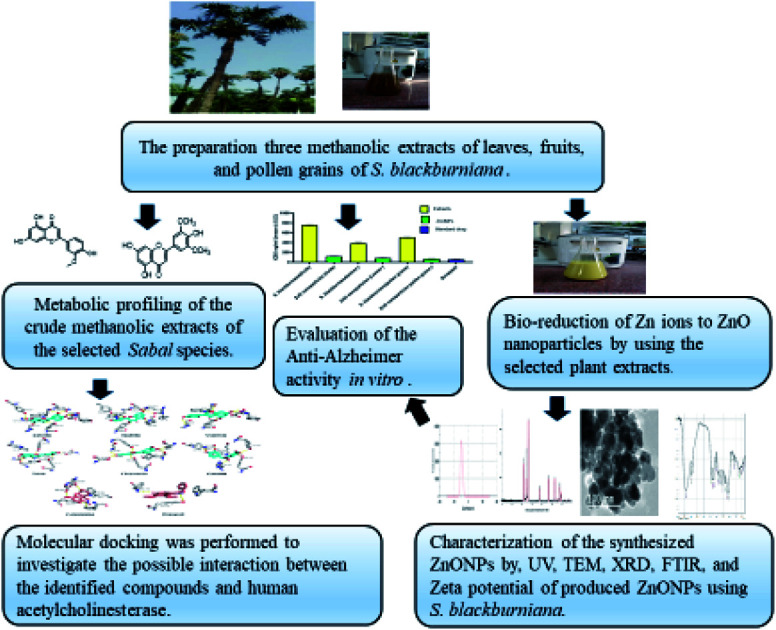
1. Introduction
Recently, nanotechnology has been used in the development and improvement of techniques for the diagnosis of several diseases and played an important role in the treatment of Alzheimer's disease1 Alzheimer's disease is considered a primary degenerative disease of the central nervous system resulting in several regarding problems such as memory, behaviour and cognition impairments, and it can ultimately lead to dementia.2 AChE inhibition is an important target for the management of Alzheimer's disease. The decrease in the level of acetylcholinesterase (AChE (leads to reduced hydrolysis of the neurotransmitter acetylcholine (ACh), and can improve the cognitive function. AChE inhibitors are the most common drugs used for management, diagnosis and treatment of Alzheimer's disease in addition to other diseases such as glaucoma, myasthenia gravis, and bladder distention diseases. Many nanoparticles such as silica dioxide, silver, titanium dioxide and zinc oxide nanoparticles have been used for the treatment of neurological diseases.3–5 Nanotechnology points out the regulating matter at a dimension of roughly 1 to 100 nanometers, which is quite different from those of larger particles; nanoparticles have unique properties and specific characteristics such as size, shape, distribution, chemical reactivity and potential applications in catalysis (as they offer high surface areas).6 Several efforts have been made to develop cheaper techniques and novel methods for the synthesis of nanoparticles.7 They exhibit unique properties, namely, high chemical stability, low dielectric constant, and good photoelectric capacity.8–10 Moreover, they are promising for various technical applications, such as nanogenerators, biosensors, solar cells, varistors, photo catalysts and photo detectors.11 They are preferred due to their non-toxic nature. Recently, the green synthesis of ZnO nanoparticles using plant extracts is simple. It offers several advantages such as timesaving (fast) processes, cost-effectiveness compared to others as well as eco-friendly (environmentally non-toxic) nature. Nanoparticles have tremendous potential applications such as antimicrobial activity.12–15 The potential use of ZnO nanoparticles in various industries such as food, pharmacy, agriculture, and cosmetics for antibacterial activity and safety is remarkable. The U.S. Food and Drug Administration classified ZnO as a generally recognized as safe (GRAS) compound.16 The genus Sabal belongs to the Palmae family (Arecaceae). It includes around twenty species and is native to the South East of the United States, to the North of South America and the West Indies. Sabal is unarmed palms with bisexual flowers.17 The aim of this study is to obtain green-synthesized ZnO nanoparticles using the extracts of leaves, fruits, and pollen grains of S. blackburniana. In addition to their characterization by UV-vis spectroscopy, transmission electron microscopy, Fourier transform infra-red spectroscopy and zeta potential studies were conducted to confirm their synthesis. Then, the in vitro investigation of the possible acetylcholinesterase enzyme (AChE) inhibitory activity of the methanolic extracts of different parts (leaves, fruits, and pollen grains) of S. blackburniana and green-synthesized ZnO nanoparticles was conducted with donepezil as the standard. Along with this, metabolomics profiling was performed using an Acquity Ultra Performance Liquid Chromatography system equipped with a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer, in order to identify the phytochemical composition of the selected extracts. Then, molecular docking studies of annotated secondary metabolites were carried out to demonstrate and explore the chemical compounds. This might contribute to the anti-Alzheimer potential. The main objective of this study is to evaluate the anticholinesterase potential of the green-synthesized ZnO nanoparticles and extracts of leaves, fruits and pollen grains of S. blackburniana in vitro. Moreover, the identification of the top and interesting metabolites can help in achieving the best binding modes inside the acetylcholinesterase's active site. This may be responsible for the effectiveness in controlling Alzheimer's disease by molecular docking of the dereplicated metabolites against human acetylcholinesterase (AChE). These can be a primary step to aid the discovery of the significantly promising anti-Alzheimer potential, which may have less serious side effects.
2. Materials and methods
2.1. Plant material
Fresh parts (leaves, fruits, and pollen grains) of S. blackburniana family (Arecaceae) had been collected during summer (2019) from Al-Orman Botanical garden, Giza, Egypt, and were authenticated by senior botanist Mrs Therris Labib, Head Specialist for plant taxonomy, El-Orman Botanical Garden, Giza, Egypt. A voucher specimen (N.O.19.6.2019 (1)) of the selected plant was deposited in the herbarium of Pharmacognosy Department, Faculty of pharmacy, Cairo University, Egypt. The plant parts were washed with tap water, and the surface washed with distilled water until no impurities remained. Then, they were shade dried at room temperature to remove moisture. The dried parts were pulverized in a clean electric blender to obtain a fine powder, which was stored in a tightly closed amber-colored glass container. Fig. 1 displays the overall scheme of materials and methods.
Fig. 1. Scheme of green synthesis of ZnO nanoparticles using Sabal blackburniana.
2.1.1. Preparation of methanolic extracts for metabolomics
The dried samples, leaves, fruits, and pollen grains (200 g), of S. blackburniana were separately extracted with 90% methanol by cold maceration till exhaustion. The total methanolic extracts of the samples were evaporated under reduced pressure using a vacuum rotary evaporator (Buchi-210, Switzerland).
2.1.2. Sample preparation for anti-Alzheimer study
Dilute purified AChE to 400 U l−1 using assay buffer. Dissolve the dried methanolic extracts of the tested samples in DMSO, and it is prudent to first test the tolerance of DMSO by the enzyme AChE; the DMSO concentration of 5 μl of test compounds added to the reaction should be 40 v% DMSO or less.
2.2. Chemicals and material for anticholinesterase activity of the selected Sabal species
Purified AChE (e.g. Sigma Aldrich cat# C3389), AChE inhibitor (e.g.), clear flat-bottom 96-well plates (e.g. VWR cat# 82050-760), pipetting devices and accessories (e.g. multi-channel pipettor), and a plate reader (Bioline Elisa Plate Reader, India (were used in the experiments.
2.3. Chemicals for LC-HRMS analysis
Methanol used for LC-HRMS analysis was supplied by Merck Co., Darmstadt, Germany. HPLC-grade acetonitrile and formic acid were obtained from Baker (The Netherlands) and Milli-Q water (Thermo Fisher Scientific Inc., Dublin, Ireland) were used for UPLC analysis. Other chemicals used in the present study were of analytical grade and supplied from Sigma-Aldrich Chemical Co. (Ireland).
2.4. Chemicals for ZnO nanoparticles
Absolute ethanol (CH3CH2OH), sodium hydroxide (NaOH) and all the reagents purchased were of analytical grade and used without any further purification. Zinc acetate dihydrate (Zn(CH3COO)2·2H2O) was purchased from Sigma-Aldrich, Germany. Distilled water (deionized water) was employed for the preparation of aqueous solution for all experiments.
2.5. Synthesis of ZnO nanoparticles using the extract of selected Sabal species
ZnO nanoparticles were synthesized by a modified method of Bala et al.11,18 First, 100 mg of the dried methanolic extracts of the selected samples were dissolved in absolute ethanol and filtered using a Whatman no. 1 filter paper. Then, 5 ml of the obtained plant extract was used as a reducing and capping agent and added dropwise to the solution containing 1 g of zinc acetate dehydrate, which was dissolved in 50 ml distilled water. The resulting mixture was stirred and kept in an ultrasonic water bath at 60 °C for 30 min, and the pH values were adjusted to 12, by adding an ammonia solution dropwise. Then, the mixture was left for 30 min until complete reduction of zinc acetate to zinc oxide nanoparticles (formation of white precipitate), and the resulting suspension was centrifuged at 12 000 rpm for 10 min. The resulting precipitate was repeatedly washed with distilled water followed by absolute ethanol to get rid of any impurities present on them. Finally, the precipitate was dried at 60 °C in an oven.
2.6. Characterization of the synthesized ZnO nanoparticles by UV-visible spectroscopy
ZnO nanoparticles were characterized and monitored by recording the UV-vis spectrum of the reaction medium using a UV spectrophotometer model Shimadzu UV-1601 (Shimadzu Corporation, Japan) in the wavelength range from 200 to 500 nm. ZnONPs samples were dissolved in methanol and the solution was placed in a quartz cuvettes with 1 cm path length.
2.7. Characterization of the synthesized ZnO nanoparticles (ZnONPs) by TEM
The characterization of the green-synthesized ZnONPs was carried out using a transmission electron microscope (TEM; Jeol model JEM-1011, Japan). Few drops of ZnONP suspension were located on a copper grid, coated with a carbon support film, and the excess liquid was removed with a filter paper and then kept at room temperature until complete solvent evaporation. The TEM image investigated the morphology and particle size of ZnO nanoparticles.
2.8. X-ray diffraction (XRD) analysis
X-ray diffraction (XRD) of dry powders of the green-synthesized ZnONPs was performed using an X-ray diffractometer (Shimadzu Lab, XRD-6000, Japan) equipped with a Cu Kα radiation source (λ = 1.5406 Å) to record the XRD spectra, and their corresponding size was calculated using Scherer's equation. The diffracted intensities were recorded from 4° to 90° at 2θ angles.
Scherer equation:D = Kλ/β cos θwhere D is the crystallite size of the particle, K represents the Scherrer constant, which is equal to 0.9, λ is the wavelength of light used for diffraction (λ = 1.54 Å), β is the full width at half maximum of the diffraction peak and θ is the angle of reflection.
2.9. Characterization of the synthesized ZnO nanoparticles by Fourier transform infra-red (FT-IR) spectroscopy
The spectral properties of zinc oxide nanoparticles were observed by Fourier transform infrared spectroscopy (FTIR-6100 spectrometer Jasco, Japan) for the characterization of the functional group attached to the surface of ZnONPs. Fourier transform infrared (FTIR) spectra of all samples were recorded, and the scanning was performed at 4 cm−1 resolution in the spectral range of 400–4000 cm−1 at room temperature.
2.10. Determination of ZnO nanoparticles by zeta potential
The nanoparticle size distribution was studied using a Zeta-sizer Ver.6.12: MAL10444595 (Malvern instruments) in a disposable cell at 25 °C.
2.11. Metabolic profiling of the methanolic extracts of selected Sabal species
Metabolic profiling was carried out on 2 mg of each crude methanolic extract of different parts (leaves, fruits, and pollen grains) of S. blackburniana (it was dissolved separately in 1 ml MeOH and then filtered using a 0.2 μm membrane filter) using an Acquity Ultra Performance Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, MA, USA).19 Chromatographic separation was performed using a BEH C18 column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, USA) with a guard column (2.1 × 5 mm, 1.7 μm particle size) and a linear binary solvent gradient of 0–100% eluent B over 6 min at a flow rate of 0.3 ml min−1, using 0.1% formic acid in water (v/v) as solvent A and acetonitrile as solvent B. The column temperature was 40 °C and the injection volume used was 2 μl. The MSConvert software was used to transfer and convert the raw data into separate positive and negative ionization files, and the resulting files were imported to the data mining software MZmine 2.10 for peak picking, deconvolution, deisotoping, alignment and formula prediction. In order to annotate the compounds, the processed data set was subjected to molecular formula prediction and peak identification via dereplication using online METLIN25 and Dictionary of Natural Products (DNP) databases.20
2.12. Anticholinesterase activity of the selected Sabal species evolution
2.12.1. Assay of AChE enzyme activity by spectrophotometry
The assessment of the acetylcholinesterase activity was carried out according to an improved Ellman method, in a 96-well plate reader using a Quanti-Chrome assay kit (USA).21 The enzyme hydrolyses the substrate acetylthiocholine resulting in the product thiocholine, which reacts with 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) to produce 2-nitrobenzoate-5-mercaptothiocholine and 5-thio-2-nitrobenzoate (a yellow color). The intensity of chromogen yellow color and its absorbance can be detected at 412 nm, proportionate to the enzyme activity in the tested samples.
2.12.2. Ninety-six-well plate reaction preparation22
First, 45 μl of AChE was transferred into separate wells and 45 μl of assay buffer was transferred into one well, which will be the no enzyme control well used as a 100% inhibition control. Then, 5 μl of 40 v% DMSO was added, following which 5 μl of the tested extracts at concentrations 10, 100, 1000, and 10 000 ng ml−1 was added to the remaining wells containing AChE, and the plate mixture was incubated at 37 °C for 15 min. Next, 154 μl Tris–HCl buffer was mixed with 1 μl acetylthiocholine iodide substrate and 0.5 μl DTNB and 150 μl of them was added to each selected extract sample, sample blank, and no-inhibitor control wells. Different concentrations of donepezil were prepared in serial concentrations as same as test extracts and the absorbance OD was measured at 412 nm at 0 min and at 10 min using a plate reader23,24 The half-maximal inhibition concentration (IC50) required to produce 50% inhibition of the enzymatic reaction (inhibition of the hydrolysis of acetylcholine) was determined.25 IC50 values were calculated by a linear approximation regression analysis of the inhibition percentage versus the extract concentration (serial dilution in methanol in concentrations (10, 100, 1000, and 10 000 ng ml−1) using the Excel program. The enzyme inhibition (%) was calculated from the rate of absorbance change with time (V = Abs/Δt), and the calculation is as follows:Inhibition (%) = (1 − ΔOD test cpd/ΔOD no-inhibitor) × 100where ΔOD test cpd is the absorbance OD at 412 nm of a test compound well at 0 min subtracted from the absorbance OD at 412 nm of a test compound well at 10 min. ΔOD no-inhibitor is the absorbance OD at 412 nm of the no-inhibitor control well at 0 min subtracted from the absorbance OD at 412 nm of the no-inhibitor control well at 10 min.
2.13. Docking experiments
Molecular docking experiments were performed using the Autodock Vina software26 The reported target proteins (human AChE) were downloaded from the protein databank website (https://www.rcsb.org/), (PDB codes: 4EY6 (crystal structure of recombinant human acetylcholinesterase in complex with (−)-galantamine) and 4EY7 (crystal structure of recombinant human acetylcholinesterase in complex with Donepezil)),27 and then they were prepared using the Autodock tools software.20 The binding sites of the co-crystalized ligands were selected for the docking experiments, and the coordinates of these binding sites were also determined using the Autodock tools software. Top-scoring binding poses with RMSD values less than 2 Å were then selected and visualized using the Pymol software.28
2.14. Statistical analysis
All experiments and analyses in the present study were carried out in triplicates. The results were expressed as mean ± S.D (mean ± standard error) of the indicated number of experiments (n ≥ 3). Statistical analysis was evaluated using one-way analysis of variance (ANOVA), followed by Tukey-Kramer's multiple comparison, using GraphPad Prism 8 and Microsoft Excel 2007. The level of statistical significance was taken at p < 0.05. The value of probability less than 0.05 was considered statistically significant (P < 0.05).
3. Results and discussion
3.1. UV-visible characterization of the synthesized ZnO nanoparticles of the selected Sabal species
They were early detected by forming a white precipitate and its nanostructures were further confirmed by UV-visible spectroscopy, transmission electron microscopy (TEM), Fourier transform infrared (FTIR) spectroscopy and zeta potential studies. Green-synthesized ZnO nanoparticles are known to show an absorption band in the range of 300–550 nm. The result of UV-vis spectroscopy revealed that all samples exhibited a strong absorption band with maxima around (354–368 nm), which is characteristic of ZnO nanoparticles29 due to their large excitation binding energy at room temperature and the surface plasmon resonance (SPR). The notable blue shift absorption for the synthesized ZnO nanoparticles can be due to a high decrease in the particle size. The position of the SPR was attributed to the degree of charge transfers between the medium, the particles, and interaction with the medium, and the particle size and shape.30 Another excitonic absorption band was located in the range of 260–280 nm, in addition to the absorption band located at 500–550 nm, as shown in Fig. S1C.† The absorption band that ranges towards longer wavelengths might be owing to the movement of the electronic cloud on the overall skeleton of the ZnO nanoparticles. All of them exhibited (Fig. S1A–C†) UV bands corresponding to the near–band gap excitonic emission and broad green emission bands due to oxygen vacancy.10 The slight variations in the values of absorbance intensity are due to the change in the particle size.31,32 UV-vis spectra exhibit significant absorption of ZnO nanoparticles33,34 with a large sample population.
3.2. TEM characterization of the synthesized ZnO nanoparticles of the selected Sabal species
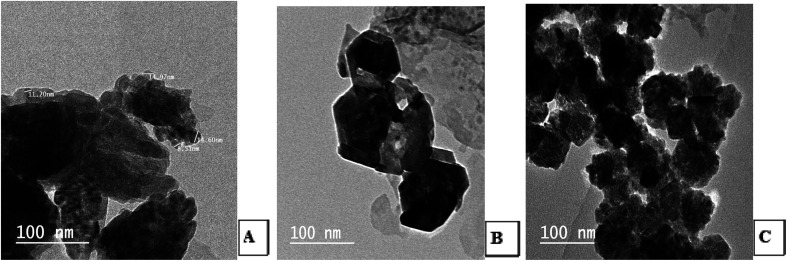
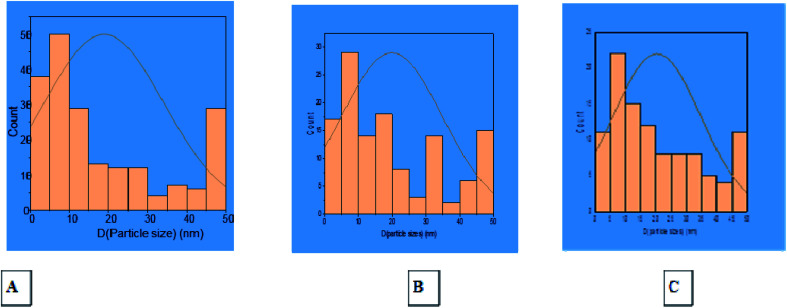
Transmission Electron Microscopic (TEM) image initially characterized the morphology and dimensions of the biosynthesized ZnO nanoparticles, and the estimated ZnO nanoparticles size from (Fig. 2A–C) TEM pictures showed that they had different shapes, predominantly irregular small spherical particles included in hexagonal structures, rod-like shaped, cubic structures, which accumulate like bullets, in addition the presence of clusters as depicted by the TEM image. The larger nanoparticle sizes observed in the samples were due to the agglomeration of smaller nanoparticles; this agglomeration may be due to van der Waals forces, polarity and electrostatic attraction of ZnO nanoparticles.35 The mean particle size distribution histogram was 18.80–20.32 nm, which was calculated from the TEM graph using the ImageJ software and determined the particle size range from 2.18 to 49.56 nm as shown in Fig. 3A–C (with range from 2.84 ± 1.53 to 48.81 ± 1.53, 2.18 nm ± 1.43 to 49.15 ± 1.43 and 2.23 ± 1.36 to 49.56 ± 1.36) for (A), (B) and (C) respectively.
Fig. 2. TEM photos for the shape and size of the synthesized ZnO nanoparticles using S. blackburniana: (A) leaves, (B) fruits, and (C) pollen grains.
Fig. 3. Histogram of particle size distribution from TEM photos of ZnO nanoparticles using S. blackburniana: (A) leaves, (B) fruits, and (C) pollen grains.
3.3. X-ray diffraction (XRD) analysis
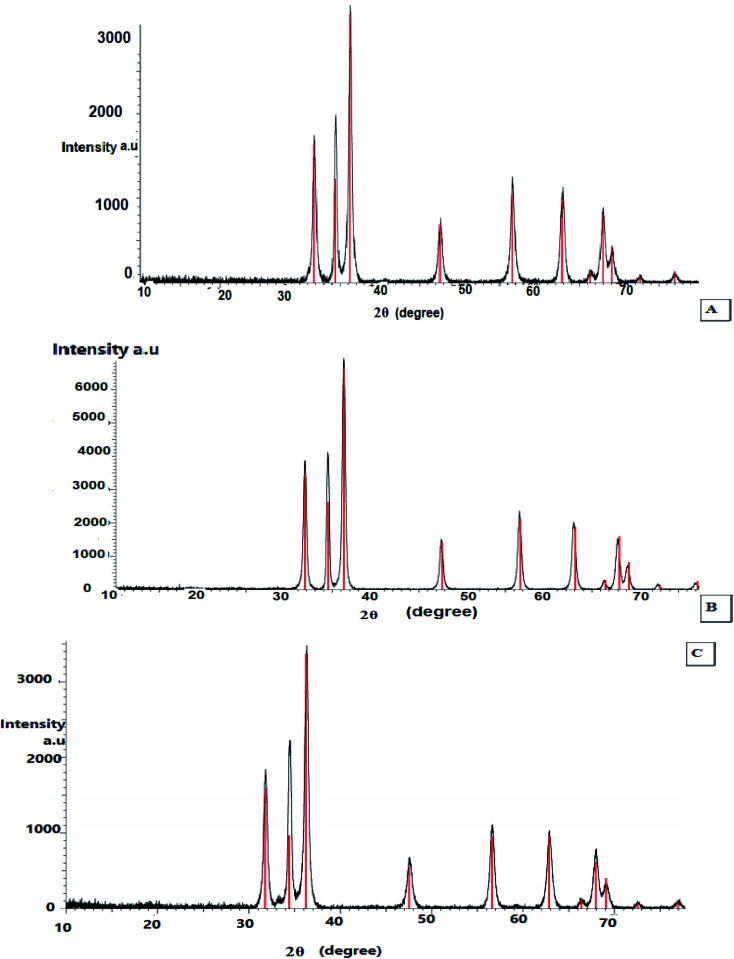
X-ray diffraction was performed to examine the textural properties such as size, crystal structure and shape of the materials, as the peak positions of the diffraction pattern provide information of translational symmetry size and shape of the unit cell. Therefore, the green-synthesized dried powder of ZnO nanoparticles using the extracts of leaves, fruits, and pollen grains of S. blackburniana was investigated by X-ray diffraction to confirm the crystalline nature of them. The results indicated that XRD spectral analysis displays the number of strong diffraction peaks corresponding to Bragg reflections with 2θ values: 31.77°, 34.43°, 36.36°, 47.55°, 56.60°, 62.87°, 66.39°, 67.96°, 69.10° and 72.88° (Fig. 4A), 31.85°, 34.55°, 36.35°, 47.70°, 56.75°, 63.09°, 66.57°, 68.17° and 72.87° (Fig. 4B), 31.78°, 34.42°, 36.26°, 47.54°, 56.61°, 62.86°, 66.39°, 67.96° and 72.57° (Fig. 4C); these peaks are in good agreement with the literature report,36–38 thus implying that the synthesized nanoparticles are identical to the hexagonal phase of zinc oxide.36 The green-synthesized ZnO nanoparticles using the above-mentioned extracts are crystalline with hexagonal wurtzite phase, and it can be stated that the obtained crystallite of ZnO nanoparticles show high purity as shown in the X-ray peak profile; the patterns were exhibited in Fig. 4. The average particle crystallite sizes obtained by the three ZnO nanoparticles using the extracts of leaves, fruits, and pollen grains synthesis were estimated using the Scherrer equation39 and found to be 47.21, 47.67, and 47.8 nm respectively from high-intensity peaks.
Fig. 4. X-ray diffraction analysis (XRD) of the synthesized ZnO nanoparticles using S. blackburniana: (A) leaves, (B) fruits, and (C) pollen grains.
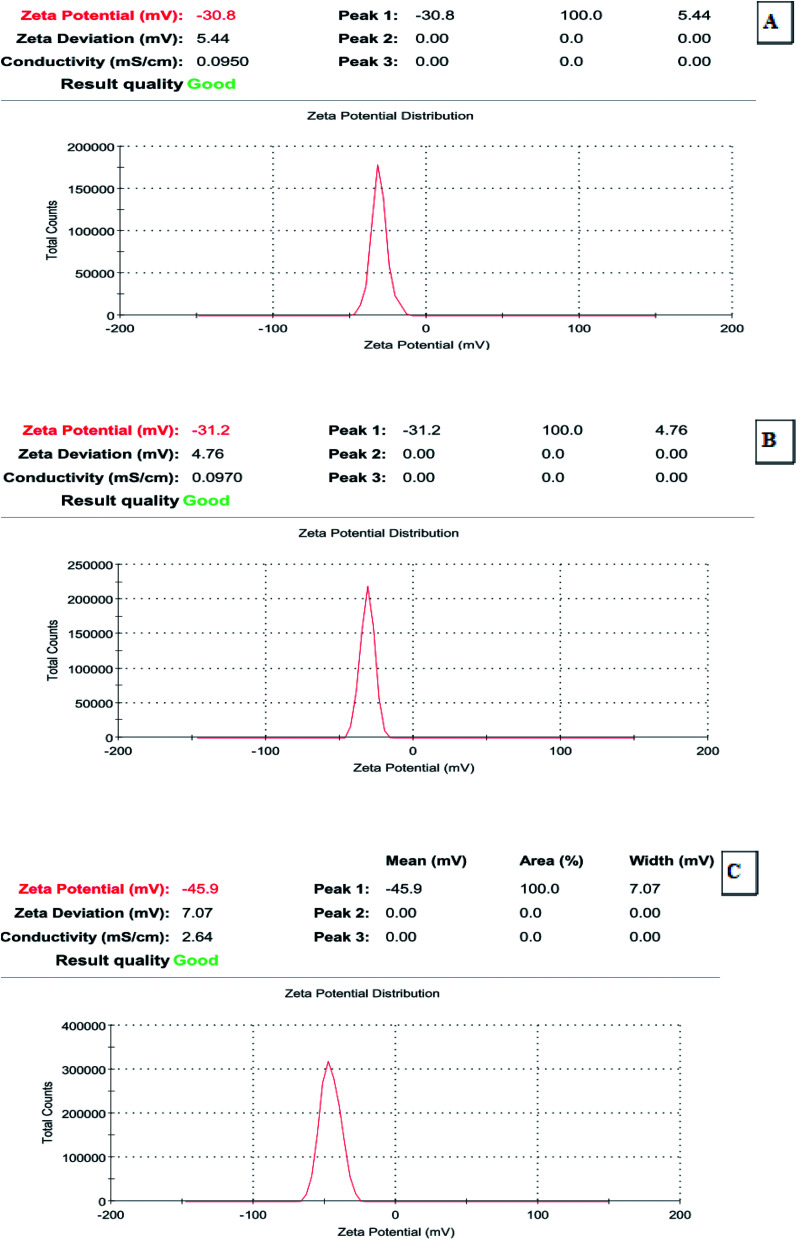
3.4. Determination of the ZnO nanoparticles by zeta potential
Zeta potential is important for the characterization and evaluation of the stability of the synthesized ZnO nanoparticles; it was determined that during electrophoresis (Zetasizer Nano), NaOH was used to adjust the pH. Recently, it has been accepted that zeta potentials do not drop below 30 mV (regardless of the sign) for the stabilization of nanoparticle preparation. The zeta potential determination demonstrated that all of the representative selected synthesized ZnO nanoparticles exhibited acceptable ZP values (−30.8 to −45.9 mV), which indicated their good stability (Fig. 5A–C).
Fig. 5. Zeta potential of the synthesized ZnO nanoparticles using S. blackburniana: (A) leaves, (B) fruits and (C) pollens grains.
3.5. Fourier transform infrared (FT-IR) spectroscopy for the characterization of the selected Sabal species-synthesized ZnO nanoparticles
FT-IR analysis is an important tool to understand and identify the characteristic functional groups present on the surface of ZnO and ascertain the purity and nature of the nanoparticles; it also demonstrated the presence of phytochemicals in the extract. The FTIR spectra (Fig. S2–S4†) show the main bands and different major peaks positions corresponding to the synthesized ZnO nanoparticles and the extracts: 3387, 3668.61, 3383.11, 3414.3, 3390.86 and 3414.01.49 cm−1, indicating the broad peak of the OH stretching vibrations. While the peaks at 2931.8, 2978.09, 2904.8 and 2978.09 cm−1 correspond to the CH3 stretching vibration, 1612.49 and 1631.78 cm−1 indicate the C C bond stretching vibration. While the peaks at 2090.84 and 2067.69 cm−1 are relevant to triple bond stretching and 1975.11 and 1917.24 cm−1 indicate aromatic overtones of ring bending. The peaks at 1496.76, 1419.61, 1415.75 and 1454.33 cm−1 correspond to polyphenol OH and the presence of an aromatic group, where the peaks at 1381.03, 1342.42, 1342.46 and 1396.46 cm−1 indicate the C–O–H bending vibration. The presence of tertiary amide, C–N stretching and N–H bending 1265.30, 1230.58, and 1296.16 cm−1 corresponds to C–O stretching in aromatic alcohols, while the peak at 1026.13 is relevant to the presence of C–O and aliphatic C–N bond stretching, and the peaks at 443.63, 462.92, 478.36, 416.62 and 451.34 cm−1, were characteristic bands of the zinc oxide, nanoparticles. The FTIR spectrum, absorption at 400 cm−1 to 600 cm−1 identifies the presence of zinc oxide nanoparticles. The peaks that were observed at 1573.91 and 1631.78 cm−1 and 478.35, 416.62, 446.77, 443.63 and 451.34 cm−1 correspond to the Zn–O stretching and deformation vibration, respectively. These further confirms the formation of zinc oxide nanoparticles by using the extracts of leaves, fruits and pollen grains of S. blackburniana (Fig. 4–6).The former observed peaks indicate the presence of functional groups such as ketones, aldehydes, hydroxyls, alcohols, phenolics, and carboxylic acid compounds with strong hydrogen bonds and stretching of the N–H group, which could be mainly attributed to flavonoids, terpenoids, glycosides, and phenols; in addition to conformed formation of green-synthesized ZnO nanoparticles, the previous results indicated that the plant extract contains various metabolites that work as capping and stabilizing agents for the synthesized ZnO nanoparticles and also prevent aggregation and pairing of the nanoparticles. The reducing capacity of plants depends on the presence of polyphenols and other chelating agents present in plants that have critical effects on the reduction potential of ions and the amounts of nanoparticle production.40 The similarity between the spectra which had some slight shifts in peak position indicated the presence of the residual plant extract in the sample as a capping agent to the ZnO nanoparticles. These may identify the biomolecules, which are responsible for efficient stabilization and capping of the metal nanoparticles.41
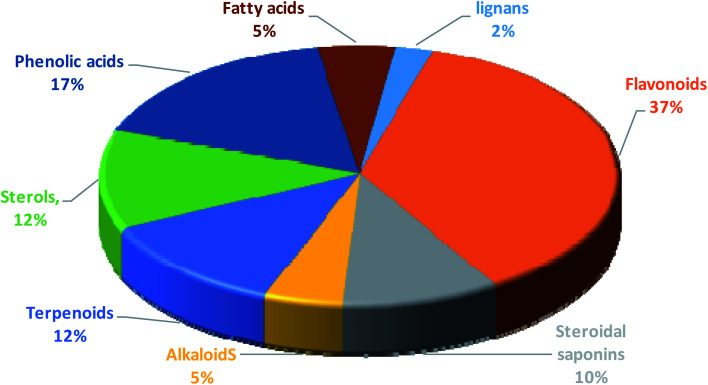
Fig. 6. Percentage of different classes of metabolites distributed in the methanolic extracts of tested Sabal species.
3.6. Metabolic profiling of the crude methanolic extracts of the selected Sabal species
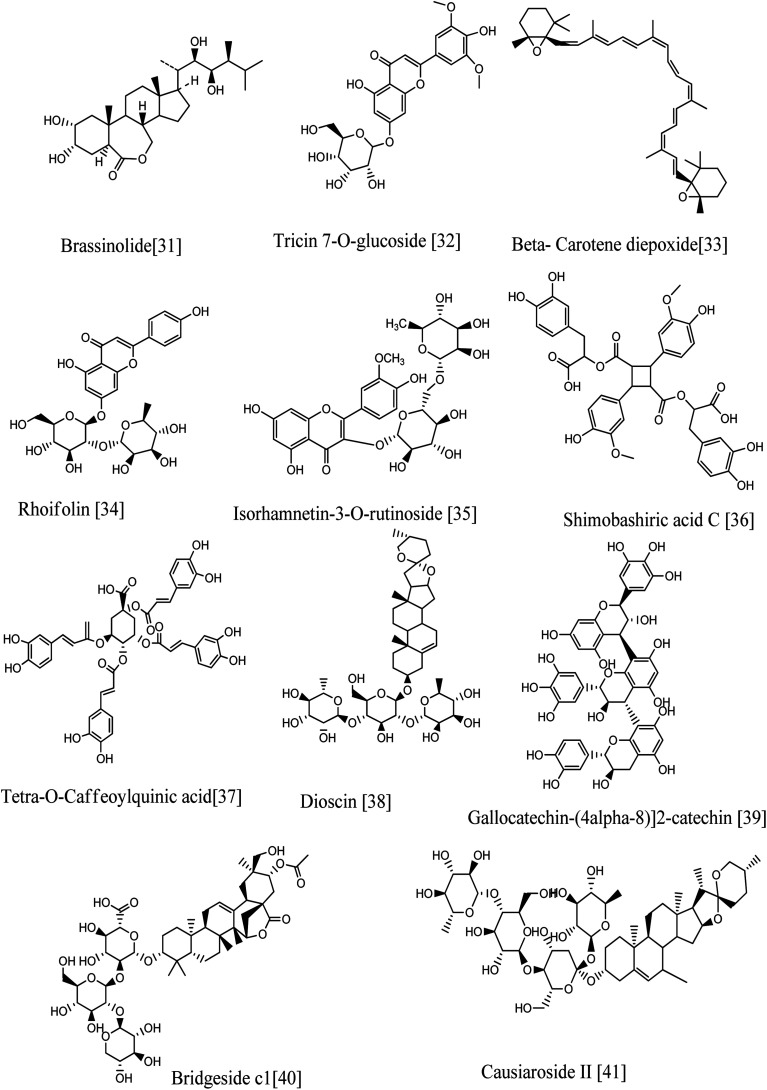
Annotation of the secondary metabolites from the crude methanolic extract of the selected plant using LC-MS.19 These dereplication studies relied on a precise molecular formula prediction along with a chemotaxonomic filtration to minimize the number of hits per molecular formula (depending on chemotaxonomic categorization).42 From DNP database (Table S1,†Fig. 7–10), forty-one metabolites were annotated belonging to certain chemical classes (Fig. 6): flavonoids, steroidal saponins, terpenoids, sterols, phenolics, alkaloids, fatty acids, and lignans.
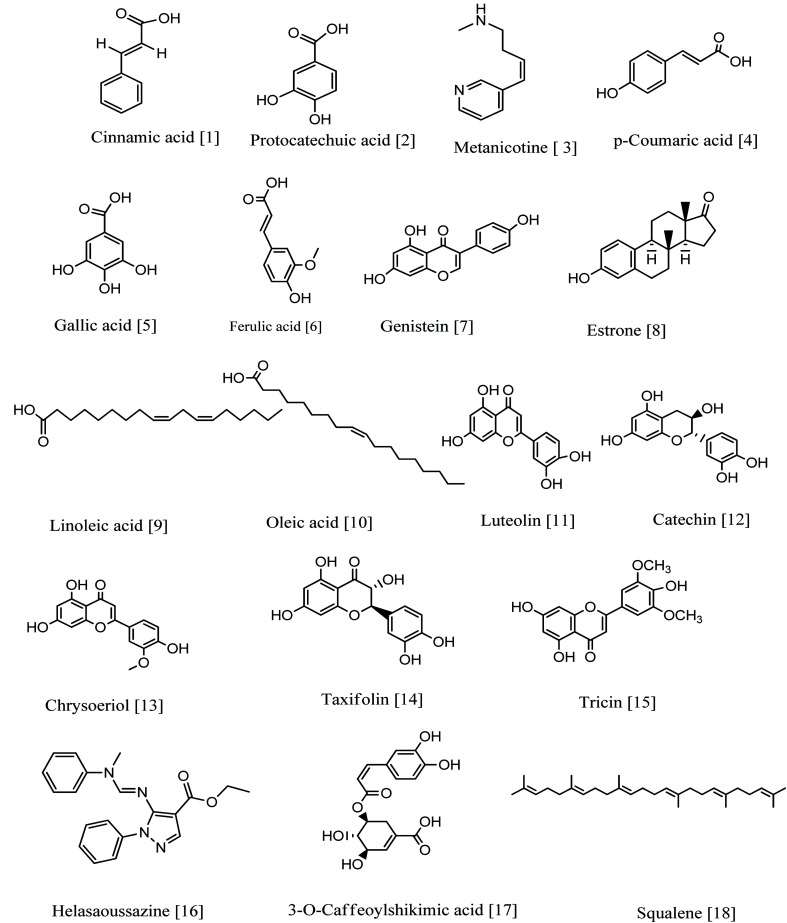
Fig. 7. Compounds annotated from the methanolic extract of the selected Sabal species.
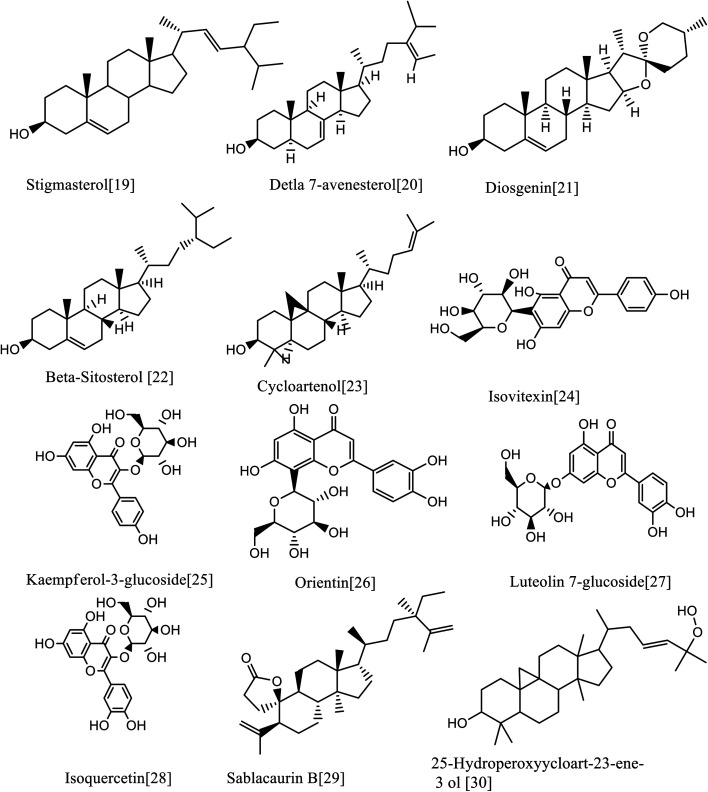
Fig. 8. Compounds annotated from the methanolic extract of the selected Sabal species.
Fig. 9. Compounds annotated from the methanolic extract of the selected Sabal species.
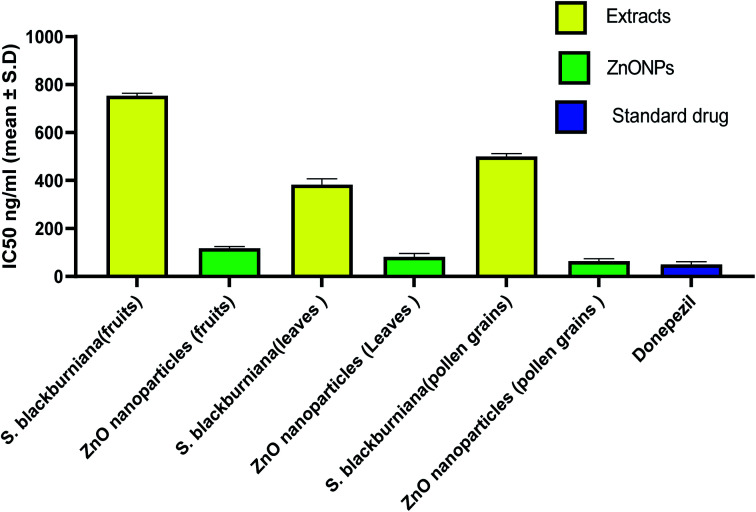
Fig. 10. IC50 values expressed in ng ml−1 for AChE inhibition by plant extracts, ZnO nanoparticles and donepezil.
3.6.1. Polyphenolics
3.6.1.1. Phenolic acids and phenolic acid derivatives
Seven phenolics (six phenolic acids and one phenolic acid derivative) were identified from the DNP database. The mass ion peak at m/z 149.1278 [M + H]+ (RT, 2.84 min), for the predicted molecular formula, C9H8O2, was dereplicated as cinnamic acid earlier isolated from palm.43 While metabolite with the molecular formula C7H6O4 for the peak at m/z 155.1043 [M + H]+ (RT, 2.32 min) was characterized as Protocatechuic acid, which was reported from Phoenix dactylifera.44 The mass ion peaks at m/z165.0900 [M + H]+ (RT, 1.78 min), in accordance to the molecular formula C9H8O3, was recognized as p-coumaric acid that was isolated from Phoenix canariensis leaves.45 In addition, other metabolites with the molecular formulae C7H6O5, and C10H10O4 for the mass ion peaks at m/z 171.1018 [M + H]+ (RT, 4.64 min) and m/z 195.1800 [M + H]+ (RT, 3.93 min) respectively were annotated as gallic acid and ferulic acid respectively, and they were earlier isolated from palm.46,47 In addition, the mass ion peak at m/z 337.1639 [M + H]+ (RT, 2.48 min) for the predicted molecular formula C16H16O8, was dereplicated as 3-O-caffeoylshikimic acid.48 On the other hand, the metabolite with the molecular formula C43H36O18 for the mass ion peak at m/z 839.17393 [M−H]− (RT, 6.53 min) was characterized as a polyphenolic compound, tetra-O-caffeoylquinic acid (phenolic acids derivatives). It was isolated from Pluchea symphytifolia.46
3.6.1.2. Flavonoids
Furthermore, fifteen flavonoids were characterized: the metabolite with the mass ion peak at m/z 271.1194 [M + H]+ (RT, 3.55 min), in accordance with the molecular formula C15H10O5, was recognized as Genistein, that is an isoflavone formerly obtained from Sabal palm seeds47 and showed significant antioxidant and antitumor activities;50,51 the metabolite with the mass ion peaks at m/z 287.1167 [M + H]+ (RT, 2.75 min) and 291.14869 [M + H]+ (RT, 2.3 min) for the predicted molecular formulae C15H10O6 and C15H14O6 respectively were annotated as luteolin (tetrahydroxyflavone) and catechin (flavanol), respectively, which were formerly obtained from palm; leaves seeds, fruits.52–54 In addition, other metabolites with the molecular formulae, C16H12O6, C15H12O7 and C17H14O7 for the mass ion peaks at m/z 301.1357 [M + H]+ (RT, 3.70 min), 305.1692 [M + H]+ (RT, 3.18 min), and 331.15 [M + H]+ (RT, 3.76 min) respectively were characterized as chryseriol (flavone), taxifolin (flavonol) and tricin (an O-methylated flavone), respectively, which were previously obtained from palm (leaves and fruits) and Sabal palm seeds.49,54,55 Moreover, the metabolites with the mass ion peaks at m/z 433.2045[M + H]+ (RT, 2.78 min) and 449.2024 [M + H]+ (RT, 2.66 min) with the suggested molecular formulas C21H20O10 and C21H20O11 were identified as isovitexin and orientin respectively, which were the two C-glycosyl flavones previously obtained from S. blackbumiana and S. causiarum.57 Similarly, the mass ion peaks at m/z 625.2084 [M + H]+ (RT, 3.05 min), 579.2576 [M + H]+ (RT, 8.72 min), 493.2376 [M + H]+ (RT, 3.04 min), 465.202 [M + H]+ (RT, 2.69 min), 449.2030 [M + H]+ (RT, 2.50 min) and 449.2029 [M + H]+ (RT, 2.54 min) respectively corresponding to the molecular formulas C28H32O16, C27H30O14, C27H30O14, C21H20O12, C21H20O11 and C21H20O11, respectively were distinguished as isorhamnetin-3-O-rutinoside, rhoifolin, tricin 7-O-glucoside, isoquercetin, luteolin 7-glucoside and kaempferol-3-glucoside respectively, which were the six glycosyloxyflavones earlier reported from S. serrulata.57,58 In addition, the metabolite with the molecular formula C45H38O20 for the mass ion peak at m/z 897.0992 [M−H]− (RT, 3.02 min) was characterized as gallocatechin-(4-alpha-8)2-catechin (flavan-3-ols) having good inhibitory activity of lipoxygenase.59 The flavonoids are the most predominant ones.
3.6.2. Alkaloids
Two alkaloids were identified: the metabolite with the mass ion peaks that at m/z 163.0739 [M + H]+ (RT, 2.93 min) and 332.3094 [M−H]− (RT, 4.74 min), corresponding to the proposed molecular formulae C10H14N2 and C19H13N4O2 respectively were identified as metanicotine and helasaoussazine respectively. metanicotine has nicotine CNS-selective, nicotine agonist activities, and is also used as a potential agent in treatment of Alzheimer's and ulcerative colitis; it was previously obtained from the leaves of Duboisia Hopwoodii.60,61 Helasaoussazine was previously obtained from P. canariensis and palm.62
3.6.3. Lignans
One neolignan was characterized. The metabolite with the mass ion peak at m/z 747.1913 [M−H]− (RT, 3.02 min) for the molecular formula C38H36O16 was dereplicated as Shimobashiric acid D.63
3.6.4. Fatty acids
Two metabolites with the molecular formulae C18H32O2 and C18H34O2 for the mass ion peaks at m/z 279.4756 [M–H]− (RT, 6.85 min) and, [M + H]+ RT, 3.85 min) respectively, were annotated as linoleic acid and oleic acid respectively, which were earlier obtained from S. serrulata.64
3.6.5. 5. Sterols
Furthermore, five sterols were annotated. The metabolites with the mass ion peaks at, m/z 271.3016 [M + H]+ (RT, 5.4 min), 413.3919 [M + H]+ (RT, 4.17 min), 415.4083 [M + H]+ (RT, 5.59 min), 413.3925 [M + H]+ (RT, 3.44 min), and 481.4135 [M + H]+ (RT, 5.9 min) in agreement with the molecular formulae C18H22O2, C30H52, C29H50O, C29H48O and C28H48O6 respectively were distinguished and dereplicated as estrone,65 stigmasterol,49, β-sitosterol,66 Δ7-avenasterol,67 and brassinolide68 respectively, which were formerly isolated from Phoenix dactylifera, Serenoa repens (W. Bartram), Sabal palm, S. causiarum and P. canariensi.69,70
3.6.6. Terpenoids
Five terpenoids were dereplicated. The metabolites with the mass ion peaks at m/z 411.377 [M + H]+ (RT, 3.18 min), 427.3924[M + H]+ (RT, 6.64 min), 459.4498 [M + H]+ (RT, 5.7 min), 569.521 [M + H]+ (RT, 6.15 min) and 469.5029 [M + H]+ (RT, 6.03 min) corresponding to the molecular formulae C30H50, C30H50O, C30H50O3, C40H56O2 and C32H52O2, respectively were annotated as squalene (hydrocarbon triterpene),67 cycloartenol (pentacyclic triterpenoid),66 25-hydroperoxyycloart-23-ene-3-ol (triterpenoid), β-carotene-2,2′-diepoxide (tetraterpene; a oxygenated carotenoid) and sablacaurin B (19-triterpenoid; nor-3,4-seco-lanostane-type) respectively that were previously isolated from S. blackburniana, S. serrulata, and S. causiarum.71,72
3.6.7. Steroidal saponins
Finally, four steroidal saponins were characterized. The metabolite with the mass ion peak at m/z 415.4092 [M + H]+ (RT 5.57 min) for the molecular formula C27H42O3, was dereplicated as diosgenin (sapogenin steroidal saponins), which was characterized from Sabal palm seeds49 and had an antitumor effect on different cancer cell lines; diosgenin is safe and effective as a chemopreventive agent against several human cancers.73 In addition, the metabolites with the mass ion peaks at m/z, 869.6766 [M + H]+ (RT, 3.47 min), 999.4723 [M + H]+ (RT 6.025 min) and 1048.7724 [M + H]+ (RT 3.10 min) for the molecular formulae C45H72O16, C51H83O22 and C49H74O21 respectively were dereplicated as dioscin,57 bridgeside C1 and causiaroside II (= p, 16P-dihydroxypregn-5-en-20-one 3-0-[a-L3 rhamnopyranosyl (1-+4)-P-d-glucopyranosyl (1-+4)] [a.-l-rhamnopyranosyl (1-+2)]-P-d-glucopyranosyl 16-o-[β-d-glucopyranosyloxy-γ-methyl]-0-[valerate]) respectively that were early obtained from S. causiarum.74,75
3.7. Anticholinesterase activity
In the last decade, botanical drugs have been currently approved in therapy with specific indications. Recently, it has been reported that biogenically synthesized nanoparticles from medicinal plants have less harmful effects than that of chemically synthesized nanoparticles.76 Therefore, medicinal plants and green synthesized nanoparticles are a vital focus of researchers as alternative therapies for the treatment of the Alzheimer and other diseases.5,77–80 In this study, the acetyl cholinesterase enzyme (AChE) inhibitory activity (anti-Alzheimer) of the crude methanolic extracts of the selected Sabal species and green-synthesized ZnO nanoparticles in vitro were evaluated and compared with that of the standard donepezil drug (AChE inhibitor). The increase in acetyl cholinesterase enzyme (AChE) inhibitory activity was the most characteristic change that occurs in Alzheimer's disease. AChE is the enzyme responsible for acetylcholine hydrolysis, from both cholinergic and noncholinergic neurons of the brain. The increase in acetylcholine levels can be achieved by the inhibition of AChE, which can help in the treatment of Alzheimer's disease (AD).3 The results indicated that all three samples of methanolic extracts of leaves, pollen grains and fruits, of S. blackburniana respectively showed a significant activity (<10 μg ml−1) with IC50 values of 382.87 ± 6.86, 500.08 ± 12.22 and 754.08 ± 9.93, ng ml−1, respectively, however, the green-synthesized ZnO nanoparticles using the pollen grains and leaves extracts respectively exhibited the highest (very potent) and promising anti-acetylcholinesterase activity with IC50 values of 63.78 ± 10.0471 and 81.99 ± 13.08 ng ml−1 respectively, followed by ZnO nanoparticles using fruit extracts with an IC50 value of 117.95 ± 6.86 ng ml−1 in comparison to the standard donepezil' IC50 value 50.7 ± 5.77 ng ml−1, as shown in (Fig. 10). Moreover, ZnO nanoparticles using pollen grain extracts exerted the highest percentage inhibition (38.30, 51.10, 69.85, and 82.56 at concentrations of 10, 100, 1000, and 10 000 ng ml−1 respectively) compared to donepezil (39.04, 56.78, 71.97, and 82.87 at concentrations of 10, 100, 1000, and 10 000 ng ml−1 respectively) on AChE enzyme activity, followed by ZnO nanoparticles using leaf extracts, and finally, ZnO nanoparticles using fruit extracts (Table 1). Overall, the former results indicated that a considerable anti-Alzheimer activity of the selected extracts could be due to the fact that they are rich source of useful metabolites such as flavonoids, which are reported as a promising lead class of compounds for the efficient design and development of multipotent anti-Alzheimer's disease drugs, and to suppress the progression of pathologies of Alzheimer's disease (AD); this is due to their ability to crush the cognitive deficits in numerous normal and transgenic preclinical animal models.81–84 Moreover, the results showed that green-synthesized ZnO nanoparticles possessed the most potent acetylcholinesterase inhibitory activity, which may be due to their large surface area. ZnO nanoparticles have been successfully exploited as drug carriers for loading and transporting drugs to target sites, thereby reducing unwanted toxicity and off-target effects, and resulting in amplified synergistic effects.85 These may be effective and useful in treating Alzheimer's disease. Further study should be done in vivo to explain the exact mechanism of inhibitory action.
Percentage inhibition of S. blackburniana extracts, ZnO nanoparticles and donepezil (concentration, ng ml−1) on AChE enzyme activity.
| Sample | % inhibition of AChE | |||
|---|---|---|---|---|
| Conc ng ml−1 | ||||
| 10 | 100 | 1000 | 10 000 | |
| S. blackburniana (fruits) | 19.16 | 31.19 | 43.22 | 78.18 |
| ZnO nanoparticles (fruits of S. blackburniana) | 36.59 | 48.48 | 60.48 | 79.63 |
| S. blackburniana (leaves) | 20.28 | 41.99 | 49.94 | 80.12 |
| ZnO nanoparticles (leaves of S. blackburniana) | 39.81 | 48.48 | 63.51 | 82.15 |
| S. blackburniana (pollen grains) | 16.31 | 37.61 | 52.80 | 77.37 |
| ZnO nanoparticles (pollen grains of S. blackburniana) | 38.30 | 51.10 | 69.85 | 82.56 |
| Donepezil | 39.04 | 56.78 | 71.97 | 82.87 |
3.8. Molecular docking
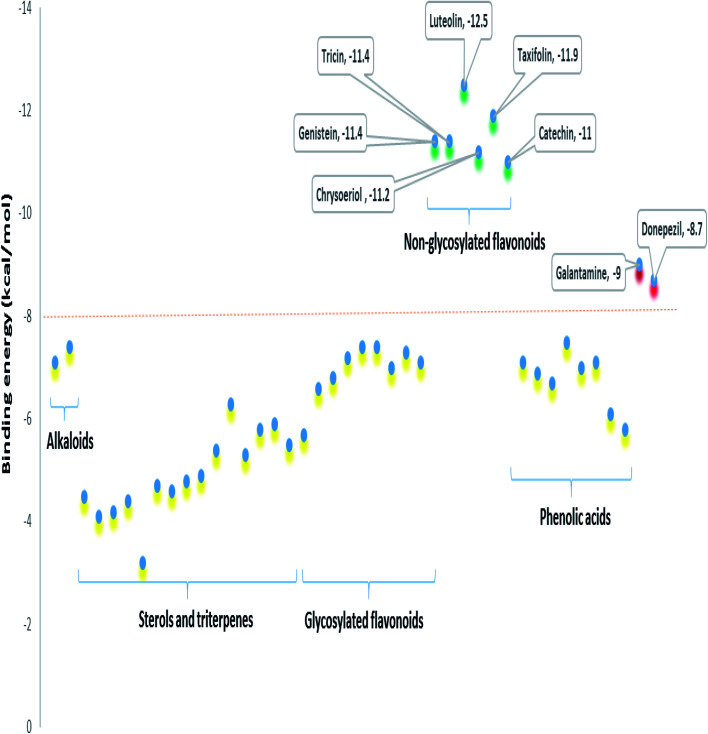
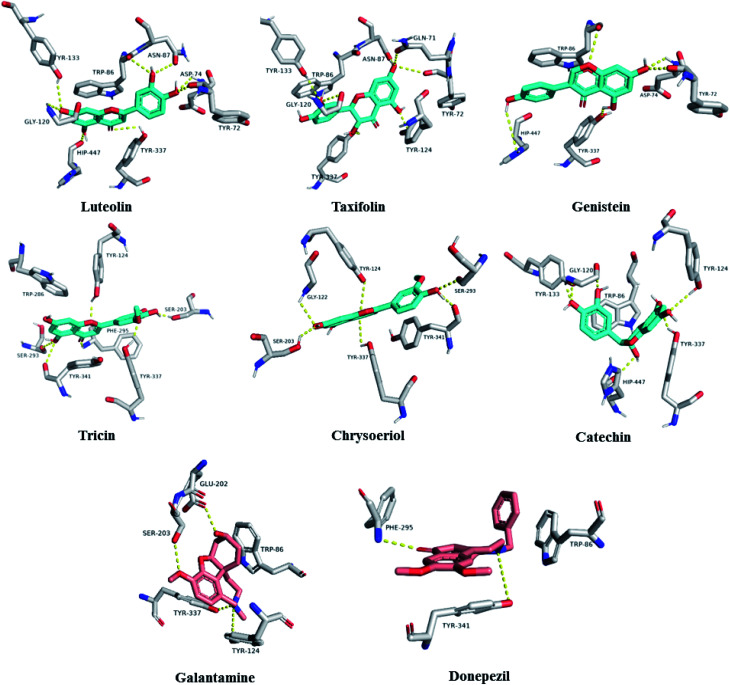
To further highlight the probable metabolites that may be associated with the anti-AChE activity of Sabal-derived extracts, all dereplicated metabolites (Table S1† and Fig. 7–11) were subjected for flexible docking analysis against human AChE (PDB: 4EY6). The application of such protocol in natural product-based drug discovery can potentially facilitate targeting of the drug candidates in complex plant extracts. As shown in Fig. 12, the non-glycosylated flavonoids such as luteolin, taxifolin, tricin, genistein, chrysoeriol and catechin (i.e. flavones, isoflavones, and flavanols) were the top-scoring metabolites (binding energy: −11 to 12.5 kcal mol−1). Additionally, they showed binding poses comparable with the co-crystalized ligands, galantamine and donepezil with more molecular interactions (H-bonds and hydrophobic interactions) and higher binding energies (Fig. 12). The key interactions of the co-crystalized ligands inside the active site were H-bonds with TYR-124, GLU-202, SER-203, PHE-295, TYR-337, and TYR-341 along with one hydrophobic interaction with TRP-86 (Fig. 12). Besides, these reported key amino acids, top-scoring flavonoids were able to interact through H-bonding with extra amino acid residues such as TYR-72, ASP-74, ASN-87, GLY-120, SER-293, and HIP-447 (Fig. 12). In conclusion, these top-scoring Sabal-derived flavonoids showed comparable binding mode with galantamine (i.e. co-crystallized ligand) inside the human AChE's active site, and this may provide a molecular explanation for the anti-AChE activity of Sabal extracts.
Fig. 11. Binding energies of the metabolites dereplicated in Sabal-derived extracts. All metabolites (yellow color) except for the non-glycosylated flavonoids (green color, more than 10) showed binding energies less than −8 kcal mol−1. Galantamine and donepezil (i.e. the co-crystalized ligands of AChE, red color) showed binding energies of −9.0 and −8.7 kcal mol−1.
Fig. 12. Binding modes of the top-scoring metabolites inside AChE's active site (cyan color). Galantamine and donepezil are the co-crystalized ligands (red color).
4. Conclusions
The biological green pathway comprises the use of the plant extracts as reducing agents, which was more preferable for the development of nanoparticles with optimal features. All the obtained results confirm that Zn ions had been reduced by the extracts of S. blackburniana under the reaction conditions. The FT-IR analysis results illustrated that there are many functional groups that were adsorbed and present on the surface of ZnO nanoparticles such as OH and C O groups, which may have an important role in bio-reduction. FT-IR spectroscopy further confirms the formation of zinc oxide nanoparticles. The estimation of acetylcholinesterase inhibitory activity of all green-synthesized ZnO nanoparticles using the three methanolic extracts of leaves, fruits, and pollen grains of S. blackburniana concluded that they showed a very potent and significant inhibitory activity, and the extracts may have several metabolomics, which possess potential neuroprotective activity. Metabolic profiling of the crude methanolic extract of selected plants using LC-MS has resulted in the annotation of forty-one metabolites, which belong to various classes, and a molecular modelling study has also been performed to provide insights into the most probable metabolites associated with the AChE inhibition. The docking results suggested that luteolin, taxifolin, tricin, genistein, chrysoeriol and catechin possessed the best binding affinity, as these compounds have been able to achieve binding modes better than co-crystalized ligands; hence, they may be the main contributors to the observed anti-AChE activity of the tested Sabal extracts, and the application of such protocols in natural product-based drug discovery can potentially facilitate the targeting of the drug candidates in complex plant extracts. In this study, for the first time, we demonstrated the green synthesis of ZnO nanoparticles using S. blackburniana and their anti-Alzheimer potential. Based on these work, the anti-acetylcholinesterase activity of the selected extracts, which are rich sources of useful metabolites, and green-synthesized ZnO nanoparticles of S. blackburniana in vitro was demonstrated. Moreover, the docking results will help discover the production of novel drugs from Sabal and suggest that they may be used for the treatment of neurodegenerative disorders such as Alzheimer's disease. Future studies should be exerted in vivo focusing possible potential neuroprotective activity for accepted anti-Alzheimer natural drugs and to evaluate that they have no serious or undesirable side effects.
Abbreviations
- ZnO NPs
Zinc oxide nanoparticles
- TEM
Transmission electron microscope
- XRD
X-ray diffraction
- FTIR
Fourier transform infra-red spectroscopy
- ZP
Zeta potential
- UPLC-MS
Acquity Ultra Performance Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer
Author contributions
Conceptualization: Seham S. El-hawwary, Usama Ramadan Abdelmohsen, Fatema R. Saber, Hanan A. Elimam, Mohamed A. El Raey, Ahmed M. Sayed, Hanan M. Abd Almaksoud. Data curation: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen, Mohamed A. El Raey. Formal analysis: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen, Fatema R. Saber, Ahmed M. Sayed. Investigation: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen, Hanan A. Elimam. Methodology: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen, Hanan A. Elimam, Mohamed A. El Raey. Ahmed M. Sayed. Project administration: Seham S. El-hawwary, Usama Ramadan Abdelmohsen, Fatema R. Saber, Hanan A. Elimam. Resources: Hanan M. Abd Almaksoud, Seham S. El-hawwary. Software: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen, Ahmed M. Sayed. Writing-Original draft: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen. Writing-review& editing: Hanan M. Abd Almaksoud, Usama Ramadan Abdelmohsen.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
We thank, Dr Omnia Magdy Hendawy, also we would like to thank M. Müller and M. Krischke for helping in LC-MS measurement.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01725j
References
- Nazem A. Mansoori G. Insci. J. 2011;4:169–193. doi: 10.5640/insc.0104169. [DOI] [Google Scholar]
- Hajipour J. Santoso R. Rezaee F. Aghaverdi H. Mahmoudi M. Perry G. Trends Biotechnol. 2017;35:937–953. doi: 10.1016/j.tibtech.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Nazıroğlu M. Muhamad S. Pecze L. Expert Rev. Clin. Pharmacol. 2017:751–2441. doi: 10.1080/17512433.2017.1324781. [DOI] [PubMed] [Google Scholar]
- Saraiva C. Praça C. Ferreira R. Santos T. Ferreira L. Bernardino L. J. Controlled Release. 2016;235:34–47. doi: 10.1016/j.jconrel.2016.05.044. [DOI] [PubMed] [Google Scholar]
- Youssif K. A. Haggag E. G. Elshamy A. M. Rabeh M. A. Gabr N. M. Seleem A. Salem M. A. Hussein A. S. Krischke M. Mueller M. J. Abdelmohsen U. R. PLoS One. 2019;14(11):e0223781. doi: 10.1371/journal.pone.0223781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalwa H., Handbook of Nanostructured Biomaterials and Their Applications in Nanobiotechnology, American Scientific Publishers, 2007, vol. 2 [Google Scholar]
- Jayaseelan C. Rahuman A. A. Kirthi A. V. et al. . Spectrochim. Acta, Part A. 2012;90:78–84. doi: 10.1016/j.saa.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Anlovar A. Kogej K. Crnjak Orel Z. Zigon M. eXPRESS Polym. Lett. 2011;5:604. doi: 10.3144/expresspolymlett.2011.59. [DOI] [Google Scholar]
- Cullity B. D., Elements of X-Ray Diffraction, Addison-Wesley, Reading, MA, 3rd edn, 1967 [Google Scholar]
- Srinivasa rao B. Rajesh kumar B. Rajagopal reddy V. Subba rao T. Chalcogenide Lett. 2011;8:177–185. [Google Scholar]
- Rao C. N. R. and Govindaraj A., Nanotubes and Nanowires (The RSC Nanoscience and Nanotechnology Series); Kroto H., and Craighead H., Royal Society of Chemistry, London, 2005 [Google Scholar]
- Fouda A. Hassan S. D. Salem S. S. Microb. Pathog. 2018;125:252–261. doi: 10.1016/j.micpath.2018.09.030. [DOI] [PubMed] [Google Scholar]
- Gouda H. Attia H. Alyami H. S. Orabi M. A. A. Gaara A. H. El Raey M. A. Int. J. Pharmacol. 2020;16(3):236–243. [Google Scholar]
- Diallo A. Ngom B. Park E. Maaza M. J. Alloys Compd. 2015;646:425–430. doi: 10.1016/j.jallcom.2015.05.242. [DOI] [Google Scholar]
- Ogunyemi S. O. Abdallah Y. Zhang M. Fouad M. H. Hong X. Ibrahim E. Mater. Res. Express. 2018;6(10):341–352. [Google Scholar]
- FDA (Food and Drug Administration), Electronic Code of Federal Regulations, Title 21, Chapter I, Subchapter E, Part 582, Subpart F, §582.5991, Last seen: October 10, 2016, from http://www.ecfr.gov/cgi-bin/text-idx
- Bailey L. B., Hortus Third, A Concise Dictionary of Plants Cultivated in The United States and Canada.Staff of the L. H. Bailey Hortorium, Cornell University, 1976 [Google Scholar]
- Bala N. Saha S. Chakraborty M. Maiti M. Das S. Basu R. Nandy P. RSC Adv. 2015;5:4993–5003. doi: 10.1039/C4RA12784F. [DOI] [Google Scholar]
- Abdelmohsen U. Cheng C. Viegelmann C. Zhang T. Grkovic T. Ahmed S. Quinn R. Hentschel U. Edrada-Ebel R. Mar. Drugs. 2014;12:1220–1244. doi: 10.3390/md12031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle M. Willett P. Klaffke W. Van Noort P. J. Chem. Inf. Comput. Sci. 2003;43:449–457. doi: 10.1021/ci025591m. [DOI] [PubMed] [Google Scholar]
- Ellman G. L. Diane C. K. Andresjr V. Featherstone R. M. Biochem. Pharmacol. 1961;7(2):88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Magnotti Jr R. A. Eberly J. P. Quarm D. E. McConnell R. S. Clin. Chem. 1987;33/10:1731–1735. doi: 10.1093/clinchem/33.10.1731. [DOI] [PubMed] [Google Scholar]
- Refaey M. S. Abdelhamid R. A. Elimam H. Ellshaier Y. A. Ali A. A. Bioorg. Chem. 2021;108:104643. doi: 10.1016/j.bioorg.2021.104643. [DOI] [PubMed] [Google Scholar]
- Bakr R. O. El-Behairy M. F. Elissawy A. M. Elimam H. Fayed M. A. A. Molecules. 2021;26(5):1198. doi: 10.3390/molecules26051198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykul S. Martinez-Hackert E. Anal. Biochem. 2016;508:97–103. doi: 10.1016/j.ab.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D. L de Groot B. J. Comput.-Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J. Rudolph M. J. Burshteyn F. Cassidy M. S. Gary E. N. Love J. Height J. J. J. Med. Chem. 2012;55(22):10282–10286. doi: 10.1021/jm300871x. [DOI] [PubMed] [Google Scholar]
- DeLano W. L., Pymol: An open-source molecular graphics tool. CCP4 Newsletter on protein crystallography, 2002, 40(1), pp. 82–92 [Google Scholar]
- Santhoshkumar J. Kumar S. V. Rajeshkumar S. Resour.-Effic. Technol. 2017;3:459–465. [Google Scholar]
- Mohammadi F. M. Ghasemi N. J. Nanostruct. Chem. 2018;8:93–102. doi: 10.1007/s40097-018-0257-6. [DOI] [Google Scholar]
- Obaid A. Al-Thabaiti S. Al-Harbi L. M. Khan Z. Global Advanced Research Journal of Microbiology. 2015;3(8):119–126. [Google Scholar]
- Tripathy A. Raichur A. Chandrasekaran N. Prathna T. Mukherjee A. J. Nanopart. Res. 2010;12(1):237–246. doi: 10.1007/s11051-009-9602-5. [DOI] [Google Scholar]
- Heath J. Shiang J. J. Chem. Soc. Rev. 1998;27:65. doi: 10.1039/A827065Z. [DOI] [Google Scholar]
- Phosphor Handbook, ed. S. Shionoya, and W. M. Yen, 1998, CRC, Boca Raton, FL [Google Scholar]; Berger L. I., Semiconductor Materials, 1997, CRC, Boca Raton, FL: T. Satyanarayana, K [Google Scholar]
- Guo H. Y. He X. M. Hu C. G. et al. . Electrochim. Acta. 2014;120:23–29. doi: 10.1016/j.electacta.2013.12.099. [DOI] [Google Scholar]
- Kumar H. Rani R. Int. Lett. Chem., Phys. Astron. 2013;19:26–36. [Google Scholar]
- Sharmila Devi R. Gayathri R. Int. J. Curr. Eng. Technol. 2014;4:2444–2446. [Google Scholar]
- Hamedani N. F. Farzaneh F. J. Sci., Islamic Repub. Iran. 2006;17:231–234. [Google Scholar]
- Jacob R. Isac J. Int. J. Chem. Stud. 2015;2:12–21. [Google Scholar]
- Siavash I. Green synthesis of metal nanoparticles using plants. Green Chem. 2011;13:2638. doi: 10.1039/C1GC15386B. [DOI] [Google Scholar]
- Erjaee H. Rajaian H. Nazifi S. Advances in Natural Sciences. Nanosci. Nanotechnol. 2017;8:1–9. [Google Scholar]
- Raheem D. Tawfike A. Abdelmohsen U. Edrada-Ebel R. Fitzsimmons-Thoss V. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-38940-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amira E. A. Behija S. E. Beligh M. Lamia L. Manel I. Mohamed H. et al. . J. Agric. Food Chem. 2012;60:10896–10902. doi: 10.1021/jf302602v. [DOI] [PubMed] [Google Scholar]
- Mansouri A. Embarek G. Kokkalou E. Kefalas P. Food Chem. 2005;89:411–420. doi: 10.1016/j.foodchem.2004.02.051. [DOI] [Google Scholar]
- Benmeddour Z. Mehinagic E. Le Meurlay D. Louaileche H. J. Funct. Foods. 2013;5:346–354. doi: 10.1016/j.jff.2012.11.005. [DOI] [Google Scholar]
- Garcia B. Marco J. A. J. Nat. Prod. 1981;44(1):111–113. doi: 10.1021/np50013a020. [DOI] [Google Scholar]
- Farag M. A. Mohsen M. Heinke R. Wessjohann L. A. Food Res. Int. 2014;64:218–226. doi: 10.1016/j.foodres.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Scholz E. Heinrich M. Hunkler D. Planta Med. 1994;60(4):360–364. doi: 10.1055/s-2006-959501. [DOI] [PubMed] [Google Scholar]
- Liu L. Xiong C. Zhu Y. Zhou H. Ma S. J. Beijing Univ. Chem. Technol. 2015;42(6):78–83. [Google Scholar]
- Dixon R. A. Ferreira D. Genistein. Phytochemistry. 2002;60:205–211. doi: 10.1016/S0031-9422(02)00116-4. [DOI] [PubMed] [Google Scholar]
- Hazato T. H. Naganawa M. Kumagai T. Aoyagi T. Umezawa H. J. Antibiot. 1979;32:217–222. doi: 10.7164/antibiotics.32.217. [DOI] [PubMed] [Google Scholar]
- Messaoudi R. Abbeddou S. Mansouri A. Calokerinos A. C. Kefalas P. Int. J. Food Prop. 2013;08:29. [Google Scholar]
- Ouafi S. Gaceb-Terrak R. Bounaga N. Lebreton P. C. R. Math. Acad. Sci. Paris. 1988;III306:399–404. [Google Scholar]
- Hong Y. J. Tomas-Barberan F. A. Kader A. A. Mitchell A. E. J. Agric. Food Chem. 2006;54:2405–2411. doi: 10.1021/jf0581776. [DOI] [PubMed] [Google Scholar]
- Michael H. N. Salib J. Y. Eskander &E. F. Phytother. Res. 2013;27(5):699–704. doi: 10.1002/ptr.4777. [DOI] [PubMed] [Google Scholar]
- Harborne C. Williams A. Greenham J. Moyna P. Phytochemistry. 1974;13:1557–1559. doi: 10.1016/0031-9422(74)80327-4. [DOI] [Google Scholar]
- Idaka K. Hirai Y. Shoji J. Chem. Pharm. Bull. 1988;36(5):1783–1790. doi: 10.1248/cpb.36.1783. [DOI] [Google Scholar]
- Hiermann A. Arch. Pharm. 1989;322(2):111–114. doi: 10.1002/ardp.19893220211. [DOI] [PubMed] [Google Scholar]
- Goupy P. Hugues M. Boivin P. Josèphe Amiot M. J. Sci. Food Agric. 1999;79(12):1625–1634. doi: 10.1002/(SICI)1097-0010(199909)79:12<1625::AID-JSFA411>3.0.CO;2-8. [DOI] [Google Scholar]
- Papke R. L. Porter Papke J. K. Br. J. Pharmacol. 2002;137(1):49–61. doi: 10.1038/sj.bjp.0704833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luanratana O. Griffin W. J. Phytochemistry. 1982;21:449–451. doi: 10.1016/S0031-9422(00)95286-5. [DOI] [Google Scholar]
- Hammami S. Jarraya H. Ben Salem S. Hamdi B. Ben Salah A. Devi P. Nefzi A. Mighri Z. J. Soc. Chim. Tunis. 2010;12:105–108. [Google Scholar]
- Murata T. Miyase T. Yoshizaki F. Hyaluronidase inhibitors from Keiskea japonica. Chem. Pharm. Bull. 2012;60:121–128. doi: 10.1248/cpb.60.121. [DOI] [PubMed] [Google Scholar]
- Catchpole O. J. Perry N. B. da Silva B. M. Grey J. B. Smallfield B. M. J. Supercrit. Fluids. 2002;22:129–138. doi: 10.1016/S0896-8446(01)00110-3. [DOI] [Google Scholar]
- Zaki A. K. Schmidt J. Hammouda F. M. Adam G. Planta Med. 1993;59:613–614. doi: 10.1055/s-2006-959837. [DOI] [Google Scholar]
- Fabre P. S. A. Jpn. Kokai Tokkyo Koho JP. 1983;58 67:625. [Google Scholar]
- El-Dib R. Kaloga M. Mahmoud L. Soliman H. Moharram F. Kolodziej H. Phytochemistry. 2004;65:1153–1157. doi: 10.1016/j.phytochem.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Yang X. H. Cai Z. Y. Cai X. Xu X. J. Li H. J. Qin Y. Chin. J. New Drugs. 2015;24(24):2854–2864. [Google Scholar]
- Hong Y. J. Tomas-Barberan F. A. Kader A. A. Mitchell A. E. J. Agric. Food Chem. 2006;54:2405–2411. doi: 10.1021/jf0581776. [DOI] [PubMed] [Google Scholar]
- Nehdi I. Omrib S. Khalil M. I. Al-Resayes S. I. Ind. Crops Prod. 2010;32(3):360–365. doi: 10.1016/j.indcrop.2010.05.016. [DOI] [Google Scholar]
- Kato T. Haller B. F. Heinrich M. Sticher O. Phytochemistry. 1996;41(4):1191–1195. doi: 10.1016/0031-9422(95)00765-2. [DOI] [PubMed] [Google Scholar]
- Almeida M. V. . Penteado C. J. Food Compos. Anal. 1988;1(4):341–352. doi: 10.1016/0889-1575(88)90034-8. [DOI] [Google Scholar]
- Raju J. Mehta R. Nutr. Cancer. 2009;61(1):27–35. doi: 10.1080/01635580802357352. [DOI] [PubMed] [Google Scholar]
- Asami A. Hirai Y. Shoji J. Chem. Pharm. Bull. 1991;39:2053–2056. doi: 10.1248/cpb.39.2053. [DOI] [Google Scholar]
- Okazaki S. Kinoshita K. Ito S. Koyama K. Yuasa H. Takahashi K. Phytochemistry. 2011;72(1):136–146. doi: 10.1016/j.phytochem.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Muhammad W. Ullah N. Haroona M. H Abbasi B. RSC Adv. 2019;9:29541–29548. doi: 10.1039/C9RA04424H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagajyothi P. C. Sreekanth T. V. Tettey C. O. Jun Y. I. Mook S. H. Medi. Chem. Lett. 2014;24:4298–4303. doi: 10.1016/j.bmcl.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Ali M. Muhammad S. Shah M. R. Khan A. Rashid U. Farooq U. et al. . Front. Pharmacol. 2017;8:327. doi: 10.3389/fphar.2017.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz M. Junaid M. Ahmed J. Ullah F. Sadiq A. Ahmad S. et al. . BMC Complementary Altern. Med. 2014;14:145. doi: 10.1186/1472-6882-14-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq A. Zeb A. Ullah F. Ahmad S. Ayaz M. Rashid U. et al. . Front. Pharmacol. 2018;9:623. doi: 10.3389/fphar.2018.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakoyiannis L. Daskalopoulou A. Pergialiotis V. Perrea D. Biomed. Pharmacother. 2019;109:1488–1497. doi: 10.1016/j.biopha.2018.10.086. [DOI] [PubMed] [Google Scholar]
- Bakhtiari M. Panahi Y. Ameli J. Darvishi B. Biomed. Pharmacother. 2017;93:218–229. doi: 10.1016/j.biopha.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Macready A. L. Kennedy O. B. Ellis J. A. Williams C. M. Spencer J. P. Butler L. T. Genes Nutr. 2009;4:227–242. doi: 10.1007/s12263-009-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socci V. Tempesta D. Desideri G. De Gennaro L. Ferrara M. Front. Nutr. 2017;4:19. doi: 10.3389/fnut.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. K. Mishra H. Ekielski A. Talegaonkar S. Vaidya B. Drug Discovery Today. 2017;22:1825–1834. doi: 10.1016/j.drudis.2017.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.